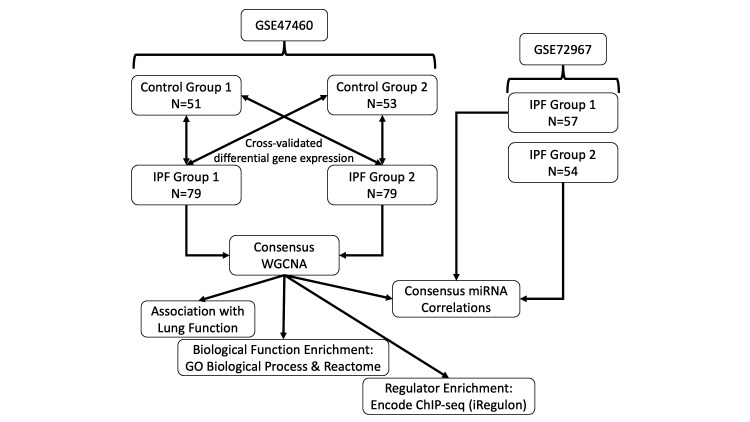
Figure 1.
Flow chart of methodologies used in this study. GSE47460 was divided into two non-overlapping datasets for cross-validated differential gene expression analysis. This was followed by applying a consensus WGCNA on the two IPF groups using the differentially expressed genes as input. Identified modules were then analysed by association with lung function and enrichment analyses for biological function and transcription factor regulators. Samples in GSE72967 were matched to the IPF groups and correlated with the module eigengene to determine association with that module. GO, gene ontology; IPF, idiopathic pulmonary fibrosis; WGCNA, weighted gene coexpression network analysis.