Abstract
Evolutionary dynamics in laboratory microbial evolution experiments can be surprisingly complex. In the last two decades, observations of these dynamics have challenged simple models of adaptation, and have shown that clonal interference, hitchhiking, ecological diversification, and contingency are widespread. In recent years, advances in high-throughput strain maintenance and phenotypic assays, the dramatically reduced cost of genome sequencing, and emerging methods for lineage barcoding have made it possible to observe evolutionary dynamics at unprecedented resolution. These new methods can now begin to provide detailed measurements of key aspects of fitness landscapes and of evolutionary outcomes across a range of systems. These measurements can highlight challenges to existing theoretical models, and guide new theoretical work towards the complications that are most widely important.
Keywords: clonal interference, ecological diversification, pleiotropy, epistasis, contingency
Surprising Complexity in Simple Experiments
For many decades, evolutionary adaptation in microbial populations was thought to proceed by "periodic selection," where individual beneficial mutations arise sequentially and either go extinct or fix in independent selective sweeps [1]. In this picture, evolution is relatively simple: mutations arise randomly and then fix or go extinct at a rate that is commensurate with their individual selective effect. Our ability to predict how a population should evolve is then only limited by our knowledge of the biological details of that specific system (i.e. the potential mutations and their corresponding mutation rates and selective effects, the biological environment in Box 1).
Box 1: Key Determinants of Evolutionary Dynamics.
Evolutionary dynamics are influenced by a number of different factors. One class of factors involves the physiology of specific organisms in particular environmental contexts. We refer to these as the biological environment; they determine how selection acts on different genotypes. Another class of factors determine how genetic variation arises and how it is inherited. We refer to these as the population genetic environment; they determine how genetic drift operates, constrain how mutations move between haplotypes, and determine which organisms compete and interact. Of course, the distinction between the biological and population genetic environment is somewhat arbitrary.
-
A:Examples of factors in the population genetic environment include:
- population size, N
- mutation rate, U
- recombination rate, R, and the physical structure of the genome
- spatial structure
-
B:Examples of factors in the biological environment include:
- the “local” distribution of mutational effects on fitness, ρ(s)
- the ruggedness of the landscape (how ρ(s) changes as a result of epistasis)
- pleiotropic effects of mutations
- statistics of environmental change
- ecological opportunities
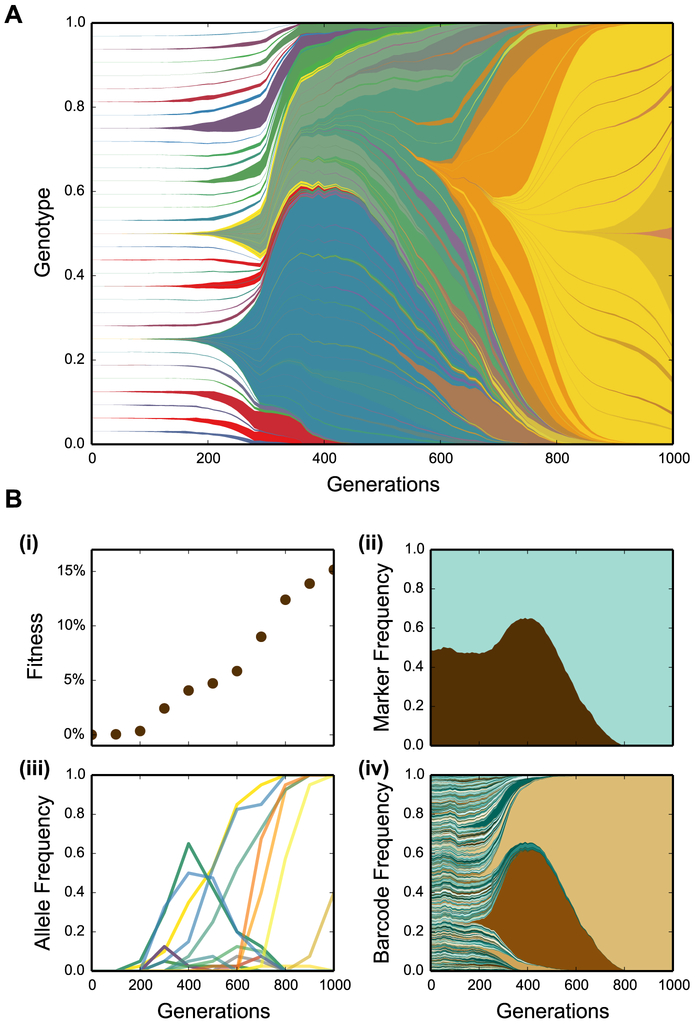
Beginning in the late 1990s, however, observations of surprising complexity in microbial evolution experiments provided convincing evidence rejecting this standard "periodic selection" picture. Instead, careful observations of rates of fitness increase [2-4] and changes in the frequencies of genetic markers over time [5-9] pointed to widespread signatures of clonal interference (see Glossary) and hitchhiking. These complications make it much harder to predict how evolution will act: we are limited not only by our knowledge of the biological details, but also by our lack of understanding of the evolutionary dynamics themselves. The basic difficulty is that many interacting loci across the genome are hopelessly intertwined -- evolution cannot change the frequencies of alleles at one locus without simultaneously affecting alleles at many other linked loci (Figure 1, Key Figure). In these settings, we cannot rely on well-established models of evolution at individual loci to predict evolutionary dynamics.
Figure 1 (Key Figure):
(A) Simulated evolutionary dynamics in an asexually evolving population, with parameter values typical in a laboratory evolution experiment. Mutations arise often enough that they cannot be selected on individually. Instead, between the appearance of a new mutation in a population and its eventual extinction or fixation, many other mutations arise in the population, either on the same genetic background or in a competing lineage. As a result, the fate of each mutation is not determined only on its own merits, but is intertwined with all other mutations in the population. Most beneficial mutations are outcompeted by fitter clones before they are able to rise to substantial frequencies. We note that these evolutionary dynamics have never been directly observed at the resolution shown here. (B) These evolutionary dynamics can be studied in laboratory settings using a range of methods: (i) Fitness assays. The relative increase in fitness of the evolving population compared to the ancestor offers a coarse view of the underlying evolutionary dynamics. (ii) The frequencies of pre-introduced genetic markers through time. As with fitness assays, changes in marker frequencies reflect the aggregate effects of multiple evolutionary events. These methods cannot resolve the effects of individual mutations. (iii) Population metagenomic sequencing offers a view of individual mutations that arise during evolution. However, only mutations that reach substantial frequencies (typically at least ~5% or more) are observable. Thus only a tiny and biased subset of all the mutations occurring in the population is visible. (iv) Newer barcoding methods make it possible to observe lineage dynamics at much higher resolution. Up to the resolution limits imposed by the evolutionary process itself (i.e. genetic drift), these lineage dynamics can be used to infer when beneficial mutations occur and their effects on fitness. However, because barcode diversity is lost as the population evolves, these methods are currently limited to studying short timescales.
Inspired in large part by these experiments, there is now a thriving theoretical community bringing methods from statistical physics and applied mathematics to the study of evolutionary dynamics in these "rapidly evolving" populations. This has led to many advances in our analytical understanding of the effects of clonal interference and other forms of linked selection [10]. However, increasingly high-resolution observations of evolutionary dynamics in laboratory evolution experiments have continued to reveal unexpected complexities that appear to be crucial to evolutionary dynamics in these systems, and call for still further theoretical work. In this article, we review these recent developments.
Studying Evolution Without Phenotype
Many studies of adaptation in both natural and laboratory populations are focused primarily on understanding phenotypes: the goal is to characterize adaptive changes and to identify the evolutionary processes by which they arose as well as their genetic and molecular basis. Experimental studies of evolutionary dynamics focus instead on understanding evolution as a stochastic algorithm. That is, given a particular set of biological details (i.e. the set of mutations that can arise and their corresponding fitness effects in all relevant environments and genetic backgrounds, often referred to as the fitness landscape), what will evolution actually do? What mutations will fix with what probabilities? How repeatable is the process, and what patterns of genetic diversity will a population display? The focus is on the role of the dynamics in determining evolutionary outcomes, and not on the nature of the adaptive phenotypes or their genetic and molecular basis. That is, we aim to study evolution as a process, without reference to the specific phenotypes in question.
In principle, given a specific landscape and set of population genetic parameters, we can address any questions about how evolution acts by implementing computational simulations of the dynamics. However, we cannot possibly measure the fitness landscape in every system we wish to understand. Instead, we hope to be able to identify key principles of evolutionary dynamics that help us understand what general features of landscapes are important and how these features influence evolution across a wide range of systems. With this goal in mind, many theoretical studies have focused on simple analytically tractable models.
Since evolutionary dynamics are inherently random, testing these models involves quantifying the probabilities of different outcomes. This requires highly controlled and replicated experiments, which make it possible to identify deviations from existing theoretical predictions that point to important new processes that future theory must account for. For example, classic results from population genetics tell us that the fixation probability of a beneficial mutation that is at frequency x within a population of constant size N and provides a fitness advantage s should be pfix(x,s) = (1-e−2Nsx)/(1-e−2Ns) [11]. This has led to the widespread view of s=1/N as a drift barrier: newly arising beneficial mutations with fitness effect less than this fix with probability approximately equivalent to a neutral mutation, while beneficial mutations with larger effects fix with probability of about 2s. Yet numerous experimental studies have shown dramatic differences from this prediction in adapting microbial populations, with beneficial mutations fixing much less often than this formula would predict [12-17]. Analysis of the evolutionary dynamics in these experiments showed that this discrepancy arises because beneficial mutations are much more common than previously appreciated in these large populations (often with sizes ranging from 106-1010), leading to widespread clonal interference that reduces the efficiency of selection and hence the fixation probability of any individual beneficial mutation. This in turn led to further theoretical analysis of these effects of clonal interference.
We now know that clonal interference tunes the characteristic effect size of evolutionarily relevant mutations, favoring larger-effect mutations and dramatically suppressing the importance of smaller-effect mutations [18-20]. This characteristic effect size depends sensitively on the overall size of a population, the mutation rate, and the evolutionary conditions. Thus, these dynamical aspects of adaptation tune the spectrum of mutations that have a chance at fixing in populations, which in turn feed back to affect evolutionary dynamics. On long enough timescales, this feedback between the raw evolutionary material and evolutionary dynamics ultimately shapes entire genomes. In recent years, there has been significant theoretical interest in characterizing how evolutionary dynamics can alter mutation rates [21, 22], the spectrum of available mutations in a genome [23], as well as expected patterns of epistasis [24, 25]. However, often too little is known about the fitness landscape to model genomic evolution over long evolutionary timescales meaningfully. In these cases, experimental evolution can be used to measure the distribution of this raw evolutionary material, which can guide future theoretical work.
As technological developments (particularly in sequencing) have recently made it possible to observe evolutionary dynamics in laboratory populations with ever increasing resolution and replication, many theoretical expectations have come under challenge. Over the coming few years, there are likely continue to be many insights derived from taking a purely observational approach, by simply watching evolutionary dynamics using these new tools in a variety of settings and asking whether we can explain what we see. The answer is often no, which can then spur new theoretical directions. Similarly, these new techniques can now allow high-throughput measurements of quantities such as distributions of pleiotropic or epistatic effects in genomes, which were not possible to make at scale using previous methods.
Technological Advances in Observing Evolutionary Dynamics
There are three key challenges in observing evolutionary dynamics. First, the underlying events are mutations, and we ultimately want to track the frequencies of all the genotypes they produce. These are typically difficult to observe directly. Second, evolutionary dynamics are fundamentally stochastic, so we typically wish to quantify the probabilities of different events. This often requires extensive replication. Finally, new mutations arise in single individuals. Their dynamics while they remain at very low frequencies within the population are typically both critically important and very difficult to observe.
Because it is difficult to observe the underlying mutations directly, early work attempted to measure phenotypic changes through time. For instance, many studies measured the competitive fitness of evolving lines through time (Figure 1 B.i). This work provided many insights into how quickly populations adapt [26], how this depends on population size and other parameters [2, 3, 27, 28], and how repeatable these phenotypic changes are across replicate populations [29, 30]. However, these phenotypic changes are a coarse view of the underlying genetic changes, and hence can only provide limited insight into the evolutionary dynamics at the sequence level.
An alternative approach has been to engineer strains in such a way that certain specific mutations lead to easily measurable phenotypic changes. For example, one can construct yeast strains in which loss-of-function mutations in the gene CAN1 lead to resistance to the drug canavanine; the frequency of these mutations can then be precisely tracked by plating on media containing this drug [31]. Other studies built on this idea to introduce other drug or fluorescent markers that become active when certain classes of mutations arise [13, 32, 33]. However, while these approaches allow us to track the frequencies of specific mutations (often at high resolution), they typically only allow us to observe a very small fraction of the genetic changes that occur within the population. We must infer something about the larger majority of genetic changes that we cannot observe from the dynamics of the small fraction we can see. Closely related to this approach, other studies have introduced neutral (or sometimes non-neutral) genetic markers to distinguish different lineages within evolving populations [1, 5, 6, 9, 12]. By tracking the frequencies of these markers through time, we can infer something about the underlying dynamics [34]. However, since these studies have typically only tracked the frequencies of two or three markers, they are only sensitive to major shifts in the composition of the population, and cannot provide any insight into dynamics at lower frequencies (Figure 1 B.ii).
These earlier methods for observing evolutionary dynamics were limited not only in resolution but also in scale. The experiments themselves were typically conducted in test tubes, flasks, or chemostats. This required substantial physical space as well as labor, which limited replication. In addition, measuring phenotypic changes such as fitness or the frequencies of drug markers was relatively laborious, so it was only practical to track evolution in at most a few dozen populations at once. More recently, it has become common to maintain populations in microplates and to maintain them using robotic liquid handling [35]. These methods make it possible for a single experimenter to maintain thousands of microbial populations in parallel. By using fluorescent proteins rather than drug or nutrient markers, it has similarly become possible to analyze some aspects of the dynamics in these populations at high throughput using flow cytometry.
More recently, advances in sequencing technology have now made it possible to track evolution at the sequence level directly, using either whole-population “metagenomic” sequencing or by sampling and sequencing individual clones [14, 15, 17, 36-41]. While these approaches involve substantial bioinformatics challenges, this makes it possible to identify individual mutations and track their frequencies through time (Figure 1 B.iii). Though it was initially not possible to do this at scale, reductions in sequencing costs now make it possible to sequence hundreds of clones or whole microbial populations samples to a depth of 20-100x on a single sequencing lane. Sample preparation, which was once a limiting factor, has also become possible to do at scale for minimal cost [42]. Thus by combining extensive sequencing with the robotic liquid handling methods described above, it is now feasible to track evolutionary dynamics at the sequence level in hundreds of replicate populations in parallel.
However, a fundamental limit of these sequencing approaches is frequency resolution. Sequencing errors and other bioinformatics challenges make it very difficult to identify and track mutations below a few percent frequency. Yet in microbial populations that often consist of millions or billions of cells, the fates of mutations are often determined by the competition of high-fitness clones at frequencies that are many orders of magnitude lower than this. While these challenges can be mitigated to some extent by increasing sequencing depth or by using approaches such as circle sequencing [43] or Duplex sequencing [44] to reduce error rates, this can dramatically increase costs. Thus this is likely to remain a major limitation of whole-genome sequencing approaches for the foreseeable future.
To circumvent this resolution problem, a new approach is to label individual cells with unique DNA barcodes at the outset of an experiment [45]. This approach exploits the same principles as older marker tracking methods, but does so using millions of unique barcodes, rather than a few fluorescent or drug markers (Figure 1B.iv). By sequencing the barcode locus, one can track the number of descendants of all individuals in the population over time at extremely high resolution. Sequencing errors are no longer limiting because barcodes can be designed to differ at several sites. As with other marker-based methods, this approach does not directly identify individual mutations. However, since the barcodes measure frequencies at very high resolution, changes in their frequencies are much more sensitive to the effects of individual mutations. Thus it is possible to infer when adaptive mutations occur, their effects on fitness, and their frequency trajectories even at very low frequencies.
These barcoding methods are promising, but do suffer from two key limitations. First, to realize the benefits of increased resolution, one must sequence the barcode locus at depths comparable to microbial population sizes (106-1010). This requires 106-1010 reads per timepoint and population sequenced, and the corresponding costs limit the extent of the replication that can be achieved. Thus far, this approach has only been used to track dynamics in a few populations in parallel [46, 47]. Second, as time progresses, barcode diversity declines as some lineages go extinct and others increase in size. Therefore this method has been limited to offering high-resolution views of only the earliest phases of clonal evolution. In principle, this second limitation could be circumvented either by “re-barcoding” the population at periodic intervals or by adapting methods to continually add diversity to existing barcodes [48], though either approach presents some technical challenges.
Which complications are important?
Theoretical studies of very simple models have provided a great deal of insight that underlies much intuition in evolutionary dynamics and population genetics. For example, many studies have analyzed how a population climbs a single fitness peak in the strong-selection-weak-mutation (SSWM) approximation where only one mutation is ever present in the population at a time. Similarly, models of neutral mutation accumulation and the balance between deleterious mutations and selection are often used to explain evolution in a “well-adapted” population that is at a local fitness peak. The implicit assumption that natural populations are typically in such a well-adapted state underlies many practical methods in population genetics.
Of course, no one believes that evolution is ever actually this simple. Nevertheless, these idealized models have widespread influence because there are countless complications that could in principle matter, and it is impossible to model all of them at once (Box 1). A key question is thus: which complications are widespread and of general importance, and what simplifications can we get away with? Experimental studies of evolutionary dynamics have played an important role in answering this question. Many of these experiments are explicitly designed to be as simple as we can make them. Thus complications that routinely arise even in these very artificially restricted settings may be to some extent genuinely widespread and unavoidable. Of course this does not rule out the possibility that other effects are important in other specific settings, but it does help point to key factors that any theoretical picture needs to grapple with.
For example, over the past two decades it has become clear that clonal interference and hitchhiking are of widespread importance. Early tests of the SSWM picture focused on very large populations, often using strains engineered to have higher than normal mutation rates, in order to probe what was imagined to be an idiosyncratic regime where the widely-used SSWM approximations might begin to break down [2]. Instead, it soon became clear that clonal interference and hitchhiking were unavoidable even in modestly-sized microbial and viral populations with wild-type mutation rates. Qualitatively similar effects of linked selection have also been observed in recombining outbred populations adapting on standing variation, where selection acts simultaneously on many sites across the genome, and recombination can only slowly decouple the effects of linked beneficial and deleterious alleles (a version of the Hill-Robertson effect) [49-53].
More recent work has also begun to challenge the assumption that populations that have evolved in a constant environment for a long period of time can be described using the standard picture of a “well-adapted” population on a fitness peak. As far as we are aware, there are no examples that fit this picture, including the long-term experiment in E. coli through at least 60,000 generations [17, 41, 54, 55]. Instead, even very large populations evolved for long periods in as constant an environment as is experimentally feasible continue to increase in fitness and to rapidly accumulate adaptive mutations. While experiments in smaller populations do sometimes reach fitness plateaus, this is not necessarily because they have reached an optimum [56]. Instead, these populations may have reached a balance between adaptation and the stochastic accumulation of deleterious mutations [57], with molecular evolution continuing at a rapid pace. These results suggest that we should question the standard picture of natural populations in a "well-adapted" state characterized by neutral evolution and deleterious mutation-selection balance.
Another widespread assumption of many models of evolution and population genetics is that evolutionary and ecological dynamics can be separated. Instead, coexisting types often spontaneously arise in laboratory evolution experiments and are maintained for long periods due to negative frequency-dependent selection [16, 17, 37, 38, 58-61]. These ecological interactions arise via a variety of different mechanisms, despite the fact that many of these experiments were explicitly designed to minimize the opportunities for ecological diversification. These ecological interactions are then often further modified as evolution continues within each coexisting type, leading to shifts in the frequencies of the types [17, 37, 38, 59, 60]. Thus evolution and ecology are fundamentally intertwined.
Other types of complex frequency-dependent interactions are also sometimes observed. For example, there are some reports of positive frequency-dependent and non-transitive (or “red queen”) fitness interactions [62-65]. However, within the limits of current resolution, these more complex effects appear to be relatively rare in microbial evolution experiments [66].
On the other hand, the effects of individual mutations can strongly depend on the genetic background in which they occur [67]. These epistatic effects of course include specific interactions involving mutations within an individual protein or pathway [68-70]. However, some experiments have shown more widespread effects, where individual mutations can often alter the future evolutionary potential of their descendants. This can occur both due to global fitness-mediated effects [30, 71-77] (e.g. higher-fitness genotypes can be generically less “adaptable” and less “robust”) and due to more idiosyncratic mechanisms [78-80]. This widespread epistasis and contingency may help explain why experimentally evolving populations do not ever appear to reach a fitness peak.
Similarly, the structure of pleiotropic effects of mutations for fitness across varying environmental conditions can be complex. While we might expect simple tradeoffs between fitness in different conditions to routinely arise from physiological constraints, the reality is often more subtle [81-87]. There are often multiple distinct ways a population can adapt to a given environmental condition, which may have a variety of effects across other environments [88-90]. The details of the population genetic environment and the statistics of fluctuating conditions can therefore play a critical role in determining the extent to which adaptation tends to lead to specialization [91].
Concluding Remarks and Future Perspectives
It could be argued that studies of evolutionary dynamics in artificial and highly simplified laboratory conditions (which often lack spatial structure, temporal variability, interactions with other species, and other complexities) are unrepresentative of evolution in natural systems. However, we view this simplicity instead as a major strength of experimental evolution, which is a powerful tool precisely because complications can be introduced in a controlled, replicable way (see Outstanding Questions). Nevertheless, one could argue that conclusions from artificial laboratory environments are simply not representative of those relevant in more “natural” settings. Testing this will ultimately require more detailed direct observations of evolution in natural environments. Some recent work has moved in this direction by using laboratory evolution techniques in more complex and realistic environments, such as by studying E. coli that are experimentally passaged through mouse guts [39] or V. fischeri living in symbiosis with squid [92]. A complementary direction will be to begin to use these general techniques and analysis frameworks to study evolution directly in natural systems, such as evolving pathogens [93-95], the immune system [96], and host-associated microbial and viral communities [97-99].
OUTSTANDING QUESTIONS.
How rugged are fitness landscapes? What do the statistical patterns of epistasis between mutations look like? How do these in turn impact evolutionary dynamics?
How common are modifiers of fitness landscapes (e.g. mutator alleles or evolvability loci) and what are their effects on evolutionary dynamics?
How does recombination affect these patterns? What do genome-wide patterns of epistatic effects look like in recombining populations? Are closely linked sites more or less likely to have epistatic interactions than distant sites?
How often do ecological interactions between closely related individuals arise? How often do they lead to robust equilibria, and how often are these ecological interactions broken by future mutations?
How different is adaptation in different environments? In what ways is laboratory adaptation similar to adaptation in more “natural” settings, and in what ways do they differ?
How does variability in the evolutionary environment impact evolutionary dynamics and the predictability of evolution? What are the pleiotropic effects of mutations across a range of environments, and how does this affect evolution in conditions that fluctuate in time or space?
It is also unclear how evolutionary dynamics in microbial populations relate to other systems. Numerous studies have analyzed evolution in other laboratory model organisms, including complex multicellular organisms such as Drosophila [51, 53] or C. elegans [100]. In these systems, standing genetic variation, differences in genome organization and ploidy, and other complications can all influence the dynamics. These factors may affect which parameter regimes are typically relevant, and what complications theoretical models must grapple with. However, many of the technical methods described here cannot be directly applied to these systems. Thus an important future direction will be to develop tools that make it possible to study evolution in these more complex organisms at higher throughput and resolution, and comparing the results to what we have learned by studying microbial systems.
HIGHLIGHTS.
Laboratory evolution experiments show that microbial populations continue to rapidly adapt even over long timescales in simple constant environments. Beneficial mutations remain abundant, and populations typically do not reach a fitness peak.
Regular patterns of "global" epistatic interactions as well as idiosyncratic interactions between individual mutations lead to evolutionary contingency. It is unclear to what extent this contingency affects the predictability and repeatability of evolution.
Spontaneous ecological diversification is common, even in simple environmental conditions that were explicitly designed to minimize this possibility. Ecological and evolutionary dynamics often occur on similar timescales.
Measurements of the pleiotropic effects of individual mutations on fitness across a range of environmental conditions reveal diverse statistical patterns of pleiotropy in different systems. These patterns interact with evolutionary dynamics to determine how populations specialize as they adapt.
ACKNOWLEDGEMENTS
M.M.D. acknowledges support from the Simons Foundation (Grant 376196), the National Science Foundation (DEB-1655960), and the National Institutes of Health (R01-GM104239).
Glossary Box
- Clonal interference
Competition between multiple different (and typically beneficial) mutations that are segregating simultaneously within the population.
- DNA barcode
A DNA sequence that "barcodes" a strain. This often refers to a naturally occuring sequenced used for species identification in ecological applications. In laboratory evolution, barcodes are sometimes instead random sequences (often ~10-30 base pairs) that are integrated by the experimenter into a specific genomic location.
- Epistasis
The dependence of phenotypic effects of mutations on the genetic background.
- Flow cytometry
A technique to measure the fluorescence profiles of individual cells in high throughput. Often used to count differently labeled cells in a population for applications such as fitness measurements.
- Fitness landscape
A general mapping between genotype and fitness in a specific environmental condition.
- Hitchhiking
The process by which a neutral or deleterious allele increases in frequency due to linkage to a beneficial mutation. Can also refer to a weakly beneficial mutation increasing in frequency due to linkage to a more strongly beneficial one.
- Pleiotropy
The effect of a mutation on multiple different phenotypes. Here, the phenotypes discussed are often fitness effects in different environments.
REFERENCES CITED
- 1.Atwood KC, Schneider LK, and Ryan FJ, Periodic selection in Escherichia coli. Proceedings of the National Academy of Sciences, 1951. 37: p. 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Visser J, et al. , Diminishing returns from mutation supply rate in asexual populations. Science, 1999. 283(5400): p. 404–406. [DOI] [PubMed] [Google Scholar]
- 3.Desai MM, Fisher DS, and Murray AW, The Speed of Evolution and Maintenance of Variation in Asexual Populations. Current Biology, 2007. 17(5): p. 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miralles R, Moya A, and Elena SF, Diminishing returns of population size in the rate of RNA virus adaptation. J. Virology, 2000. 74(8): p. 3566–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao KC and Sherlock G, Molecular Characterization of Clonal Interference During Adaptive Evolution in Asexual Populations of Saccharomyces cerevisiae. Nature Genetics, 2008. 40: p. 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegreness M, et al. , An equivalence principle for the incorporation of favorable mutations in asexual populations. Science, 2006. 311(5767): p. 1615–1617. [DOI] [PubMed] [Google Scholar]
- 7.Imhof M and Schlotterer C, Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA, 2001. 98: p. 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miralles R, et al. , Clonal interference and the evolution of RNA viruses. Science, 1999. 285(5434): p. 1745–1747. [DOI] [PubMed] [Google Scholar]
- 9.De Visser JAGM and Rozen DE, Clonal Interference and the Periodic Selection of New Beneficial Mutations in Escherichia coli. Genetics, 2006. 172: p. 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neher RA, Genetic Draft, Selective Interference, and Population Genetics of Rapid Adaptation. Annual Review of Ecology, Evolution, and Systematics, 2013. 44(1): p. 195–215. [Google Scholar]
- 11.Kimura M, On the probability of fixation of mutant genes in a population. Genetics, 1962. 47: p. 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenkel EM, Good BH, and Desai MM, The Fates of Mutant Lineages and the Distribution of Fitness Effects of Beneficial Mutations in Laboratory Budding Yeast Populations. Genetics, 2014. 196(4): p. 1217–1226. PMCID: PMC3982683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang GI, Botstein D, and Desai MM, Genetic variation and the fate of beneficial mutations in asexual populations. Genetics, 2011. 188(3): p. 647–61. PMCID: PMC3176544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang GI, et al. , Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature, 2013. 500: p. 571–574. PMCID: PMC3758440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvitek DJ and Sherlock G, Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment. PLoS Genetics, 2013. 9(11): p. e1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madamsetti R, Lenski RE, and Barrick JE, Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics, 2015. 200(2): p. 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good BH, et al. , The dynamics of molecular evolution over 60,000 generations. Nature, 2017. 551(7678): p. 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good BH, et al. , Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proceedings of the National Academy of Sciences, 2012. 109: p. 4950–4955. PMCID: PMC3323973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallatschek O, The noisy edge of traveling waves. Proceedings of the National Academy of Sciences, 2011. 108(5): p. 1783–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher DS, Asexual evolution waves: fluctuations and universality. Journal of Statistical Mechanics: Theory and Experiment, 2013. 2013(01): p. P01011. [Google Scholar]
- 21.Sung W, et al. , Drift-barrier hypothesis and mutation-rate evolution. Proceedings of the National Academy of Sciences, 2012. l09(45): p. 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good BH and Desai MM, Evolution of mutation rates in rapidly adapting asexual populations. Genetics, 2016. 204(3): p. 1249–1266. PMCID: PMC5105855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice DP, Good BH, and Desai MM, The Evolutionarily Stable Distribution of Fitness Effects. Genetics, 2015. 200(1): p. 321–329. PMCID: PMC4423373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neher RA and Shraiman BI, Statistical genetics and evolution of quantitative traits. Reiews of Modern Physics, 2011. 83: p. 1283–1300. [Google Scholar]
- 25.de Visser JAGM, Cooper TF, and Elena SF, The causes of epistasis. Proceedings of the Royal Society B: Biological Sciences, 2011. 278(1725): p. 3617–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenski R, et al. , Long-term experimental evolution in Escherichia coli I. Adaptation and divergence during 2,000 generations. American Naturalist, 1991. 138: p. 1315–1341. [Google Scholar]
- 27.Kryazhimskiy SK, Rice DP, and Desai MM, Population Subdivision and Adaptation in Asexual Populations of Saccharomyces cerevisiae. Evolution, 2012. 66: p. 1931–1941. [DOI] [PubMed] [Google Scholar]
- 28.Nahum JR, et al. , A tortoise–hare pattern seen in adapting structured and unstructured populations suggests a rugged fitness landscape in bacteria. Proceedings of the National Academy of Sciences, 2015. 112(24): p. 7530–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Travisano M, et al. , Experimental tests of the roles of adaptation, chance, and history in evolution. Science, 1995. 267(5194): p. 87–90. [DOI] [PubMed] [Google Scholar]
- 30.Kryazhimskiy S, et al. , Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science, 2014. 344(6191): p. 1519–1522. PMCID: PMC4314286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paquin C and Adams J, Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature, 1983. 302: p. 495–500. [DOI] [PubMed] [Google Scholar]
- 32.MacLean RC, Perron GG, and Gardner A, Diminishing Returns From Beneficial Mutations and Pervasive Epistasis Shape the Fitness Landscape for Rifampicin Resistance in Pseudomonas aeruginosa. Genetics, 2010. 186(4): p. 1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baym M, et al. , Spatiotemporal microbial evolution on antibiotic landscapes. Science, 2016. 353(6304): p. 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al. , Estimation of the rate and effect of new beneficial mutations in asexual populations. Theoretical Population Biology, 2012. 81(2): p. 168–178. [DOI] [PubMed] [Google Scholar]
- 35.Desai MM, Statistical questions in experimental evolution. Journal of Statistical Mechanics: Theory and Experiment, 2013. 2013(01): p. P01003. [Google Scholar]
- 36.McDonald MJ, Rice DP, and Desai MM, Sex speeds adaptation by altering the dynamics of molecular evolution. Nature, 2016. 531(7593): p. 233–236. PMCID: PMC4855304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traverse CC, et al. , Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proceedings of the National Academy of Sciences, 2013. 110: p. E250–E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herron MD and Doebeli M, Parallel Evolutionary Dynamics of Adaptive Diversification in Escherichia coli. PLOS Biology, 2013. 11(2): p. e1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barroso-Batista J, et al. , The First Steps of Adaptation of Escherichia coli to the Gut Are Dominated by Soft Sweeps. PLOS Genetics, 2014. 10(3): p. e1004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrick JE, et al. , Genome Evolution and Adaptation in a Long-Term Experiment with Escherichia Coli. Nature, 2009. 461: p. 1243–1247. [DOI] [PubMed] [Google Scholar]
- 41.Tenaillon O, et al. , Tempo and mode of genome evolution in a 50,000-generation experiment. Nature, 2016. 536(7615): p. 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baym M, et al. , Inexpensive Multiplexed Library Preparation for Megabase-Sized Genomes. PLOS ONE, 2015. 10(5): p. e0128036. PMCID: PMC4441430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lou DI, et al. , High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proceedings of the National Academy of Sciences, 2013. 110(49): p. 19872–19877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt MW, et al. , Detection of ultra-rare mutations by next-generation sequencing. Proceedings of the National Academy of Sciences, 2012. 109(36): p. 14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blundell JR and Levy SF, Beyond genome sequencing: Lineage tracking with barcodes to study the dynamics of evolution, infection, and cancer. Genomics, 2014. 104(6): p. 417–430. [DOI] [PubMed] [Google Scholar]
- 46.Levy SF, et al. , Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature, 2015. 519(7542): p. 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blundell JR, et al. , The dynamics of adaptive genetic diversity during the early stages of clonal evolution. bioRxiv, 2017: p. 170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalhor R, Mali P, and Church GM, Rapidly evolving homing CRISPR barcodes. Nature Methods, 2016. 14: p. 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosheleva K and Desai MM, Recombination alters the dynamics of adaptation on standing variation in laboratory yeast populations. Molecular Biology and Evolution, 2018. 35(1): p. 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parts L, et al. , Revealing the genetic structure of a trait by sequencing a population under selection. Genome Research, 2011. 21(7): p. 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burke MK, et al. , Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature, 2010. 467: p. 587. [DOI] [PubMed] [Google Scholar]
- 52.Burke MK, Liti G, and Long AD, Standing Genetic Variation Drives Repeatable Experimental Evolution in Outcrossing Populations of Saccharomyces cerevisiae. Molecular Biology and Evolution, 2014. 31(12): p. 3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orozco-terWengel P, et al. , Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Molecular Ecology, 2012. 21 (20): p. 4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiser MJ, Ribeck N, and Lenski RE, Long-Term Dynamics of Adaptation in Asexual Populations. Science, 2013. 342(6164): p. 1364. [DOI] [PubMed] [Google Scholar]
- 55.Lenski RE, et al. , Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli. Proceedings of the Royal Society B: Biological Sciences, 2015. 282(1821). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silander OK, Tenaillon O, and Chao L, Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol, 2007. 5(4): p. e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal S, et al. , Dynamic Mutation Selection Balance as an Evolutionary Attractor. Genetics, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rainey PB and Travisano M, Adaptive Radiation in a Heterogeneous Environment. Nature, 1998. 394: p. 69–72. [DOI] [PubMed] [Google Scholar]
- 59.Frenkel EM, et al. , Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proceedings of the National Academy of Sciences, 2015. 112(36): p. 11306–11311. PMCID: PMC4568650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plucain J, et al. , Epistasis and Allele Specificity in the Emergence of a Stable Polymorphism in Escherichia coli. Science, 2014. 343(6177): p. 1366. [DOI] [PubMed] [Google Scholar]
- 61.Behringer MG, et al. , Escherichia coli cultures maintain stable subpopulation structure during long-term evolution. Proceedings of the National Academy of Sciences, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rendueles O, Amherd M, and Velicer Gregory J. Positively Frequency-Dependent Interference Competition Maintains Diversity and Pervades a Natural Population of Cooperative Microbes. Current Biology, 2015. 25(13): p. 1673–1681. [DOI] [PubMed] [Google Scholar]
- 63.Chao L and Levin BR, Structured habitats and the evolution of anticompetitor toxins in bacteria. Proceedings of the National Academy of Sciences, 1981. 78(10): p. 6324–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paquin CE and Adams J, Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature, 1983. 306: p. 368. [DOI] [PubMed] [Google Scholar]
- 65.Kerr B, et al. , Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature, 2002. 418: p. 171–174. [DOI] [PubMed] [Google Scholar]
- 66.de Visser JA and Lenski RE, Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evol Biol, 2002. 2: p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jerison ER and Desai MM, Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Current Opinion in Genetics & Development, 2015. 35: p. 33–39. PMCID: PMC4710057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinreich DM, et al. , Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science, 2006. 312: p. 111–114. [DOI] [PubMed] [Google Scholar]
- 69.de Vos MGJ, et al. , Environmental Dependence of Genetic Constraint. PLOS Genetics, 2013. 9(6): p. e1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenaillon O, et al. , The Molecular Diversity of Adaptive Convergence. Science, 2012. 335(6067): p. 457–461. [DOI] [PubMed] [Google Scholar]
- 71.Jerison ER, et al. , Genetic variation in adaptability and pleiotropy in budding yeast. eLife, 2017. 6: p. e27167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moore F, Rozen D, and Lenski R, Pervasive compensatory adaptation in Escherichia coli. Proc Roy Soc, London B, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perfeito L, et al. , Rates of fitness decline and rebound suggest pervasive epistasis. Evolution, 2014. 68(1): p. 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrick JE, et al. , Escherichia coli rpoB Mutants Have Increased Evolvability in Proportion to Their Fitness Defects. Molecular Biology and Evolution, 2010. 27(6): p. 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanjuan R, et al. , Epistasis and the adaptability of an RNA virus. Genetics, 2005. 170: p. 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan AI, et al. , Negative Epistasis Between Beneficial Mutations in an Evolving Bacterial Population. Science, 2011. 332(6034): p. 1193–1196. [DOI] [PubMed] [Google Scholar]
- 77.Chou H-H, et al. , Diminishing Returns Epistasis Among Beneficial Mutations Decelerates Adaptation. Science, 2011. 332(6034): p. 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woods RJ, et al. , Second-Order Selection for Evolvability in a Large Escherichia coli Population. Science, 2011. 331(6023): p. 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blount ZD, Borland CZ, and Lenski RE, Historical Contingency and the Evolution of a Key innovation in an Experimental Population of Escherichia coli. PNAS, 2008. 105: p. 7899–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blount ZD, et al. , Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature, 2012. 489(7417): p. 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cooper VS and Lenski RE, The population genetics of ecological specialization in evolving Escherichia coli populations. Nature, 2000. 407(6805): p. 736–739. [DOI] [PubMed] [Google Scholar]
- 82.Leiby N and Marx CJ, Metabolic Erosion Primarily Through Mutation Accumulation, and Not Tradeoffs, Drives Limited Evolution of Substrate Specificity in Escherichia coli. PLOS Biology, 2014. 12(2): p. e1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.MacLean RC, Bell G, and Rainey PB, The evolution of a pleiotropic fitness tradeoff in Pseudomonas fluorescens. Proceedings of the National Academy of Sciences of the United States of America, 2004. 101(21): p. 8072–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duffy S, Turner PE, and Burch CL, Pleiotropic costs of niche expansion in the RNA bacteriophage Phi6. Genetics, 2006. 172: p. 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jasmin J-N and Zeyl C, Evolution of pleiotropic costs in experimental populations. Journal of Evolutionary Biology, 2013. 26(6): p. 1363–1369. [DOI] [PubMed] [Google Scholar]
- 86.Li Y, et al. , Hidden Complexity of Yeast Adaptation under Simple Evolutionary Conditions. Current Biology, 2018. 28(4): p. 515–525.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer JR, Guldelj I, and Beardmore R, Biophysical mechanisms that maintain biodiversity through trade-offs. Nature Communications, 2015. 6: p. 6278. [DOI] [PubMed] [Google Scholar]
- 88.Rodriguez-Verdugo A, et al. , Different tradeoffs result from alternate genetic adaptations to a common environment. Proceedings of the National Academy of Sciences, 2014. 111(33): p. 12121–12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bennett AF and Lenski RE, An experimental test of evolutionary trade-offs during temperature adaptation. Proceedings of the National Academy of Sciences, 2007. 104(suppl 1): p. 8649–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ostrowski EA, et al. , Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution, 2005. 59(11): p. 2343–2352. [PubMed] [Google Scholar]
- 91.Turner P and Elena S, Cost of host radiation in an RNA virus. Genetics, 2000. 156(4): p. 1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soto W, Punke EB, and Nishiguchi MK, Evolutionary perspectives in a mutualism of sepiolid squid and bioluminescent bacteria: combined usage of microbial experimental evolution and temporal population genetics. Evolution, 2012. 66(5): p. 1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luksza M and Lassig M, A predictive fitness model for influenza. Nature, 2014. 507(7490): p. 57–61. [DOI] [PubMed] [Google Scholar]
- 94.Zanini F, et al. , Population genomics of intrapatient HIV-1 evolution. eLife, 2015. 4: p. e11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lieberman TD, et al. , Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet, 2011. 43(12): p. 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horns F, et al. , Signatures of selection in the human antibody repertoire: selective sweeps, competing subclones, and neutral drift. bioRxiv, 2017: p. 145052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao S, et al. , Adaptive evolution within the gut microbiome of individual people. bioRxiv, 2018: p. 208009. [Google Scholar]
- 98.Garud NR, et al. , Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. bioRxiv, 2018: p. 210955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Minot S, et al. , Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America, 2013. 110(30): p. 12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teotónio H, et al. , Experimental Evolution with Caenorhabditis Nematodes. Genetics, 2017. 206(2): p. 691–716. [DOI] [PMC free article] [PubMed] [Google Scholar]