Abstract
The concentrations of high- and low-density lipoprotein cholesterol and triglycerides are influenced by smoking, but it is unknown whether genetic associations with lipids may be modified by smoking. We conducted a multi-ancestry genome-wide gene-smoking interaction study in 133,805 individuals with follow-up in an additional 253,467 individuals. Combined meta-analyses identified 13 novel loci, some of which were detected only because the association differed by smoking status. Additionally, we demonstrated the importance of including diverse populations, particularly in studies of interactions with lifestyle factors, where genomic and lifestyle differences by ancestry may contribute to novel findings.
Editorial summary:
A multi-ancestry genome-wide gene-smoking interaction study identifies 13 new loci associated with serum lipids.
Serum lipids, such as triglycerides and high- and low-density lipoprotein cholesterol (HDL and LDL), are influenced by both genetic and lifestyle factors. Over 250 lipid loci have been identified,1–6 yet, it is unclear to what extent lifestyle factors modify the effects of these variants, or those yet to be identified. Smoking is associated with an unfavorable lipid profile,7,8 warranting its investigation as a lifestyle factor that potentially modifies genetic associations with lipids. Identifying interactions using traditional 1 degree of freedom (1df) tests of SNP x smoking terms may have low power, except in very large sample sizes. To enhance power, a 2 degree of freedom (2df) test that jointly evaluates the interaction and main effects was developed.9
The Gene-Lifestyle Interactions Working Group, under the aegis of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium10, was formed to conduct analyses of lifestyle interactions in the genetic basis of cardiovascular traits. As both genetic and lifestyle factors differ across populations with different ancestry backgrounds, and to address the underrepresentation of non-European populations in genomic research, great effort went into creating a large, multi-ancestry resource for these investigations.11 Here, we report a genome-wide interaction study that uses both the 1df test of interaction and the 2df joint test of main and interaction effects to test the hypothesis that genetic associations of serum lipids differ by smoking status.
Results
Novel Loci
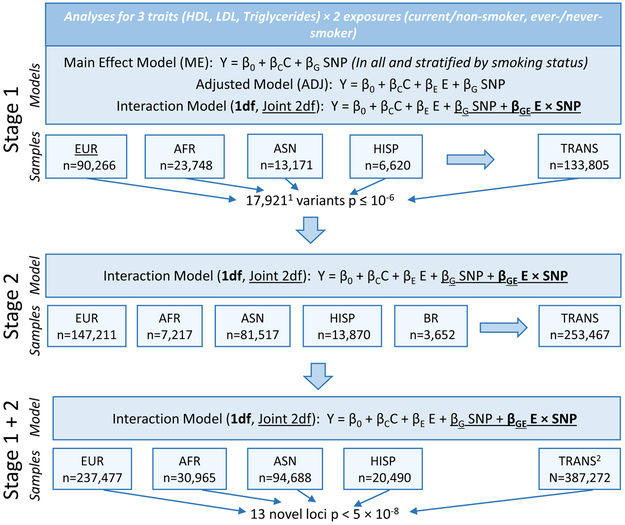
We conducted genome-wide interaction meta-analyses for current and ever-smoking status in up to 133,805 individuals of European (EUR), African (AFR), Asian (ASN) and Hispanic (HISP) ancestries (Supplementary Tables 1–3), with follow-up of 17,921 variants with p ≤ 10−6 (not pruned for linkage disequilibrium [LD]) in an additional 253,467 individuals of EUR, AFR, ASN, HISP, and Brazilian (BR) ancestries (Supplementary Tables 4–6), as described in Figure 1. Of these, 16,389 variants (487 loci, defined by +/− 1 MB) passed filters and were included in stage 2 analyses. Ninety percent of variants (14,733) and 22% of loci (109) replicated in stage 2 (variants: p <0.05/16,389, loci: p <0.05/487). We conducted meta-analyses of stage 1 and 2 results (Manhattan Plots Supplementary Figure 1; QQ Plots, Supplementary Figure 2) and identified 13 novel loci with p < 5 × 10−8 that were at least 1 MB away from previously reported lipid loci (Table 1; results by stage: Supplementary Table 7; forest plots: Supplementary Figures 3 and 4; regional association plots: Supplementary Figure 5). These loci had low false discovery rate (FDR) q-values (all q < 3 × 10−4; Supplementary Table 8). We report novel loci with p < 5 × 10−8 as well as those passing a more stringent threshold (p < 6.25 × 10−9), adjusting for 2 smoking exposures, 2 interaction tests, and ancestry-specific and trans-ancestry tests. The patterns observed in these results are described below and illustrated using output from stage 1 meta-analyses, where results from a main effect model (in all and stratified by smoking exposure) and a smoking-adjusted main effect model were also available (Figure 1; Supplementary Table 9).
Figure 1. Study Overview:
Summary of data included in this study. 116,389 variants passed filtering criteria and were included in stage 2 analyses. 2Trans-ancestry (TRANS) stage 1 and 2 combined meta-analyses were meta-analyses of stage 1 TRANS and stage 2 TRANS meta-analyses, and not meta-analyses of ancestry-specific stage 1 and stage 2 combined meta-analyses.
Table 1:
Statistically Significant (p < 5×10−8) Results in Stage 1 and 2 Meta-Analysis
| Index Variant (Nearest Gene)1 |
Bld 37 Chr:Position |
1000 Genomes Freq2 AFR/AMR/ASN/EUR |
Tested Allele: Freq |
Ancestry | Trait/ Exposure3 |
Stage 1 + 2 | Stage 1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Effect | SE | Int. Effect |
SE | 1df Int. P-value4 |
2df Joint P-value4 |
n | Adj. Main Effect P-value5 |
|||||||
| Loci with Evidence for Interaction | |||||||||||||||
| rs12740061 (LOC105378783) |
1:69407810 | 0.01/0.17/0.02/0.22 | T: 0.05 | AFR | HDL/CS | 16,606 | 0.02 | 0.0082 | −0.11 | 0.019 | 7.40E-09 | 2.4E-08 | 15,499 | 0.98 | |
| rs77810251 (PTPRZ1) |
7:121504149 | 0.02/0.22/0.34/0.11 | A: 0.04 | AFR | HDL/ES | 24,253 | 0.052 | 0.0083 | −0.06 | 0.012 | 9.50E-07 | 1.2E-9* | 23,146 | 1.60E-04 | |
| rs73453125 (CNTNAP2) |
7:146084573 | 0.09/0.02/0/0 | A: 0.07 | TRANS, AFR | LDL/CS | 40,566 | 1.9 | 0.69 | −8.3 | 1.4 | 1.70E-07 | 2.0E-08 | 24,668 | 0.76 | |
| rs56167574 (PRKAG2) |
7:151245975 | 0.13/0.01/0/0 | A: 0.12 | AFR | LDL/ES | 25,778 | 1.9 | 0.8 | −6.1 | 1.1 | 1.50E-08 | 8.4E-08 | 23,353 | 0.08 | |
| rs79950627 (MIR4686) |
11:2233790 | 0.06/0.01/0/0 | A: 0.05 | TRANS, AFR | LDL/CS | 38,272 | −0.1 | 0.79 | −8.4 | 1.6 | 1.40E-06 | 7.2E-09 | 23,348 | 0.25 | |
| rs60029395 (ZNF729) |
19:22446748 | 0.15/0.01/0.03/0 | A: 0.13 | AFR | TRIG/CS | 19,048 | 0.041 | 0.0092 | −0.097 | 0.018 | 3.30E-08 | 8.2E-08 | 15,747 | 0.17 | |
| rs7364132 (DGCR8) |
22:20096172 | 0.19/0.02/0/0 | A: 0.16 | AFR, TRANS | TRIG/ES | 23,935 | 0.012 | 0.0091 | −0.066 | 0.013 | 8.80E-07 | 2.5E-08 | 21,834 | 0.0055 | |
| Probable Main Effect Loci (No Evidence of Interaction) | |||||||||||||||
| rs12144063 (EYA3) |
1:28406047 | 0.35/0.28/0.53/0.30 | T: 0.37 | TRANS | HDL/CS, ES | 375,418 | −0.004 | 0.00069 | −0.00033 | 0.0016 | 0.75 | 1.3E-10* | 131,057 | 4.70E-07 | |
| rs10937241 (ETV5) |
3:185822774 | 0.30/0.31/0.58/0.19 | A: 0.17 | EA, TRANS | HDL/CS, ES | 230,919 | −0.008 | 0.0012 | 0.0021 | 0.0026 | 0.65 | 4.2E-12* | 90,266 | 4.50E-07 | |
| rs34311866 (TMEM175) |
4:951947 | 0.01/0.07/0.12/0.20 | C: 0.17 | TRANS, EA | HDL, TRIG/CS | 351,489 | −0.006 | 0.00097 | 0.0014 | 0.0022 | 0.61 | 1.6E-9* | 115,640 | 2.10E-06 | |
| rs73729083 (CREB3L2) |
7:137559799 | 0.11/0.04/0.02/0 | C: 0.05 | TRANS, AFR | LDL/ES, CS | 84,091 | −3.7 | 0.66 | −0.37 | 0.95 | 0.53 | 1.3E-14* | 35,909 | 2.00E-10 | |
| rs10101067 (EYA1) |
8:72407374 | 0.04/0.07/0.13/0.06 | C: 0.08 | TRANS | TRIG/CS | 317,809 | 0.014 | 0.0025 | −0.0092 | 0.0053 | 0.069 | 4.1E-08 | 102,263 | 2.10E-06 | |
| rs4758675 (B3GNT4) |
12:122691738 | 0.02/0/0/0 | C: 0.02 | AFR | TRIG/CS | 12,982 | −0.13 | 0.025 | −0.029 | 0.057 | 0.85 | 1.3E-08 | 11,875 | 3.60E-08 | |
Abbreviations: African ancestry (AFR), Current Smoking (CS), European ancestry (EUR), Ever-Smoking (ES), Trans-ancestry (TRANS), Triglycerides (TRIG).
Listed variants represent the lead associations within 1 MB region for the 2 and 1 degree of freedom tests of the variant × smoking interaction after excluding variants within 1 MB of known lipids loci. If variant is in/within 2 KB of a gene, that gene name is listed;
Frequency of the tested allele in 1000 Genomes data by ancestry: Asian (ASN), Americas (AMR), African (AFR), and European (EUR)
If the region was associated with the trait in more than one meta-analysis, the most statistically significant result is listed first and described in table;
Bolding indicates genome-wide statistical significance;
P-values in this column come from a smoking-adjusted main effect model (available in Stage 1 cohorts only, see Figure 1);
Findings with an asterisk are statistically significant using a stricter p-value threshold, after Bonferroni correction for 2 smoking traits, 2 interaction tests, and ethnic and trans-ethnic testing (p < 5 × 10−8/8=6.25 × 10−9).
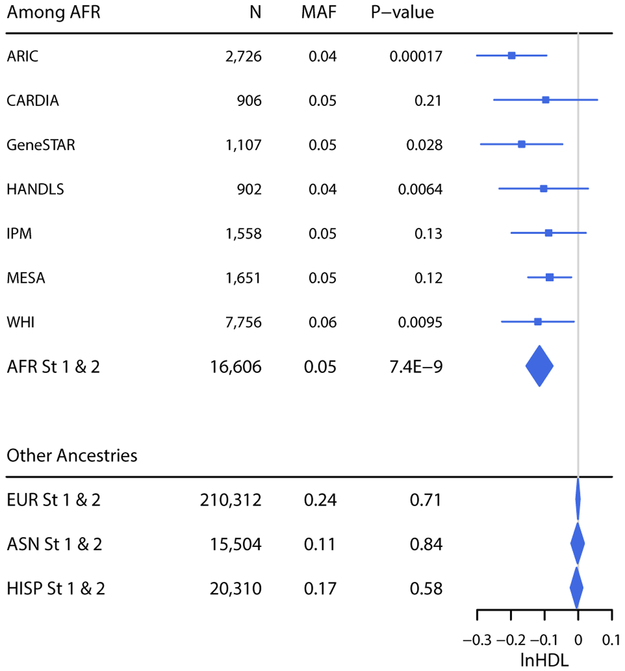
Notably, many novel loci were statistically significant only in AFR meta-analyses. For 7 of the 13 novel loci, the minor allele frequencies (MAF) of the index variants were highest in AFR, and inter-ancestry differences in MAF and/or LD may explain the failure to detect similar associations in other ancestries. However, some AFR-only associations were unlikely to be due to diminished power in non-AFR meta-analyses. For instance, the effect of rs12740061 (NC_000001.10:g.69407810C>T; LOC105378783) on HDL was significantly modified by current smoking status among AFR (p1df = 7.4 × 10−9; Figure 2, Table 1), such that the genetic effect was stronger among current smokers than non-smokers (Supplementary Table 9). In contrast, there was virtually no evidence for association in any other ancestry, despite higher MAF (Figure 2). The potential influence of under-adjustment for principal components (PCs) on these results was evaluated by excluding the 6 studies adjusting for only 1 PC (the average number of PCs among AFR studies was 4.2); effect estimates were similar and p-values were increased or similar, consistent with a ~20% reduction in sample size (Supplementary Table 10).
Figure 2.
Interaction of rs12740061 (LOC105378783) and Current Smoking (1df). A forest plot showing the betas (95% confidence intervals) and p values (1df) for the rs12740061 × Current Smoking interaction term in linear regression models of HDL adjusted for age, sex, study-specific covariates (if applicable), smoking status, and principal components. Results for each AFR study are shown, as well as the ancestry-specific combined stage 1 and 2 meta-analyses.
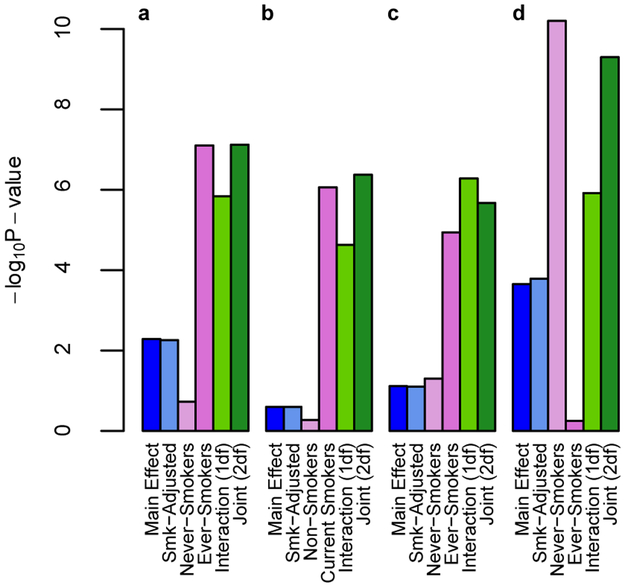
We observed interactions where notable associations were only found among current or ever-smokers, with effect sizes close to zero among non- or never-smokers, including a statistically significant association for the 2df joint test of main and interaction effects for rs7364132 (NC_000022.10:g.20096172G>A; DGCR8) × ever-smoking on triglycerides (p2df = 2.5 × 10−8; Table 1). Main effect models stratified by smoking status showed a strong genetic association with triglycerides among ever-smokers (difference in mean ln triglycerides per A allele β = −0.05, p = 7.9 × 10−8), with a negligible association among never-smokers (β = 0.01, p = 0.19; Figure 3a). This association was not significant in a non-stratified main effect model (Table 1; Supplementary Table 9), and was only detectable when modeling permitted a different association across smoking strata. Similar results were observed for rs79950627 (NC_000011.9:g.2233790G>A; MIR4686) × current smoking on LDL (Figure 3b), and rs56167574 (NC_000007.13:g.151245975G>A; PRKAG2) × ever-smoking on LDL (Figure 3c, Supplementary Table 9).
Figure 3.
Associations Observed Primarily Among One Smoking Stratum. For selected variants for which an association was primarily observed only in one smoking stratum, a comparison of the p values for stage 1 linear association models, including a main effect model adjusted for age, sex, principal components, and study-specific covariates (as appropriate) in all individuals and stratified by smoking exposure; a model additionally adjusted for smoking exposure; and a model that also includes a smoking exposure × SNP interaction term, from which a 1df test of interaction and a 2df joint test of main effect and interaction were calculated. a.) rs7364132 (DGCR8) × ever-smoking on triglycerides (n = 21,834 [11,113 never smokers; 10,725 ever-smokers]), b.) rs79950627 (MIR4686) × current smoking on LDL (n = 23,348 [18,384 non-smokers; 4,973 current smokers]), c.) rs56167574 (PRKAG2) × ever smoking on LDL (n = 23,353 [11,700 never smokers; 11,649 ever-smokers]), and d.) rs77810251 (PTPRZ1) × ever smoking on HDL (n = 23,146 [11,560 never smokers; 11,592 ever-smokers]).
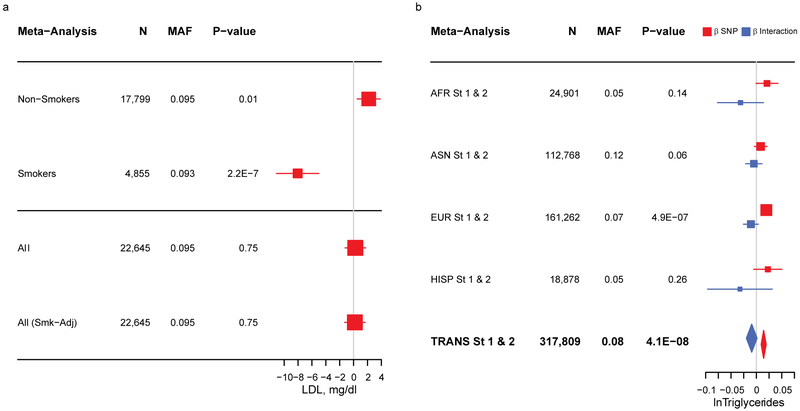
We also observed interactions where the association was in opposite directions in the exposed vs. unexposed stratum, with a larger, more statistically significant association among smokers. For instance, current smoking modified the association between rs73453125 (NC_000007.13:g.146084573G>A; CNTNAP2) and LDL (Table 1). In stratified main effect models, the A allele was associated with lower LDL among current smokers (β = −8.1 mg/dL, p = 2.2 × 10−7), but higher LDL among non-smokers (β = 2.18 mg/dL, p = 0.01; Figure 4a, Supplementary Table 9). In a non-stratified smoking-adjusted main effects model, no association between rs73453125 and LDL was detected (β = 0.3 mg/dL, p = 0.98). Similar results were observed for rs12740061 (LOC105378783) (Supplementary Table 9).
Figure 4.
Forest Plots of Selected Associations. (a.) Plot showing the association between rs73453125 and LDL among AFR in stage 1 (where a series of models were available). Variant betas (95% confidence intervals) and p values are drawn from main effect linear regression models of Non-Smokers, Smokers, all individuals, and all individuals with adjustment for smoking status. (b.) Plot showing the association between rs10101067 (EYA1) and triglycerides in ancestry-specific and combined analysis from stages 1 and 2. Variant main and interaction betas (95% confidence intervals) are drawn from linear regression models that include a current smoking × SNP term and p values are for the 2df joint test of main effect and interaction.
Although many interactions manifested as associations significant only, or more strongly, in smokers, for rs10937241 (NC_000003.11:g.185822774A>G; ETV5), rs34311866 (NC_000004.11:g.951947T>C; TMEM175), rs10101067 (NC_000008.10:g.72407374G>C; EYA1), and rs77810251 (NC_000007.13:g.121504149G>A; PTPRZ1), the associations observed among non- or never-smokers were more statistically significant. Notably, in stratified main effect models, rs77810251 was associated with increased HDL among never-smokers (β = 0.05 lnHDL, p = 6.3 × 10−11) with no significant association among ever-smokers (β = −0.005 lnHDL, p = 0.56; Figure 3d; Supplementary Table 9). In a smoking-adjusted main effect model of never- and ever-smokers together, the association was markedly reduced (β = 0.02 lnHDL, p = 1.6 × 10−4).
The 2df joint test simultaneously evaluates main and smoking interaction effects; some of our results appear to capture a main effect of the variant. For instance, the 2df test for rs12144063 (EYA3) detected an association (p = 1.3 × 10−10), while the 1df test of interaction does not (p = 0.75). The minor alleles for this and three other variants (rs10937241 [ETV5], rs34311866 [TMEM175], and rs10101067 [EYA1]) were common across populations, and their effects were small in magnitude and yet reached genome-wide statistical significance (rs10101067 [EYA1]; Figure 4b), consistent with expectations for novel main effect loci in well-studied populations. There are two findings, however, for which the relatively large sample size in the AFR meta-analyses appeared to facilitate detection. The MAF for rs73729083 (NC_000007.13:g.137559799T>C; CREB3L2) was much greater among AFR than in HISP and ASN (not present among EUR), and the variant effect estimates were large and consistent across ancestries, while the interaction effect estimates were inconsistent, with wide confidence intervals (Supplementary Figure 3f). The minor allele for rs4758675 (NC_000012.11:g.122691738C>A; B3GNT4) was only present in AFR (Supplementary Figure 3k), but variant effect estimates were consistent across AFR studies, with interaction effect estimates approaching the null (Supplementary Figure 4e). In total, 6 of the 13 novel loci that we identified appear to be driven by main effects of the variant while the remainder show some evidence of interaction.
There were 16 additional novel loci identified in stage 1 meta-analyses (p1df or p2df < 5 × 10−8) for which the variants were unavailable for analysis in stage 2 cohorts. These loci were identified only in AFR meta-analyses (many were AFR-specific variants; Table 2). Due to the relatively small number and size of available AFR cohorts in stage 2 (total n = 7,217; n < 2,000 per cohort), these relatively low frequency variants did not pass filters for minor allele count within exposure groups. Nevertheless, these associations had low FDR q-values (all q < 2.4 × 10−4) in stage 1, and some appear worthy of further investigation. One particularly interesting candidate is rs17150980 (NC_000007.13:g.78173734T>C; MAGI2) × ever-smoking on triglycerides (p2df = 1.4 × 10−9), for which consistent effects for both the variant and the interaction were observed across AFR studies, but not in other ancestries (Supplementary Figure 6).
Table 2:
Statistically Significant (p < 5×10−8) Results in Stage 1 Meta-Analysis Unavailable in Stage 21
| Index Variant (Nearest Gene)2 |
Bld 37 Chr:Position |
1000 Genomes Freq3 AFR/AMR/ASN/EUR |
Tested Allele: Freq |
Ancestry | Trait/ Exposure |
Stage 1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Effect | SE | Int. Effect |
SE | 1df Interaction P-value3 |
2df Joint P-value4 |
Adj. Main Effect P-value5 |
||||||
| rs140602625 (EXOC6B) |
2:72849325 | 0.01/0/0/0 | C: 0.02 | AFR | LDL/CS | 7,755 | −3.4 | 3.1 | −35 | 7.1 | 1.0E-6 | 1.5E-8 | 0.018 |
| rs114138886 (LOC107985905) |
2:84428024 | 0.02/0/0/0 | T: 0.02 | AFR | LDL/CS | 7,755 | 2.4 | 2.9 | −29 | 5.4 | 9.3E-8 | 4.4E-8 | 0.47 |
| rs149776574 (REEP1) |
2:86472455 | 0.01/0.08/0/0.06 | G: 0.02 | AFR | TRIG/CS | 7,756 | −0.048 | 0.033 | 0.40 | 0.069 | 4.2E-10* | 5.1E-10* | 0.88 |
| rs143396479 (LOC105374426/TMEM33) |
4:41911366 | 0.02/0/0/0 | A: 0.01 | AFR | LDL/ES | 10,912 | −16.0 | 2.6 | 15 | 4.5 | 0.022 | 6.8E-9 | 0.0094 |
| rs148187465 (MARCH1) |
4:164639694 | 0.01/0/0/0 | C: 0.01 | AFR | LDL/CS | 7,755 | −2.1 | 3.0 | −32 | 6.2 | 3.7E-7 | 4.9E-9* | 0.032 |
| rs76687692 (G3BP1) |
5:151189283 | 0.03/0/0/0 | A: 0.01 | AFR | LDL/CS | 9,418 | 2.7 | 3.2 | 25 | 5.5 | 0.0013 | 4.8E-9* | 0.0016 |
| rs73339842 (LINC01938) |
5:164967406 | 0.02/0.01/0/0 | G: 0.02 | AFR | TRIG/CS | 7,756 | 0.046 | 0.033 | −0.41 | 0.071 | 8.5E-9 | 3.3E-8 | 0.96 |
| rs115580718 (BMP6) |
6:7880037 | 0.02/0/0/0 | G: 0.01 | AFR | TRIG/CS | 7,756 | −0.12 | 0.036 | −0.29 | 0.082 | 0.00045 | 1.2E-9* | 1.6E-6 |
| rs17150980 (MAGI2) |
7:78173734 | 0/0.12/0.45/0.01 | C: 0.03 | AFR | TRIG/ES | 12,972 | −0.17 | 0.028 | 0.24 | 0.044 | 7.5E-8 | 1.4E-9* | 0.085 |
| rs116592443 (LYZL2) |
10:30884890 | 0.02/0/0/0 | A: 0.01 | AFR | TRIG/CS | 7,756 | 0.073 | 0.038 | −0.46 | 0.081 | 1.8E-8 | 1.2E-7 | 0.76 |
| rs115628664 (UNC5B) |
10:72899880 | 0.03/0/0/0 | G: 0.01 | AFR | TRIG/CS | 7,756 | 0.027 | 0.040 | −0.39 | 0.071 | 4.7E-8 | 6.7E-9* | 0.44 |
| rs183911507 (TP53I11) |
11:44978366 | 0.01/0/0/0 | G: 0.02 | AFR | TRIG/CS | 10,287 | −0.043 | 0.029 | 0.33 | 0.059 | 1.7E-8 | 6.5E-8 | 0.82 |
| rs199771018 (STOML3) |
13:39507838 | 0.02/0/0/0 | T: 0.02 | AFR | HDL/CS | 7,756 | −0.019 | 0.019 | 0.23 | 0.037 | 1.2E-9* | 6.3E-10* | 0.55 |
| rs190976513 (LOC105370255) |
13:71114207 | 0.02/0.01/0/0 | A: 0.02 | AFR | LDL/CS | 10,234 | −5.1 | 2.6 | −20 | 5.2 | 9.3E-5 | 3.2E-8 | 1.1E-4 |
| rs182600360 (LOC105370531) |
14:63607120 | 0.02/0/0/0 | A: 0.02 | AFR | LDL/CS | 7,755 | 6.6 | 3.3 | −39 | 7.1 | 4.4E-8 | 3.3E-7 | 0.56 |
| rs62064821 (CCT6B) |
17:33280904 | 0.01/0.04/0/0.06 | T: 0.01 | AFR | LDL/CS | 10,234 | 8.5 | 3.3 | −30 | 5.5 | 3.1E-8 | 6.0E-7 | 0.17 |
Abbreviations: African ancestry (AFR), Current Smoking (CS), Ever-Smoking (ES), Triglycerides (TRIG).
All loci have some evidence for interaction (p<0.05 for 1df test of interaction); thus, results not categorized into “Loci with Evidence for Interaction” or “Probable Main Effects (without evidence for interaction)”;
Listed variants represent the lead associations within 1 MB region for the 2 and 1 degree of freedom tests of the variant × smoking interaction after excluding variants within 1 MB of known lipids loci. If variant is in/within 2 KB of a gene, that gene name is listed;
Frequency of the tested allele in 1000 Genomes data by ancestry: Asian (ASN), Americas (AMR), African (AFR), and European (EUR);
Bolding indicates genome-wide statistical significance;
P-values in this column come from a smoking-adjusted main effect model (available in Stage 1 cohorts only, see Figure 1).
Findings with an asterisk indicate statistical significance using a stricter p-value threshold, after Bonferroni correction for 2 smoking traits, 2 interaction tests, and ethnic and trans-ethnic testing (5 × 10−8/8 = 6.25 × 10−9).
As we ran analyses for both current and ever-smoking status, we evaluated novel associations across smoking exposures to further characterize those loci (Supplementary Table 11). For the 6 probable main effect loci (EYA3, ETV5, TMEM175, CREB3L2, EYA1, B3GNT4), an association of similar statistical significance was observed across smoking status definitions for the 2df joint test, with similar lack of effect for the 1df test of the interaction, consistent with the interpretation that smoking status was unimportant, with the main effect driving the association. For the locus in which a stronger association was observed among non-smokers (PTPRZ1), the 1df interaction p value was dramatically reduced (from 9.5 × 10−7 for ever-smoking to 0.011 for current smoking), consistent with any smoke exposure altering the association between this variant and HDL, and including former smokers with the never smokers (as in the current smoking analysis) diluting the observed association among never smokers. For the reported interactions with current smoking, all the effect estimates were greatly reduced in the ever-smoking analysis, suggesting that active smoking is the relevant exposure. For the reported interactions with ever-smoking, markedly reduced statistical significance was observed in the current smoking analysis, likely reflecting a drop in power from excluding former smokers from the exposed group.
We conducted a secondary analysis of smoking dose in two of our AFR cohorts with measured cigarettes per day for four interaction loci (see methods for selection criteria): rs12740061 (LOC105378783), rs73453125 (CNTNAP2), rs79950627 (MIR4686), and rs7364132 (DGCR8). For each of these variants, a stronger association was observed with increasing smoking dose (Supplementary Table 12), and the interaction was statistically significant for all variants but rs7364132, which was just over our threshold for statistical significance (p = 0.0035 vs. p < 0.0021).
Conditional analysis showed no evidence that the novel associations were driven by variants at known lipids loci (Supplementary Table 13). Imputation quality for novel variants was high (minimum 0.75), with sample-size weighted average imputation quality of 0.90 and minor allele frequencies that match publicly-available datasets (Supplementary Table 14).
Interactions at Known Loci
We examined interactions at known lipid loci. Since results for the 2df test at known lipid loci are expected to predominantly reflect previously identified main effects, we exclusively evaluated the 1df test of interaction. No interactions within known loci were statistically significant (p1df < 0.05/269 known loci in our data). To evaluate whether the proportion of known variants with p1df < 0.05 was higher than would be expected by chance (5%), we conducted binomial tests for each trait-exposure combination (p-values Bonferroni-corrected for multiple tests). There was significant enrichment of known variants with 1df interaction p < 0.05: HDL-current smoking p = 9.6 × 10−12, HDL-ever smoking p = 5.9 × 10−7, LDL-current smoking p = 8.4 × 10−15, LDL-ever smoking p = 3.1 × 10−5, triglycerides-current smoking p = 4.0 × 10−3, triglycerides-ever smoking p = 3.1 × 10−4. We conducted power calculations under different interaction scenarios to determine the conditions under which an interaction analysis and a main effect analysis would both be sufficiently powered to detect the same locus (i.e. when an interaction could be detected in a locus previously identified in a main effect analysis; Supplementary Table 15). At current trans-ancestry meta-analyses sample sizes and assuming a large effect size, there was limited power to detect either a main effect or an interaction when an association was larger or only present among smokers (main effect <1%; interaction 77%), or when associations differed in magnitude but not direction (main effect >99%; interaction <1%); thus, making it unlikely to detect an interaction at a known locus. We were well-powered for both interaction and main effect analyses to detect smoking interactions for which smoking eliminates or drastically reduces a large association among non- or never-smokers. We identified one such interaction in our data, for PTPRZ1 in AFR only, which may not have been previously identified in a main effect analysis because of limited power of AFR main effect analyses thus far.
Proportion Variance Explained by Identified Loci
Ten studies from four ancestries were used to calculate the proportion of the variance in lipid traits explained by the genome-wide statistically significant novel loci: 13 loci from stage 1 and 2 combined meta-analyses (Table 1), and 16 loci from stage 1 that were not available in stage 2 analyses (Table 2). Two different methods were used (Online Methods), and the range of findings across these methods are presented (Supplementary Table 16). In AFR, novel variants and their interactions explained 1.0–2.7% of HDL, 0.7–2.6% of LDL, and 1.3–3.2% of triglycerides. The proportion explained was smaller among EUR (0.06–0.14% of HDL, 0.01–0.07% of LDL, and 0.10–0.19% of triglycerides), ASN (0.27–0.86% of HDL, 0.09–0.82% of LDL, and 0.8–1.5% of triglycerides), and HISP (0.2–0.4% of HDL, 0.2–0.5% of LDL, and 0.2–0.4% of triglycerides). These results should be considered in the context of the inter-ancestry MAF differences: the proportion of novel variants that could be evaluated varied by ancestry, with 94–97% among the AFR cohorts, but only 32–39% among the EUR and ASN cohorts, and 55% in the HISP cohort. In contrast, each of the cohorts investigated had similar proportions of the requested known variants (83–96%).
Reproducing Known Lipids Associations
We evaluated the degree to which our data reproduce previously reported lipid loci. Given that approximately 81% of cohorts in stage 1 were included both in this and in previous efforts, this analysis is not a formal replication. For comparability with traditional GWAS, we evaluated results from stage 1 main effect models. Of the 356 previously reported associations for 279 variants (compiled from1–6,12), there were 236 associations for 189 variants that were confirmed in our data (consistent direction and p < 0.05/356), for a 66.3% concordance rate (Supplementary Table 17).
Bioinformatics
To characterize the potential impact of our novel associations for chronic disease risk and to investigate biological mechanisms, we conducted a series of follow-up analyses and annotations. We performed extensive bioinformatics annotation on variants within the 29 novel loci (Tables 1 and 2). These loci included 78 associated variants that were in or near 33 unique genes (Supplementary Table 18). We conducted look-up of these variants in previously conducted GWAS for related traits (Supplementary Tables 19–24), the Genotype-Tissue Expression (GTEx v7.0) portal and Regulome DB (Supplementary Table 25), HaploReg v4.1 (Supplementary Table 26), and an analysis of cis- and trans- expression quantitative trait loci (eQTL) in whole blood from Framingham Heart Study participants (Supplementary Table 27). Additionally, for each trait we performed DEPICT gene prioritization (Supplementary Tables 28–30), gene set enrichment (Supplementary Tables 31–33), and tissue or cell type enrichment analyses13 (Supplementary Tables 34–37), using both novel and known loci. Notable findings from these follow-up analyses are summarized below by locus.
Consistent with our observations of an association of the C allele for rs10101067 (EYA1) with higher triglycerides, this allele was associated with increased risk of coronary artery disease (β = 0.036, p= 0.03; Supplementary Table 19), ischemic stroke (β = 0.11, p= 0.04; Supplementary Table 20), and higher waist to hip ratio adjusted for BMI (β = 0.029 units, p= 6.5 × 10−4, with similar results observed for waist circumference adjusted for BMI; Supplementary Table 21).
We found an association of the T allele of rs12144063 (NC_000001.10:g.28406047G>T; EYA3) with lower HDL. This allele was associated with increased risk of all stroke types (β = 0.05, p = 0.04), as well as stroke subtypes (Supplementary Table 20). rs7529792 (NC_000001.10:g.28306250C>T), a variant in LD with rs12144063 (r2 = 0.97) regulates gene expression of EYA3 and has a high Regulome DB score (1b; Supplementary Table 25). Haploreg also shows regulatory features for rs12144063, including being in a promoter location expressed in liver and brain, in enhancer histone marks, and at DNAse marks for EYA3 (Supplementary Table 26). DEPICT predicted a role for these variants in regulating EYA3 and XKR8 (Supplementary Table 28), which encodes a phospholipid scramblase important in apoptotic signaling14.
We report an interaction between smoking and rs77810251 (PTPRZ1) with the minor allele associated with higher HDL only among never-smokers. While this variant was not available in look-up data for GIANT, a variant in this locus with a similar association, rs740965 (NC_000007.13:g.121513561T>G), was associated with lower BMI among EUR (β = −0.01 kg/m2, p= 0.01, similar results for trans-ancestry analysis). This variant was also associated with lower waist circumference adjusted for BMI among EUR women (β = −0.016, p = 0.04; Supplementary Table 21). PTPRZ1 was shown to be downregulated in cells treated with an acute dose of nicotine15, which supports our observation of a lack of an association of PTPRZ1 variants among ever-smokers.
We report a main effect of rs34311866 on HDL and triglycerides. rs34311866 is a missense variant in TMEM175, which has been associated with Parkinson’s disease16 and type 2 diabetes17. This variant contributes to the regulation of DGKQ (p = 5.3 × 10−21) and is an eQTL of DGKQ in adipose, artery, lung, nerve and thyroid tissue (Supplementary Table 25). The expression of DGKQ is more strongly regulated by another significantly associated variant in this locus, rs4690220 (NC_000004.11:g.980464A>G), which is located upstream of IDUA and in an intron of SLC26A1. This variant had a high score in the RegulomeDB (1f), supporting a potential functional effect (Supplementary Table 25). Importantly, DGKQ has been implicated in studies of cholesterol metabolism18, bile acid signaling, glucose homoeostasis in hepatocytes19, primary biliary cirrhosis20, and Parkinson’s disease21–24. DGKQ interacts with the key lipid enzymes LPL, LIPG, and PNPLA3 (Supplementary Figure 7). These results suggest that the observed association with HDL and triglycerides could act on cholesterol metabolism through regulation of DGKQ. Also, rs34311866 is a trans-eQTL for GNPDA1 (Supplementary Table 27); expression of this gene has been associated with a set of traits, including hyperlipidemia25.
In our data, there was a significant rs12740061 (LOC105378783) × smoking interaction, such that the minor allele was associated with decreased HDL only among current smokers. This variant is a trans-eQTL for TAS1R1 (Supplementary Table 27). Variants in this gene have been found to influence taste receptors, notably affecting cigarette smoking habits26.
Discussion
In this study, we evaluated gene-smoking interactions in large, multi-ancestry, meta-analyses of serum lipids, using varying associations among smoking subgroups to improve the ability to detect novel lipid loci. We report 13 novel loci for serum lipids from stage 1 and 2 meta-analyses. Sixteen additional statistically significant novel loci were found in stage 1 but were unavailable in stage 2. All 29 novel associations had a low q-value (p < 3 × 10−4). Using both the 1df test of interaction and the 2df joint test of main and interaction effects in this study allowed us to improve our inferences based on the results: the 2df test bolstered the power to detect interactions, while the 1df test could discriminate between associations that predominantly reflected main effects vs. interactions.
Our results provide support for future efforts to evaluate lifestyle interactions with complex traits. We identified loci for which an association with serum lipids was only observed in one smoking stratum. In main effect models at these loci, the signal from one subgroup was not detected when all individuals were evaluated together (regardless of adjusting for smoking). These loci could only be observed by an analysis that was either smoking-stratified or contained an interaction term, highlighting the importance of considering potential effect modification in association studies. Additionally, through use of the joint 2df test, we identified six loci that appear to show novel main effects. Consistent with this characterization, five of these loci were within 500 KB of variants identified in recent large-scale association studies using main effect models: ETV27–29, TMEM17528, EYA128, EYA328, and B3GNT428.
With 23,753 AFR individuals in the Stage 1 analyses and 30,970 AFR individuals overall, this work represents one of the largest studies of serum lipids in AFR. It is therefore unsurprising that two of our novel lipid loci (CREB3L2 and B3GNT4) appear to be driven primarily by genetic main effects. Importantly, these associations could not have been detected in EUR, as the tested allele for both rs4758675 (B3GNT4) and rs73729083 (CREB3L2) are absent in EUR.
In addition to these probable main effect loci, the prominence of novel loci that were statistically significant only in AFR meta-analyses deserves further discussion. Some findings could not be effectively evaluated in other ancestry groups because of inter-ancestry MAF differences: the minor alleles for half of the variants were much more frequent in AFR. More puzzling, however, is the discovery of loci with evidence of strong interactions in AFR but not in meta-analyses in other ancestries, despite comparable or higher allele frequencies, such as were observed with rs12740061 (LOC105378783; Figure 2) or rs17150980 (MAGI2; Supplementary Figure 6). This phenomenon suggests inter-ancestry differences in either genomic or environmental context. There are variants in LD (r2 > 0.2) among AFR for rs12740061 (LOC105378783) and rs17150980 (MAGI2) that are not in LD with these variants in other ancestries30, but these variants were directly tested in our study with no evidence of an association in non-AFR analyses. Thus, it is unlikely that inter-ancestry LD differences explain these results, although unmeasured causal variants are a possibility. Inter-ancestry differences in smoking are also a potential explanation. In addition to known differences in smoking patterns31, there are pronounced ancestry differences in preferred cigarette type, with over 85% of AFR smokers using menthol cigarettes compared to 29% of EUR smokers (in the US)32. Menthol cigarettes are thought to facilitate greater absorption of harmful chemicals because of deeper inhalation31,33 through desensitization of nicotinic acetylcholine receptors that cause nicotine-induced irritation34. Evidence for an excess risk of cardiovascular disease associated with mentholated cigarettes, however, is equivocal35–39. Ancestry differences in smoking-related metabolites and carcinogens have been reported40–43, and differential metabolism of key compounds may underlie observed differences by ancestry. Some behaviors/conditions that co-occur with smoking may also differ by ancestry, and this additional factor may modify the observed genetic associations with serum lipids.
The biological mechanisms through which smoking influences the observed genetic associations will require further investigation, as the myriad components of cigarette smoke and their downstream consequences (including oxidative stress and inflammation) affect pathways throughout the body44. However, there is evidence for differential expression of PTPRZ115, LPL15 and LDLR45 in cells exposed to an acute dose of nicotine. Also, concentrations of CETP46, ApoB47, and LPL48 are associated with smoking status.
The sample size attained for diverse ancestries is a key strength of our study, particularly among AFR. As a result, we were able to identify loci that had not been previously detected in meta-analyses of ancestries that are better represented in genomic research. Additionally, our use of nested models in our stage 1 analyses allowed us to more fully characterize loci. Despite these strengths, however, a smaller number of AFR studies were available for stage 2, resulting in an inability to follow up on some of our stage 1 low frequency findings.
In conclusion, this large, multi-ancestry genome-wide study of gene-smoking interactions on serum lipids identified 13 novel loci based on combined analysis of stages 1 and 2, and an additional 16 novel loci based on stage 1 that were unavailable in stage 2. Some loci were detected only in analyses stratified by smoking status or with a smoking interaction term, thus motivating further study of gene × environment interactions with other lifestyle factors to identify new loci for lipids and other complex traits. We demonstrate the importance of including diverse populations, reaching a sufficient sample size in these analyses for discovery of novel main effect lipid loci for AFR. Careful consideration of ancestry may be of particular importance for gene × environment interactions, as ancestry may be a proxy for both genomic and environmental context.
URLs
1000 Genomes Project: http://www.internationalgenome.org/
dbGaP: https://www.ncbi.nlm.nih.gov/gap
dbSNP: http://ncbi.nlm.nih.gov/snp/
DEPICT: http://data.broadinstitute.org/mpg/depict/
EasyQC: http://www.genepi-regensburg.de/easyqc
EasyStrata: http://www.genepi-regensburg.de/easystrata
ENCODE: https://www.encodeproject.org/
forestplot: http://cran.r-project.org/web/packages/forestplot/
GCTA: http://cnsgenomics.com/software/gcta
geepack: http://cran.r-project.org/web/packages/geepack/
GenABEL: https://github.com/cran/GenABEL
Gene Ontology: http://www.geneontology.org/
GTEx: https://gtexportal.org/home/
HaploReg: http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php
KEGG: http://www.genome.jp/kegg/
LocusZoom: http://locuszoom.sph.umich.edu/
METAL: http://genome.sph.umich.edu/wiki/METAL
NCBI Entrez gene: ncbi.nlm.nih.gov/gene/
ProbABEL: https://github.com/GenABEL-Project/ProbABEL
Reactome: http://bioconductor.org/packages/release/data/annotation/html/reactome.db.html
RegulomeDB: http://www.regulomedb.org/
Roadmap Epignomics: http://www.roadmapepigenomics.org/
sandwich: http://cran.r-project.org/web/packages/sandwich/index.html
STRING database: http://string-db.org/
Online Methods
Details regarding motivation and methodology of this and other projects of the CHARGE Gene-Lifestyle Interactions Working Group are available in our recently published methods paper11, and detailed information on study design can be found in the Life Sciences Reporting Summary.
Participants
Analyses included men and women between 18 and 80 years of age of European (EUR), African (AFR), Asian (ASN), Hispanic (HISP), and (in stage 2 only) Brazilian (BR) ancestry. Participating studies are described in Supplementary Materials, with further details of sample sizes, trait distribution, and data preparation available in Supplementary Tables 1–6. Considerable effort was expended to engage as many studies of diverse ancestry as possible. This work was approved by the Washington University in St. Louis Institutional Review Board and complies with all relevant ethical regulations. Each study obtained informed consent from participants and received approval from the appropriate institutional review boards.
Phenotypes
Analyses evaluated the concentrations of high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), and triglycerides. LDL could be either directly assayed or derived using the Friedewald equation (if triglycerides ≤ 400 mg/dL and individuals were fasting for at least 8 hours). Lipid-lowering drug use was defined as any use of a statin drug or any unspecified lipid-lowering drug after 1994 (when statin use became common). If LDL was directly assayed, adjustment for lipid-lowering drug was performed by dividing the LDL value by 0.7. If LDL was derived using the Friedewald equation, total cholesterol was first adjusted for lipid-lowering drug use (total cholesterol/0.8) before calculation of LDL by the Friedewald equation. No adjustments were made for any other lipid medication, nor were adjustments made to HDL or triglycerides for medication use. If samples were from individuals who were non-fasting (fasting ≤ 8 hours), then neither triglycerides nor calculated LDL were used. Both HDL and triglycerides were natural log-transformed, while LDL remained untransformed. In the event that multiple measurements of lipids were available (i.e. in a longitudinal study), analysts selected the visit for which data were available for the largest number of participants, and the measurement from that visit was included in analyses.
Environmental Exposure Status
Smoking variables evaluated were current smoking status (yes/no) and ever smoking status (yes/no). Current smokers were included in the exposed group for both of these variables, and never smokers were included in the unexposed group for both of these variables. Former smokers were included in the unexposed group for the current smoking variable and the exposed group for the ever-smoking variable. Smoking variables were coded as 0/1 for unexposed/exposed groups.
Genotype Data
Genotyping was performed by each participating study using genotyping arrays from either Illumina (San Diego, CA, USA) or Affymetrix (Santa Clara, CA, USA). Each study conducted imputation using various software. The cosmopolitan reference panel from the 1000 Genomes Project Phase I Integrated Release Version 3 Haplotypes (2010–11 data freeze, 2012–03-14 haplotypes) was specified for imputation and used by most studies, with some using the HapMap Phase II reference panel instead. Only variants on the autosome and with MAF of at least 0.01 were considered. Specific details of each participating study’s genotyping platform and imputation software are described (Supplementary Tables 3 and 6). Genotype was coded as the dosage of the imputed genetic variant, coded additively (0,1,2).
Stage 1 Analysis
Stage 1 genome-wide interaction analyses included 29 cohorts contributing data from 51 study/ancestry groups and up to 133,805 individuals of EUR, AFR, ASN, and HISP ancestry (Supplementary Tables 1–3). All cohorts ran three models in all individuals: a main effect model, a model adjusted for smoking, and an interaction model that included a multiplicative interaction term between the variant and smoking status (Figure 1). Additionally, the main effect model was run stratified by smoking exposure. All models were run for 3 lipids traits (HDL, LDL, and triglycerides) and 2 smoking exposures (current smoking and ever smoking). Thus, each study/ancestry group completed 30 GWAS (using 5 models × 3 traits × 2 exposures).
All models were adjusted for age, sex, and field center (as appropriate). Principal components derived using genotyped SNPs were included based on the study analyst’s discretion. All AFR cohorts were requested to include at least the first principal component, and 71% of AFR cohorts used multiple PCs (with 25% using 10 PCs). The average number of PCs used was 4.2. Additional cohort-specific covariates could be included if necessary to control for other potential confounding factors. Studies including participants from multiple ancestry groups conducted and reported analyses separately by ancestry. Participating studies provided the estimated genetic main effect and robust estimates of standard error for all requested models. In addition, for the models with an interaction term, studies also reported the interaction effects and robust estimates of their standard errors, and a robust estimate of the corresponding covariance matrix between the main and interaction effects. To obtain robust estimates of covariance matrices and robust standard errors, studies with only unrelated participants used R packages; either sandwich or ProbABEL. If the study included related individuals, either generalized estimating equations (R package geepack) or linear mixed models (GenABEL, MMAP, or R) were used. Sample code provided to studies to generate these data has been previously published (see Supplementary Materials 11).
Extensive quality control (QC) was performed using EasyQC49 on study-level (examining the results of each study individually), and then on ancestry-level (examining all studies within each ancestry group together). Study-level QC consisted of exclusion of all variants with MAF < 0.01, extensive harmonization of alleles, and comparison of allele frequencies with ancestry-appropriate 1000 Genomes reference data. Ancestry-level QC included the compilation of summary statistics on all effect estimates, standard errors and p-values across studies to identify potential outliers, and production of SE-N and QQ plots to identify analytical problems (such as improper trait transformations)50. Variants were excluded from ancestry-specific meta-analyses for an imputation score < 0.5; the same threshold was implemented regardless of imputation software, as imputation quality measures are shown to be similar across software51. Additionally, variants were excluded if the minimum of the minor allele count in the exposed or unexposed groups × imputation score was less than 20. To be included in meta-analyses, each variant had to be available from at least 3 studies or 5,000 individuals contributing data.
Meta-analyses were conducted for all models using the inverse variance-weighted fixed effects method as implemented in METAL. We evaluated both a 1 degree of freedom test of interaction effect (1df) and a 2 degree of freedom joint test of main and interaction effects (2df), following previously published methods9. A 1df Wald test was used to evaluate the 1df interaction, as well as the main effect and the smoking-adjusted main effect in models without an interaction term. A 2df Wald test was used to jointly test the effects of both the variant and the variant x smoking interaction52. Meta-analyses were conducted within each ancestry separately, and then trans-ancestry meta-analyses were conducted on all ancestry-specific meta-analyses. Genomic control correction was applied before all meta-analyses.
Variants that were associated in any analysis at p ≤ 10−6 were carried forward for analysis in Stage 2. A total of 17,921 variants from 519 loci (defined by physical distance +/− 1 MB) were selected for Stage 2 analyses.
Stage 2 Analysis
Variants selected for Stage 2 were evaluated in 50 cohorts, with data from 75 separate ancestry/study groups totaling up to 253,467 individuals (Supplementary Tables 4–6). In addition to the 4 ancestry groups listed above, stage 2 analyses also included studies of Brazilian (BR) individuals. BR were considered only in the trans-ancestry meta-analyses, since there were no stage 1 BR results for meta-analysis. In stage 2, variants were evaluated only in a model with the interaction term (Figure 1).
Study- and ancestry-level QC was carried out as in stage 1. In contrast to stage 1, no additional filters were included for the number of studies or individuals contributing data to stage 2 meta-analyses, as these filters were implemented to reduce the probability of false positives, and were less relevant in stage 2. Stage 2 variants were evaluated in all ancestry groups and for all traits, no matter what specific meta-analysis met the p-value threshold in the stage 1 analysis. Genomic control was not applied to stage 2 meta-analyses, given the expectation of association. To ensure quality of analyses, all quality control and meta-analyses of replication data were completed independently by analysts at two different institutions (ARB and JLB [NIH], EL, XD, and CTL [Boston University]), with differences resolved through consultation.
Meta-Analyses of Stages 1 and 2
Given the increased power of combined meta-analysis of stage 1 and 2 results compared with a discovery and replication strategy53, combined stage 1 and 2 meta-analyses were carried out for all the selected variants . We report variants significant at 5 × 10−8 as well as those significant at Bonferroni correction for 2 smoking traits, 2 interaction tests, and ancestry-specific and trans-ancestry testing, with p-value of 6.25 × 10−9 (5 × 10−8/8). Loci that are significant at the stricter p-value are identified in main tables. Loci were defined based on physical distance (+/− 1 MB) and are described by the index variant (i.e. the most statistically significant variant within each locus). Novelty was determined by physical distance (+/− 1 MB) from known lipids loci compiled from large meta-analyses1–5,12. False Discovery Rate q values were determined using EasyStrata to implement the Benjamini-Hochberg method of calculation. Results were visualized using R 3.1.0, including the package ‘forestplot’ (Supplementary Figures 3 and 4), and LocusZoom v1.4 (Supplementary Figure 5) for regional association plots.
Smoking Dose Analysis
To further characterize these associations, we evaluated an interaction between smoking dose and a few of the observed novel loci. While smoking dose data was not available for many of the included studies, we conducted secondary analysis on smoking dose interaction in a subset of loci in our two largest AFR studies: WHI-SHARE and ARIC. We identified 4 loci from our main results (LOC105378783, CNTNAP2, MIR4686, DGCR8) for follow-up based on the following criteria: an interaction locus (as opposed to a probable main effect), stronger association observed among smokers compared to non-/never-smokers, the presence of contributing cohort(s) with smoking dose variables available and with p < 0.05 for reported result (to ensure sufficient power for analysis). We investigated these 4 loci using 3 methods of characterizing cigarettes per day: a quantitative variable, a categorical variable based on meaningful dose levels (less than a half a pack, between a half a pack and a pack, and more than a pack per day), and binary variable defined by the median of cigarettes per day in that cohort. Dose variables were defined separately by smoking status, such that cigarettes per day for former smokers were set to 0 for variables defined for current smokers, while the cigarettes per day for both current and former smokers were quantified when defined for ever smokers. Statistical significance was set at p < 0.0021, Bonferroni correction for investigation of 4 loci, 3 smoking dose variables, and 2 smoking status exposures.
Conditional Analyses
To assess independence of novel loci from established lipids loci, we conducted conditional analyses using GCTA. GCTA’s conditional and joint analysis option (COJO) calculates approximate conditional and joint association analyses based on summary statistics from a GWAS meta-analysis and individual genotype data from an ancestry-appropriate reference sample (for LD estimation). For novel loci from predominantly AFR meta-analyses, the LD reference set included unrelated AFR from HUFS, CFS, JHS, ARIC, and MESA (total N = 8,425). For novel loci from predominantly EUR meta-analyses, the LD reference set included unrelated EUR from ARIC (total N = 9,770). Excluding HUFS, these data were accessed through dbGaP (ARIC phs000280.v2.p1, phs000090.v2.p1; CFS phs000284.v1.p1; JHS phs000286.v4.p1, phs000499.v2.p1; and MESA phs000209.v13.p1, phs000420.v6.p3) and imputed to 1000 Genomes phase 1 v. 3 using the Michigan Imputation Server54 For loci with a p < 5 × 10−8 for the 1df test of interaction, results from stage 1 and 2 meta-analyses were adjusted for all known lipids loci. A method for running conditional analyses for 2df tests has not been implemented within GCTA, therefore we evaluated loci with a p < 5 × 10−8 for the 2df joint test of main and interaction effects by conditioning stage 1 stratified analyses on known lipids loci (stratified analyses were not conducted in stage 2 studies). The conditioned 2df joint test of main and interaction effects was then calculated using EasyStrata50 on the conditioned stratified results.
Power Calculations for Detecting Interactions at Known Lipids Loci
To better contextualize our lack of detection of an interaction at a known locus, we conducted power calculations under a variety of scenarios. We explored the power to detect both an interaction and a main effect, making assumptions based on our data, as the sample sizes achieved in this project are comparable to the largest main effect GWAS for lipids1,5. Using previously developed analytical power formulas55, we evaluated three interaction scenarios: a pure interaction effect (no effect in non-smokers and a positive effect in current smokers), a quantitative interaction (effects in the same direction across strata, but of different magnitude), and a qualitative interaction (effects in opposite directions and of different magnitude). We assumed stage 1 + 2 sample sizes and 19% prevalence of smoking (as in our data). For the purposes of illustration, we assumed relatively large effects which explain 0.06% of the variance in the lipid trait; the median variance explained from known lipid loci, as estimated from a previous publication (their Supplemental Table 1)2, is 0.04%.
Proportion of Variance Explained
To evaluate the proportion of the variance explained by our novel associations, we conducted additional analyses of our variants of interest in cohorts of diverse ancestries (Supplementary Table 16). In each of 10 studies from 4 ancestries (EUR, AFR, ASN, and HISP), we ran a series of nested regression models to determine the relative contribution of each set of additional variables. The first model included only standard covariates (age, sex, center, principal components, etc.). The second model additionally included smoking status (both current and ever smoking). The third added known variants1–5,12. The fourth model added all novel variants, and the last model also included interaction terms for novel variants. For the purposes of this analysis, novel variants included the lead variant for each genome-wide significant locus in the meta-analyses of stages 1 and 2 (Table 1) and that were significant but only available in stage 1 meta-analyses (Table 2). By subtracting the r2values from each of these nested regression models, the proportion of variance explained by the additional set of variables was determined. We conducted these analyses using two approaches. In Approach 1, all variants with MAF ≥ 0.01 and imputation quality ≥ 0.3 were included in regression models. While the imputation quality threshold used for the main analyses (≥ 0.5) was higher in order to reduce the risk of spurious associations, we selected a lower threshold for this secondary analysis to maximize the number of variants of interest included. In Approach 2, to avoid possible overfitting, stepwise regression was used for variant selection, such that only variants that were associated (p < 0.05) were retained in the model. All variants were considered in models for each trait and ancestry, regardless of the trait or ancestry in which the association was identified.
Reproducing Previously Reported Lipids Associations
To evaluate the degree to which our data confirmed previous associations, we evaluated statistically significant associations reported from recent large meta-analyses1–5,12. In the event of overlap between reports, the most statistically significant variant-trait association was considered, for a total of 346 unique associations for 269 variants. Output from our main effect models (stage 1) was extracted for all ancestries for each previously reported variant-trait combination. Reproducibility was determined by p < 0.05 in any ancestry and a consistent direction of effect (Supplementary Table 17).
Functional Inference
To evaluate the degree to which our novel variants might influence other cardiometabolic traits, we extracted our novel variants (Tables 1 and 2) from previous studies. Supplementary Tables 19–24 present the association of these variants with coronary artery disease and myocardial infarction, using data from the CARDIoGRAM consortium56; neurological traits, using data from the Neurology Working Group of the CHARGE Consortium; anthropometry, using data from the GIANT consortium.57-59 adoptive smoking interaction, using data from the GIANT consortium 60; diabetes and related traits, using data from MAGIC61, AAGILE62, and DIAGRAM63, 64; and kidney outcomes, using data from the COGENT-Kidney consortium65.
To conduct functional annotation of our novel variants (Supplementary Tables 18, 25–27), we used NCBI Entrez gene (see URLs) for gene information, dbSNP to translate positions to human genome build 38, HaploReg (v4.1) and RegulomeDB for gene expression and regulation data from ENCODE and RoadMap projects, and GTEx v7.0 for additional gene expression information. We also investigated our novel variants in cis- and trans-eQTL data based on analysis of the whole blood of Framingham Heart Study participants66.
Pathway and Gene Set Enrichment Analyses
We conducted DEPICT analyses13 based on genome-wide significant (p< 5 × 10−8) variants separately for the three traits HDL, LDL and triglycerides (Supplementary Tables 28–37). To obtain input for the prioritization and enrichment analyses, DEPICT first created a list of non-overlapping loci by applying a combined distance and LD based threshold (500 KB flanking regions and LD r² > 0.1) between the associated variants and the 1000 Genomes reference data. DEPICT then obtained lists of overlapping genes by applying an LD based threshold (r2 > 0.5) between the non-overlapping variants and known functional coding or cis-acting regulatory variants for the respective genes. Finally, the major histocompatibility complex region on chromosome 6 (base position 25,000,000 – 35,000,000) was removed from further analyses. DEPICT prioritized genes at associated regions by comparing functional similarity of genes across associated loci using a gene score that was adjusted for several confounders, such as gene length. Utilizing lead variants from 500 pre-compiled null GWAS the scoring step was repeated 50 times to obtain an experiment-wide FDR for the gene prioritization. Second, DEPICT conducted gene-set enrichment analyses based on a total of 14,461 pre-compiled reconstituted gene sets. The reconstituted gene sets involve 737 Reactome database pathways, 2,473 phenotypic gene sets (derived from the Mouse Genetics Initiative)67, 184 Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathways, 5,083 Gene Ontology database terms, and 5,984 protein molecular pathways (derived from protein-protein interactions68). Third, DEPICT conducted tissue and cell type enrichment analyses based on expression data in any of the 209 MeSH annotations for 37,427 microarrays of the Affymetrix U133 Plus 2.0 Array platform. In addition, we used STRING database for identifying protein x protein interactions.
Data Availability
All summary results will be made available in dbGaP (phs000930.v7.p1).
Supplementary Material
Acknowledgments
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Human Genome Research Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. This project was largely supported by a grant from the US National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (R01HL118305) and by the Intramural Research Program of the National Human Genome Research Institute of the National Institutes of Health through the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (Z01HG200362). Additional and study-specific acknowledgments appear in the Supplementary Note.
Appendix
Author Contributions
All authors reviewed and approved the manuscript. Study concept and design: A.B.Z., A.C.M., A.C.P., A.J.O., A.R., A.R.B., A.R.W., B.I.F., B.L.H., C.A.M.K., C.Ballantyne, C.Bouchard, C.C.K., C.C.L., C.D.L., C.H., C.Langenberg, C.M.van D., C.M.K., C.N.R., C-T.L., C.Y., C-Y.C., D.C.R., D.I.C., D.M.B., D.R.W., D.W.B., E.B., E.P.B., E.R.F., E.S.T., F.R.R., G.W., H.A., H.J.de S., H.Watkins, I.G., I.J.D., I.K., J.B.J., J.Ding, J.Divers, J.D.F., J.E.Hixson, J.E.K., J.I.R., J.K., Jianjun Liu, J.M.C., J.M.S., J-M.Y., K.C., K.K.L., K.L.M., L.A.C., Lifelines Cohort Study, L.E.W., L.J.L., M.A.I., M.A.P., M.Brown, M.Boehnke,M.Farrall, M.Fornage, M.He, M.K., M.K.E., M.Laakso, M.S., N.G.F., N.J.S., N.J.W., N.K., N.L.P., N.P., N.S., O.P., O.T.R., P.F., P.G., P.H., P.K., P.K.E.M., P.M.R., P.S., R.A.S., R.M.D., R.R., R.S.C., S.C., S.K.M., S.L.R.K., S.R., S.T.T., T.A., T.A.L., T.B.H., T.F., T.K.R., T.Lehtimäki, T.N.K., T.R., T.W., T.Y.W., U.de F., V.G., W.B.W., W.P.K., X.G., Y.K., Y.Liu, Y.W., Y.X.W., and Y.Y.T. Phenotype data acquisition and/or quality control: A.B.Z., A.C., A.C.P., A.D.M., A.G., A.J.O., A.K., A.Metspalu, A.P., A.P.R., A.R.B., A.R.V.R.H., A.R.W., A.W.M., B.E.C., B.G., B.I.F., B.L.H., B.M.P., B.O.T., B.Penninx, C.A.M.K., C.Ballantyne, C.Bouchard, C.D.L., C.E.L., C.Gieger, C.H., C.J., C.Langenberg, C.Li, C.M.K., C.M.van D., C.N.R., C.O.S., C.P.N., C.Y., D.C.R., D.H., D.M.B., D.R.J., D.R.W., D.W.B., E.E., E.P.B., E.S.T., F.R., F.R.R., F-C.H., G.J.P., G.R.B., G.W., H.G., H.J.de S., H.J.G., H.M.S., H.Tiemeier, H.Wang, I.J.D., I.K., I-T.L., J.A.S., J.B.J., J.Ding, J.Divers, J.D.F., J.E.K., J.H.Z., Jian’an Luan, Jingjing Liang, J.M.C., J.M.S., J-M.J.J., J-M.Y., J-S.W., K.C., K.K.L., K.Leander, K.Liu, K.Schwander, K-H.L., L.A.C., Lifelines Cohort Study, L.F.B., L.J.B., L.M., L.M.R., L.R.Y., M.Alver, M.Amini, M.A.P., M.Brown, M.Boissel, M.C., M.F.F., M.He, M.Hirata, M.K., M.K.E., M.K.W., M.N., M.P.C., M.S., M.W., N.F., N.G.F., N.J.S., N.J.W., N.L.P., N.P., N.S., N.Y.Q.T., O.H.F., O.P., O.T.R., P.A.P., P.H., P.J.S., P.K., P.K.E.M., P.M.R., P.S., P.W.F., R.A.S., R.M., R.M.D., R.R., R.S.C., S.E.H., S.L.R.K., S.S., S.S.R., S.T.T., T.A.L., T.E., T.F., T.K., T.K.R., T.Lehtimäki, T.M., T.N.K., T.R., T.S., T.W., T-D.W., U.de F., Understanding Society Scientific Group, W.B.W., W.P.K., Y.C.T., Y.Liu, and Y.Lu. Genotype data acquisition and/or quality control: A.B.Z., A.C.P., A.G., A.G.U., A.L., A.Metspalu, A.R.B., A.R.V.R.H., A.T.K., A.V.S., B.E.C., B.G., B.I.F., B.L.H., B.M.P., B.O.T., B.Prins, C.Bouchard, C.C.K., C.C.L., C.Gao, C.K., C.Langenberg, C.Li, C.M.K., C.N.R., C.P.N., C-K.H., C-T.L., D.C.R., D.E.A., D.I.C., D.M.B., D.O.M-K., E.B., E.B.W., E.E., E.L., E.P.B., E.R.F., E.S.T., E.Z., F.G., F.P.H., F.R., F.R.R., F-C.H., H.G., H.Wang, I.J.D., I.K., I.M.N., J.A.S., J.E.Hixson, J.E.Huffman, J.E.K., J.F.C., J.H.Z., J.I.R., Jian’an Luan, Jingjing Liang, Jianjun Liu, Jingmin Liu, J.M.C., J.M.S., K.C., K.D.T., K.K.L., K.Leander, K.Schwander, K.Strauch, L.A.C., Lifelines Cohort Study, L.M., L.M.R., L.R.Y., Lan Wang, L-P.L., M.Alver, M.Amini, M.A.N., M.A.P., M.Boissel, M.C., M.Fornage, M.F.F., M.K., M.K.E., M.P., M.P.C., N.A., N.D.P., N.J.S., N.J.W., N.K., N.L.P., N.S., O.P., P.B.M., P.H., P.J.V.M., P.K.E.M., P.W.F., R.A.S., R.D., R.J.F.L., R.M., R.N.E., S.E.H., S.H., S.K.M., S.L.R.K., S.S.R., S.T.T., T.E., T.K.R., T.Lehtimäki, T.N.K., T.R., U.de F., Understanding Society Scientific Group, W.Zhao, X.D., X.S., X.Z., Y.F., Y.H., Y.Liu, Y.Momozawa, Y.Y.T., Y-D.I.C., and Z.A. Data analysis and interpretation: A.B.Z., A.C.M., A.C.P., A.G., A.Mahajan, A.P.M., A.P.R., A.R., A.R.B., A.R.V.R.H., A.S., A.U.J., A.V.S., B.I.F., B.K., B.M.P., B.O.T., B.Prins, C.A.W., C.Bouchard, C.D.L., C.Gao, C.Gieger, C.Li, C.N.R., C.P.N., C-T.L., C-Y.C., D.C.R., D.H., D.I.C., D.M.B., D.O.M-K., D.V., E.B.W., E.E., E.L., E.R.F., E.S.T., F.G., F.P.H., F.T., F-C.H., G.C., G.W., H.G., H.S., I.G., I.M.N., I.N., J.A.S., J.B.J., J.Divers, J.E.Hixson, J.E.Huffman, J.F.C., J.H.Z., Jian’an Luan, Jingmin Liu, J.S.F., J.Y., J.Z., K.Leander, K.R., L.A.C., Lifelines Cohort Study, L.F.B., L.M.R., L.R.Y., Lan Wang, Lihua Wang, L-P.L., M.Amini, M.A.N., M.A.R., M.A.S., M.Fornage, M.Farrall, M.F.F., M.K., M.K.E., M.P., M.R., M.R.B., M.S., N.D.P., N.F., N.J.S., N.M., P.A.P., P.B.M., P.H., P.J.V.M., P.S.V., R.D., R.J.F.L., R.N., R.N.E., R.S.C., S.A.G., S.B.K., S.E.H., S.H., S.K.M., S.L., S.L.R.K., S.M.T., T.K.R., T.Louie, T.M.B., T.N.K., T.R., T.S., T.V.V., T.W.W., T.Y.W., W.B.W., W.Zhao, X.C., X.D., X.G., X.S., Y.H., Y.J., Y.K., Y.Lu, and Y.X.W. Performed Look-ups: A.E.J., A.Mahajan, A.P.M., A.R.B., COGENT-Kidney Consortium, D.I.C., K.Y., M.G., N.F., and T.W.W.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests except for the following. Oscar H Franco received grants from Metagenics (on women’s health and epigenetics) and from Nestle (on child health); Jost Bruno Jonas serves as a consultant for Mundipharma Co. (Cambridge, UK); Patent holder with Biocompatibles UK Ltd. (Franham, Surrey, UK) (Title: Treatment of eye diseases using encapsulated cells encoding and secreting neuroprotective factor and / or anti-angiogenic factor; Patent number: 20120263794), and Patent application with University of Heidelberg (Heidelberg, Germany) (Title: Agents for use in the therapeutic or prophylactic treatment of myopia or hyperopia; Europäische Patentanmeldung 15 000 771.4); Mike A. Nalls’ participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, National Institutes of Health, Bethesda, MD, USA, as a possible conflict of interest Dr. Nalls also consults for Illumina Inc, the Michael J. Fox Foundation and University of California Healthcare among others; Neil Poulter has received financial support from several pharmaceutical companies that manufacture either blood pressure-lowering or lipid lowering agents or both, and consultancy fees; Peter Sever has received research awards from Pfizer Inc.; Bruce M Psaty serves on the DSMB of a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson; and Laura J Bierut is listed as an inventor on Issued U.S. Patent 8,080,371,”Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
References
- 1.Willer CJ et al. Discovery and refinement of loci associated with lipid levels. Nature genetics 45, 1274–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Do R et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nature genetics 45, 1345–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peloso GM et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. American journal of human genetics 94, 223–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spracklen CN et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Human Molecular Genetics 26, 1770–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med 358, 1240–9 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Kar D et al. Relationship of cardiometabolic parameters in non-smokers, current smokers, and quitters in diabetes: a systematic review and meta-analysis. Cardiovascular Diabetology 15, 158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong C et al. Cigarette smoke exposure impairs reverse cholesterol transport which can be minimized by treatment of hydrogen-saturated saline. Lipids in Health and Disease 14, 159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning AK et al. Meta-analysis of Gene-Environment interaction: joint estimation of SNP and SNP×Environment regression coefficients. Genetic Epidemiology 35, 11–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Psaty BM et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from five cohorts. Circulation. Cardiovascular genetics 2, 73–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao DC et al. Multiancestry Study of Gene–Lifestyle Interactions for Cardiovascular Traits in 610 475 Individuals From 124 Cohorts. Design and Rationale 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanktree MB et al. Genetic meta-analysis of 15,901 African Americans identifies variation in EXOC3L1 is associated with HDL concentration. Journal of Lipid Research 56, 1781–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pers TH et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nature Communications 6, 5890–5890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki J, Imanishi E & Nagata S Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proceedings of the National Academy of Sciences of the United States of America 113, 9509–9514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J et al. Genome-Wide Expression Analysis Reveals Diverse Effects of Acute Nicotine Exposure on Neuronal Function-Related Genes and Pathways. Frontiers in Psychiatry 2, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Parkinson Disease Genomics, C. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 377, 641–649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng MCY et al. Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes. PLOS Genetics 10, e1004517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai K, Lucki NC & Sewer MB Silencing diacylglycerol kinase-theta expression reduces steroid hormone biosynthesis and cholesterol metabolism in human adrenocortical cells(). Biochimica et biophysica acta 1841, 552–562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai K & Sewer MB Diacylglycerol kinase θ couples farnesoid X receptor-dependent bile acid signalling to Akt activation and glucose homoeostasis in hepatocytes. The Biochemical journal 454, 267–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordell HJ et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nature Communications 6, 8019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards TL et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Annals of human genetics 74, 97–109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lill CM et al. Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson’s Disease Genetics: The PDGene Database. PLOS Genetics 8, e1002548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nalls MA et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nature genetics 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankratz N et al. Meta-analysis of Parkinson disease: Identification of a novel locus, RIT2. Annals of Neurology 71, 370–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J et al. Phlegm-Dampness Constitution: Genomics, Susceptibility, Adjustment and Treatment with Traditional Chinese Medicine. The American Journal of Chinese Medicine 41, 253–262 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Choi J-H et al. Variations in TAS1R taste receptor gene family modify food intake and gastric cancer risk in a Korean population. Molecular Nutrition & Food Research 60, 2433–2445 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann TJ et al. A large electronic-health-record-based genome-wide study of serum lipids. Nature Genetics 50, 401–413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klarin D et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu DJ et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nature Genetics 49, 1758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward LD & Kellis M HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research 40, D930–D934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.U.S. Department of Health and Human Services. Tobacco Use Among U.S. Racial/Ethnic Minority Groups—African Americans, American Indians and Alaska Natives, Asian Americans and Pacific Islanders, and Hispanics: A Report of the Surgeon General ( U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Atlanta, GA, 1998). [Google Scholar]
- 32.Villanti AC et al. Changes in the prevalence and correlates of menthol cigarette use in the USA, 2004–2014. Tobacco Control 25, ii14 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ross KC, Dempsey DA, Helen G, Delucchi K & Benowitz NL The influence of puff characteristics, nicotine dependence, and rate of nicotine metabolism on daily nicotine exposure in African American smokers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 25, 936–943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ton HT et al. Menthol Enhances the Desensitization of Human α3β4 Nicotinic Acetylcholine Receptors. Molecular Pharmacology 88, 256–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander LA et al. Why We Must Continue to Investigate Menthol’s Role in the African American Smoking Paradox. Nicotine & Tobacco Research 18, S91–S101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones MR, Tellez-Plaza M & Navas-Acien A Smoking, Menthol Cigarettes and All-Cause, Cancer and Cardiovascular Mortality: Evidence from the National Health and Nutrition Examination Survey (NHANES) and a Meta-Analysis. PLoS ONE 8, e77941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munro HM, Tarone RE, Wang TJ & Blot WJ Menthol and Nonmenthol Cigarette Smoking: All-Cause Deaths, Cardiovascular Disease Deaths, and Other Causes of Death Among Blacks and Whites. Circulation 133, 1861–1866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray RP, Connett JE, Skeans MA & Tashkin DP Menthol Cigarettes and Health Risks in Lung Health Study Data. Nicotine & Tobacco Research 9, 101–107 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Vozoris NT, Mhsc, Md & Frcpc. Mentholated cigarettes and cardiovascular and pulmonary diseases: A population-based study. Archives of Internal Medicine 172, 590–593 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Stable EJ, Herrera B, Jacob IP & Benowitz NL Nicotine metabolism and intake in black and white smokers. JAMA 280, 152–156 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Khariwala SS et al. Cotinine and Tobacco-Specific Carcinogen Exposure Among Nondaily Smokers in a Multiethnic Sample. Nicotine & Tobacco Research 16, 600–605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain RB Distributions of selected urinary metabolites of volatile organic compounds by age, gender, race/ethnicity, and smoking status in a representative sample of U.S. adults. Environmental Toxicology and Pharmacology 40, 471–479 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Benowitz NL, Dains KM, Dempsey D, Wilson M & Jacob P Racial Differences in the Relationship Between Number of Cigarettes Smoked and Nicotine and Carcinogen Exposure. Nicotine & Tobacco Research 13, 772–783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U.S. Department of Health and Human Services. The Health Consequences of Smoking: 50 Years of Progress A Report of the Surgeon General. ( U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Atlanta, GA, 2014). [Google Scholar]
- 45.Ito S et al. Nicotine-Induced Expression of Low-Density Lipoprotein Receptor in Oral Epithelial Cells. PLoS ONE 8, e82563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dullaart RP, Hoogenberg K, Dikkeschei BD & van Tol A Higher plasma lipid transfer protein activities and unfavorable lipoprotein changes in cigarette-smoking men. Arteriosclerosis, Thrombosis, and Vascular Biology 14, 1581–1585 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Frondelius K et al. Lifestyle and Dietary Determinants of Serum Apolipoprotein A1 and Apolipoprotein B Concentrations: Cross-Sectional Analyses within a Swedish Cohort of 24,984 Individuals. Nutrients 9, 211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onat A et al. Preheparin serum lipoprotein lipase mass interacts with gender, gene polymorphism and, positively, with smoking. in Clinical Chemistry and Laboratory Medicine Vol. 47 208 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nature protocols 9, 1192–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler TW et al. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics 31, 259–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchini J & Howie B Genotype imputation for genome-wide association studies. Nat Rev Genet 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Kraft P, Yen YC, Stram DO, Morrison J & Gauderman WJ Exploiting Gene-Environment Interaction to Detect Genetic Associations. Human Heredity 63, 111–119 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Skol AD, Scott LJ, Abecasis GR & Boehnke M Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38, 209–213 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Das S et al. Next-generation genotype imputation service and methods. Nature Genetics 48, 1284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler TW et al. Approaches to detect genetic effects that differ between two strata in genome-wide meta-analyses: Recommendations based on a systematic evaluation. PLoS One 12, e0181038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikpay M et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47, 1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood AR et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Locke AE et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shungin D et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Justice AE et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nature Communications 8, 14977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manning AK et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature genetics 44, 659–669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu CT et al. Trans-ethnic Meta-analysis and Functional Annotation Illuminates the Genetic Architecture of Fasting Glucose and Insulin. Am J Hum Genet 99, 56–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaulton KJ et al. Genetic fine-mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nature genetics 47, 1415–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DIAbetes Genetics Replication Meta-analysis Consortium, Asian Genetic Epidemiology Network Type Diabetes Consortium, South Asian Type Diabetes Consortium Mexican American Type Diabetes Consortium & Type Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples Consortium. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature genetics 46, 234–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahajan A et al. Trans-ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity. American Journal of Human Genetics 99, 636–646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joehanes R et al. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biology 18, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blake JA et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res 42, D810–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lage K et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol 25, 309–16 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All summary results will be made available in dbGaP (phs000930.v7.p1).