Summary
Sporulation in Bacillus subtilis is a paradigm of bacterial development, which involves the interaction between a larger mother cell and a smaller forespore. The mother cell and the forespore activate different genetic programs, leading to the production of sporulation-specific proteins. A critical gap in our understanding of sporulation is how vegetative proteins, made before sporulation initiation, contribute to spore formation. Here we present a system, spatiotemporally regulated proteolysis (STRP), that enables the rapid, developmentally-regulated degradation of target proteins, providing a suitable method to dissect the cell- and developmental stage-specific role of vegetative proteins. We have used STRP to dissect the role of two major vegetative sigma factors, σH and σA, during sporulation. Our results suggest that σH is only required in predivisional cells, where it is essential for sporulation initiation, but that it is dispensable during subsequent steps of spore formation. However, we provide evidence that σA plays different roles in the mother cell, where it replenishes housekeeping functions, and in the forespore, where it plays an unexpected role in promoting spore germination and outgrowth. Altogether, our results demonstrate that STRP has the potential to provide a comprehensive molecular dissection of every stage of sporulation, germination and outgrowth.
Keywords: Bacillus subtilis, sporulation, inducible protein degradation, ssrA, sigma factors
Graphical Abstract
Spore formation in Bacillus subtilis represents a simple developmental process that involves the interaction between only two cells. Here we present a method, called spatiotemporally regulated proteolysis (STRP), to rapidly deplete target proteins in a cell- and developmental stage-specific manner during sporulation. We show that STRP has the potential to provide a comprehensive molecular dissection of every stage of sporulation, germination and outgrowth.
Introduction
Cellular differentiation is a pivotal step in every developmental process, from human ontogeny to spore formation in certain bacteria. Sporulation in the bacterium Bacillus subtilis has become a paradigm for cell differentiation and development in bacteria (Errington, 2003; Hilbert and Piggot, 2004; Higgins and Dworkin, 2012; Tan and Ramamurthi, 2014; Narula et al., 2016). Triggered by nutrient starvation, spore formation involves the close interaction between two cells that differ in both size and developmental fate: the smaller forespore, which becomes the dormant and highly resilient spore following maturation, and the larger mother cell, which lyses after contributing to the formation of the mature spore.
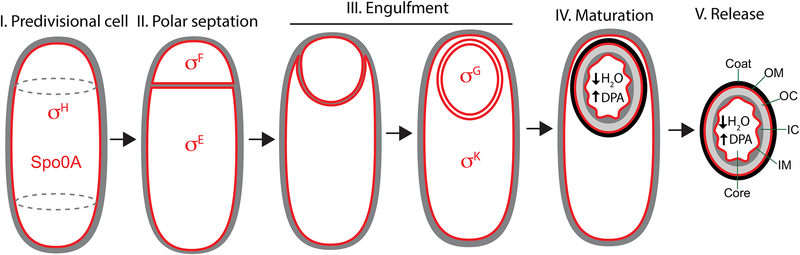
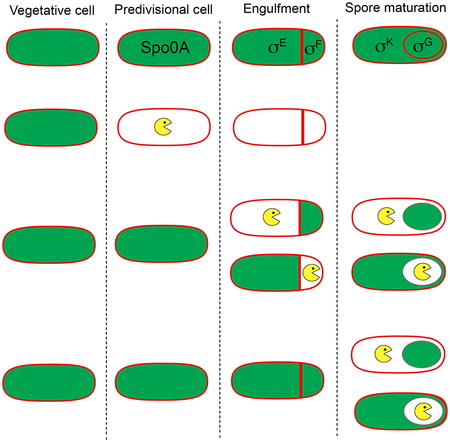
The sporulation process is depicted in Fig. 1. The forespore and the mother cell are the result of an asymmetrically positioned cell division event in which the septum is formed close to a single cell pole, in a process called polar septation. After polar septation, the membrane of the mother cell engulfs the forespore in a phagocytosis-like process that culminates with the forespore surrounded by two membranes and enclosed within the mother cell cytoplasm. The forespore subsequently matures inside the mother cell, where it is protected from the external medium. Spore maturation involves the formation of a peptidoglycan layer called the cortex between the inner and outer forespore membranes (Popham and Bernhards, 2015), the assembly of a proteinaceous coat around the outer membrane (McKenney et al., 2013), and the dehydration of the forespore cytoplasm, which endows the spore with a bright appearance under phase-contrast microscopy. Once maturation is complete, the mother cell lyses and the spore is released into the environment, where it can remain dormant for years. Mature spores are resistant to a variety of environmental challenges (Setlow, 2006), including high temperature, desiccation, and a variety of chemicals such as antibiotics, to which spores are impermeable. Spore germination is triggered in response to specific nutrients and other stimuli (Setlow, 2014). During germination the spore core is rehydrated, leading to the resumption of metabolic activities and vegetative growth.
Figure 1. The sporulation pathway in B. subtilis.
Membranes are in red, and peptidoglycan in gray. Transcription factors active in different cells and stages are shown red. I. In predivisional cells, the division sites (dotted ovals) are shifted to polar positions. II. Polar septation produces a small forespore and a large mother cell. III. After polar septation, the mother cell engulfs the forespore in a phagocytosis-like process. After engulfment, the forespore is enclosed within the mother cell cytoplasm and delimited by two membranes. IV. The forespore matures through the formation of a peptidoglycan cortex (gray) between the inner and outer forespore membranes, the assembly of a proteinaceous coat around the outer membrane (black), and the dehydration of the forespore core, which accumulates dipicolinic acid (DPA). V. Once maturation is completed, the mother cell lyses and the spore is released into the environment, where it remains dormant until conditions are appropriate for germination. OM, outer membrane; OC, outer cortex; IC, inner cortex; IM, inner membrane.
The regulatory program that controls sporulation is relatively well understood (Errington, 2003; Hilbert and Piggot, 2004). Several transcriptional regulators are sequentially activated, which orchestrate distinct programs of gene expression in the mother cell and forespore at different developmental stages (Fig. 1). The entry into sporulation is controlled by the stationary phase-specific σ factor, σH, and the phosphorelay response regulator, Spo0A, which together govern gene expression in the predivisional cell. After polar septation, a cascade of cell-specific σ factors become active in the forespore and the mother cell. Immediately after polar septation, σF is activated in the forespore, followed by the activation of σE in the mother cell. Upon engulfment completion, σG becomes active in the forespore and σK in the mother cell. Together, the cell-specific σ factors control the transcription of ~560 genes (Eichenberger et al., 2004; Wang et al., 2006; Eichenberger, 2012), most of which are sporulation-specific and not transcribed in vegetative cells.
The study of sporulation has traditionally focused on deciphering the function of genes transcribed by sporulation-specific σ factors. Although there are still many questions to be answered, thanks to the concerted efforts of thousands of researchers over the past fifty years, we have achieved a relatively comprehensive understanding of the function of many sporulation-specific proteins. However, most B. subtilis proteins are produced during vegetative growth, before polar septation, and the manner in which they contribute to sporulation remains largely unknown. This critical gap in our knowledge of sporulation is mainly due to the lack of suitable genetic tools to inhibit the function of specific proteins in a precise, cell- and developmental stage-specific manner during spore formation. The precisely regulated inactivation of target proteins is critical because many such proteins are important for growth, so null mutations may be nonviable or unable to enter sporulation. Furthermore, because sporulating cells do not grow or divide following polar septation, methods based on inhibition of transcription or translation to deplete specific proteins have limited utility. Indeed, the average half life of bacterial proteins is ~8–20 h in growing and stationary phase cells (Koch and Levy, 1955; Borek et al., 1958; Mandelstam, 1958; Kock et al., 2004), which, due to the absence of growth during sporulation, would produce a negligible reduction in the amount of protein in the ~3 h between polar septation and forespore maturation (at 37°C). Thus, there is a need for new systems to rapidly deplete proteins, preferably in a cell- and developmental stage-specific manner.
In 2008, Griffith and Grossman developed an inducible, targeted protein degradation system in B. subtilis (Griffith and Grossman, 2008), which provides an opportunity to circumvent these limitations. The system is based on the addition of a modified ssrA tag from E. coli (hereafter ssrA*) to the C-terminus of the target protein, and the expression of the E. coli SspB (SspBEc) from inducible promoters. When SspBEc is produced, it binds to the ssrA* tag and delivers the target protein to the endogenous B. subtilis protease, ClpXP, for degradation. This system supports the degradation of target proteins within minutes after the induction of sspBEc expression (Griffith and Grossman, 2008; Eswaramoorthy et al., 2014; Yen Shin et al., 2015; Lamsa et al., 2016; Lopez-Garrido et al., 2018), allowing for the rapid depletion of proteins in non-dividing cells.
We have previously shown that the expression of sspBEc from sporulation-specific promoters dependent on σF and σE supports the efficient degradation of ssrA*-tagged proteins in a cell-specific manner during sporulation (Yen Shin et al., 2015; Lopez-Garrido et al., 2018). Here, we expand on our previous study to build a comprehensive framework, called spatiotemporally regulated proteolysis (STRP), to rapidly deplete target proteins in a cell- and developmental stage-specific manner. We demonstrate that our framework is well suited to dissect the cell- and stage-specific requirement of target proteins during sporulation. First, we provide evidence that degradation of abundant target proteins does not affect the turnover of native ClpXP substrates. Second, we show that degradation of control proteins with well-documented roles in sporulation produce phenotypes equivalent to those of the null mutants. Third, we demonstrate that proteins produced before polar septation are rapidly degraded in a spatially- and temporally-controlled manner.
We have applied STRP to dissect the spatiotemporal requirement of the stationary-phase σ factor σH, and the essential vegetative σ factor σA, during sporulation. Our results indicate that σH is only required in predivisional cells, and does not play a major role once polar septation occurs. However, we present evidence that σA plays different roles in mother cell and forespore: in the mother cell, it may replenish housekeeping functions needed to complete sporulation; in the forespore, it is required early in sporulation to produce spores that germinate efficiently and it is essential for spore outgrowth.
Results
Degradation of abundant ssrA*-tagged proteins does not saturate ClpXP protease in B. subtilis
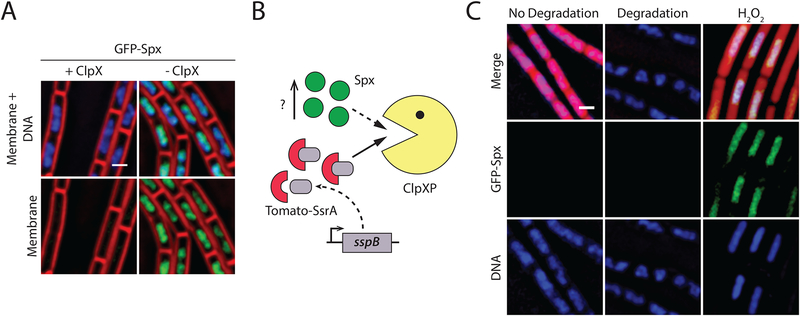
Targeted protein degradation systems based on modified ssrA tags have emerged as a powerful genetic tool in different bacteria, including B. subtilis (McGinness et al., 2006; Griffith and Grossman, 2008; Wei et al., 2011). Because those systems rely upon endogenous proteases (ClpXP in B. subtilis) to degrade target proteins, a potential caveat is that inducing the degradation of an abundant target protein could saturate the protease, preventing processing of its natural substrates (Cookson et al., 2011). To test this possibility, we sought to develop an in vivo reporter for ClpXP saturation in B. subtilis. Towards this end, we constructed an N-terminal fusion of the green fluorescent protein (GFP) to the stress-related transcriptional regulator, Spx. A strain producing GFP-Spx as the only source of Spx was resistant to diamide (data not shown), suggesting that the fusion protein is functional (Nakano et al., 2003). Under standard laboratory conditions, Spx is maintained at low levels due to proteolysis by ClpXP (Nakano et al., 2002). Accordingly, no GFP-Spx is observed by fluorescence microscopy (Fig. 2A, +ClpXP). However, when the amount of ClpXP becomes limiting, GFP-Spx accumulates, yielding a clear fluorescent signal that co-localizes with the nucleoid (Fig 2A, -ClpXP). We reasoned that if inducing the degradation of ssrA*-tagged proteins saturated ClpXP, we would observe an accumulation of GFP-Spx signal that co-localizes with the nucleoid. To test this idea, we induced the degradation of an ssrA*-tagged derivative of the red fluorescent protein (Tomato-ssrA*) produced at high levels from a strong constitutive promoter, and looked for the appearance of GFP-Spx (Fig 2B). As a positive control, this strain was also treated with hydrogen peroxide, which has previously been shown to evoke the Spx response (Leelakriangsak et al., 2007). Degradation of Tomato-ssrA* did not result in the accumulation of GFP-Spx, whereas hydrogen peroxide treatment did (Fig 2C). These data suggest that targeting abundant proteins for degradation by ClpXP has little or no effect on ClpXP housekeeping activity. Thus, the phenotypes observed upon degradation of target proteins are likely the result of target protein inactivation rather than a consequence of ClpXP saturation.
Figure 2. Degradation of an abundant protein does not affect turnover of a native ClpXP substrate.
A. Fluorescence microscopy of a strain carrying a GFP-Spx fusion that expresses ClpXP under the control of an IPTG-inducible promoter (BER0552), grown in the presence (left, +ClpXP) or absence (right, -ClpXP) of 1mM IPTG. When ClpX is limiting, GFP-Spx accumulates. Membranes are stained with FM4–64 (red) and DNA with DAPI (blue). Scale bar, 1μm.
B. Inducing degradation of abundant ssrA*-tagged proteins should cause accumulation of GFP-Spx if ClpXP is saturated.
C. Fluorescence microscopy of a strain carrying a GFP-Spx fusion, producing the red fluorescent protein, tomato, from a strong promoter and SspBEc from a xylose-inducible promoter (BER0577), grown in the absence (left, no degradation) or in the presence (middle, degradation) of 1% xylose, or with 2mM H2O2 (right) as a positive control. Imaging was performed 2 hours after treatment. Inducing degradation does not yield an accumulation of GFP-Spx, but treatment with hydrogen peroxide, which is known to evoke the Spx response, does. Membranes are stained with FM4–64 (red) and DNA with DAPI (blue). Scale bar, 1 μm.
Development of a cell-specific protein degradation system during B. subtilis sporulation
To adapt targeted protein degradation to the study of sporulation in B. subtilis, we explored the possibility of using developmentally-regulated promoters to drive sspBEc expression. We have previously employed σF- and σE-dependent promoters to produce SspBEc and degrade the SpoIIIE DNA translocase after polar septation (Yen Shin et al., 2015; Lopez-Garrido et al., 2018). σF and σE are rapidly inactivated once engulfment is completed (Li and Piggot, 2001), which limits the ability of this approach to study proteins that are required throughout sporulation. Thus, we decided to expand the set of developmentally-regulated promoters to increase the versatility of the protein degradation system.
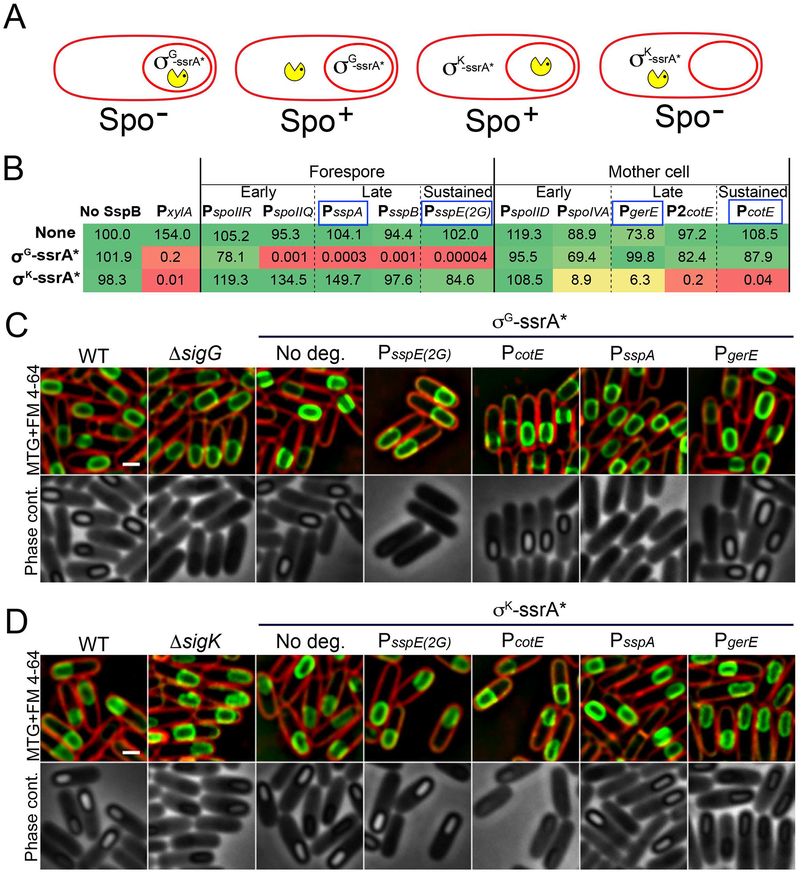
We designed a genetic strategy to screen for cell-specific promoters that triggered the efficient degradation of target proteins when driving the expression of sspBEc (Fig. 3A). Specifically, we constructed strains producing ssrA*-tagged versions of the essential sporulation proteins, σG and σK, which orchestrate cell-specific transcription after engulfment in the forespore and mother cell, respectively (Fig. 1). B. subtilis mutants lacking σG or σK are unable to form spores (Table S1). However, the addition of the ssrA* tags did not produce any observable defect in spore morphogenesis or titer (Fig. 3B–D; Table S1), suggesting that both tagged proteins are fully functional. Expression of sspBEc from a xylose-inducible promoter, however, yielded a dramatic reduction in spore titer for both strains (Fig. 3B; Table S1), indicating that σG-ssrA* and σK-ssrA* were efficiently degraded. Note that the addition of 1% of xylose alone to sporulating cultures did not reduce spore titers (Fig. 3B) nor affect the progression of sporulation (Fig. S1). We then chose sporulation cell-specific promoters to drive sspBEc expression and tested if they triggered the efficient degradation of σG-ssrA* and σK-ssrA*. We selected promoters belonging to three different temporal classes:
Early promoters, active immediately after polar septation in the forespore (σF-dependent) or in the mother cell (σE-dependent), but inactive after engulfment. We selected the σF-dependent promoters PspoIIR (Karow et al., 1995) and PspoIIQ (Londoño-Vallejo et al., 1997), and the σE-dependent promoters PspoIID (Clarke et al., 1986; Rong et al., 1986) and PspoIVA (Roels et al., 1992).
Late promoters, active in the forespore (σG-dependent) or the mother cell (σK-dependent) after engulfment. Here we used the σG-dependent promoters PsspA and PsspB (Nicholson et al., 1989) and the σK-dependent promoters PgerE (Cutting et al., 1989) and P2cotE (Zheng and Losick, 1990) for late forespore and mother cell expression, respectively. Note that PsspB drives the expression of the Bacillus sspB gene, which encodes the major β-type small acid-soluble protein and is unrelated to the degradation adaptor protein SspBEc.
Sustained promoters, continuously active in the forespore (σF- and σG-dependent) or the mother cell (σE- and σK-dependent) during and after engulfment. The synthetic PsspE(2G) (Sun et al., 1991), which is recognized by both σF and σG, was used to continuously transcribe sspBEc in the forespore. To continuously transcribe sspBEc in the mother cell, we used the two cotE promoters, the first of which is activated by σE (P1cotE), and the second by σK (P2cotE) (Zheng and Losick, 1990).
Figure 3. Optimization of cell- and stage-specific protein degradation system.
A. Diagram representing the genetic strategy used to screen for cell-specific promoters that trigger the efficient degradation of target proteins when driving the expression of sspBEc. We constructed ssrA*-tagged versions of the σG and σK, which are essential for sporulation and required in the forespore and mother cell, respectively. Degradation of either protein in the cell in which they are required should yield a strong sporulation defect (Spo−), while expression of sspBEc in the opposite cell should have no effect (Spo+).
B. Relative spore titers of strains expressing sspBEc from different cell-specific promoters (columns) in the absence of ssrA*-tagged protein (none), when σG is tagged with ssrA* (σG-ssrA*) or when σK is tagged with ssrA* (σK-ssrA*). To facilitate data comparison, the spore titer of the wild type strain (no ssrA*, no SspBEc) was relativized to 100. The actual values are shown in Table S1. The color indicates the severity of the spore titer defect (green, no defect; yellow, moderate defect; red, severe defect). Expression from PxylA was induced by the addition of 1 % xylose. Sporulation-specific promoters promoters were classified in three temporal classes according to their expression pattern: early promoters are activated shortly after polar septation, and inactivated after engulfment; late promoters activated after engulfment; sustained promoters are continuously active throughout engulfment and forespore maturation. The promoters selected for further characterization are indicated by a blue box.
C. Micrographs of wild type sporangia (WT), σG mutant sporangia (ΔsigG) and sporangia in which σG is tagged with ssrA* (σG-ssrA*) in the absence of SspBEc (No deg.) or when SspBEc is produced from the sustained promoters PsspE(2G) and PcotE or the late promoters PsspA and PgerE. The top row shows membranes stained with FM 4–64 (red) and Mitotracker green (green). The bottom row shows phase contrast pictures. Pictures were taken six hours after sporulation induction (t6). Scale bar, 1 μm.
D. Micrographs of wild type sporangia (WT), σK mutant sporangia (ΔsigK) and sporangia in which σK is tagged with ssrA* (σK-ssrA*) in the absence of SspBEc (No deg.) or when SspBEc is produced from the sustained promoters PsspE(2G) and PcotE or the late promoters PsspA and PgerE. The top row shows membranes stained with FM 4–64 (red) and Mitotracker green (green). The bottom row shows phase contrast pictures. Pictures were taken six hours after sporulation induction (t6). Scale bar, 1 μm.
As shown in Fig. 3B, forespore expression of sspBEc from PspoIIQ, PsspE(2G), PsspA and PsspB resulted in significant spore titer defects in strains expressing σG tagged with ssrA*. Similarly, mother cell expression of sspBEc from PspoIVA, PcotE, PgerE and P2cotE significantly reduced the spore titer of the σK-ssrA* strain. Importantly, no spore titer defects were observed in the σG-ssrA* and σK-ssrA* strains when sspBEc was expressed in the cell in which the transcription factor is not active. Only the early promoters PspoIIR and PspoIID failed to trigger efficient degradation of σG-ssrA* and σK-ssrA*, respectively. Both promoters are only transiently active during engulfment (Clarke et al., 1986; Karow et al., 1995; Wu and Errington, 2000; Eichenberger et al., 2004). In addition, PspoIIR is particularly weak (Sharp and Pogliano, 2002; Ojkic et al., 2016). Thus, it is possible these promoters do not yield enough SspBEc to mediate the complete and/or sustained degradation of σG-ssrA* and σK-ssrA* once engulfment is complete. To avoid complications due to inactivation of promoters after engulfment, we focused on sustained and late promoters for further characterization. We specifically selected the sustained promoters PsspE(2G) and PcotE and the late promoters PsspA and PgerE. Although P2cotE produced a stronger spore titer defect than PgerE when σK was tagged with ssrA* (Fig. 3B), we observed that its pattern of expression was variable, leading to premature degradation of target proteins in a fraction of the sporangia (not shown). We therefore decided to exclude P2cotE from further studies. The sustained and late promoters that we selected for use in our system are indicated by a blue box in Figure 3B.
Characterization of cell-specific degradation from sustained and late promoters
We next performed experiments to confirm that the selected promoters conferred the expected temporal and spatial regulation on sspBEc expression, and that they yielded enough SspBEc to completely degrade target proteins. First, we made transcriptional fusions to gfp to confirm the expression pattern from the different promoters using fluorescence microscopy (Fig. S2). As expected PsspE(2G) and PcotE conveyed sustained forespore- and mother cell-specific gfp expression that could be visualized shortly after polar septation, whereas PsspA and PgerE conveyed late expression in these cells. Expression of sspBEc from these promoters had no detectable effect on sporulation (Fig. 3B; Table S1). In addition, we compared the expression levels from the selected sustained or late promoters with the expression level of a xylose-inducible promoter (PxylA) induced with a xylose concentration (0.01 %) that we determined was enough to completely degrade abundant ssrA* tagged proteins during vegetative growth (Fig. S3). As shown in Fig. S3E, expression from sustained and late promoters yielded intracellular GFP concentrations that were at least 20 fold higher than the concentration achieved when GFP was expressed from PxylA induced with 0.01 % of xylose, suggesting that, when produced from the selected sustained and late promoters, SspBEc is not limiting for degradation. Finally, we performed microscopy to determine if the cytological profiles obtained upon cell-specific degradation of σG-ssrA* and σK-ssrA* matched those of the σG- and σK- mutants. We used a combination of fluorescence and phase contrast microscopy to assess the completion of two developmental milestones: engulfment membrane fission, which depends on early cell-specific gene expression, and forespore dehydration, which depends on late cell-specific gene expression. To assess the completion of engulfment, we simultaneously stained sporangia with two membrane dyes: FM 4–64, which fluoresces red and is membrane-impermeable, and Mitotracker Green (MTG), which fluoresces green and is membrane-permeable (Sharp and Pogliano, 1999). During engulfment, the membrane of the forespore is exposed to the culture medium, and is accessible to both FM 4–64 and MTG. After engulfment culminates with membrane fission, the forespore is completely enclosed within the mother cell cytoplasm and the forespore membranes can only be stained by the membrane-permeable dye MTG, but not FM 4–64. This allows sporangia that have completed engulfment membrane fission to be easily distinguished from those that have not (Sharp and Pogliano, 1999). To assay forespore dehydration, we used phase-contrast microscopy and observed the appearance of phase-bright spores.
Degradation of σG-ssrA* from the sustained promoter PsspE(2G) or the late promoter PsspA produced cytological profiles that phenocopied that of the σG- null mutant, with completion of engulfment membrane fission, but no forespore dehydration (Fig. 3C). Production of SspBEc from the mother cell-specific promoters PcotE or PgerE in strains producing σG-ssrA*, however, resulted in wild type profiles (Fig. 3C). Similarly, degradation of σK-ssrA* from PcotE or PgerE allowed completion of engulfment membrane fission, but yielded less dehydrated, phase-gray forespores, similar to the σK- null mutant (Fig. 3D). As expected, production of SspBEc from forespore-specific promoters did not trigger σK-ssrA* degradation and resulted in wild-type cytological profiles (Fig. 3D). To confirm that σG and σK were efficiently degraded, we tagged each protein with GFP and ssrA*. A clear GFP fluorescence signal was observed in the forespore and the mother cell in strains producing σG-GFP-ssrA* and σK-GFP-ssrA*, respectively (Fig. S4). The signal completely disappeared when sspBEc was expressed in the appropriate compartment from either sustained or late promoters (Fig. S4), indicating that both proteins were efficiently degraded. The cytological profiles and spore titers obtained after degradation of σG-GFP-ssrA* and σK-GFP-ssrA* were equivalent to those obtained after of σG-ssrA* and σK-ssrA* degradation (Fig. S4; Table S1).
It is interesting to note that degradation of σG and σK from late promoters produced cytological profiles equivalent to those of the null mutants (Fig. 3C, D; Fig. S4). Because transcription from late promoters depends on σG and σK, those σ factors must be initially activated, allowing a burst of expression of the genes under their control, including sspBEc. Once enough SspBEc has accumulated, it will trigger the degradation of σG-ssrA* and σK-ssrA*. Thus, these results suggest that σG and σK activity is required continuously during forespore maturation, and not just transiently after engulfment has been completed.
Overall, the above results demonstrate that production of SspBEc from developmentally-regulated promoters can be used to trigger the cell-specific degradation of sporulation-specific target proteins, yielding cytological profiles equivalent to the null mutants.
Cell- and stage-specific degradation of vegetative proteins
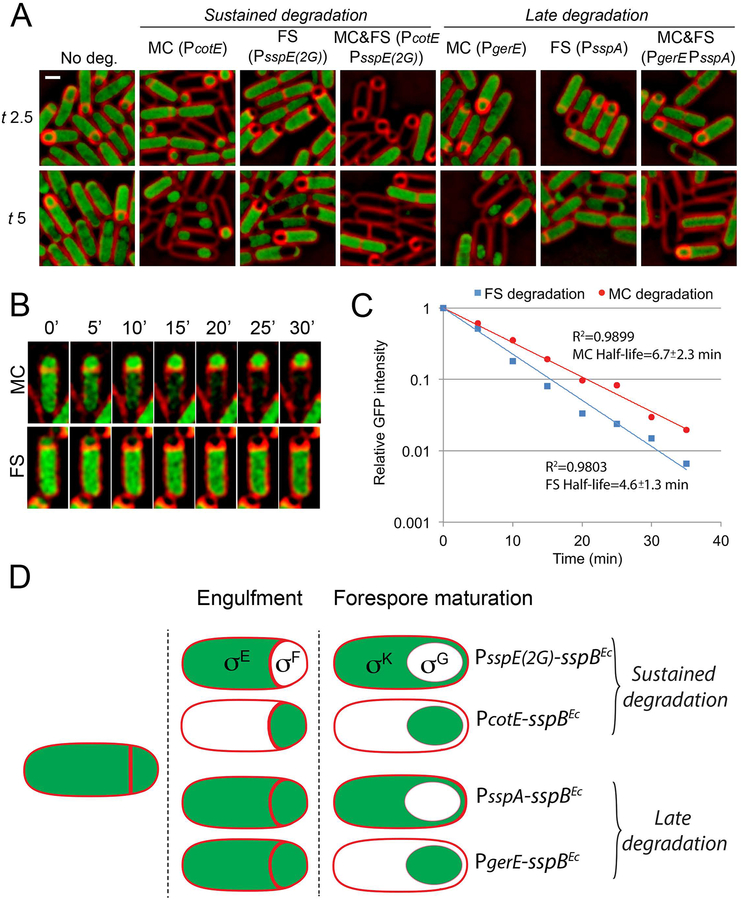
To assess whether expression of sspBEc from the selected promoters led to an efficient, spatiotemporally-regulated degradation of target proteins made before polar septation, we studied the degradation of a YtsJ-GFP-ssrA* fusion protein (Lamsa et al., 2016). YtsJ is an abundant, non-essential cytoplasmic protein that is not required for sporulation. We visually evaluated degradation based on the disappearance of GFP fluorescence (Fig. 4A). The results obtained can be summarized as follows: (i) When sspBEc was not expressed, a clear fluorescent signal was observed filling the cytoplasm of the mother cell and the forespore, as well as the cytoplasm of predivisional cells. (ii) Production of SspBEc from sustained promoters triggered the continuous degradation of YtsJ-GFP-ssrA* in the forespore (PsspE(2G)) or the mother cell (PcotE), starting shortly after polar septation. We also triggered degradation in both cells simultaneously by producing SspBEc from PsspE(2G) and PcotE at the same time; in this case, fluorescence was only observed in cells before polar septation. (iii) Production of SspBEc from the late promoters triggered degradation in the forespore (PsspA), in the mother cell (PgerE) or in both (PsspA PgerE) only after engulfment completion. In every case, no fluorescence was detected in the cell types in which YtsJ-GFP-ssrA had been degraded, indicating that SspBEc production led to the rapid and efficient degradation of the target protein.
Figure 4. Cell- and stage-specific degradation of vegetative proteins.
A. Fluorescence microscopy showing the cell-specific degradation of YtsJ-GFP-ssrA*. SspBEc was produced from sustained promoters (indicated in parentheses) in the mother cell (MC), forespore (FS), or both (MC&FS) to induce the sustained degradation of YtsJ-GFP-ssrA*. To induce degradation only after engulfment (late degradation), sspBEc was expressed from late promoters. Membranes were stained with FM 4–64. Scale bar, 1 μm.
B. Timelapse microscopy showing the disappearance of YtsJ-GFP-ssrA* fluorescence signal when degradation is induced in the mother cell (MC, sspBEc expressed from PcotE) or in the forespore (FS, sspBEc expressed from PsspE(2G)). Images taken every 5 minutes are shown. Membranes were stained with FM 4–64.
C. YtsJ-GFP-ssrA* degradation kinetics when SspBEc is produced in the forespore (blue squares and line) or in the mother cell (red circles and line). In both cases, the disappearance of the GFP signal follows an exponential decay. The coefficient of determination of the regression curves (R2), and the half-lives of YtsJ-GFP-ssrA* when degradation is induced in the mother cell or the forespore are indicated in the graph. Data represent the average of 17 and 14 sporangia for forespore and mother cell degradation, respectively.
D. STRP setup. The use of sustained promoters (PsspE(2G) and PcotE) allows the sustained degradation of target proteins in the mother cell or the forespore. Expression of sspBEc from late promoters (PsspA and PgerE), however, triggers degradation of target proteins only after engulfment is completed. The combination of both classes of cell-specific promoters allows the evaluation of the spatial and temporal requirement of proteins made before polar septation.
Next, we performed timelapse microscopy to determine how quickly YtsJ-GFP-ssrA* was degraded upon induction of sspBEc expression. We specifically used sustained promoters, which are active immediately after polar septation, to drive the expression of sspBEc, since polar septation provides a clear marker for the onset of degradation. We stained sporangia with the membrane dye FM 4–64 and monitored the disappearance of YtsJ-GFP-ssrA* fluorescence signal upon induction of sspBEc expression in the mother cell or the forespore from PcotE and PsspE(2G), respectively (Fig. 4B). To correct for GFP photobleaching due to continuous imaging, we calculated the ratio between the GFP intensity in the compartment in which degradation was induced and the other compartment. Once degradation started, the decrease in GFP fluorescence followed an exponential decay. Degradation was slightly faster in the forespore (protein half-life 4.6±1.3 min) than in the mother cell (protein half-life 6.7±2.3 min) (Fig. 4C). It has been reported that ClpXP is more active in the mother cell than in the forespore (Kain et al., 2008). It seems therefore unlikely that the faster degradation kinetics in the forespore is due to differences in ClpXP activity between both cells, in which case we would expect degradation to happen faster in the mother cell. Instead, the difference might reflect a slower accumulation of SspBEc in the mother cell due to its larger volume or differences in promoter strength. The kinetic data from cell-specific degradation are in good agreement with degradation speeds estimated at the population level in vegetative cells (Griffith and Grossman, 2008; Eswaramoorthy et al., 2014; Lamsa et al., 2016). Importantly, according to the estimated degradation rates, cell-specific expression of sspBEc would allow complete depletion of target proteins within ~20–30 min, which is significantly less time than required for engulfment under these conditions (~90 min, Ojkic et al. 2016).
These results show that expression of sspBEc from sustained and late promoters allow rapid degradation of proteins made before polar septation in a spatially and temporally controlled manner. Sustained promoters remain active in the forespore or mother cell throughout sporulation, while late promoters are active only after engulfment. Therefore, the use of both temporal classes of promoters to trigger degradation of target proteins, in combination with various assays to assess the completion of developmental milestones (Harwood et al., 1990), allows for the dissection of the temporal and cell-specific requirement of target proteins during sporulation (Fig. 4D). We have named this experimental approach spatiotemporally regulated proteolysis (STRP). In the next sections, we have used STRP to dissect the cell-specific role that two key vegetative sigma factors, σH and σA, play during sporulation.
The stationary phase sigma factor σH is only required in predivisional cells
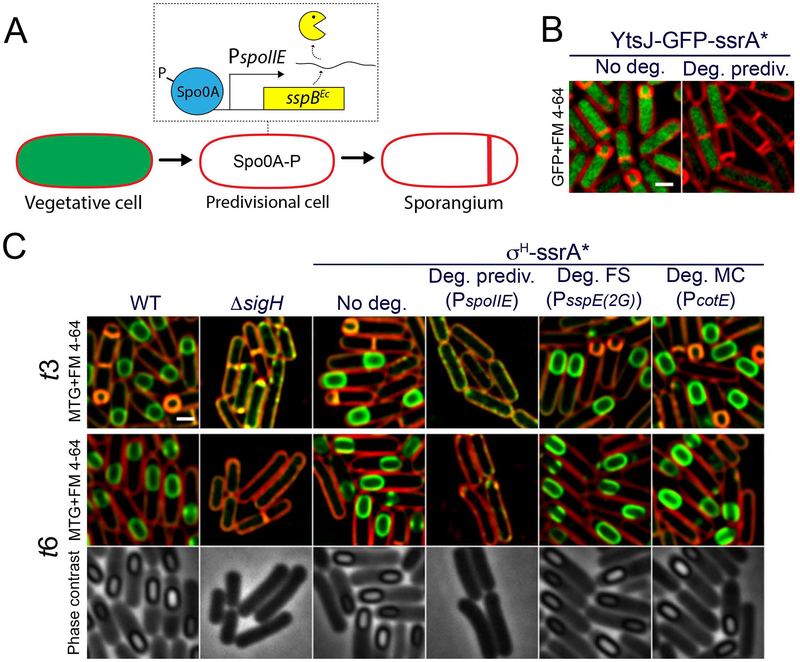
The initiation of sporulation requires the activation of the stationary-phase sigma factor, σH, which is required for polar septation. Accordingly, σH- mutants are unable to undergo polar septation and do not initiate sporulation (Fig. 5C, ΔsigH). While its importance in sporulation initiation is well established, it is unclear if σH activity is required in the mother cell or forespore after polar septation. To evaluate this possibility, we constructed a σH-ssrA* fusion protein, which supports sporulation at levels comparable to the wild type (Table S1). First, we validated that degradation of σH-ssrA* prior to polar septation suppressed the formation of the polar septum. For this purpose we used two different strategies. First, we induced degradation of σH-ssrA* by expressing sspBEc from a xylose-inducible promoter (Fig. S5), which triggers σH-ssrA* degradation continuously during the growth of the culture. Second, we expressed sspBEc from the Spo0A-dependent promoter PspoIIE, which becomes active in predivisional cells, shortly before polar septation (Fig. 5A). This second strategy restricts degradation to predivisional cells that are about to commit to sporulation, a point that we confirmed by monitoring the disappearance of YtsJ-GFP-ssrA* (Fig. 5B). In both cases, degradation of σH-ssrA* prior to polar septation resulted in the inhibition of polar septum formation (Fig. 5C; Fig. S5), confirming the requirement of σH-dependent transcription in predivisional cells to start sporulation. We next determined the cytological profiles obtained upon cell-specific degradation of σH-ssrA* from forespore- and mother cell-specific sustained promoters. As shown in Fig. 5C, degradation of σH-ssrA* in the forespore or mother cell yielded cytological profiles equivalent to the control strain. These results suggest that σH does not play a critical role in either the forespore or the mother cell. Altogether, our results indicate that σH-dependent transcription is required in predivisional cells until the polar septum is formed, but that it is dispensable after polar septation.
Figure 5. σH activity is only required in predivisional cells.
A. Diagram representing the strategy used to induce the degradation of target proteins in predivisional cells about to undergo polar septation. Before polar septation, Spo0A becomes phosphorylated and controls the expression of several genes required for entry into sporulation. Expression of sspBEc from promoters activated by Spo0A would trigger degradation in predivisional cells, before polar septation occurs. We specifically used the promoter of spoIIE to drive sspBEc expression.
B. Degradation of YtsJ-GFP-ssrA* in predivisional cells. Membranes were stained with FM 4–64. Scale bar, 1 μm.
C. Micrographs of wild type sporangia (WT), σH mutant sporangia (ΔsigH) and sporangia in which σH is tagged with ssrA* (σH-ssrA*) in the absence of SspBEc (No deg.) or when SspBEc is produced in predivisional cells from PspoIIE (Deg. prediv.), in the forespore from PsspE(2G) (Deg. FS) or in the mother cell from PcotE (Deg. MC). Pictures were taken 3 (t3) and 6 (t6) hours after sporulation induction. Membranes stained with FM 4–64 (red) and Mitotracker green (green) are shown for t3 and t6. Phase contrast pictures are only shown for t6. Scale bar, 1 μm.
Effect of cell-specific degradation of σA on spore formation
σA is the only essential sigma factor in B. subtilis, and it controls the transcription of housekeeping genes in vegetative cells (Haldenwang, 1995). During sporulation, σA is present in both the mother cell and the forespore (Yen Shin et al., 2015), and it is bound to a fraction of the core RNA polymerase (Ju et al., 1999; Fujita, 2000). In addition, it has been shown that σA is active and that it mediates gene transcription in both cells (Li and Piggot, 2001). However, the actual relevance of σA-mediated transcription during sporulation is not known.
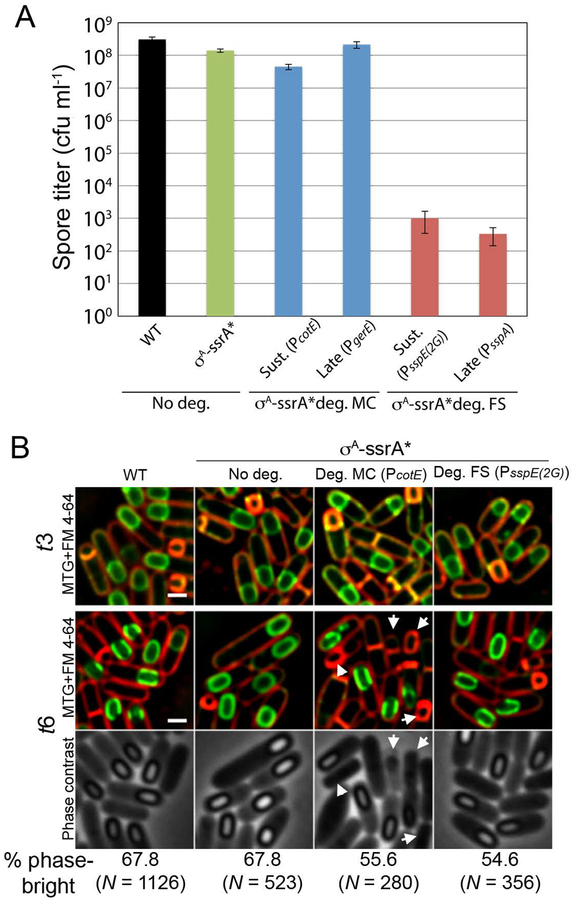
To address this question, we used a σA-ssrA* version that supported a vegetative growth rate close to that of wild type in the absence of degradation, but did not support growth when sspBEc expression was induced from a xylose-dependent promoter (Lamsa et al., 2016). First, we studied the effect of cell-specific degradation of σA-ssrA* from sustained and late promoters on spore titers, measured as the ability of heat-treated spores to form colonies in the presence of nutrients. In the absence of degradation, a strain containing σA-ssrA* showed a mild spore titer reduction of ~2–3 fold compared to the wild type (Fig. 6A), suggesting that the functionality of the tagged protein was somewhat impaired, but still enough to support spore formation. When degradation was induced in the mother cell from the sustained promoter PcotE, there was a modest additional 3-fold reduction in spore titer. No additional reduction was observed when SspBEc was produced from the late mother cell-specific promoter PgerE (Fig. 6A). These results suggest that σA-dependent transcription does not play a major role in the mother cell during sporulation, although it might contribute to replenish the pool of housekeeping proteins early during development. When degradation was induced in the forespore from sustained (PsspE(2G)) or late (PsspA) promoters, however, there was a dramatic reduction in the spore titer of ~five orders of magnitude (Fig. 6A). A similar reduction was observed when σA-ssrA* was degraded simultaneously in the mother cell and in the forespore (Table S1).
Figure 6. Cell-specific degradation of σA.
A. Spore titers, measured as the number of colony forming units per ml (cfu ml−1) in heat-killed sporulating cultures, of wild type (WT) and σA-ssrA* strains, and strains in which σA-ssrA* degradation is triggered in the mother cell (σA-ssrA* deg. MC) or in the forespore (σA-ssrA* deg. FS) from sustained (Sust.) or late promoters. The promoters used to drive the expression of sspBEc are indicated in parentheses. Data represent the average and standard deviation of at least three independent experiments.
B. Micrographs of wild type sporangia (WT) and σA-ssrA* sporangia in the absence of degradation (No deg.) or when degradation is induced in the forespore (Deg. FS), mother cell (Deg. MC) or both (Deg. both) from sustained promoters (in parentheses). Pictures were taken 3 (t3) and 6 (t6) hours after sporulation induction. Lysed cells upon mother cell degradation at t6 are indicated by arrows. The percentage of sporangia containing phase-bright spores at t6 is shown at the bottom, including the umber of sporangia (N) analyzed for each strain. Lysed sporangia were excluded from the quantification. Membranes stained with FM 6–64 (red) and Mitotracker green (green) for each time point. Phase contrast pictures are shown in the bottom row for t6. Scale bar, 1 μm.
To determine the specific sporulation steps affected by cell-specific degradation of σA-ssrA*, we performed microscopy and observed the cytological profiles upon degradation of σA-ssrA* from sustained promoters. As shown in Fig. 6B, tagging σA with ssrA* had no detectable effect on engulfment or on the formation of phase bright spores, compared to the wild type. When degradation was induced in the mother cell, we observed occasional cell lysis, which might explain the mild spore titer defect obtained upon σA-ssrA* degradation in the mother cell (Fig. 6A; Table S1). Nevertheless, a significant fraction of the forespores were phase bright by t6. Surprisingly, when σA-ssrA* was degraded in the forespore there was no detectable defect in spore formation, indicating that σA is not required in the forespore for the production of phase-bright spores. Altogether, these results indicate that σA-dependent transcription is dispensable in both the forespore and the mother cell for the production of apparently mature spores.
Forespore σA is required for spore germination and outgrowth
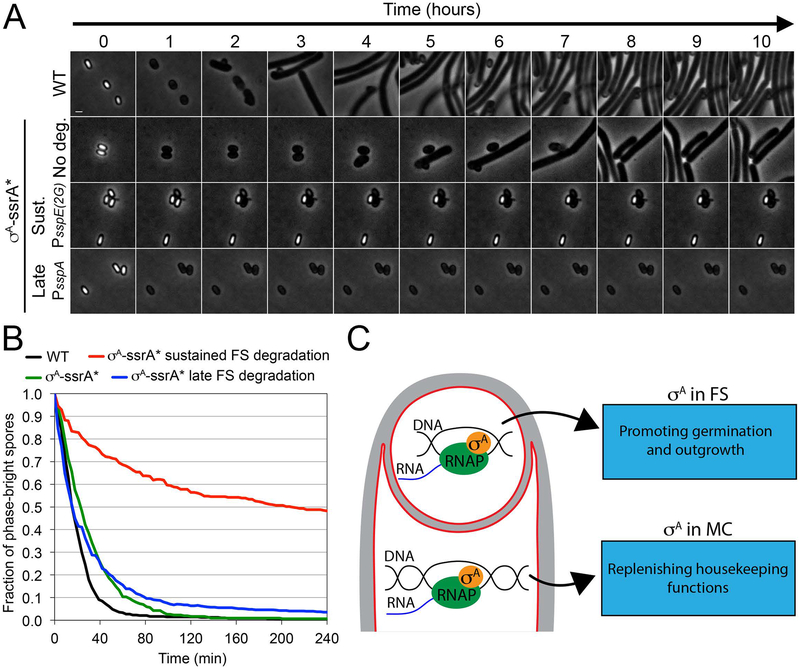
The above results suggest that the dramatic spore titer reduction observed when σA-ssrA* is degraded in the forespore is not due to defects in forespore development. We noted that when σA-ssrA* degradation was induced in the forespore from either sustained or late promoters, the few colonies formed by the germinating spores after overnight incubation at 30°C were remarkably small (not shown). We therefore reasoned that the spore titer defects observed after degradation of σA-ssrA* in the forespore might be the result of deficient germination or outgrowth of spores devoid of σA, rather than the consequence of compromised spore development. In agreement with this idea, we observed the appearance of an increasing number of colonies after longer incubation of the plates at 30°C (Fig. S6).
To further explore this possibility, we monitored germination and outgrowth of individual spores over time using phase-contrast timelapse microscopy. During germination, the spore core is rehydrated, which causes the spores to transition from phase-bright to phase-dark. Simultaneously, the spore cortex is degraded and the coat is detached, allowing the germinated spore to resume vegetative growth during a process called outgrowth. We purified spores from the wild type strain, σA-ssrA* (no degradation) strain, and from strains in which σA-ssrA* degradation was induced in the forespore from either sustained or late promoters. We induced germination by placing the spores on an LB agarose pad supplemented with 10 mM of L-alanine, and used timelapse phase-contrast microscopy to monitor germination and outgrowth over time (Fig. 7A). We also estimated germination efficiency at the population level by monitoring the loss in optical density of spore suspensions upon addition of 10 mM of L-alanine (Fig. S7). σA-ssrA* spores exhibited germination kinetics similar to that of wild-type spores (Movie S1 and S2; Fig. S7), with more than 90% phase-dark spores 80 minutes after imaging onset (Fig. 7B). Outgrowth was slightly delayed, however, compared to wild type (Fig. 7A), consistent with the idea that the σA-ssrA* fusion protein is not completely functional. Spores from a strain in which σA-ssrA* was degraded in the forespore from the late forespore promoter PsspA (Fig. 7A–B; Movie S4; Fig. S7) germinated normally, but showed no outgrowth, demonstrating that σA is required for the resumption of vegetative growth after germination, as expected. Surprisingly, spores from a strain in which σA-ssrA* was degraded from the sustained promoter PsspE(2G) showed a dramatic germination defect, with only ~50% of the spores transitioning to phase-dark four hours after adding the germinant L-alanine (Fig. 7A–B; Movie S3; Fig. S7). Those spores that germinated showed no outgrowth. This suggests that σA activity is required early in sporulation to produce spores that germinate efficiently, and that σA activity in the spore is necessary for outgrowth. Potential models to explain these results are discussed below.
Figure 7. Forespore σA contributes to spore germination and outgrowth.
A. Phase-contrast timelapse microscopy of germinating spores from wild type (WT), σA-ssrA* (No deg.), σA-ssrA* sustained forespore degradation (Sust.) and σA-ssrA* late forespore degradation (Late) strains. Images taken every hour for 10 hours are shown. Complete time series of images taken every 3 minutes for 10 hours are provided in movies S1–S4.
B. Fraction of phase-bright spores over time after germination induction, calculated from timelapse microscopy shown in B. Black line, wild type spores (N = 547); green line, σA-ssrA* spores (N = 318); red line, σA-ssrA* sustained forespore degradation spores (N = 434); blue line, σA-ssrA* late forespore degradation spores (N = 668). The fraction of phase bright spores at the onset of imaging was relativized to 1 for every strain. More than 84% of the spores were phase-bright initially.
C. σA-dependent transcription in the mother cell might contribute to replenish housekeeping functions during sporulation. In the forespore, σA-dependent transcription makes critical contributions to the germination and outgrowth of mature spores.
Discussion
Here we present a method, called spatiotemporally regulated proteolysis (STRP), to rapidly deplete target proteins in a cell- and developmental stage-specific manner during sporulation in B. subtilis. STRP allows the first systematic evaluation of the cell- and developmental-stage specific requirement of proteins made before polar septation, and has the potential to complete the list of the cellular machineries required to assemble a spore. It also provides a versatile new tool to study proteins that play critical roles at several stages of sporulation, such as the peptidoglycan biosynthetic machinery, which is required for polar septation, engulfment, and cortex biosynthesis.
A key feature of STRP is that it is based on the direct degradation of target proteins, rather than on the inhibition of transcription or translation. Our data indicate that target proteins are depleted within minutes after polar septation in the mother cell and in the forespore. This is critical to study the role of specific proteins during sporulation, when the lack of growth prevents the dilution of proteins and mRNAs that is required by other methods such as CRISPR interference.
An essential requirement to achieve the spatiotemporally controlled degradation of target proteins is the use of tightly regulated promoters to drive the expression of sspBEc. The sporulation regulatory program provides an excellent genetic framework for this purpose, since it includes a plethora of promoters that become active in specific cell types and developmental stages. Because these promoters show no ectopic expression, target protein degradation is only triggered in the cell and developmental stage in which the promoters are active. We have explored cell-specific promoters that are continuously active in the mother cell or the forespore after polar septation, which support the sustained degradation of target proteins throughout sporulation, and promoters that become active in either cell only when engulfment is completed, which trigger the late degradation of target proteins. This design provides a systematic method to determine the requirement of target proteins in the mother cell or the forespore at different developmental stages. Proteins required only during engulfment would cause sporulation defects only when subjected to sustained degradation, but not when subjected to late degradation. However, proteins required after engulfment would also produce sporulation defects upon late degradation. Altogether, our results show that STRP is well suited to dissect the spatiotemporal role of proteins made before polar septation, including essential proteins, during development. This opens new avenues in the study of sporulation, since it provides the possibility to investigate how vegetative cellular processes are compartmentalized and coordinated between different cell-types.
We here focus our efforts on the transcriptional machinery of sporulation, which provides an ideal system to test our methodology. In addition, there are key outstanding questions concerning the role played by vegetative transcription factors late in sporulation. We have used STRP to dissect the spatiotemporal requirement of two vegetative σ factors, σH and σA, during sporulation. σH controls gene expression upon entry into stationary phase, and some of the genes in the σH regulon are essential for sporulation initiation. Degradation of σH-ssrA* in predivisional cells by expressing sspBEc from a high-threshold Spo0A~P dependent promoter that is activated shortly before polar septation (PspoIIE; Fujita et al., 2005) blocks polar septation (Fig. 5C). This suggests that the initial σH activity after entry into stationary phase is not sufficient for initiation of sporulation. However, degradation of σH-ssrA* after polar septation in the mother cell, forespore or both does not produce any sporulation defects (Fig, 5C). Altogether, these results indicate that σH activity is required in predivisional cells until the polar septum is formed, but that after polar septation, σH activity is dispensable.
σA is required for the transcription of housekeeping genes in vegetative cells, and is the only essential σ factor in B. subtilis. During sporulation, σA is found in both mother cell and forespore (Yen Shin et al., 2015), and at least a fraction is bound to the core RNA polymerase (Ju et al., 1999; Fujita, 2000). Accordingly, σA-dependent transcription has been demonstrated in both cells (Li and Piggot, 2001). However, our results show that σA activity is asymmetrically required in mother cell and forespore. The sustained degradation of σA in the mother cell produces only a mild spore titer defect, and no spore titer defect is observed after late degradation (Fig. 6A). These results suggest that σA plays a minor role in the mother cell, perhaps to replenish housekeeping functions in sporangia in which they become limiting. It is therefore possible that the σA-dependent functions provided before polar septation are enough to support sporulation in most sporangia, or that transcription of key genes under σA-control can be mediated by either σE or σK during sporulation, relieving the need for σA-dependent transcription in the mother cell. In support of this latter hypothesis, it has been shown that the transcription of genes involved in the synthesis of peptidoglycan precursors becomes dependent on mother cell-specific σ factors during sporulation (Eichenberger et al., 2003; Eichenberger et al., 2004), which leads to the accumulation of peptidoglycan precursors for cortex synthesis after engulfment (Vasudevan et al., 2007).
Degradation of σA in the forespore produces a dramatic spore titer reduction (Fig. 6A) that our data suggest is the result of germination and outgrowth defects (Fig. 6; Fig. 7). Germination of dormant, dehydrated spores is triggered by specific germinants that are recognized by receptors in the inner spore membrane. This triggers the release of dipicolinic acid, probably through channels made of SpoVA proteins, which leads to a partial rehydration of the spore core. Subsequently, cell wall lytic enzymes that degrade the cortex are activated, allowing the full expansion and rehydration of the spore core, in preparation for outgrowth (Setlow, 2014). During outgrowth, the germinated spore is transformed into a rod-shaped, growing, vegetative cell. Outgrowth is completely abolished when σA is degraded from either sustained or late promoters (Fig. 7; Movies S3 and S4). This is expected, since σA-mediated gene expression is likely required to resume growth after exiting dormancy. It seems reasonable to think that degradation of σA-ssrA* in the forespore results in mature spores devoid of σA. Therefore, when transcription restarts after germination, σA-dependent housekeeping transcripts will not be produced, preventing the growth of the reviving spore. Even if new σA-ssrA* molecules are produced during outgrowth, they will be quickly degraded due to the presence of residual SspBEc in the spores, which will target σA-ssrA* for degradation by ClpXP.
The germination defect is only observed when σA is continuously degraded in the forespore, starting shortly after polar septation. We conceive two potential models to explain this observation: First, it is possible that σA-dependent transcription is required in germinating spores to mediate the transition from phase-bright to phase-dark spores, which is consistent with recent results suggesting that translation happens during germination and that it is required to complete the germination process (Sinai et al., 2015). However, we consider this possibility unlikely, since late degradation of σA in the forespore completely blocks outgrowth, but does not produce a significant germination defect (Fig. 7). Of course, it is possible that some intact σA remains in the spores after late degradation, and that this supports germination. However, the outgrowth defect after late σA-ssrA* degradation argues against this possibility.
The second model to explain the germination defect observed when σA is continuously degraded in the forespore is that σA-dependent transcription is required in the developing forespore, early during spore formation, to produce germination-proficient spores. In this context, it is possible that specific factors produced in the forespore under σA-control are required for the germination of mature spores. Candidates for such factors are translation proteins that are required for efficient germination, which must be present in mature spores or quickly synthesized at the onset of germination (Sinai et al., 2015). It is also possible that σA-dependent transcription plays a more general role in developing forespores, by providing housekeeping functions that confer to the forespore an optimal metabolic or structural state that promotes the formation of a functional germination machinery. Nevertheless, further experiments are required to clarify the requirement of σA in the forespore for germination.
Altogether, these results outline a potential application of STRP to identify cellular pathways involved in spore germination and outgrowth. This is particularly appealing, since it is unclear whether specific biosynthetic activities are required in the dehydrated, dormant spore to mediate germination (Sinai et al., 2015; Boone and Driks, 2016; Korza et al., 2016; Sinai and Ben-Yehuda, 2016).
As discussed above, the use of targeted degradation to deplete proteins has key advantages compared to other methods. However, it also has limitations that are important to consider. Perhaps the most apparent limitation is that not all proteins are suitable for degradation. The ssrA*/SspBEc degradation system requires that target proteins have an accessible cytoplasmic C-terminus for degradation, which excludes all extracellular proteins and membrane proteins with extracytoplasmic C-termini as potential targets, as well as cytoplasmic proteins in which the tag is sterically inaccessible due to the folding of the protein. The use of degradation systems based on N-terminal tags might provide an alternative to degrade some of the membrane proteins with extracytoplasmic C-termini (Sekar et al., 2016). Second, the inducible protein degradation system entails the modification of the target protein by adding the fifteen amino acid ssrA* tag, which might significantly affect protein functionality. However, ~95% of the ~200 proteins we have tagged so far (see below) tolerate the addition of the tag, suggesting that the ssrA* tag does not tend to significantly interfere with protein folding nor function.
Altogether, our results show that STRP holds great potential for providing a comprehensive molecular dissection of not only spore development, but also germination and outgrowth. We have undertaken a genome-wide approach to tag with ssrA* every essential protein with cytoplasmic C-termini in B. subtilis. The systematic exploration of our collection will provide a comprehensive overview of the spatiotemporal role of essential vegetative cellular pathways during the different developmental stages in B. subtilis.
Experimental procedures
Strain construction
All the strains used in this study are derivatives of B. subtilis PY79, and are listed in Table S2. A list of plasmids and primers as long with detailed descriptions of plasmid construction, is provided in supplementary information (Tables S3 and S4). In every case, ssrA* was added to the appropriate gene in its native locus. The amino acid sequence of the ssrA* used in our constructs was AANDENYSENYALGG, and was previously described by Griffith and Grossman (2008). sspBEc was inserted in different ectopic loci in B. subtilis chromosome, depending on the cell in which expression was taking place. For forespore-specific expression, we selected the amyE locus, which is trapped in the forespore after polar septation (Wu and Errington, 1998), allowing immediate expression before the forespore chromosome is fully translocated. For mother cell-specific expression, we inserted sspBEc in the terminus-proximal pelB locus, which encodes a pectate lyase involved in polygalacturonic acid degradation. Because pelB (174°) is located in the chromosome close to the dif site (172°) (Sciochetti et al., 2001), it is one of the last loci to be translocated to the forespore (Ptacin et al., 2008), which can transiently boost sspBEc expression in the mother cell after polar septation. We previously inserted sspBEc in the ectopic thrC locus for mother cell-specific expression. This creates a threonine auxotrophy, and therefore, the sporulation medium has to be supplemented with threonine. Under those conditions sporulation progress normally through engulfment, but we have observed that forespore maturation is delayed compared to threonine prototroph strains (not shown). For expression in predivisional cells, sspBEc was inserted in the lacA locus. Xylose inducible expression was achieved by inserting sspBEc in the xylA locus (Lamsa et al., 2016), which simultaneously creates a mutant unable to use xylose as a carbon source, preventing the depletion of the inducer.
Culture conditions
For microscopy experiments, sporulation was induced by resuspension (Sterlini and Mandelstam, 1969), except that the bacteria were grown in 25% LB prior to resuspension, rather than CH medium. Cultures were grown at 37°C for batch culture microscopy experiments, and at 30°C for timelapse fluorescence microscopy experiments.
For determination of heat-resistant spore titers and spore purification, cultures were grown and sporulated in DSM broth (Schaeffer et al., 1965).
To study YtsJ-GFP-ssrA* degradation and PxylA expression in vegetative cells (Fig. S3), cultures of the appropriate strains were grown in LB at 37°C until O.D.600 0.2. The cultures were than split into 2 ml aliquots and incubated for 45 minutes in LB or LB supplemented with different concentrations of xylose, at 37°C. Then 12 μl aliquots were used for microscopy, and 1 ml aliquots were used for protein extraction for Western blot.
Spore titer assay
Strains were cultured in 2 ml of DSM for 24 h at 37°C. Cultures were then heated at 80°C for 20 min, serially diluted, and plated on LB. Spore titers were calculated based on colony counts.
Spore purification
Sporulation was induced by exhaustion in 250 ml of DSM. Strains were grown for 72 h and then collected and washed with 4°C sterile water. The spores were incubated in sterile water at 4°C to lyse any remaining vegetative cells. Spores were further purified over a phosphate-polyethylene glycol aqueous biphasic gradient (Harrold et al., 2011). The top organic phase containing the spores was removed and extensively washed in at least 50 vol of 4°C sterile water. The spores were pelleted, resuspended in sterile water, and stored at 4°C. Sample purity was checked using phase contrast microscopy. Spore samples of >95% purity (phase-bright spores) were used in the studies.
Batch culture microscopy
Samples from sporulation cultures were taken at the indicated time points and transferred to 1.2% agarose pads for imaging. When appropriate, membranes were stained with 0.5 μg ml−1 FM 4–64 (Life Technologies) and 1 μg ml−1 Mitotracker green (Life Technologies). FM 4–64 was added to the agarose pad, whereas Mitotracker green was mixed with the cells before transferring them to the pad. Cells were visualized on an Applied Precision DV Elite optical sectioning microscope equipped with a Photometrics CoolSNAP-HQ2 camera. Pictures were deconvolved using SoftWoRx v5.5.1 (Applied Precision). The median focal plane is shown.
Timelapse fluorescent microscopy to visualize YtsJ-GFP-ssrA* degradation
Sporulation was induced at 30°C. 1.5 h after sporulation induction, 0.5 μg ml−1 FM 4–64 was added to the culture and incubation continued for another 1.5 h. Seven-μl samples were taken 3 h after resuspension and transferred to agarose pads prepared as follows: 2/3 volume of supernatant from the sporulation culture; 1/3 volume 3.6% agarose in fresh A+B sporulation medium; 0.17 μg ml−1 FM 4–64. Pads were partially dried, covered with a glass slide, and sealed with petroleum jelly to avoid dehydration during timelapse imaging. Pictures were taken in an environmental chamber at 30°C every 5 min for 5 h. To visualize FM 4–64 stained membranes, excitation/emission filters were TRITC/CY5, excitation light transmission was set to 5%, and exposure time was 0.1 s. To visualize GFP signal, excitation/emission filters were FITC/FITC, excitation light transmission was set to 50%, and exposure time was 0.5 s.
Quantification of YtsJ-GFP-ssrA* degradation kinetics
To calculate YtsJ-GFP-ssrA* degradation kinetics, we measured the mean cytoplasmic GFP fluorescence in the mother cell and the forespore over time by drawing polygons encompassing either cell. After subtracting the mean background fluorescence, we calculated the mean fluorescence ratio between the cell in which degradation was triggered and the other cell of the same sporangium. This allowed us to correct for GFP photobleaching. We defined time zero as the time frame immediately before GFP degradation was first observed, and relativized the fluorescence ratios of subsequent time points with respect to time zero, that was arbitrarily given the value of one. The decrease in the fluorescence ratio over time followed an exponential decay curve. We fitted the data for each sporangia to an exponential decay function and determined the protein half-life upon degradation induction for individual sporangia. The reported half-lives are the averages ± standard deviations of 17 and 14 sporangia for forespore and mother cell degradation, respectively. The graph in Fig. 4C shows the average fluorescence ratio of the different sporangia over time.
Timelapse phase contrast microscopy to visualize spore germination and outgrowth
Purified spores were diluted in water to an O.D.600 of 0.3, and 10 μl of the spore suspension were placed on 1.2% agarose pads prepared in LB and supplemented with the germinant L-alanine (10 mM). Pads were partially dried, covered with a glass slide, and sealed with petroleum jelly to avoid dehydration during timelapse imaging. Phase contrast pictures were taken at 30°C every 3 minutes during 10 hours. Light transmission was set to 32% and exposure time was 0.1 s. Note that, after placing the spores in the agarose pads, there was a time lag of ~15 minutes until we started taking pictures. To minimize the number of spores that germinated during this time, spores were not heat-activated before timelapse microscopy. We quantified the number of spores that transitioned from phase-bright to phase-dark during the first four hours of imaging, focusing only on spores that were phase bright at the onset of imaging.
Quantification of spore germination by loss of optical density
Germination assays by loss of O.D.580 were performed as described in Harwood et al. 1990. Briefly, spores were heat activated at 70°C for 20 min in water, and resuspended in 10 mM Tris-HCl, pH 8.4, to obtain an O.D.580 of 0.3. The resuspended spores were incubated at 37°C for 20 min, and then germination was induced by the addition of L-alanine 10 mM. O.D.580 was measured every 5 min for an hour.
Protein extracts and Western blot analysis
Bacterial cultures were grown as described in “Culture conditions”. Bacterial cells present in 1 ml aliquots of the cultures were collected by centrifugation (5,000 × g, 2 min) and suspended in 60 μl of Tris-sucrose buffer [33 mM Tris-HCl, pH8.0; 40% sucrose; 1 mM EDTA] containing 1 mg ml−1 lysozyme, 160 μg ml−1 PMSF, and 1:100 diluted Sigma P2714 protease inhibitor cocktail, and incubated at 37°C for 10 min. Then, membranes were dissolved by adding 2 μl of Triton X-100, samples were mixed with 60 μl of Laemmli loading buffer [1.3% SDS; 10% (v/v) glycerol; 50 mM Tris-HCl; 1.8% β-mercaptoethanol; 0.02% bromophenol blue, pH 6.8] and boiled for 10 min. Proteins present in 5-μl aliquots of each sample were resolved by Tris-Tricine-PAGE, using 12% gels. Conditions for protein transfer have been described elsewhere. Optimal dilutions of anti-SspBEc and anti-σA primary antibodies were 1:5,000. Goat anti-rabbit horseradish peroxidase-conjugated antibody (1:20,000) was used as secondary antibody. Proteins recognized by the antibodies were visualized by chemoluminescence using the luciferin-luminol reagents, in a BioRad Gel Doc XR+ System. For quantification, the intensity of the bands was determined using ImageJ. σA was used as loading control.
Supplementary Material
Acknowledgements
We are thankful to Prof. Tania Baker for providing anti-SspBEc antibody and to Prof. David Rudner for providing pDR110. This work was supported by NIH Grant R01-GM57045. JLG was supported by an EMBO Long Term Fellowship (ALTF 1274–2011) during part of the duration of this project. AT, MLR and MD were part of the Magistère de Génétique Graduate Program, (Université Paris Diderot, 75013 Paris, France), which supported their stay in the lab. K. Pogliano is a co-founder of Linnaeus Bioscience Inc (La Jolla, CA), a shareholder, and a member of the Advisory Board. The terms of this arrangement have been reviewed and are managed by the University of California, San Diego in accordance with its conflict of interest policies.
References
- Boone T, and Driks A (2016) Protein synthesis during germination: Shedding new light on a classical question. J Bacteriol 198: 3251–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek E, Ponticorvo L, and Rittenberg D (1958) Protein turnover in micro-organisms. Proc Natl Acad Sci U S A 44: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S, Lopez-Diaz I, and Mandelstam J (1986) Use of lacZ gene fusions to determine the dependence pattern of the sporulation gene spoIID in spo mutants of Bacillus subtilis. J Gen Microbiol 132: 2987–2994. [DOI] [PubMed] [Google Scholar]
- Cookson NA, Mather WH, Danino T, Mondragón-Palomino O, Williams RJ, Tsimring LS, and Hasty J (2011) Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol Syst Biol 7: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S, Panzer S, and Losick R (1989) Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol 207: 393–404. [DOI] [PubMed] [Google Scholar]
- Eichenberger P (2012) Genomics and cellular biology of endospore formation In Bacillus : cellular and molecular biology. Graumann PL (ed.). Caister Academic Press, Norfolk, UK: pp. 319–350. [Google Scholar]
- Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, Ooij C Van Silvaggi J, Gonzalez-Pastor JE, et al. (2003) The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327: 945–972. [DOI] [PubMed] [Google Scholar]
- Errington J (2003) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1: 117–126. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Winter PW, Wawrzusin P, York AG, Shroff H, and Ramamurthi KS (2014) Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet 10: e1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M (2000) Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5: 79–88. [DOI] [PubMed] [Google Scholar]
- Fujita M, González-Pastor JE, and Losick R (2005) High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187: 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, and Grossman AD (2008) Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol 70: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, and Stragier P (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61. [DOI] [PubMed] [Google Scholar]
- Haldenwang WG (1995) The sigma factors of Bacillus subtilis. Microbiol Rev 59: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrold ZR, Hertel MR, and Gorman-Lewis D (2011) Optimizing Bacillus subtilis spore isolation and quantifying spore harvest purity. J Microbiol Methods 87: 325–329. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM, Chambert R, Galizzi A, Gally D, Gruss AD, et al. (1990) Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, New York, Brisbane, Toronto, Singapore. [Google Scholar]
- Higgins D, and Dworkin J (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, and Piggot PJ (2004) Compartmentalization of gene expression during Bacillus subtilis spore formation. 68: 234–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, and Grossman AD (1993). Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev 7: 283–294. [DOI] [PubMed] [Google Scholar]
- Ju J, Mitchell T, Peters H, and Haldenwang WG (1999) Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J Bacteriol 181: 4969–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain J, He GG, and Losick R (2008) Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol 190: 6749–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow LM, Glaser P, and Piggot PJ (1995) Identification of a gene, spoIIR, which links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 92: 2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL, and Levy HR (1955) Protein turnover in growing cultures of Escherichia coli. J Biol Chem 217: 947–957. [PubMed] [Google Scholar]
- Kock H, Gerth U, and Hecker M (2004) The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J Bacteriol 186: 5856–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korza G, Setlow B, Rao L, Li Q, and Setlow P (2016) Changes in Bacillus spore small molecules, rRNA, germination and outgrowth after extended sub-lethal exposure to various temperatures: evidence that protein synthesis is not essential for spore germination. J Bacteriol 198: 3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa A, Lopez-Garrido J, Quach D, Riley EP, Pogliano J, and Pogliano K (2016) Rapid Inhibition Profiling in Bacillus subtilis to identify the mechanism of action of new antimicrobials. ACS Chem Biol 11: 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelakriangsak M, Kobayashi K, and Zuber P (2007) Dual negative control of spx transcription initiation from the P 3 promoter by repressors PerR and YodB in Bacillus subtilis. J Bacteriol 189: 1736–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, and Piggot PJ (2001) Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc Natl Acad Sci U S A 98: 12538–12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garrido J, Ojkic N, Khanna K, Wagner F, Villa E, Endres R and Pogliano K (2018) Chromosome translocation inflates Bacillus forespores and impacts cellular morphology. Cell. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, Fréhel C, and Stragier P (1997) spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24: 29–39. [DOI] [PubMed] [Google Scholar]
- Mandelstam J (1958) Turnover of protein in growing and non-growing populations of Escherichia coli. Biochem J 69: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker T. a, and Sauer RT (2006) Engineering controllable protein degradation. Mol Cell 22: 701–707. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, and Eichenberger P (2013) The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Küster-Schöck E, Grossman AD, and Zuber P (2003) Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A 100: 13603–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Zheng G, Nakano MM, and Zuber P (2002) Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J Bacteriol 184: 3664–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula J, Fujita M, and Igoshin OA (2016) Functional requirements of cellular differentiation: lessons from Bacillus subtilis. Curr Opin Microbiol 34: 38–46. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Sun DX, Setlow B, and Setlow P (1989) Promoter specificity of sigma G-containing RNA polymerase from sporulating cells of Bacillus subtilis: identification of a group of forespore-specific promoters. J Bacteriol 171: 2708–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, López-Garrido J, Pogliano K, and Endres RG (2016) Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. Elife 5: e18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, and Bernhards CB (2015) Spore Peptidoglycan. Microbiol Spectr 3: TBS-0005–2012. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, and Bustamante C (2008) Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol 15: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels S, Driks A, and Losick R (1992) Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol 174: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S, Rosenkrantz MS, and Sonenshein AL (1986) Transcriptional control of the Bacillus subtilis spoIID gene. J Bacteriol 165: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygus T, and Hillen W (1991). Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl. Microbiol. Biotechnol 35: 594–599. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, and Aubert JP (1965) Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A 54: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti S, Piggot P, and Blakely G (2001) Identification and characterization of the dif site from Bacillus subtilis. J Bacteriol 183: 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar K, Gentile AM, Bostick JW, and Tyo KEJ (2016) N-terminal-based targeted, inducible protein degradation in Escherichia coli. PLoS One 11: e0149746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101: 514–525. [DOI] [PubMed] [Google Scholar]
- Setlow P (2014) Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, and Pogliano K (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci U S A 96: 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, and Pogliano K (2002) Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295: 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L, and Ben-Yehuda S (2016) Commentary: Changes in Bacillus spore small molecules, rRNA, germination, and outgrowth after extended sublethal exposure to various temperatures: Evidence that protein synthesis is not essential for spore germination. Front Microbiol 7: 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L, Rosenberg A, Smith Y, Segev E, and Ben-Yehuda S (2015) The molecular timeline of a reviving bacterial spore. Mol Cell 57: 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M, and Richter R (1994). Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142: 79–83. [DOI] [PubMed] [Google Scholar]
- Sterlini JM, and Mandelstam J (1969) Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Fajardo-Cavazos P, Sussman MD, Tovar-Rojo F, Cabrera-Martinez RM, and Setlow P (1991) Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J Bacteriol 173: 7867–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, and Ramamurthi KS (2014) Spore formation in Bacillus subtilis. Environ Microbiol Rep 6: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, and Popham DL (2007) Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol 65: 1582–1594. [DOI] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, et al. (2006) The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358: 16–37. [DOI] [PubMed] [Google Scholar]
- Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, et al. (2011) Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci U S A 108: 4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (1998) Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol 27: 777–786. [DOI] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (2000) Identification and characterization of a new prespore-specific regulatory gene, rsfA, of Bacillus subtilis. J Bacteriol 182: 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Shin J, Lopez-Garrido J, Lee S-H, Diaz-Celis C, Fleming T, Bustamante C, and Pogliano K (2015) Visualization and functional dissection of coaxial paired SpoIIIE channels across the sporulation septum. Elife 4: e06474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, and Losick R (1984). A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet 195: 424–433. [DOI] [PubMed] [Google Scholar]
- Zheng L, and Losick R (1990) Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol 212: 645–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.