Abstract
β-Lactamases are the resistance enzymes for β-lactam antibiotics, of which four classes are known. β-lactamases hydrolyze the β-lactam moieties of these antibiotics, rendering them inactive. It is shown herein that the class D OXA-10 β-lactamase depends critically on an unusual carbamylated lysine as the basic residue for both the enzyme acylation and deacylation steps of catalysis. The formation of carbamylated lysine is reversible. Evidence is presented that this enzyme is dimeric and carbamylated in living bacteria. High-resolution x-ray structures for the native enzyme were determined at pH values of 6.0, 6.5, 7.5, and 8.5. Two dimers are present per asymmetric unit. One monomer in each dimer was carbamylated at pH 6.0, whereas all four monomers were fully carbamylated at pH 8.5. At the intermediate pH values, one monomer of each dimer was carbamylated, and the other showed a mixture of carbamylated and non-carbamylated lysines. It would appear that, as the pH increased for the sample, additional lysines were “titrated” by carbamylation. A handful of carbamylated lysines are known from protein crystallographic data, all of which have been attributed roles in structural stabilization (mostly as metal ligands) of the proteins. This paper reports a previously unrecognized role for a noncoordinated carbamylate lysine as a basic residue involved in mechanistic reactions of an enzyme, which indicates another means for expansion of the catalytic capabilities of the amino acids in nature beyond the 20 common amino acids in development of biological catalysts.
In the presence of compelling physiological needs, nature aptly develops catalysts to meet the given requirement of the organism. Such is clearly the case in evolution of determinants for resistance to antibiotics (1, 2). The organisms that evolve or acquire such mechanisms are capable of surviving the antibiotic challenge. Among the multiple mechanisms for resistance to antibiotics that nature has devised (1, 2), resistance enzymes for β-lactam antibiotics are the best understood. β-Lactamases catalyze hydrolysis of the β-lactam moieties of these antibacterial agents; thereby, the drug is made ineffective. Because of the extensive clinical use of β-lactam antibiotics over the past 50 years, a strong selection power has favored organisms that harbor the genes for these resistance enzymes. Over 340 distinct β-lactamases are known (3, 4) that fall into four enzyme classes: A, B, C, and D. It is now accepted that β-lactamases have descended from a family of bacterial enzymes referred to as penicillin-binding proteins (5, 6). Furthermore, each class of β-lactamases has evolved from a distinct penicillin-binding protein (6). It would appear that, in the course of these evolutionary processes, four distinct catalytic machineries have developed (7–9).
Class B enzymes are zinc dependent, whereas the remaining three classes have pursued an active-site-serine strategy. The enzymes of classes A, C, and D undergo acylation by the substrate at a serine residue, and the acylated enzyme species undergoes deacylation in the second step of the catalytic process. Although the acylation event has been handed down from the parental penicillin-binding proteins and appears to be shared among these enzymes, the mechanisms for deacylation are different for all three classes, including factors such as the directions of the approach of the hydrolytic water molecule, as well as the details of the catalytic facilitation by the enzymes (3, 6, 7, 9).
Enzymes of class D are the least understood. The structural information for these enzymes has emerged only recently (8, 10–12). A feature for the catalytic events of class D enzymes appears to be the use of the same residues for both the enzyme acylation and deacylation steps (8). Furthermore, the residue of central importance for this enzyme appears to be a carbamylated lysine. As will be discussed herein, carbamylated Lys-70 is the basic residue that promotes the serine hydroxyl for the acylation step. This carbamylated lysine is also in a suitable position to activate the incoming hydrolytic water for the deacylation event. Hence, class D enzymes enjoy symmetry in their catalytic events. We provide evidence in this report that carbamylation of lysine is indispensable for the activity of the OXA-10 class D β-lactamase, and that it provides a means for expansion of the catalytic capabilities of the amino acids in proteins beyond the 20 common amino acids in development of this biological catalyst.
Materials and Methods
Antibiotics and other reagents were purchased from Sigma, unless otherwise stated. The growth medium was purchased either from Difco or Fisher Scientific. The chromatography media were from Bio-Rad Laboratories, and NaH13CO3 (99% enriched) was purchased from Cambridge Isotope Laboratories (Cambridge, MA). NaH14CO3 was purchased from Amersham Pharmacia. Escherichia coli BL21(DE3) was from GIBCO/BRL; E. coli DH5α and plasmid pET24a+ were from Novagen.
Isolation and Purification of the OXA-10 Wild-Type and Lys-70-Ala Mutant β-Lactamases.
The Lys-70-Ala mutant was generated by using the QuickChange Site-Directed mutagenesis protocol (Stratagene). Specifically, the oxa-10 gene in the pET24a+ vector (8) was amplified by using Turbo PfuI DNA polymerase and two mutagenic primers: Lys70AlaD, CCAGCATCAATTTGCGATCCCCAACGC; and Lys70AlaR, GCGTTGGGGATCGCAAATGTTGATGCTGG. The PCR product was further treated with DpnI restriction enzyme to digest the methylated DNA, and E. coli DH5α was transformed by plasmid DNA. The nucleotide sequence of the entire oxa gene was verified, and E. coli BL21(DE3) was retransformed with it. Both the wild-type and mutant β-lactamases were purified from the growth medium and the periplasmic space according to a published protocol (8). We followed the procedure of Neu and Heppel in liberation of the periplasmic content (13).
Modification of the OXA-10 β-Lactamase by 14CO2.
The OXA-10 β-lactamase (1.3 mg) was dialyzed against degassed 25 mM sodium acetate buffer (pH 4.5), and then against degassed 100 mM sodium phosphate buffer (pH 7.5). The solution was supplemented with 20 mM NaH14CO3 with specific activity of 1.0 mCi/mmol and was subjected to a Sephadex G-25 (1 × 20 cm) desalting column preequilibrated with 100 mM sodium phosphate buffer (pH 7.5), under an atmosphere of argon. Elution was performed with the same buffer. Aliquots of 1 ml were collected, and radioactivity was measured. This experiment was performed three times. The entire experiment was also carried out with nonradioactive sodium bicarbonate (a total of four times) to measure the protein yield and the activity after the column.
13C NMR Experiments.
The OXA-10 β-lactamase (10 mg) was dialyzed against several changes of degassed 25 mM sodium acetate buffer (pH 4.5), and then against degassed 10 mM sodium phosphate buffer, 0.1 mM EDTA (pH 7.5). Finally, the enzyme was dialyzed against the last buffer containing 10% D2O and 20 mM NaH13CO3 and then concentrated to give a concentration of 1 mM. The 13C NMR spectrum of the OXA-10 β-lactamase modified by 13C-labeled carbon dioxide was collected at 25°C. After determination of the 13C NMR spectrum for the OXA-10 enzyme, the enzyme was dialyzed against 25 mM sodium acetate buffer (pH 4.5), followed by 10 mM sodium phosphate buffer, 0.1 mM EDTA (pH 7.5), and 10% D2O to allow for the removal of the carbamate from the enzyme, and the 13C NMR spectrum was determined again. The Lys-70-Ala mutant variant was treated similarly, and its 13C NMR spectrum was determined at 25°C.
Determination of Carbon Dioxide Binding Constants.
Unless otherwise stated, all of the buffers were degassed under vacuum and were subsequently purged by argon. The enzyme (2.9 μM, 100 μl) was decarbamylated by incubation in 25 mM sodium acetate buffer (pH 4.5), under vacuum for 8–10 min. The activity of the enzyme (23 nM) was measured by monitoring hydrolysis of penicillin G (200 μM) in 100 mM sodium phosphate buffer (pH 7.5) in a total volume of 500 μl. Carbon dioxide was provided by the addition of concentrated NaHCO3, prepared in the assay buffer. The carbon dioxide concentration in solution was calculated as described before (14). The ionic strength was kept constant at 0.3 M by the addition of Na2SO4. Fitting of the results was carried out by the grafit software to a one-site-binding equation. Fluorometric measurements were performed in a Spex Industries (Metuchen, NJ) Fluoromax luminescence spectrophotometer to determine the binding constant of carbon dioxide to the OXA-10 β-lactamase. The protein was excited at 295 nm, and emission was recorded at 340 nm. The slit width was kept at 0.4 or 0.5 mm. Experiments were carried out at 25°C in 100 mM sodium phosphate buffer (pH 7.5). Aliquots of concentrated NaHCO3, prepared in the same buffer, were added to the enzyme solution (2 μM) to provide the desired carbon dioxide concentrations.
Kinetic Studies of the OXA-10 β-Lactamase.
All kinetics measurements were performed on a Hewlett-Packard 8453 diode array spectrophotometer, at room temperature in 100 mM sodium phosphate buffer (pH 7.0) with and without supplements of sodium bicarbonate. The enzyme concentration in the assays was varied from 10 nM to 70 nM depending on the substrate. Hydrolysis of the substrates was monitored at the corresponding wavelengths: ampicillin at 240 nm (Δɛ = 538 M−1⋅cm−1), carbenicillin at 240 nm (Δɛ = 400 M−1⋅cm−1), penicillin G at 240 nm (Δɛ = 570 M−1⋅cm−1), oxacillin at 260 nm (Δɛ = 258 M−1⋅cm−1), cloxacillin at 260 nm (Δɛ = 140 M−1⋅cm−1), cephaloridine at 267 nm (Δɛ = 1,000 M−1⋅cm−1), cephalothin at 262 nm (Δɛ = 2,830 M−1⋅cm−1), nitrocefin at 482 nm (Δɛ = 17400 M−1⋅cm−1). Kinetic parameters, kcat, Km, and kcat/Km, were determined from Lineweaver-Burk plot. Substrate concentrations were varied such that Km values were flanked by six data points. In cases of biphasic turnover behavior, both phases were characterized.
Dependence of Kinetic Parameters on pH.
The pH dependence of the hydrolytic activity of the OXA-10 β-lactamase for turnover of penicillin G and nitrocefin (a chromogeneic cephalosporin that undergoes monophasic turnover) was investigated. All assays were performed at room temperature. The individual reactions were buffered (pH range of 4.5 to 9.5) by a three-component buffer system containing 50 mM acetic acid, 50 mM Mes, and 100 mM Tris, which gave an ionic strength of 0.1 M (15).
Circular Dichroism Spectra.
The CD-spectra of the wild-type OXA-10 and the Lys-70-Ala mutant β-lactamases (6 μM) were recorded in a Jasco (Easton, MD) spectrophotometer at 25°C in 10 mM sodium phosphate buffer (pH 7.0).
Conformational Change of OXA-10 β-Lactamase.
Experiments with different concentrations of enzyme and substrates were performed. Oxacillin or cloxacillin (10 or 1.0 mM) was incubated with the enzyme (2 or 0.5 μM) in a total volume of 100 μl, respectively. Aliquots (2 μl or 8 μl) were removed and diluted 250-fold into a solution of 200 μM penicillin G in 100 mM sodium phosphate (pH 7.0), at 1-min intervals up to 10 min. The activity of the enzyme was measured by monitoring hydrolysis of penicillin G immediately. We paid attention to the time when a first-order recovery of activity was noted, which was seen after 4–6 min of incubation with the substrate.
Crystallization.
Single crystals of the OXA-10 β-lactamase were obtained by the hanging drop method as previously described (10). Briefly, 1 μl of a protein solution at 10 mg/ml in 20 mM sodium potassium phosphate buffer (pH 7.8) was mixed with 1 μl of reservoir solution [2.0 M ammonium sulfate, either in 100 mM Tris⋅HCl (pH 8.5) or 100 mM Na/Hepes (pH 7.5)] at 4°C. Crystals of average size 300 × 300 × 50 μm (3) were obtained after 2 weeks.
The influence of pH on carbamylation of Lys-70 in the protein crystals was assessed in the following manner. The drop containing the crystals grown at pH 7.5 was transferred above a 1-ml reservoir made of 2.2 M ammonium sulfate, 100 mM Na/Mes, at pH 6.0 or 6.5, and equilibration was allowed to proceed. With volatile precipitating agents, such as ammonium sulfate, the pH of the drop at equilibrium is determined by the pH of the reservoir solution (16).
Protein-inhibitor complexes were prepared by a 5-min soaking of the crystal grown at pH 7.5 in a solution of sodium 6β-hydroxyisopropylpenicillanate (125 mM), in ammonium sulfate (2.2 M), 100 mM Na/Hepes (pH 7.5). Crystals were cryoprotected by a 45-s immersion in the reservoir solution complemented with 20% (vol/vol) ethylene glycol, before crycooling in a gaseous nitrogen flux at 100 K.
Data Collection and Processing.
Diffraction data were collected at the European Synchrotron Radiation Facility (ESRF; Grenoble, France), at beamline ID14EH1 (native structures at pH 6.0, pH 6.5, and pH 7.5), beamline ID14 EH2 (inhibited enzyme derivative), or beamline BM30A (native structure at pH 8.5). Crystals at all pH values were isomorphous with typical cell parameters of a = 67.3 Å, b = 82.4 Å, c = 101.2 Å, and β = 95.9°. Data were processed with mosflm (17), scaled and merged with scala (18), and converted to amplitudes with truncate, from the ccp4 (19) suite of programs (Table 1). About 1,700–2,000 reflections from each data set were randomly extracted for the calculation of the free R factor.
Table 1.
Crystallographic data processing and refinement statistics
| Native pH 7.5 | Native pH 6.5 | Native pH 6.0 | Soaked crystals | |||||
|---|---|---|---|---|---|---|---|---|
| Resolution, Å | 53.16–1.39 | 27.40–1.70 | 33.77–1.90 | 44.77–1.70 | ||||
| Overall | 1.47–1.39 | Overall | 1.79–1.70 | Overall | 2.00–1.90 | Overall | 1.79–1.70 | |
| No. observations | 841,487 | 95,038 | 307,069 | 44,375 | 215,405 | 26,849 | 432,971 | 45,364 |
| No. reflections | 216,019 | 29,650 | 118,305 | 17,487 | 84,615 | 11,824 | 119,631 | 16,474 |
| Multiplicity | 3.9 | 3.2 | 2.6 | 2.5 | 2.5 | 2.3 | 3.6 | 2.8 |
| Completeness, % | 99.0 | 93.3 | 98.3 | 99.7 | 97.8 | 94.0 | 99.1 | 93.8 |
| Rsym | 0.082 | 0.373 | 0.068 | 0.341 | 0.085 | 0.404 | 0.064 | 0.289 |
| I/σ | 11.8 | 3.3 | 12.2 | 4.1 | 9.7 | 3.0 | 12.9 | 3.2 |
| R factor | 0.15311 | 0.18344 | 0.17941 | 0.17688 | ||||
| Rfree | 0.18090 | 0.22068 | 0.22234 | 0.21043 | ||||
| rms deviation | ||||||||
| Bond length, Å | 0.013 | 0.018 | 0.023 | 0.006 | ||||
| Bond angle, ° | 1.581 | 1.686 | 1.931 | 1.191 | ||||
| Dihedrals, ° | 15.744 | 16.034 | 16.642 | 14.341 | ||||
Refinement.
The structure of the OXA-10 enzyme, refined to 1.8 Å (10), was used as a starting model for rigid-body refinement, after removal of all solvent molecules and of the carbamyl group on Lys-70. The sigmaA weighted electron density maps were displayed with turbo-frodo. They clearly indicated the carbamylation and the acylation status of Lys-70 and Ser-67, respectively, in each subunit of the two dimers in the asymmetric unit. Carbamylated lysines and modified serines were introduced accordingly at this stage. Further refinement was performed by using the maximum likelihood method as implemented in refmac (20), including a bulk solvent correction. In the last steps of refinement, water molecules were introduced by using arp (21). Anisotropic B-factor refinement was applied in the last refinement cycle except for the native structure at pH 6.0.
Results and Discussion
The x-ray crystal structure of the native OXA-10 β-lactamase at pH 8.5 indicated that Lys-70 in both subunits of the dimeric enzyme was carbamylated (10). We presented an argument that the active site environment around this lysine would appear to have evolved to accommodate the carbamylated residue (10). For example, the side chain of Lys-70 is sequestered in a hydrophobic environment within the active site, created by five invariant residues. It is likely that this hydrophobic environment would favor the free base form of the lysine side chain, facilitating its carbamylation in the presence of carbon dioxide. Furthermore, three specific electrostatic interactions stabilize the carbamate group on Lys-70 (10).
Modification of the OXA-10 β-Lactamase by 14CO2 and 13CO2.
Carbamylation of the OXA-10 enzyme is reversible. The enzyme that had been incubated with 25 mM NaHCO3 at pH 7.5 lost 50% of its activity when run through a desalting Sephadex G-25 column. However, it regained full activity when replenished with NaHCO3. In another experiment, the enzyme was incubated at pH 4.5 to facilitate full decarbamylation. Then the pH was adjusted to 7.5, and the solution was supplemented with radio-labeled NaHCO3 (25 mM). We found that the degree of carbamylation of the enzyme by 14CO2 correlated with the same degree of recovery of activity.
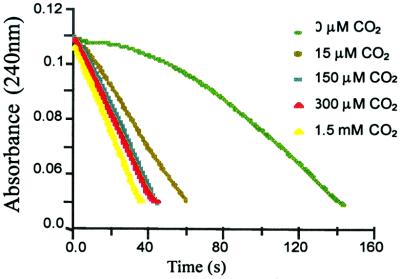
The enzyme after incubation at pH 4.5, followed by adjustment of the pH to 7.5 (in absence of bicarbonate supplement), dramatically lost activity (12% remaining; Fig. 1). The gradual increase in activity may be due to residual carbon dioxide in solution, an observation noted with rubisco, an enzyme that has a carbamylated lysine, indicating that traces of carbon dioxide are difficult to eliminate in aqueous solution (22). As supplemental carbon dioxide is added to this protein solution, the activity recovers fully (Fig. 1) with saturation (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
Figure 1.
Penicillin G (200 μM) hydrolysis progress curves with the OXA-10 β-lactamase (23 nM). The solutions were supplemented with sodium bicarbonate in quantities that would result in the given concentrations of carbon dioxide indicated.
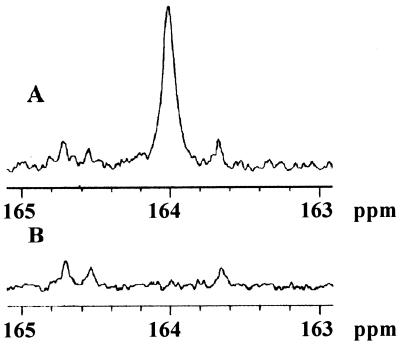
The presence of carbamylated enzyme was independently documented by a 13C NMR experiment (Fig. 2). The OXA-10 β-lactamase was decarbamylated at pH 4.5, the pH was adjusted to 7.5, and the solution was subsequently incubated with 20 mM 13C-labeled sodium bicarbonate. The 13C NMR spectrum showed the appearance of a signal at 164.0 ppm (Fig. 2A), corresponding to the carbamate residue, such as reported in the literature for the enzyme rubisco (23) and for small molecule carbamates (24). Other signals of significance were at 160.6 and 124.0 ppm (not shown), which correspond to the equilibrium between bicarbonate and carbonate and to the free carbon dioxide, respectively. When the labeled protein was incubated at pH 4.5, followed by dialysis in degassed buffer at pH 7.5, the signal at 164.0 ppm disappeared, consistent with the proposal for reversible carbamylation of Lys-70 (data not shown).
Figure 2.
The 13C NMR spectra of OXA-10 β-lactamase (A) and Lys-70-Ala OXA-10 mutant enzyme (B) in 10 mM sodium phosphate, 0.1 mM EDTA (pH 7.5) buffer supplemented with 20 mM NaH13CO3.
We subsequently prepared the Lys-70-Ala mutant variant of the OXA-10 enzyme. This enzyme is entirely inactive. Nitrocefin, the exceedingly reactive chromogeneic substrate, showed a mere 0.05% turnover in the presence of the mutant protein (5 μM protein and 100 μM nitrocefin). The extent of nitrocefin turnover indicated the formation of product (0.05 μM) substantially below the concentration of the protein used. With other more typical substrates such as penicillin G, we were not able to detect any turnover. Circular dichroic spectra of the wild-type enzyme and the Lys-70-Ala mutant variant were identical (data not shown), so mutation at position Lys-70 did not alter the secondary structural elements of the enzyme. The 13C NMR (Fig. 2B) of this mutant enzyme, which is impaired for acylation, indicated that the protein is not carbamylated (absence of the 164.0 ppm signal), as was expected.
Carbamylation of Lys-70 is reversible, as discussed above. We have evaluated the dissociation constant for carbon dioxide by two different methods. One method was based on activity assay and the other on the inherent fluorescence property of the enzyme. The activity-based measurement was carried out with penicillin G, and the nature of the measurements was such that it had to be carried out at nanomolar enzyme concentration (below the dimer/monomer dissociation constant of 1 μM). Hence, we measured a dissociation constant (Kd) for carbon dioxide of 12.4 ± 0.01 μM for the monomeric OXA-10 β-lactamase. In light of the fact that the active site has a tryptophan (Trp-154) at a distance of 3.0 Å from the carbamate, carbamylation of the enzyme was expected to affect the inherent fluorescence of the protein. A study by Chen and Barkley has shown that carboxylates of glutamate or aspartate were moderate quenchers of emission by tryptophan residues (25). We carried out the fluorescence experiments at a micromolar concentration of the protein, above the monomer/monomer dissociation constant, because the fluorescence assay was insensitive at nanomolar concentration. The value for the dissociation constant for carbon dioxide was evaluated at 0.23 ± 0.05 μM. The Hill coefficient was close to unity and indicative of a lack of cooperativity in the two carbamylation events within dimeric species. There is a 52-fold difference in these two dissociation constants, with the dimer undergoing carbamylation more readily. In light of the fact that the in vivo concentrations of carbon dioxide has been reported to be 1.3 mM (26), the OXA-10 β-lactamase is expected to be fully carbamylated in bacteria. We calculated the periplasmic concentration of the OXA-10 β-lactamase in two clinical strains of Pseudomonas (source of OXA-10) to be 4–15 μM (see supporting information). This concentration range is actually an underestimation, because we cannot expect that the procedure for liberation of the enzyme from the periplasmic space would release 100% of the enzyme contents for all bacteria. Regardless, the underestimation of 4–15 μM gives a concentration minimum of at least 4-fold above the dissociation constant for the dimer formation; hence, the OXA-10 β-lactamase is a dimer in vivo.
Steady-State Kinetics.
Because of the high activity of the enzyme, kinetics experiments could be carried out only at nanomolar concentrations of the enzyme. In these conditions, a typical feature of turnover kinetics by a number of class D β-lactamases, including the OXA-10 enzyme, is that they show at times biphasic properties. Recently, two studies attributed the biphasic nature of the kinetics to the dynamics of dimer/monomer protein equilibrium (11, 27). We, too, see biphasic kinetics for some substrates, but because we carried our experiments at enzyme concentrations that are three orders of magnitude below the dissociation constant for the dimer formation, we cannot attribute the effects of such a dimer/monomer equilibrium to our observations.
The experiments with various substrates were carried out with the purified native OXA-10 β-lactamase with and without supplementation of sodium bicarbonate as a source of carbon dioxide. In light of our discussion above that the OXA-10 enzyme is fully carbamylated in living bacteria, the results of the kinetic experiments in the presence of sodium bicarbonate are most relevant (Table 2). Under these conditions, only oxacillin and cloxacillin showed biphasic kinetic. As the results of Table 2 argue, the OXA-10 enzyme is catalytically more competent in turnover of penicillins, although it also turns over cephalosporins generally well.
Table 2.
Kinetic parameters in the presence of 50 mM NaHCO3 at pH 7.0
| Substrate | First phase
|
Second phase
|
||||
|---|---|---|---|---|---|---|
| kcat, s−1 | Km, μM | kcat/Km, M−1⋅s−1 | kcat, s−1 | Km, μM | kcat/Km, M−1⋅s−1 | |
| Penicillin G | 109 ± 3 | 23 ± 0.4 | (5 ± 1) × 106 | — | — | — |
| Ampicillin* | 143 ± 7 | 34 ± 4 | (4.2 ± 0.6) × 106 | — | — | — |
| Carbenicillin* | 112 ± 14 | 92 ± 16 | (1.2 ± 0.3) × 106 | — | — | — |
| Oxacillin | 1261 ± 27 | 29 ± 2 | (4.3 ± 0.3) × 107 | 331 ± 4 | 101 ± 21 | (3.3 ± 0.7) × 106 |
| Cloxacillin | 1533 ± 16 | 114 ± 22 | (1.3 ± 0.3) × 107 | 129 ± 1 | 110 ± 8 | (1.2 ± 0.1) × 106 |
| Cephaloridine* | 57 ± 14 | 374 ± 94 | (1.5 ± 0.5) × 105 | — | — | — |
| Cephalothin | 8.3 ± 0.1 | 32 ± 2 | (2.6 ± 0.1) × 105 | — | — | — |
In the absence of supplemental bicarbonate these substrates showed biphasic kinetics: ampicillin: kcat1 = 174 ± 34 s−1, Km1 = 27 ± 8 μM, kcat1/Km1 = (6 ± 2) × 106 M−1⋅s−1; kcat2 = 245 ± 96 s−1, Km2 = 183 ± 30 μM, kcat2/Km2 = (1.3 ± 0.6) × 106 M−1⋅s−1. Carbenicillin: kcat1 = 200 ± 8 s−1, Km1 = 191 ± 10 μM, kcat1/Km1 = (1.0 ± 0.1) × 106 M−1⋅s−1; kcat2 = 121 ± 9 s−1, Km2 = 430 ± 36 μM, kcat2/Km2 = (2.8 ± 0.3) × 105 M−1⋅s−1. Cephaloridine: kcat1 = 38 ± 7 s−1, Km1 = 344 ± 78 μM, kcat1/Km1 = (1.1 ± 0.3) × 105 M−1⋅s−1; kcat2 = 30 ± 12 s−1, Km2 = 2046 ± 804 μM, kcat2/Km2 = (1.4 ± 0.8) × 104 M−1⋅s−1.
In the absence of the supplemental bicarbonate, the OXA-10 enzyme exhibited biphasic turnover kinetics for ampicillin, carbenicillin, and cephaloridine. The data can be fitted to two distinct turnover events, for which the parameters were evaluated (Table 2). On addition of bicarbonate, the kinetics simplified to monophasic, and the parameters for the monophasic turnover kinetics approached very closely those of the first phase of turnover kinetics in the absence of bicarbonate. It is important to note that, in the case of the two phases, the effect is largely on Km, which is increased for the second phase.
The occurrence of the second phase, at pH 7.0, with these substrates may possibly arise from a decarbamylation event in the course of catalysis that would lead to inactive enzyme species, as described earlier. However, because the process of carbamylation is reversible, the active enzyme would be reconstituted by carbamylation of Lys-70, to resume the catalytic process. This hypothesis seems in line with our observations that biphasic kinetics at pH of 5.0 and 7.0, with ampicillin as substrate, could be fitted to monophasic kinetics at pH values of 7.5, 8.0, and 8.5 (data not shown). This observation would also argue for the facility of carbamylation of Lys-70 at higher pH values for the enzyme. Crystallographic evidence for this assertion will be presented later in this manuscript.
We believe that the mechanistic basis for biphasic kinetics with oxacillin and cloxacillin—which persisted despite supplementation of bicarbonate—is more distinct than the possibility described above for ampicillin, carbenicillin, and cephaloridine. These two substrates have unusual sterically encumbered side chains at their respective C6. On acylation of the active site serine, it would appear that these substrates induce a conformational change in the protein, which results in biphasic kinetics. The first phase is a burst before the conformational change, which gives rise to the second and slower phase for substrate turnover, which lasts until substrate depletion. The evidence for this statement comes from the following experiment. Oxacillin was incubated with the OXA-10 enzyme, and the enzyme was allowed to enter the second phase. An aliquot of this mixture was diluted into a solution of penicillin G. One notes a period of recovery of activity before the onset of steady-state turnover of penicillin G, presumably corresponding to the recovery of a conformation that allows for penicillin G hydrolysis. This recovery takes place with closely similar first-order rate constants of (1.0 ± 0.3) × 10−3 s−1 and (0.8 ± 0.4) × 10−3 s−1 with oxacillin and cloxacillin, respectively. These values are substantially smaller than the kcat values for these two substrates in the second phase (Table 2), implying that the recovery of the initial conformational state is slower than turnover. Hence, in the course of catalysis with oxacillin, after deacylation, the enzyme experiences reacylation by another substrate molecule and enters the catalytic process already in the conformational state for the second phase, before the enzyme has the opportunity to recover to the original conformational state responsible for the first and more rapid phase of hydrolysis. In addition to this type of conformational change, the possibility of decarbamylation/recarbamylation cannot be ruled out, but clearly the process is more complicated with oxacillin and cloxacillin. It is noteworthy that oxacillin and cloxacillin, because of their unusual C6 side chains, have been shown to cause biphasic kinetics because of such conformational change with other β-lactamases (28).
The pH dependence of the kinetic parameters for turnover of penicillin G and nitrocefin were studied. The data fit a two-pK model, with an acidic limb of 5.8–6.2 and a basic limb of 8.2–9.2 (see supporting information). These results are entirely consistent with the lower pKa describing the protonation state of carbamate and the higher pKa defining the titration of the second active site lysine, namely Lys-205. Therefore, for optimal catalysis, carbamate should be unprotonated and Lys-205 needs to be protonated. The low pKa value of 5.8 is consistent with the pKa value of 5.2 that was reported for a small molecule carbamate in the literature (29).
Structures of Native Enzymes at Different pH Values.
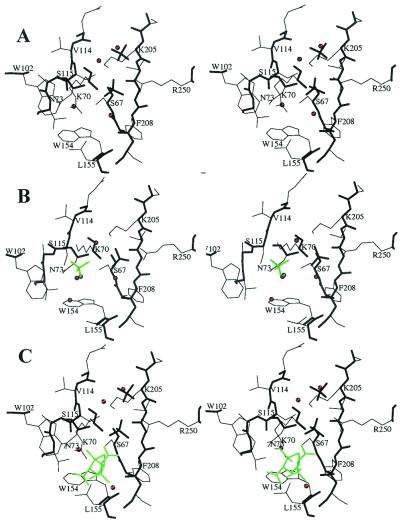
Data processing and refinement statistics are summarized in Table 1. The OXA-10 enzyme exists as a dimeric species, referred to in the text as the AC and BD dimers found in the asymmetric unit. The structures of the native enzyme were solved at four different pH values: 8.5 (1.80 Å), 7.5 (1.40 Å), 6.5 (1.70 Å) and 6.0 (1.90 Å). They revealed two geometries of the active sites. Subunits A and B displayed identical structures at all pH values whereas differences were seen for subunits C and D, depending on the pH value. At pH 8.5, the catalytic Lys-70 residue in all subunits was carbamylated (Fig. 3A). One oxygen atom of the carbamyl groups is hydrogen bonded to the hydroxyl group of Ser-67 (2.8 Å) and to the side chain nitrogen atom of Trp-154 (3.0 Å). The second oxygen atom of the carbamate group interacts with a buried water molecule (2.8 Å), itself hydrogen bonded to Asn-73OD1 (3.1 Å). The oxyanion hole is filled with a water molecule located at 3.2 Å and 2.8 Å to the Ser-67 and Phe-208 main chain nitrogen atoms, respectively. The hydroxyl group of Ser-115 interacts with a water molecule (3.1 Å) and with the amino group of Lys-205 (3.2 Å), homologous to Lys-234 in class A enzymes. The other hydrogen bonding partner of Ser-115OG is a sulfate anion (SUL1) anchored by a salt-bridge interaction to Arg-250, homologous to Arg-244 in class A enzymes.
Figure 3.
Active site structure at pH 8.5 (A), pH 6.0 (B), and when Ser-67 is acylated (C). Main chain, thick lines; side chain, thin lines. Water molecules are indicated as red spheres. The additional sulfate and water molecules observed at pH 6.0 (B) and the inhibitor (C) are depicted in green.
At pH 6.0, the active sites of monomers C and D displayed a non-carbamylated Lys-70 (Fig. 2B) that goes with a major alteration of the active site geometry. Instead of lying close to Ser-67, the amino group of the side chain of Lys-70 moves away from this catalytic residue, toward the main chain atoms of residues 111–114. The distance between the amino group of Lys-70 and the hydroxyl group of Ser-67 increases from 2.8 Å to 4.6 Å. A water molecule binds in the space previously occupied by the carbamate group, at hydrogen bond distance to the side chain nitrogen atom of Trp-154 (3.3 Å) and to the water molecule (3.3 Å) bound to the buried Asn-73OD1 (3.1 Å).
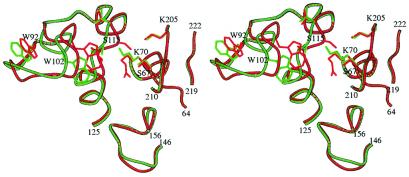
The displacement of the side chain of Lys-70 toward the main chain atoms of residues 111–114, involves a motion of residues 114 to 119 away from the active site, and a 180° rotation of the peptide bond between Val-144 and Ser-115. In these new conformations, the amino group of the Lys-70 side chain interacts with the main chain oxygen atom of Val-114 (2.8 Å), and the distance between the hydroxyl groups of Ser-115 and Ser-67 increases from 3.2 Å to 7.1 Å. A new sulfate ion (SUL2) binds at hydrogen bond distance to the side chains of Ser-115 (2.6 Å), Ser-67 (2.6 Å), and Lys-70 (2.8 Å). To accommodate the displacement of residues 114–119, residues 92–104 move away from the active site, as much as 3 Å for Trp-102 (Fig. 4).
Figure 4.
Superimposition of the OXA-10 β-lactamase structure when Lys-70 is carbamylated (red) and when it is not carbamylated (green).
At the intermediate pH values 6.5 and 7.5, monomers C and D displayed two states and conformations of Lys-70, clearly visible in the electron density maps. One of them corresponds to a carbamylated lysine, and the other to a non-carbamylated lysine. Their locations and environments were identical to those previously described in the structures solved at pH 8.5 and 6.0, respectively. Weak electron densities were found in the regions 92–104 and 114–119 because the simultaneous occurrence of the conformations observed at pH 6.0 and 8.5 leads to overlapping atomic positions.
Hence, comparing the structures of OXA-10 at four different pH values, essentially two active site geometries are observed. Monomers A and B, at all pH values, and monomers C and D, at pH 8.5, display a carbamylated Lys-70 residue located at close proximity to the hydroxyl group of Ser-67. At pH 6.0, Lys-70 in monomers C and D is not carbamylated, and the catalytic center (Ser-67, Lys-70, and Ser-115) falls apart. This result also occurred in the OXA-13 enzyme, a close relative to OXA-10, which was crystallized at pH 5.5 (12). However, in both geometries, the electrostatic potential in the vicinity of the Ser-67 hydroxyl group remains positive, binding either the carbamate group on Lys-70 or a sulfate anion (SUL2). This feature may be related to the inhibitory properties of anions on the OXA-10 enzyme. Binding of these species in the active site can, for electrostatic reasons, promote decarbamylation of Lys-70, as we indeed have observed from carbamylated crystals soaked with gold cyanide (8, 10). The enzyme lost activity, and the structure showed the non-carbamylated Lys-70 in the same location and conformation to that seen in the structure of the OXA-10 enzyme solved by Paetzel and colleagues (11) at pH 6.5 in the presence of the chloride ion.
To underscore the critical need for carbamylation of Lys-70, we observed that crystallized OXA-10 β-lactamase experienced active site acylation by a β-lactam inhibitor only at the subunits that were carbamylated (Fig. 3C). At pH 7.5, each subunit within a given dimer (AC or BD) behaved differently. Monomers A and B were carbamylated at Lys-70 and acylated at Ser-67, whereas monomers C and D were not carbamylated and not acylated. In these monomers, the conformation of the active site was similar to what was observed in the native structures at pH 6.0.
We have shown in this manuscript that the class D OXA-10 β-lactamase has evolved a distinct mechanism for its reaction compared with other β-lactamases. This enzyme depends on a critical carbamylated lysine in its active site for its catalytic mechanism. The results reported herein argue for the fact that disparate classes of β-lactamases have pursued different mechanisms to carry out the same reaction. The findings underscore the independent evolution of this reaction from distinct ancestral proteins in response to different selection factors (6).
The case of the OXA-10 β-lactamase is the first for the involvement of a non-metal-coordinate carbamylated lysine as a base in the active site of an enzyme. Carbamylated lysine may be incorporated into proteins without the need for other enzymes to create it posttranslationally. As such, it is a useful novel amino acid that would expand the versatility of the side chains of amino acids beyond the 20 common ones. It is in essence the structural counterpart to arginine, except that its side chain is negatively charged at the pH range useful for biological catalysts.
Supplementary Material
Acknowledgments
We are indebted to Dr. George Jacoby for providing us with the two Pseudomonas clinical strains. Synchrotron facilities for data collections were provided by the European Synchrotron Radiation Facility (ESRF; beam lines BM30A, ID14EH1, and ID14EH2). The work in France was funded in part by the Program de Recherche Fondamentale en Microbiologie (MENRT) and Centre National de la Recherche Scientifique (CNRS). The work in the United States was supported by a grant from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates of the enzymes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1K54, 1K55, 1K56, and 1K57).
References
- 1.Kotra L P, Golemi D, Vakulenko S, Mobashery S. Chem. Ind. 2000. 341–344. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C T. Nature (London) 2000;406:775–778. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Mobashery S. Adv Exp Med Biol. 1998;456:71–98. [PubMed] [Google Scholar]
- 4.Bush K. Clin Infect Dis. 2001;311:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 5.Kelly J A, Kuzin A P, Charlier P, Fonzé E. Cell Mol Life Sci. 1998;54:353–358. doi: 10.1007/s000180050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massova I, Mobashery S. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotra L P, Samama J-P, Mobashery S. In: Bacterial Resistance to Antimicrobials: Mechanisms, Genetics, Medical Practice and Public Health. Lewis A, Salyers A, Haber H, Wax R G, editors. New York: Dekker; 2001. pp. 123–159. [Google Scholar]
- 8.Golemi D, Maveyraud L, Vakulenko S, Tranier S, Ishiwata A, Kotra L P, Samama J-P, Mobashery S. J Am Chem Soc. 2000;122:6132–6133. [Google Scholar]
- 9.Patera A, Blaszczak L C, Shoichet B K. J Am Chem Soc. 2000;122:10504–10512. [Google Scholar]
- 10.Maveyraud L, Golemi D, Kotra L P, Tranier S, Vakulenko S, Mobashery S, Samama J-P. Structure. 2000;8:1289–1298. doi: 10.1016/s0969-2126(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 11.Paetzel M, Danel F, de Castro L, Mosimann S C, Page M G P, Strynadka N C J. Nat Struct Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 12.Pernot L, Frenois F, Rybkin T, L'Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. J Mol Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 13.Neu H C, Heppel L A. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 14.Butter J N. Carbon Dioxide Equilibria and Their Applications. Reading, MA: Addison-Wesley; 1982. [Google Scholar]
- 15.Ellis K J, Morrison J F. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 16.McPherson A. Crystallization of Biological Macromolecules. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 17.Leslie A G W. In: Computational Aspects of Protein Crystals Analysis: Proceedings of the Daresbury Study Weekend. Helliwell J R, Machin P A, Papiz M Z, editors. Daresbury, U.K.: Daresbury Laboratory; 1987. pp. 39–50. [Google Scholar]
- 18.Evans P R. In: Data Collection and Processing: Proceedings of the CCP4 Daresbury Study Weekend. Sawyer L, Isaacs N, Bayley S, editors. Daresbury, U.K.: Daresbury Laboratory; 1993. pp. 114–122. [Google Scholar]
- 19.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov G, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Perrakis A, Sixma T K, Wilson K S, Lamzin V S. Acta Crystallogr D. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 22.Lorimer G H, Badger M R, Andrews T J. Biochemistry. 1976;15:529–536. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- 23.O'Leary M H, Jaworski R J, Hartman F C. Proc Natl Acad Sci USA. 1979;76:673–675. doi: 10.1073/pnas.76.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch M, Korth H-G, Sustmann R, de Groot H. Chem Res Toxicol. 2000;13:451–461. doi: 10.1021/tx990138z. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Barkley M D. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 26.Tien M, Berlett B S, Levine R L, Chock P B, Stadtman E R. Proc Natl Acad Sci USA. 1999;96:7809–7814. doi: 10.1073/pnas.96.14.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danel F, Frère J-M, Livermore D M. Biochim Biophys Acta. 2001;1546:132–142. doi: 10.1016/s0167-4838(01)00133-9. [DOI] [PubMed] [Google Scholar]
- 28.Citri N, Samuni A, Zyk N. Proc Nat Acad Sci USA. 1976;73:1048–1052. doi: 10.1073/pnas.73.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roughton F J W, Rossi-Bernardi L. Carbon Dioxide: Chemical, Biochemical and Physiological Aspects. Washington, DC: U.S. Government Printing Office; 1970. , NASA no. SP-188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.