Most new drug treatments fail because they lack efficacy (1). In sepsis research, new therapies must contend with an additional barrier: the intractable heterogeneity of the sepsis syndrome (2). Together, these challenges have so far proved insurmountable. Hundreds of clinical trials have been conducted, at a cost of hundreds of millions of dollars, to test new agents to modulate the host response to injury in sepsis. None have succeeded (2).
The sepsis syndrome itself is simultaneously too broad and too narrow. Sepsis encompasses numerous different etiologies and pathophysiological processes, but—by definition (3)—excludes sterile injuries that lead to the same pathophysiology and organ failures, such as trauma, burns, hemorrhage, and pancreatitis.
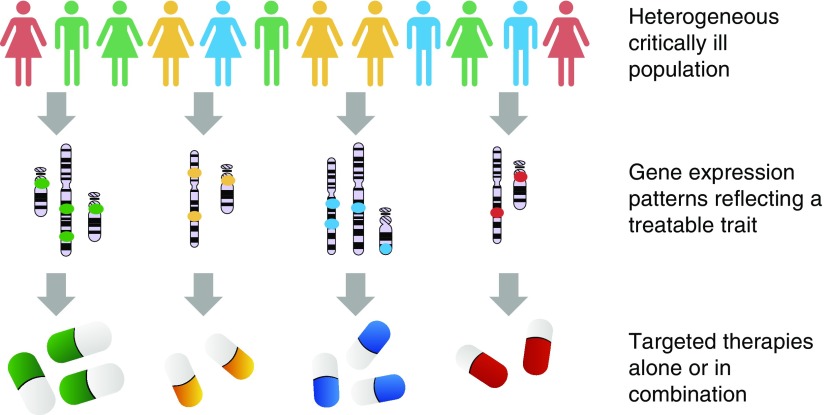
Some components of heterogeneity in sepsis are clinically apparent, such as variability in causal pathogens, comorbidities, environmental factors, and host genetics. But there is also evidence from recent studies (4–6) that important pathophysiological processes that are active in sepsis patients may vary in ways that are not directly observable at the bedside. If so, there is a chance that these processes may be amenable to different treatments (Figure 1).
Figure 1.
Deep phenotyping in practice. In a heterogeneous population, composite gene expression signals, either alone or in combination with clinical and other observations, may predict net benefit from a particular therapy. In reality, it is very likely that some patients will belong to multiple endotypes (indicated by colors on the image).
Large observational studies of blood transcriptomics applied to sepsis populations have provided several models based on molecular classification of patients with sepsis. In particular, the Genomic Advances in Sepsis (GAinS) consortium in the United Kingdom (4, 6) and the Molecular Diagnosis and Risk Stratification of Sepsis (MARS) consortium in the Netherlands detected distinct molecular endotypes in leukocyte genome-wide expression profiles from samples collected on ICU admission. The MARS consortium identified four molecular endotypes in all-cause sepsis (designated MARS 1–4) (6), whereas the GAinS consortium identified two molecular endotypes in community-acquired pneumonia (designated sepsis response signature 1 [SRS1] and SRS2) (4). More recently, in an impressive demonstration of the power of open science and data sharing (7), Sweeney and colleagues (5) identified three clinical signatures—termed inflammopathic, coagulopathic, and adaptive—using pooled data from publicly available gene expression data from other studies of patients with sepsis. Both the MARS and SRS molecular endotypes were associated with different mortality rates.
This is a necessary first step. But after these observations, the question remains as to whether the MARS/SRS signatures relate to therapeutically targetable immunopathologies. Subgroups may reflect different disease severities, or other features of the patients that are irrelevant to their care. To detect a treatment effect in these subgroups, it is necessary to acquire gene expression data from patients enrolled in randomized clinical trials.
For the first time, direct evidence of such an effect is reported by Antcliffe and colleagues (pp. 980–986) in this issue of the Journal (8). Using data from the VANISH (Vasopressin versus Norepinephrine as Initial Therapy in Septic Shock) trial, a generalized linear model based on a previously identified seven-gene SRS classifier (DYRK2, CCNB1IP1, TDRD9, ZAP70, ARL14EP, MDC1, and ADGRE3) enabled the authors to stratify 176 patients as SRS1 (47%) or SRS2 (53%). Patients stratified in this fashion did not differ in demographics and most baseline clinical characteristics (except for rates of ischemic heart disease). However, in line with the group’s previous findings (4), 28-day mortality in the placebo group was higher in SRS1 (37%) than in SRS2 (8%) patients (8). Serum lactate at baseline was also higher in SRS1 patients. Together, these observations indicate that, to some extent, the SRS classification reflects disease severity.
If severity (rather than distinct pathophysiology) underlies the difference between these groups of patients, an interaction with steroid treatment might be anticipated. Large trials of steroids in sepsis and septic shock (9, 10) have reported trends toward a treatment benefit in patients with the highest risk of death. Whether these trends are real, and if so, whether they are simply a consequence of a higher event rate in this group (heterogeneity of treatment effect), are open questions at present. Based on these studies, we would have predicted a higher probability of detectable benefit from steroids among patients classified as SRS1. In fact, an interaction was detected between hydrocortisone use and SRS2-classified patients, resulting in increased mortality estimates with an adjusted odds ratio of 8.3 (95% confidence interval, 1.4–47.8), that is, a signal consistent with harm from steroid treatment in the less-severe SRS2-classified group.
Collectively, these results and those from therapeutic trials using subclassifications of acute respiratory distress syndrome (11, 12) imply that there are divergent effects from a single intervention across and within different patient endotypes, bringing them closer to the definition of a true disease endotype (13).
The investigators in the VANISH trial are to be congratulated for having the foresight to acquire transcriptomic data within a randomized controlled trial. Although confirmatory replication will be necessary, this work brings us a step closer to the primary aim of stratified medicine research: new phenotypes with direct therapeutic consequences. It is our view that future clinical trials in critical illness should consider from the outset the probability that any new therapy may have a differential effect in a subgroup of patients, and that subgroup may only be identifiable through deep phenotyping. Among the available methodologies, preservation of whole-blood RNA is the most pragmatic way to enable future deep phenotyping.
The dichotomous SRS1/2 classification simplifies analysis, but the groupings are drawn by bisecting what appears to be a unimodal distribution (4). This suggests that the SRS classification reflects two extremes of a continuously varying underlying biological process.
This move from the identification of subgroups to the detection of continuous “treatable traits” within clinical populations has become a major focus of work in other fields (13); we, and many others, would argue that sepsis research is in particular need of these new approaches (2). Going further, it is very plausible that any physiological process that is active in a large proportion of patients with sepsis will also be active in some patients with severe sterile injury. As with other therapeutic approaches in critical care medicine, new treatable traits may be generalizable across critical illnesses.
If the information necessary to predict response to a given therapy is present in measured clinical variables, or in the whole-blood transcriptome, then detecting it becomes entirely a matter of data analysis. With current techniques, huge numbers of patients will be needed to overcome signal/noise ratios. Integration of transcriptomic signatures with genetic associations (14) may enable more efficient detection of key underlying processes. Ultimately, these approaches may identify new, specific drug targets to modulate the host response to critical injury (15), and actionable estimates of individual treatment effect for critically ill patients.
Footnotes
Originally Published in Press as DOI: 10.1164/rccm.201811-2148ED on December 12, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Arrowsmith J. Trial watch: phase II failures: 2008-2010. Nat Rev Drug Discov. 2011;10:328–329. doi: 10.1038/nrd3439. [DOI] [PubMed] [Google Scholar]
- 2.Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20:195–203. doi: 10.1016/j.molmed.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4:259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018;46:915–925. doi: 10.1097/CCM.0000000000003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. MARS Consortium. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5:816–826. doi: 10.1016/S2213-2600(17)30294-1. [DOI] [PubMed] [Google Scholar]
- 7.Dunning JW, Merson L, Rohde GGU, Gao Z, Semple MG, Tran D, et al. ISARIC Working Group 3, ISARIC Council. Open source clinical science for emerging infections. Lancet Infect Dis. 2014;14:8–9. doi: 10.1016/S1473-3099(13)70327-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antcliffe DB, Burnham KL, Al-Beidh F, Santhakumaran S, Brett SJ, Hinds CJ, et al. Transcriptomic signatures in sepsis and a differential response to steroids: from the VANISH randomized trial. Am J Respir Crit Care Med. 2019;199:980–986. doi: 10.1164/rccm.201807-1419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annane D, Renault A, Brun-Buisson C, Megarbane B, Quenot J-P, Siami S, et al. CRICS-TRIGGERSEP Network. Hydrocortisone plus fludrocortisone for adults with septic shock. N Engl J Med. 2018;378:809–818. doi: 10.1056/NEJMoa1705716. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesh B, Finfer S, Cohen J, Rajbhandari D, Arabi Y, Bellomo R, et al. ADRENAL Trial Investigators and the Australian–New Zealand Intensive Care Society Clinical Trials Group. Adjunctive glucocorticoid therapy in patients with septic shock. N Engl J Med. 2018;378:797–808. doi: 10.1056/NEJMoa1705835. [DOI] [PubMed] [Google Scholar]
- 11.Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, et al. ARDS Network. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Latent class analysis of ARDS subphenotypes: analysis of data from two randomized controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell CD, Baillie JK. Treatable traits and therapeutic targets: goals for systems biology in infectious disease. Curr Opin Syst Biol. 2017;2:139–145. doi: 10.1016/j.coisb.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baillie JK, Bretherick A, Haley CS, Clohisey S, Gray A, Neyton LPA, et al. IIBDGC Consortium. Shared activity patterns arising at genetic susceptibility loci reveal underlying genomic and cellular architecture of human disease. PLoS Comput Biol. 2018;14:e1005934. doi: 10.1371/journal.pcbi.1005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baillie JK. Translational genomics: targeting the host immune response to fight infection. Science. 2014;344:807–808. doi: 10.1126/science.1255074. [DOI] [PubMed] [Google Scholar]