Abstract
Bacteria and their bacteriophages coexist and coevolve for the benefit of both in a mutualistic association. Multiple mechanisms are used by bacteria to resist phages in a trade-off between survival and maintenance of fitness. In vitro studies allow inquiring into the fate of virus and host in different conditions aimed at mimicking natural environment. We analyse here the mutations emerging in a clinical Pseudomonas aeruginosa strain in response to infection by Ab09, a N4-like lytic podovirus and describe a variety of chromosomal deletions and mutations conferring resistance. Some deletions result from illegitimate recombination taking place during long-term maintenance of the phage genome. Phage variants with mutations in a tail fiber gene are selected during pseudolysogeny with the capacity to infect resistant cells and produce large plaques. These results highlight the complex host/phage association and suggest that phage Ab09 promotes bacterial chromosome rearrangements. Finally this study points to the possible role of two bacterial genes in Ab09 phage adhesion to the cell, rpsB encoding protein S2 of the 30S ribosomal subunit and ORF1587 encoding a Wzy-like membrane protein involved in LPS biosynthesis.
Introduction
Bacteriophages play an important role in shaping the bacterial genome by exerting a selective pressure [1]. Bacterial resistance to bacteriophage is principally mediated by modification of the phage receptors although other mechanisms can play a role by blocking the viral entry and multiplication [2]. Recently a large number of new antiphage defense systems present in multicomponent defense islands (among which restriction-modification and CRISPR-Cas systems) have been described [3,4]. In return phage counteracts the defense mechanisms by developing different strategies [5]. Long-term phage/bacteria arm-race has been extensively investigated in the laboratory, mostly using coevolution in chemostats, and a few studies have observed coevolution in natural environment [6,7]. In the environment, many factors influence the fate of bacteria and phage variants by affecting the fitness as observed with Pseudomonas fluorescens for example [8]. In P. aeruginosa the filamentous Pf4 prophage is essential for several stages of the biofilm life-cycle and contributes to the bacterial virulence [9]. Adaptation of a bacterial strain to stress or to a particular niche may be accompanied by important rearrangements of the genome. Reduction of the genomic repertoire is the result of large deletions caused by homologous and illegitimate recombination during permanent association of pathogenic bacteria with their host [10]. Recombination between IS elements or prophages have been shown to be the most frequent mechanism for inversions and deletions [11,12,13]. Another actor in phage/bacteria evolution is the RecA-dependent spontaneous prophage induction (SPI) that influence bacterial fitness [14]. This may be linked to SOS response but also results from other mechanisms.
Whereas the effect of prophage on their host has been widely described, little is known about the effect of lytic phages on the natural rate of evolution, principally due to lack of experimental systems to observe such interactions. In vitro investigations have shown that bacterial resistance to phages could modify their virulence [15]. We previously set-up an assay to analyze the mechanisms of resistance developed by the reference P. aeruginosa strain PAO1 following infection by lytic phages [16]. In this assay which allowed the analysis of a micropopulation of resistant bacteria, we found that pseudolysogeny played an important role in allowing the maintenance of phages and the emergence of mutations at a high rate (essentially single nucleotide or short deletions). Different genes involved in synthesis of phage receptors, mostly lipopolysaccharide (LPS) and type IV pili were affected, and we showed that phase variation was a major factor in inducing mutations [16]. Pseudolysogeny has been observed in different phage/bacteria systems in slowly growing or starved cells and is believed to be a mean to preserve phage and bacteria until better conditions are available [17]. In Propionibacterium acnes, establishment of pseudolysogeny by phages (all belonging to a single genus) is highly dependent on the host strain [18,19]. It is likely that such long-term maintenance of phages, which may be induced by lack of resources or by specific bacterial associations, such as in communities and biofilms, may be frequent in nature, thus favoring bacterial evolution.
Here we selected a new clinical P. aeruginosa strain, PcyII-10, susceptible to numerous phages, to further investigate the emergence of pseudolysogeny in response to infection by a lytic phage and the nature of resistant variants. We confirmed the importance of pseudolysogeny as a natural state of lytic phage/host interaction but in addition to our previous work with PAO1, we observed a high level of large chromosomal deletions arising during phage-maintenance, some being unrelated to phage resistance. The results also suggest that two unsuspected bacterial genes could be involved in phage resistance.
Material and methods
Ethics statement
The present project is in compliance with the Helsinki Declaration (Ethical Principles for Medical Research Involving Human Subjects). The bacterial strain was collected as part of the patients' usual care, without any additional sampling, as previously reported [20]. Ethic committees in each hospital providing bacterial isolates were consulted and they declared that patient informed consent was not needed: the Percy hospital “Comité d’éthique et des experimentations” and the Armand Trousseau hospital “Comité Consultatif pour la Protection des Personnes dans la Recherche Biomédicale (CCPPRB) Ile-De-France–Paris–Saint Antoine”.
Bacteria and phages
After the first steps of purification from a clinical sample, P. aeruginosa PcyII-10 strain (serotype O6) has been maintained in the laboratory for several years without colony reisolation. Its genome has been fully sequenced using PacBio and Illumina sequencing (sequence accession number LT673656) (Pourcel et al. manuscript in preparation). For the present study a single colony was picked, grown in LB and used for whole DNA genome extraction, for isolation of phage resistant variants and to prepare a stock at -80°C. The genome was resequenced (Illumina sequencing). Reference strain PAO1 was used to evaluate the number of variants resisting infection by Ab09. Strains C2-10 and PARD80 were previously characterized P. aeruginosa clinical strains [21,22].
Podovirus vB_PaeP_C2-10_Ab09 (Ab09) and myovirus vB_PaeM_PAO1_Ab27 (Ab27) were previously described [23]. Phage Ab27 was used to test the host-range of variants resisting Ab09.
Recovery of phage-resistant variants
Two “multiplicity of infection” (MOI) of 0.001 and 0.01 were used to infect PcyII-10 or PAO1 and recover resistant variants, as previously described [16,24]. Bacteria surviving infection after 72 h at 37°C were purified by three passages from a colony (passage 3 or P3) before being further characterized.
Electron Microscopy (EM) observation
To search for traces of phage multiplication in pseudolysogens the bacteria were embedded in Epon resin and sliced as described [25]. Briefly, bacteria were grown overnight from a single colony in 2 ml of LB and the bacterial pellet was recovered after centrifugation. After fixation using 2.5% glutaraldehyde, the cells were dehydrated with ethanol and acetone before being fixed in Epon “Agar low viscosity” (Oxford Instruments). The resin blocs were sliced into 90 nm sections. Before EM observation, staining was performed using uranyl acetate 2% for 15 min, followed by three 5 min washes with water.
DNA extraction, PCR and sequencing
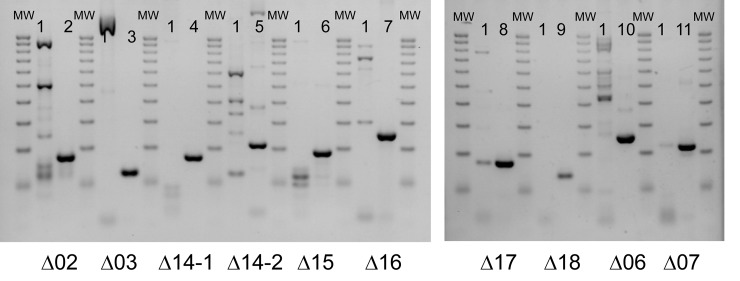
Thermolysates were produced by diluting 10 μl of overnight culture in 200 μl of water, heating at 95°C for 5 min and cooling on ice. For DNA purification, bacteria were lysed in lysis buffer (Tris 10 mM, pH 7.8, EDTA 10 mM, NaCl 10 mM, SDS 0.5%wt/vol), treated with proteinase K at 50 μg ml-1 for 2 h at 50°C, followed by one phenol and one chloroform extraction, and ethanol precipitation. PCR was performed on thermolysates or purified DNA using oligonucleotides listed in S1 Table. The expected sizes of the PCR fragment following amplification of deleted regions were: Δ02 173bp, Δ03 127bp, Δ06 260bp, Δ07 221bp, Δ14_1 171bp, Δ14_2 220bp, Δ15 191bp, Δ16 257bp, Δ17 177bp, Δ18 138bp. Sanger sequencing of PCR products was performed by Genewiz (Takeley, UK).
Gene cloning and expression
PCR amplicons were cloned into the pUCP24 plasmid, a generous gift of Dr. Schweizer, modified in order to introduce a NcoI restriction site [26]. This is a shuttle vector which replicates in E. coli and in P. aeruginosa, and contains a multiple cloning site downstream lacZα. The PcyII-10 rpsB gene and ORF1587 were PCR-amplified using oligonucleotides which included restriction sites for respectively BspLU11I and HindIII, and BspHI and HindIII (S1 Table). The amplicons were digested, ligated into the vector treated by NcoI and HindIII, and transformed into E. coli in which replication of pUCP24 is optimal [26]. The insert was Sanger-sequenced in a selected recombinant which was then used to transform P. aeruginosa variants by electroporation using the fast protocol described by Choi et al. [27]. Transformants were selected using 10 μg ml-1 Gentamycin. The colony aspect was observed under the microscope. The transformants were then tested for their susceptibility to Ab09 and its large plaque variant and to Ab27.
Whole genome sequencing
Ten μg of purified DNA was sent for draft whole genome Illumina MiSeq sequencing to the High-throughput Sequencing Platform of I2BC (CNRS, Gif sur Yvette, France). Libraries were made from sheared fragments of DNA with a mean size of 900bp, and 250bp paired-end reads were produced. One and a half million up to 2.5 million reads were obtained corresponding to an average 130 to 230 fold coverage. The mutations were identified by comparison with the parental genome (PcyII-10 or Ab09) using tools in GeneiousR11 (Biomatters, New Zealand) as described [16].
Nucleotide sequence accession number
The DNA sequence of the PcyII-10 strain has been deposited under accession number LT673656. The twelve archives of sequencing reads produced in the course of this study were deposited in the European Nucleotide Archive (ENA) under bioproject PRJEB18612 (https://www.ebi.ac.uk/ena/data/view/PRJEB18612; PcyII-10 resequenced colony (POU32), variants V02 (POU35), V03 (POU33), V06 (POU43), V07 (POU44), V10 (POU45), V14 (POU34), V15 (POU36), V16 (POU29), V17 (POU23), V18 (POU27), V19 (POU25).
Results
High frequency of deletions in PcyII-10 variants resisting phage Ab09
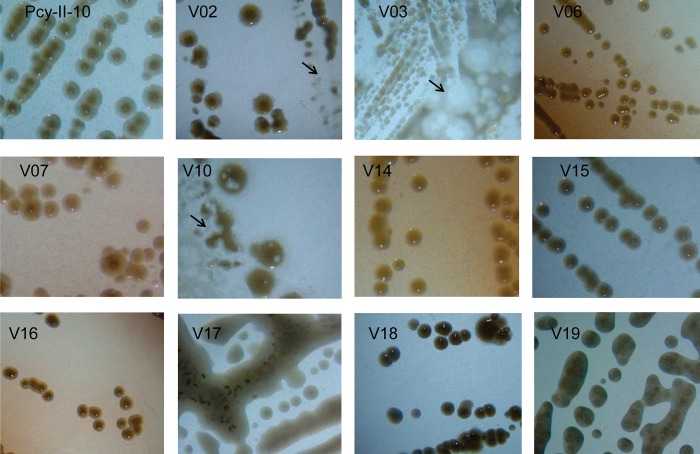
In our previous investigations we found that phage Ab09, a N4-like podovirus, used LPS as a primary receptor but was capable of infecting PAO1 mutants defective in O-Ag synthesis (although with a lower efficiency) suggesting the existence of another receptor at the cell surface [16,25]. This phage selected for mucoid mutants with a high efficiency and was capable of establishing a pseudolysogenic state. Here P. aeruginosa PcyII-10 and PAO1 were infected by Ab09 at a multiplicity of infection (MOI) of 0.001 or 0.01 before plating the mixture in soft agar on Luria-Bertani (LB) solid medium and incubating for 72h at 37°C. Interestingly, the proportion of slow-growers among PcyII-10 colonies surviving infection was much higher as compared to PAO1 whereas the amount of large mucoid colonies was ten times less with PcyII-10 (S1 Fig). Twenty PcyII-10 resistant variants displaying different phenotypes, including two mucoid variants, were recovered and purified by three colony-isolation steps (passage 3 or P3). Nine variants grew very poorly and were not further analysed. At this stage an heterogeneity in colony size and morphology was observed for many variants, a characteristic reminiscent of PAO1 pseudolysogens behavior (Fig 1) [16]. On LB solid medium, lysis could be seen in densely rich bacterial zones and in some colonies (see for example PcyII-10-V02, PcyII-10-V03 and PcyII-10-V10 in Fig 1), most likely caused by the release of phages. We tested for the presence of phages in the supernatant of P3 overnight cultures in LB and found titers ranging from 107 to 3x1010 phages per ml in all except PcyII-10-V14 and PcyII-10-V17. Four variants (PcyII-10-V06, PcyII-10-V07, PcyII-10-V14 and PcyII-10-V16) formed small reddish-brown colonies whereas both mucoid and non-mucoid colonies were produced by variant PcyII-10-V17. Finally, eleven variants were kept at P3 for storage at -80°C in glycerol, whole genome sequence (WGS) analysis and phenotypic characterization (Fig 1).
Fig 1. Colony morphology of PcyII-10 and phage-resistant variants.
Colonies formed by PcyII-10 and eleven variants plated at P3 on LB agar were photographed after 24 h at 37°C. The arrows point to lysis zones caused by phages. Variant V03 produces small colonies with irregular shapes. Variants V06, V07, V14 and V16 produce a reddish-brown pigment, the pyomelanin. Variants V17 and V19 were mucoid.
Persistence of the phage
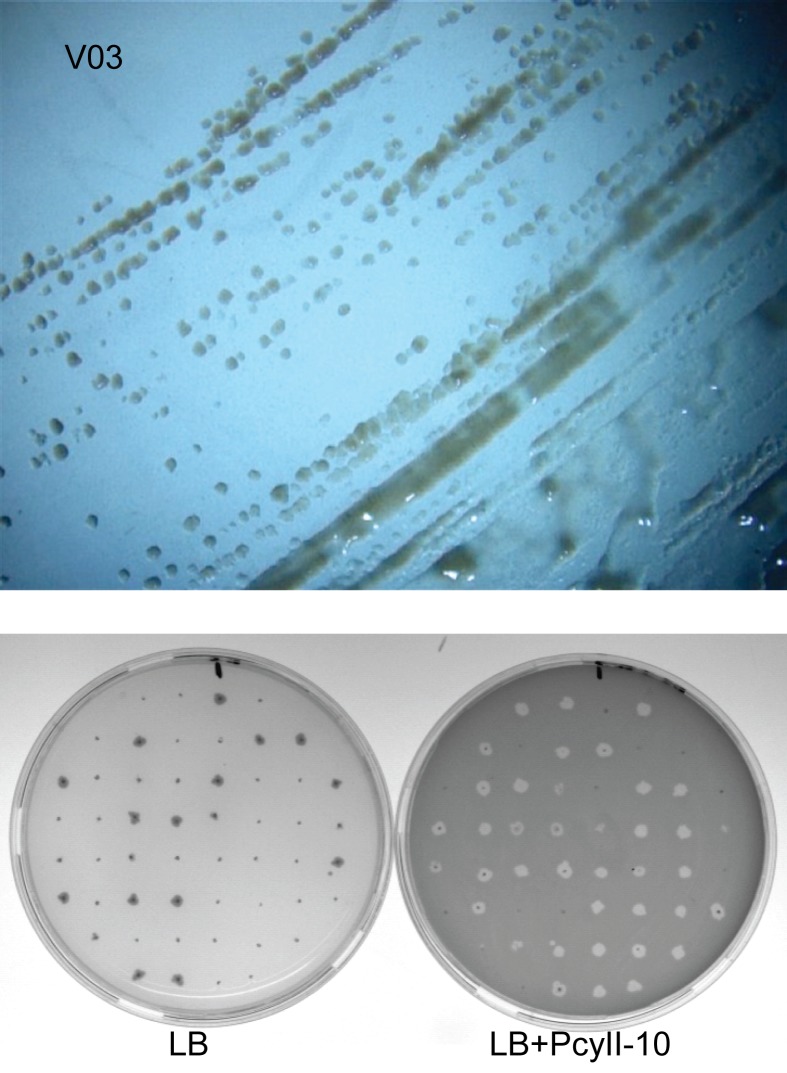
We analysed the capacity of five variants to produce phages after additional replatings by spreading the P3 stock on LB agar plates (Fig 2 top panel shows PcyII-10-V03) and inoculating 52 colonies first on an LB agar plate then on a plate covered with a lawn of soft agar containing PcyII-10 (Fig 2 bottom panel) [24]. A halo of lysis was observed around bacteria producing phages and these grew poorly on the LB plates. The percentage of P4 colonies producing phages was 40, 73, 20, 50, 80% for PcyII-10-V02, PcyII-10-V03, PcyII-10-V07, PcyII-10-V08 and PcyII-10-V10 respectively. The procedure was repeated eight times with variant PcyII-10-V03, each time selecting phage-producing bacteria and we observed that 80% of the colonies were still pseudolysogens. Occasionally larger colonies could be observed, suggesting the emergence of sub-variants as previously observed in PAO1.
Fig 2. Evaluation of Ab09 persistence in PcyII-10-V03.
(A) Colonies obtained after plating the P3 stock on a LB agar plate. (B) Fifty-two colonies recovered from the P3 stock were simultaneously plated using a sterilized pipette tip on LB agar (left) and on PcyII-10 embedded in soft agar overlay (right). The clear zone around the bacterial colony in the right panel is due to phage lysis of the indicator bacteria.
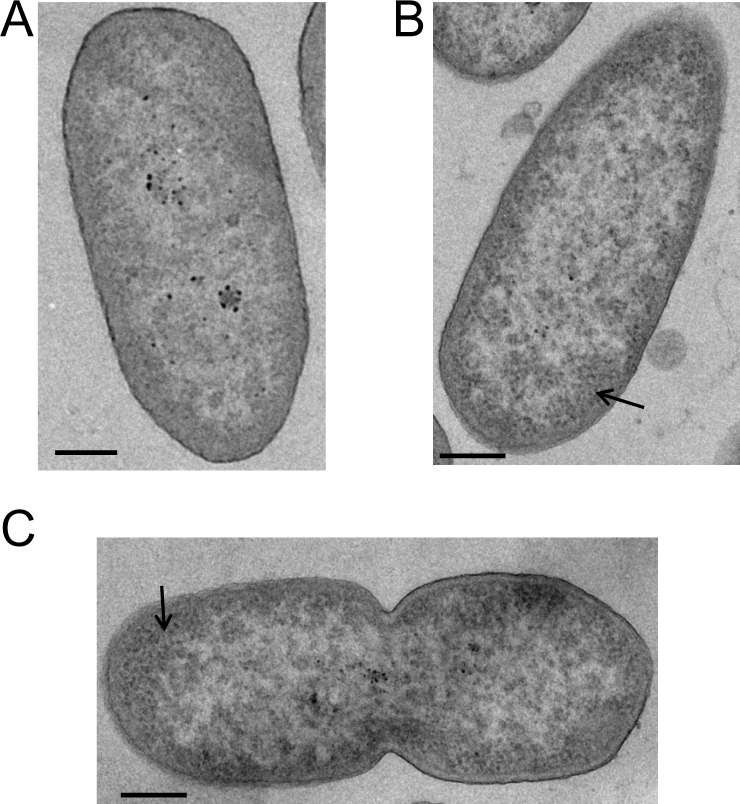
To search for the possible presence of complete virions inside pseudolysogens, variants PcyII-10-V07 and PcyII-10-V10 at P3 were cultured in LB, embedded in resin before thin sections were analysed by electron microscopy (EM). No evidence of virions could be found inside the bacteria but densely stained and tightly packed structures could be observed in the two pseudolysogens. They may represent ribonucleoproteins (RNPs) as previously described in bacteriophage infected cells [28,29], suggesting the existence of active metabolism. A large number of PcyII-10-V07 cells had clear cytoplasm and altered membranes, some showing blebs, a sign of cellular death (Fig 3 shows PcyII-10 and the variants PcyII-10-V07 and PcyII-10-V10). This suggested that there was a block in the multiplication cycle, probably at the late stage before production of virions. The observed structural alteration might be the result of the mutations as well as that of phage multiplication.
Fig 3. Observation of pseudolysogens by electron microscopy.
Transmission electron micrograph of bacterial section from (A) The parental strain PcyII-10, (B) Variant PcyII-10-V07 and (C) Variant PcyII-10-V10. The arrow in (B) and (C) points to tightly packed material that might correspond to ribonucleoprotein particles. The scale bar corresponds to 200nm.
Whole genome sequencing of Ab09-resistant variants
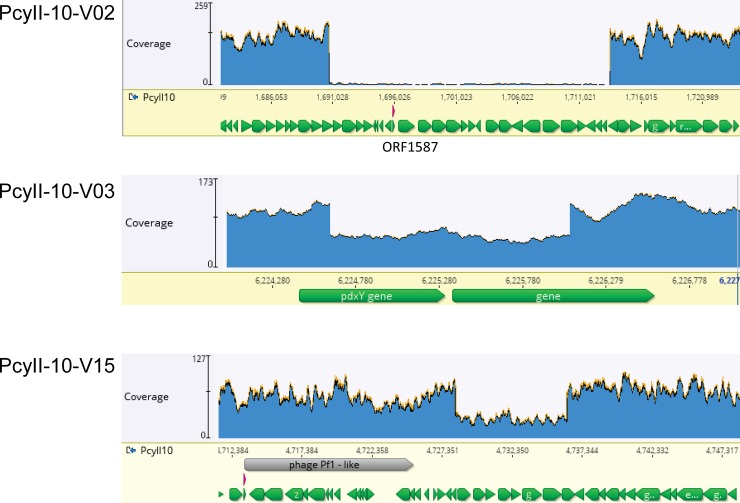
The genome of eleven Ab09-resistant variants at P3 was totally sequenced as well as that of the parental PcyII-10 culture allowing the identification in the variants of five single nucleotide changes and ten deletions (Table 1). Two variants possessed only a single nucleotide mutation (PcyII-10-V10 and PcyII-10-V19), four possessed a single deletion (PcyII-10-V02, PcyII-10-V06, PcyII-10-V16 and PcyII-10-V18) and five were double mutants. As revealed by the mapping of sequencing reads on the progenitor PcyII-10 genome, the mutations were not always present in 100% of the cells (Table 1). Whereas in three samples the whole population of cells appeared to possess a deletion, in six variants, one or two deletions were present only in a fraction of cells (Fig 4 shows three examples). Point mutations were always detected in close to 100% of the sequencing reads and in three variants a deletion was also detected in a fraction of reads (PcyII-10-V03, PcyII-10-V15, PcyII-10-V17). Four single nucleotide changes occurred in a mononucleotide stretch, causing phase-variation mutations. The existence of a mutation and/or deletion in only a fraction of the cells and the high frequency of double mutants even after three colony-purification steps suggested that new genetic modifications emerged inside pseudolysogenic bacteria, the situation previously observed with PAO1 resistant to lytic phages. The striking difference with PAO1 phage-resistant variants was the existence in PcyII-10 variants of a variety of deletions never observed in PAO1, some being very large. Ab09 reads were found in all sequenced samples, reaching relatively high levels (above 0.6%) in four variants.
Table 1. Characteristics of the mutations in the eleven phage resistant PcyII-10 variants.
| Variant (P3) | Δ | Deletion (bp) | Positions | Read % | Point mutation | Position | Read % | Phenotype | Ab09 DNA % | |
|---|---|---|---|---|---|---|---|---|---|---|
| V02 | 02 | 22,763 | 1,690,775 | 1,713,537 | 100% | 0.6 | ||||
| V03 | 03 | 1,434 | 6,224,639 | 6,226,072 | 50% | rpsB A(3)->A(2) | 1,402,765 | 100% | Blue | 1.84 |
| V06 | 06 | 535,972 | 2,930,640 | 3,465,943 | 100% | Brown | ||||
| V07 | 07 | 276,763 | 3,053,155 | 3,334,409 | 80% | Brown | ||||
| V10 | ORF1587 T(4)->T(3) | 1,697,201 | 100% | 0.19 | ||||||
| V14 | 14_1 | 2,999 | 1,941,686 | 1,944,684 | 20% | 3.69 | ||||
| 14_2 | 56,271 | 3,273,717 | 3,329,987 | 50% | Brown | |||||
| V15 | 15 | 7,920 | 4,728,358 | 4,736,277 | 40% | ORF1587 T(7)->T(8) | 1,697,063 | 95% | 0.66 | |
| V16 | 16 | 340,123 | 3,052,982 | 3,393,104 | 100% | Brown | ||||
| V17 | 17 | 11,975 | 4,713,278 | 4,725,252 | 80% | mucA C(2)->C(1) | 4,678,812 | 100% | Mucoid | |
| V18 | 18 | 64,894 | 4,713,278 | 4,778,171 | 50% | |||||
| V19 | mucA G->A | 4,678,7,60 | 100% | Mucoid | ||||||
Fig 4. Distribution of sequencing reads on the PcyII-10 genome forthree Ab09-resistant variants.
Mapping of sequencing reads to the genome of PcyII-10 revealed the existence of deleted zones. In variant PcyII-10-V02 no reads mapped to the Δ02 region, whereas in PcyII-10-V03 and PcyII-10-V15 the Δ03 and Δ15 regions were undeleted in an estimated 50% and 40% of cells respectively.
Identification of the mutations and relationship with phage resistance
Because sequencing revealed the existence of mixed populations of cells in some variants at P3, we performed further colony purifications from frozen stocks and searched for the presence of the detected single mutations or deletions by PCR analysis and Sanger sequencing. Using primers selected on both sides of the deleted region and inside this region (S1 Table) all the deletions could be confirmed by PCR analysis, on the DNA used for whole genome sequencing and in newly isolated colonies (Fig 5). Similarly, the point mutations were verified by PCR amplification and direct Sanger sequencing of the amplification product, confirming their absence at a detectable level in PcyII-10 before infection, and presence in the variants. We also checked for the presence of Ab09 and selected colonies that were free of phages in order to analyse the effect of the sole mutations. Variants were tested for their susceptibility to Ab09 and to the P. aeruginosa myovirus Ab27 which uses LPS as a primary receptor [25] (Table 2).
Fig 5. Analysis of ten deletions by PCR amplification and agarose gel electrophoresis.
For each deletion, two DNA samples were analysed, PcyII-10 and the corresponding variant. MW is for Molecular Weight marker, a 100bp fragment ruler from 100bp to 1000bp.
Table 2. Susceptibility of the PcyII-10 variants and subvariants to phages.
| Phage-free isolates | Deletion (Δ)a | Mutation | Phenotypeb | Phage susceptibilityc | |
|---|---|---|---|---|---|
| Ab09 | Ab27 | ||||
| V02 | 02 | UN | S | R | |
| V03(1) | wt | rpsB | Blue | R | S |
| V03(2) | 03 | rpsB | Blue | R | S |
| V06 | 06 | SC brown | R | R | |
| V07(1) | wt | LC pale | S | R | |
| V07(3) | 07 | SC brown | R | R | |
| V10(1) | wt | UN | S | S | |
| V10(4) | ORF1587 | UN | R | R | |
| V14(3) | 14_1 | UN | I | I | |
| V14(8) | 14_2 | SC brown | R | R | |
| V15(1) | 15 | ORF1587 | UN | R | R |
| V15(7) | wt | ORF1587 | UN | R | R |
| V16 | 16 | SC brown | R | R | |
| V17-nmucd | 17(Pf1) | UN | S | S | |
| V17-mucd | 17(Pf1) | mucA | Mucoid | I | R |
| V18(5) | 18 | SC | R | R | |
| V18(7) | wt | UN | S | S | |
| V19 | mucA | Mucoid | I | I | |
a wt, wild type
b UN, unaffected, SC, small colonies; LC, large colonies
c S susceptible, R resistant, I intermediate growth
dnmuc, non mucoid, muc, mucoid
As expected, all the tested colonies from PcyII-10-V02 possessed the 22,763bp deletion Δ02 which encompassed the aliphatic amidase operon amiEBCRS involved in motility and virulence [30], genes of the carbon-phosphorus lyase pathway and others involved in synthesis of the flagella. Notably, Δ02 included ORF1587 encoding a Wzy-like membrane protein, and found to be mutated in two other variants, PcyII-10-V10 and PcyII-10-V15. ORF1587 shows important similarities at the structural level with Wzy an oligosaccharide polymerase involved in O-Ag B-chains biosynthesis of certain serotypes, such as PAO1 O5 (Fig 6). A PcyII-10-V02 colony free of phages was susceptible to phage Ab09 and totally resistant to Ab27. Thus in PcyII-10-V02 we suppose that primary phage-resistance to Ab09 was due to immunity conferred by the presence of phage Ab09, still detected after three steps of colony purification (0.6% of reads), and not to deletion Δ02.
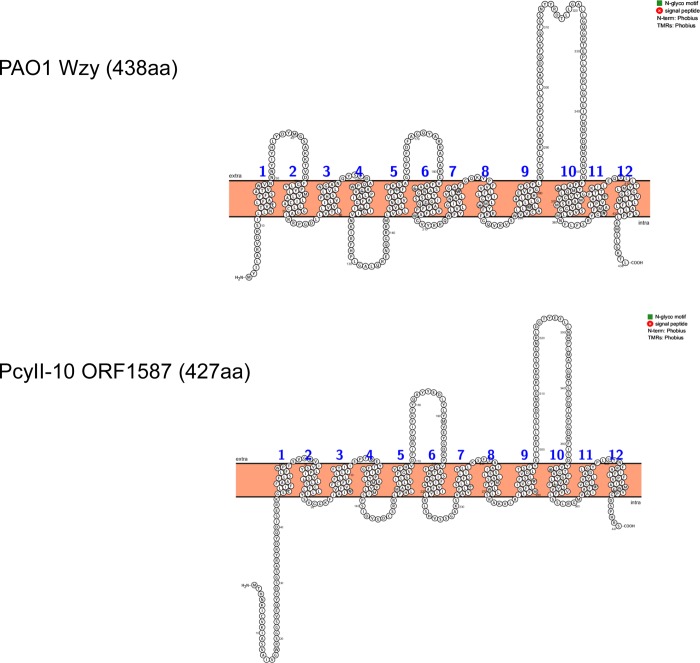
Fig 6. Comparison of Wzy and ORF1587 topologies using Protter.
The sequenced PcyII-10-V03 genome showed both a mutation in rpsB, the gene for the 30S ribosomal protein S2, and a 1,434bp deletion (deletion Δ03). The growth of variant V03 was very poor and at P3 it still produced phages at high concentration. The rpsB mutation, an A(3)->A(2) single nucleotide deletion present in 100% sequencing reads which resulted in premature stop and production of a 134 amino acids protein, could be confirmed by PCR amplification and Sanger sequencing. The rpsB gene is an essential component of the translation machinery in Escherichia coli [31]. Deletion Δ03 which encompassed two genes, pdxY a pyridoxal kinase and an ORF encoding an acyltransferase of unknown function, was originally present in half of the bacterial population, as shown by the distribution of sequencing reads. Out of eight colonies obtained from the frozen P3 stock, four possessed the deletion. Non-deleted PcyII-10-V03 bacteria such as variant PcyII-10-V03(1) were resistant to Ab09 suggesting that the rpsB mutation was sufficient to confer resistance to Ab09. Interestingly, phage Ab27 could grow normally on this mutant.
All the PcyII-10-V06 colonies were small, reddish-brown, and possessed a single large deletion of 536kb (Δ06) including the homogentisate 1,2-dioxygenase hmgA gene, the absence of which is responsible for the brown color [32], and galU a key gene for the O-Ag biosynthesis. This variant was resistant to Ab09 and Ab27.
Red and white colonies were observed upon replating of the PcyII-10-V07 P3 stock, the red ones being smaller in size and possessing the 277kb deletion Δ07 which encompassed hmgA and galU. The red colonies were resistant to Ab09 and Ab27. A subvariant, PcyII-10-V07(1), forming large white colonies was not deleted and was normally susceptible to Ab09 but resistant to Ab27, suggesting the emergence of a novel mutation.
PcyII-10-V10 possessed a T(4)->T(3) mutation in ORF1587 resulting in production of a truncated 248 aminoacids protein (instead of 427aa). PcyII-10-V10 at P3 produced a large amount of phages and upon replating, different colonies could be picked with different phenotypes. PcyII-10-V10(1) forming normal size colonies was susceptible to Ab09 and Ab27 and was not mutated in ORF1587 whereas three other tested colonies of smaller size (such as V10(4)) were mutated and resisted infection by Ab09 and Ab27.
Two derivatives of PcyII-10-V14 could be obtained containing deletion Δ14_1 or deletion Δ14_2. Variant PcyII-10-V14(3) with the 2,999bp deletion Δ14_1 formed aggregates in LB and showed reduced susceptibility to Ab09 and Ab27 (small turbid plaques). Δ14_1 included four genes (ORF1811 to ORF1814): wbpT, wbpU, wbpV and wbpL, involved in O-Antigen biosynthesis [33]. PcyII-10-V14(8) with the 56,271bp deletion Δ14_2 and making red colonies, conferred resistance to Ab09 and Ab27. Δ14_2 extended from ORF3001 corresponding to gene yejE, to part of gene hmgA and included galU.
PcyII-10-V15 also possessed a mutation in ORF1587 (T(7)->T(8)) causing a frameshift and premature stop and production of a truncated protein of 332 amino acids. The mutation was found in all tested colonies at P4. In addition sequencing revealed a 7,920bp deletion (deletion Δ15) extending from ORF4387 (lsrG gene) involved in control of quorum sensing, to ORF4395. Δ15 started 3kb away from a Pf1-like prophage copy and included the migA gene, an alpha-1,6-rhamnosytransferase involve in LPS biosynthesis, and a membrane protein of unknown function. Among 16 tested PcyII-10-V15 colonies all resisting Ab09 and Ab27, only five possessed deletion Δ15, PcyII-10-V15(1) possessing the deletion and PcyII-10-V15(7) being intact. Analysis of ORF1587 in PcyII-10-V15(7) confirmed the presence of the mutation, suggesting that alteration of this gene was responsible for resistance.
PcyII-10-V16 colonies were small, reddish-brown, and possessed a single large deletion of 340kb (Δ16), including the hmgA and galU genes. They were fully resistant to both Ab09 and Ab27.
Deletion Δ17 in variant PcyII-10-V17 corresponding to a copy of the Pf1-like prophage was confirmed in six isolated colonies out of 10. Among them, a mucoid colony had in addition a point mutation in mucA conferring resistance to Ab27 and reduced growth of Ab09. Non-mucoid colonies were susceptible to both phages.
PcyII-10-V18 possessed deletion Δ18 encompassing the Pf1-like prophage and 64,894 bp flanking genomic DNA. Genes in this region included, in addition to migA, several glycosyltransferases, a Type II secretion system and genes for iron transport. PcyII-10-V18 was resistant to both Ab09 and Ab27. A large amount of Pf1-like reads were found among the genome sequencing data showing that Pf1 multiplied actively at the third passage in this variant.
PcyII-10-V19 colonies were all mucoid due to a mutation in mucA which conferred reduced susceptibility to both Ab09 and Ab27.
Genomic distribution of deletions
The deletions were distributed into six genomic regions, three of them associated with a unique event (Δ02, Δ03, Δ14_1) and two with respectively three (region N°1) and four (region N°2) events. In region N°1 Δ17 and Δ18 included the Pf1-like prophage whereas Δ15 started 3kb after this prophage insertion site suggesting a role for the virus in the deletion. Δ06, Δ07, Δ14_2 and Δ16 encompassing the hmgA gene covered region N°2 at the replication terminus.
We searched for the presence of direct repeats flanking the deleted regions and found them in Δ02, Δ03, Δ06, Δ14_1 and Δ18, with length of respectively eight nucleotides (5’-CGGCGCCA-3’), nine nucleotides (5’-CGCTGGTGG-3’), ten nucleotides (5’-CGCCTGCTCG-3’), ten nucleotides (5’-TGGGGGATGC-3’) and seven nucleotides (5‘-AGGGTTC-3’). No significant conserved sequences were found flanking the other deletions.
Variability of phage genomes and isolation of large-plaque forming variants
Ab09 phage reads were found in all sequenced variants and in particularly significant amounts (0.6% to 3.69% Table 1) in PcyII-10-V02, PcyII-10-V03, PcyII-10-V14 and PcyII-10-V15, allowing assembly of a genome with a higher than 50X coverage, and search for possible mutations as compared to the parental Ab09 phage. The Ab09 phage genome assembled from PcyII-10-V02 and PcyII-10-V14 reads was identical to the parental genome. In variant PcyII-10-V03 the phage possessed a single G->T mutation at position 29,815 in ORF48 encoding a putative 1100 amino acids tail fiber protein, leading to a change of amino acid S289Y. In PcyII-10-V15 the assembled phage genome showed three modifications in ORF48, a G->T change in 100% of reads (position 29,815) and a A->G on about 50% reads at positions 30,159 leading to a V926A change, and a silent G->A mutation at position 32,564. This reflected an ongoing evolution of the phage in pseudolysogens.
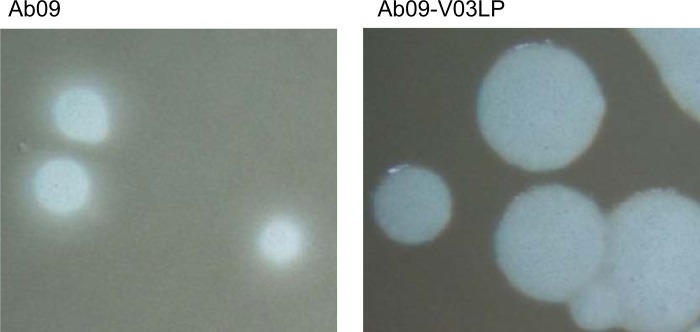
While titrating phages produced by an overnight culture of variant PcyII-10-V03 we noticed some large clear plaques among a majority of smaller plaques with a halo, the usual aspect for Ab09 plaques. We thus recovered a small (Ab09-V03SP) and a large (Ab09-V03LP) plaque, amplified the phages on PcyII-10 and performed further characterization. On PcyII-10 Ab09-V03LP plaques were 4mm in diameter, clear with sharp edges whereas the parent Ab09 and Ab09-V03SP produced 2mm plaques with irregular edges and a halo (Fig 7). On three additional strains, no difference in plaque size and morphology was seen between Ab09 and the two variants: the mean diameter was 3mm, 2mm and 4mm on P. aeruginosa strains PAO1, C2-10 and PARG80 respectively. Contrarily to progenitor Ab09 and Ab09-V03SP, Ab09-V03LP could grow on PcyII-10-V03, producing very large plaques (up to 6mm in diameter). In order to try and understand the basis for this phenotype, the complete genome of phage variant Ab09-V03LP was sequenced revealing three point mutations in ORF48. The first one in the N-terminal part, a S289Y substitution, was not specific for Ab09-V03LP as it was already present in the DNA extracted from PcyII-10-V03 pseudolysogen and also in Ab09-V03SP. The V926A substitution was observed in the phage present in the PcyII-10-V15 variant whereas the L893R substitution was specific to the large-plaque forming phage, Ab09-V03LP. This substitution in the COOH terminal region of the protein replacing an hydrophobic residue by a positively charged residue might lead to significant change in the protein secondary structure.
Fig 7. Plaque morphology of the large-plaque mutant of Ab09 produced by variant PcyII-10-V03.
Plaques produced by the wild type Ab09 phage have irregular fuzzy shapes and are surrounded by a halo whereas the Ab09-V03LP mutant forms large plaques with sharp edges and no halo.
The role of rpsB and ORF1587
Two new genes appeared to be candidates for a role in phage infection, rpsB and ORF1587 and we therefore performed additional experiments to test this hypothesis. We performed an adsorption assay on PcyII-10 and on isolated variants (PcyII-10-V03(1) for rpsB, and PcyII-10-V15(7) and PcyII-10V10(4) for ORF1587) that only possessed a mutation in these genes. On the PcyII-10 parental strain, phage adsorption was observed after 10 min whereas no adsorption was observed on the variants.
The two genes were cloned into pUCP24, an expression vector for P. aeruginosa, and the recombinants were electroporated into the mutants. Complementation of the rpsB mutation in PcyII-10-V03(1) resulted in normal colony phenotype and was fully efficient in restoring susceptibility to Ab09 (S2 Fig). Interestingly the expression of ORF1587 in PcyII-10-V10(4) restored susceptibility to Ab09, Ab09-V03LP and Ab27 but the plaques where very small, whereas the transformed PcyII-10-V15(7) cells remained resistant to both phages.
Discussion
N4-like phages are podoviruses distributed into four genera frequently isolated from different Gram-negative species and characterized by the presence of a giant virion-associated RNA polymerase [34,35,36]. In P. aeruginosa the corresponding genus “Lit1virus” includes Lit1, PA26 and Ab09 which display a rather large host spectrum, making them interesting tools for phage therapy. They form plaques with a halo, likely related to degradation of capsular exopolysaccharides by a depolymerase activity [37,38,39]. The interactions between E. coli and phage N4, and the basis for phage-resistance have been largely studied but little is known about the other members of this family. We confirm here using the clinical and minimally subcultured P. aeruginosa strain PcyII-10 that Ab09 adopts a pseudolysogenic behavior at a high rate in starving bacteria, and that this favors the emergence of mutants as previously shown using the PAO1 laboratory strain [16]. During maintenance of the phage a small proportion of bacteria can support a full multiplication cycle and die releasing new phages, whereas some cells maintain small amounts of viral genome and other are cured, becoming prey for the free phages. As opposed to the carrier state established by the levivirus LeviOr01 in which large quantities of virions could be seen inside enlarged cells [40], there is apparently no Ab09 virions accumulating in the pseudolysogens, but the bacteria appear to be deeply affected. Therefore the blockage might be at the level of phage multiplication and not lysis. This was observed with phage T4 in starved cells, a phenomenon called lysis inhibition (LIN) induced by superinfection of T4-infected cells at any time during the multiplication cycle [41,42]. Some core and variable genes in members of the N4-like family show striking similarities to those of the T4 superfamily [43]. Phage Ab09 possesses two genes related to phage T4 rIIA and rIIB which appear to play an indirect role in LIN, promoting delayed lysis but there is apparently no Ab09 equivalent of gene rI which is the real actor of LIN [44,45,46].
In this work we show that the fate of infection by phage Ab09 is dependent on the host. In PcyII-10 variants resisting Ab09 infection point mutations are rather rare as compared to PAO1 phage-resistant mutants, whereas deletions are very abundant. The rate is higher than the expected in vitro deletion rate calculated in Salmonella enterica growing in rich medium [47] and some of these deletions are very large. In addition, in a majority of variants the deleted genomes are found in less than 100% of cells after three steps of colony isolation and could be segregated by further subplatings. This shows that the deletions took place after the initial purification of the phage-resistant colonies and therefore in bacteria which continued to evolve. Curiously, some of the deletions appear not to be necessary for resistance to Ab09. We hypothesize that persistence of Ab09 in the bacteria in a pseudolysogenic state destabilizes the genome, leading to an important degree of secondary deletions.
Two deletions involve a copy of the Pf1-like prophage, and free Pf1-like phage DNA is detected in significant amounts in three additional resistant variants, suggesting that Ab09 infection leads to the prophage activation. In addition to Pf1 prophage excision, flanking chromosomal DNA is deleted in PcyII-10-V18 and the deletion in PcyII-10-V15 was 3kb away from the insertion site, suggesting that this region is a hotspot for chromosome rearrangement. The presence of a Pf1-like filamentous phage was previously proposed to be associated with the production of small colony variants (SCVs) [48]. PcyII-10 spontaneously produces SCVs but the link with Pf1-like phage activation is not established. Activation of Pf1 transcription was observed in bacteria infected with phage phiKZ although the amount of Pf1 DNA was not increased, and it was assumed that the filamentous phage was unsuccessfully attempting to escape the cells [49].
Four different deletions were found in a region (called here region N\2) in which other large deletions were already described in association with a brown/red pigmentation. This was the case for example in P. aeruginosa strains isolated in the course of an eight-years survey of a single cystic fibrosis (CF) patient (188kb deletion) [50], in P. aeruginosa CF strains [51] and in P. aeruginosa strain P1A resistant to PAP1, a PAKP1-like phage [52]. All the deletions affect galU, a key gene in LPS biosynthesis, most likely responsible for the phage-resistance. Pyomelanin production responsible for the red color is due to loss of the homogentisate gene hmgA (position 3,329,705 to 3,331,003) [32,53]. This loss was reported to favour persistence in the lung of CF patients [32], and to increase stress resistance, persistence, and virulence in some bacterial species [54,55], but it can also produce an opposite effect in other specie [56]. The regions covered by the large deletions are centered on the replication terminus. It was found that pausing of DNA replication was a factor favoring deletion formation in E. coli [57] and that in Salmonella the replication terminus is a hot-spot for transposon-mediated deletion [58].
As compared to deletions spontaneously occurring in chronic infections, we did not find a direct repeat on both side of all the large deletions containing the hmgA gene [55] in agreement with observations by Shen et al. [59]. Indeed these authors showed that MutL promotes large chromosomal deletions through non homologous end joining (NHEJ). Repeats were present flanking the other deletions implying a role of recA-mediated recombination. Illegitimate recombination was likely to be responsible for five deletions with homologous region of 6bp to 10bp in length. For bacteria it was proposed that 20 to 25bp of homologous sequence is required for RecA-dependent homologous recombination [60,61]. Mutations in MutS and SbcCD, the prokaryotic homologue of Rad50/Mre11 enhance illegitimate recombination and increase the rate of deletion [12,62]. A portion of the SbcCD subunit C gene is deleted in PcyII-10 which may have an effect on deletion rate. MutS is intact in PcyII-10 and cannot be involved in the deletion rate. We analysed additional known antimutator genes and did not find any significant differences in PcyII-10 as compared to PA14 or PAO1 reference strains [63].
All mutation/deletions except one, the rpsB mutant in V03, are likely to affect the biosynthesis of LPS. Mutations were found at two different positions in a gene of unknown function, ORF1587, both leading to the production of a truncated protein, lack of O-Ag B chains and resistance to Ab09 and Ab27. ORF1587 lies between the Arg tRNA and amiE genes, a region of heterogeneity in serotype O6 P. aeruginosa strains. As the equivalent of wzy was not found in the O-antigen biosynthesis gene cluster of O6 strains it is possible that ORF1587 functions like Wzy [33,64]. Pan et al. described at the same position in the genome, a gene that they believe is the wzy gene, Y880_03157 of O6 PAK P. aeruginosa [65]. As previously shown for Wzy proteins in different serotypes, ORF1587 encodes a protein with a similar topology but with little homology at the amino acid level (Fig 5) [64,66]. Complementation of variant PcyII-10-V15 with the wild type gene did not restore susceptibility to Ab09 and this may be due to the dominant action of the mutated gene. It is compatible with the fact that PcyII-10-V02, deleted for ORF1587 was susceptible to Ab09 and suggests that another wzy-like gene is present in PcyII-10.
Although carbohydrate chains of the LPS seem to be the first molecules to which the phage binds, it seems reasonable to think that a receptor at the membrane is important for secondary adsorption and ejection of the DNA as observed with other podoviruses [67]. Four gene products are required for the adsorption of E. coli N4 phage (NfrA, B, C, D), one of which is a cytoplasmic protein [68,69]. Both host and phage proteins are thought to constitute a channel spanning the outer and inner membranes through which the DNA and the virion-associated RNA polymerase are injected into the cell. The N4 protein gp65 which constitutes the sheath surrounding the tail tube is adsorbed on the cell surface by recognition of the outer membrane protein NfrA (+ NfrB) [70]. Interestingly, we observed a mutation in a gene that was not affected in PAO1 variants resistant to Ab09 and which may be specifically involved in binding of Ab09 to its receptor. Mutation in rpsB in variant PcyII-10-V03 leading to synthesis of a truncated protein was responsible for altered growth but did not appear to be lethal. PcyII-10-V03 resisted infection by Ab09 but was susceptible to Ab27. A derivative of Ab09 with increased growth capacity was isolated from PcyII-10-V03 P3 pseudolysogens and it showed a mutation in a putative tail fiber protein. Like other ribosomal proteins, RpsB has been shown to be exposed at the surface of Pseudomonas and other Gram- bacteria [71,72] and also found in the periplasm [73]. It is possible that the phage fiber interacts with RpsB for infection. Alternatively protein S2 of the 30S ribosomal subunit may play a role in translation of leaderless mRNAs as demonstrated for phage lambda [74]. However our results are preliminary, and additional investigations will be necessary to understand the precise role of RpsB in phage susceptibility.
As previously observed in PAO1 pseudolysogens, long-term maintenance of phages leads to emergence of phage mutants that outgrowth the parental phage. Interestingly the observed mutants are all tail fiber protein mutants. At P3, the Ab09-V03 phage variant produced two kinds of plaques, one of which was reminiscent of the large sharp-edged plaques (r-type) formed by T4 phages R rapid-lysis mutants, as compared to the wild type rough-edged plaques [75,76]. Sequencing of the large-plaque forming phage showed mutations in the putative tail fiber gene ORF48 a protein which in similar phages form a trimer and interacts with other tail protein and with carbohydrates. The morphology of phage plaque is the result of different factors, including adsorption and diffusion in the agar, but also efficient lysis [77]. The sharp-edged phenotype of Ab09-V03LP is apparently not related to lysis but rather to adsorption and is observed on PcyII-10 in which the phage evolved but not on three other susceptible strains. As demonstrated in a joint theoretical-experimental approach using E. coli and one of its phage, slow adsorption can maximize plaque size [78].
Supporting information
(TIF)
PcyII-10-V03(1) PcyII-10-V10(4) and PcyII-10-V15(7) were transformed either by the vector pUCP24 (A) or by the recombinants (B). Colonies were observed on medium containing Gentamycin and the bacteria were tested for their susceptibility to four dilutions of phage Ab09 (1), Ab09-V03LP (2) and Ab27 (3).
(TIF)
(DOCX)
Acknowledgments
We thank Lory-Anne Baker and Anthony Goudin for their help in characterizing PcyII-10 variants. This work has benefited from the facilities and expertise of the High-throughput Sequencing Platform of I2BC. The present work has benefited from the core facilities of Imagerie-Gif, (http://www.i2bc.paris-saclay.fr), a member of IBiSA (http://www.ibisa.net), supported by “France-BioImaging” (ANR-10-INBS-04-01), and the Labex “Saclay Plant Science” (ANR-11-IDEX-0003-02).
Data Availability
The DNA sequence of the PcyII-10 strain has been deposited under accession number LT673656. The twelve archives of sequencing reads produced in the course of this study were deposited in the European Nucleotide Archive (ENA) under bioproject PRJEB18612 (https://www.ebi.ac.uk/ena/data/view/PRJEB18612; PcyII-10 resequenced colony POU32, variants V02 (POU35), V03 (POU33), V06 (POU43), V07 (POU44), V10 (POU45), V14 (POU34), V15 (POU36), V16 (POU29), V17 (POU23), V18 (POU27), V19 (POU25)).
Funding Statement
GV received a grant from the Direction Générale de l’Armement (DGA) and the Agence Nationale de la Recherche (ANR, France), ‘Resisphage’ ANR-13-ASTRID-0011–01. The funder did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brockhurst MA, Buckling A, Rainey PB (2005) The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci 272: 1385–1391. 10.1098/rspb.2005.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 3.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, et al. (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, Yaakov G, et al. (2018) DISARM is a widespread bacterial defence system with broad anti-phage activities. Nature microbiology 3: 90–98. 10.1038/s41564-017-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson JE, Magadan AH, Sabri M, Moineau S (2013) Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11: 675–687. 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 6.Maura D, Debarbieux L (2012) On the interactions between virulent bacteriophages and bacteria in the gut. Bacteriophage 2: 229–233. 10.4161/bact.23557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laanto E, Hoikkala V, Ravantti J, Sundberg LR (2017) Long-term genomic coevolution of host-parasite interaction in the natural environment. Nature communications 8: 111 10.1038/s41467-017-00158-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Smet J, Hendrix H, Blasdel BG, Danis-Wlodarczyk K, Lavigne R (2017) Pseudomonas predators: understanding and exploiting phage-host interactions. Nature reviews Microbiology. [DOI] [PubMed] [Google Scholar]
- 9.Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, et al. (2009) The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J 3: 271–282. 10.1038/ismej.2008.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran NA (2002) Microbial minimalism: genome reduction in bacterial pathogens. Cell 108: 583–586. [DOI] [PubMed] [Google Scholar]
- 11.Ernst RK, D'Argenio DA, Ichikawa JK, Bangera MG, Selgrade S, Burns JL, et al. (2003) Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environmental microbiology 5: 1341–1349. [DOI] [PubMed] [Google Scholar]
- 12.Rau MH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L (2012) Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ Microbiol 14: 2200–2211. 10.1111/j.1462-2920.2012.02795.x [DOI] [PubMed] [Google Scholar]
- 13.Raeside C, Gaffe J, Deatherage DE, Tenaillon O, Briska AM, t RN, et al. (2014) Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. MBio 5: e01377–01314. 10.1128/mBio.01377-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanda AM, Thormann K, Frunzke J (2015) Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. Journal of bacteriology 197: 410–419. 10.1128/JB.02230-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosseinidoust Z, van de Ven TG, Tufenkji N (2013) Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol 79: 6110–6116. 10.1128/AEM.01421-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latino L, Midoux C, Hauck Y, Vergnaud G, Pourcel C (2016) Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology 162: 748–763. 10.1099/mic.0.000263 [DOI] [PubMed] [Google Scholar]
- 17.Los M, Wegrzyn G (2012) Pseudolysogeny. Adv Virus Res 82: 339–349. 10.1016/B978-0-12-394621-8.00019-4 [DOI] [PubMed] [Google Scholar]
- 18.Lood R, Collin M (2011) Characterization and genome sequencing of two Propionibacterium acnes phages displaying pseudolysogeny. BMC genomics 12: 198 10.1186/1471-2164-12-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Yan R, Zhong Q, Ngo S, Bangayan NJ, Nguyen L, et al. (2015) The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. The ISME journal 9: 2116 10.1038/ismej.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu-Thien H, Corbineau G, Hormigos K, Fauroux B, Corvol H, Clement A, et al. (2007) Multiple-locus variable-number tandem-repeat analysis for longitudinal survey of sources of Pseudomonas aeruginosa infection in cystic fibrosis patients. J Clin Microbiol 45: 3175–3183. 10.1128/JCM.00702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llanes C, Pourcel C, Richardot C, Plesiat P, Fichant G, Cavallo JD, et al. (2013) Diversity of beta-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: a French multicentre study. J Antimicrob Chemother 68: 1763–1771. 10.1093/jac/dkt115 [DOI] [PubMed] [Google Scholar]
- 22.Sobral D, Mariani-Kurkdjian P, Bingen E, Vu-Thien H, Hormigos K, Lebeau B, et al. (2012) A new highly discriminatory multiplex capillary-based MLVA assay as a tool for the epidemiological survey of Pseudomonas aeruginosa in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis 31: 2247–2256. 10.1007/s10096-012-1562-5 [DOI] [PubMed] [Google Scholar]
- 23.Essoh C, Latino L, Midoux C, Blouin Y, Loukou G, Nguetta SP, et al. (2015) Investigation of a Large Collection of Pseudomonas aeruginosa Bacteriophages Collected from a Single Environmental Source in Abidjan, Cote d'Ivoire. PLoS One 10: e0130548 10.1371/journal.pone.0130548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latino L, Pourcel C (2018) Recovery and Characterization of Bacteria Resisting Infection by Lytic Bacteriophage. Methods in molecular biology 1693: 85–98. 10.1007/978-1-4939-7395-8_8 [DOI] [PubMed] [Google Scholar]
- 25.Latino L, Caroff M, Pourcel C (2017) Fine structure analysis of lipopolysaccharides in bacteriophage-resistant Pseudomonas aeruginosa PAO1 mutants. Microbiology 163: 848–855. 10.1099/mic.0.000476 [DOI] [PubMed] [Google Scholar]
- 26.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ (1994) Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148: 81–86. [DOI] [PubMed] [Google Scholar]
- 27.Choi KH, Kumar A, Schweizer HP (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64: 391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 28.Bradley DE (1967) Ultrastructure of bacteriophage and bacteriocins. Bacteriological reviews 31: 230–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiebe WJ, Chapman GB (1968) Fine structure of selected marine pseudomonads and achromobacters. Journal of bacteriology 95: 1862–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clamens T, Rosay T, Crepin A, Grandjean T, Kentache T, Hardouin J, et al. (2017) The aliphatic amidase AmiE is involved in regulation of Pseudomonas aeruginosa virulence. Scientific reports 7: 41178 10.1038/srep41178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG (2011) Systematic chromosomal deletion of bacterial ribosomal protein genes. Journal of molecular biology 413: 751–761. 10.1016/j.jmb.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Rojas A, Mena A, Martin S, Borrell N, Oliver A, Blazquez J (2009) Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology 155: 1050–1057. 10.1099/mic.0.024745-0 [DOI] [PubMed] [Google Scholar]
- 33.Belanger M, Burrows LL, Lam JS (1999) Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145 (Pt 12): 3505–3521. [DOI] [PubMed] [Google Scholar]
- 34.Wittmann J, Klumpp J, Moreno Switt AI, Yagubi A, Ackermann HW, Wiedmann M, et al. (2015) Taxonomic reassessment of N4-like viruses using comparative genomics and proteomics suggests a new subfamily—"Enquartavirinae". Arch Virol 160: 3053–3062. 10.1007/s00705-015-2609-6 [DOI] [PubMed] [Google Scholar]
- 35.Ceyssens PJ, Brabban A, Rogge L, Lewis MS, Pickard D, Goulding D, et al. (2010) Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the "N4-like viruses". Virology 405: 26–30. 10.1016/j.virol.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan Y, Buchan A, Chen F (2015) Novel N4 Bacteriophages Prevail in the Cold Biosphere. Applied and environmental microbiology 81: 5196–5202. 10.1128/AEM.00832-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceyssens PJ, Lavigne R (2010) Bacteriophages of Pseudomonas. Future Microbiol 5: 1041–1055. 10.2217/fmb.10.66 [DOI] [PubMed] [Google Scholar]
- 38.Glonti T, Chanishvili N, Taylor PW (2010) Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J Appl Microbiol 108: 695–702. 10.1111/j.1365-2672.2009.04469.x [DOI] [PubMed] [Google Scholar]
- 39.Cornelissen A, Ceyssens PJ, Krylov VN, Noben JP, Volckaert G, Lavigne R (2012) Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology. [DOI] [PubMed] [Google Scholar]
- 40.Pourcel C, Midoux C, Vergnaud G, Latino L (2017) A carrier state is established in Pseudomonas aeruginosa by phage LeviOr01, a newly isolated ssRNA levivirus. The Journal of general virology 98: 2181–2189. 10.1099/jgv.0.000883 [DOI] [PubMed] [Google Scholar]
- 41.Los M, Wegrzyn G, Neubauer P (2003) A role for bacteriophage T4 rI gene function in the control of phage development during pseudolysogeny and in slowly growing host cells. Res Microbiol 154: 547–552. 10.1016/S0923-2508(03)00151-7 [DOI] [PubMed] [Google Scholar]
- 42.Golec P, Wiczk A, Los JM, Konopa G, Wegrzyn G, Los M (2011) Persistence of bacteriophage T4 in a starved Escherichia coli culture: evidence for the presence of phage subpopulations. The Journal of general virology 92: 997–1003. 10.1099/vir.0.027326-0 [DOI] [PubMed] [Google Scholar]
- 43.Chan JZ, Millard AD, Mann NH, Schafer H (2014) Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Frontiers in microbiology 5: 506 10.3389/fmicb.2014.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paddison P, Abedon ST, Dressman HK, Gailbreath K, Tracy J, Mosser E, et al. (1998) The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: a new perspective. Genetics 148: 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burch LH, Zhang L, Chao FG, Xu H, Drake JW (2011) The bacteriophage T4 rapid-lysis genes and their mutational proclivities. Journal of bacteriology 193: 3537–3545. 10.1128/JB.00138-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Young R (2016) The Last r Locus Unveiled: T4 RIII Is a Cytoplasmic Antiholin. Journal of bacteriology 198: 2448–2457. 10.1128/JB.00294-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JC, Andersson DI (2005) Bacterial genome size reduction by experimental evolution. Proceedings of the National Academy of Sciences of the United States of America 102: 12112–12116. 10.1073/pnas.0503654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb JS, Lau M, Kjelleberg S (2004) Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186: 8066–8073. 10.1128/JB.186.23.8066-8073.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ceyssens PJ, Minakhin L, Van den Bossche A, Yakunina M, Klimuk E, Blasdel B, et al. (2014) Development of giant bacteriophage varphiKZ is independent of the host transcription apparatus. Journal of virology 88: 10501–10510. 10.1128/JVI.01347-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, et al. (2006) Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proceedings of the National Academy of Sciences of the United States of America 103: 8487–8492. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tummler B (2011) Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol 13: 1690–1704. 10.1111/j.1462-2920.2011.02483.x [DOI] [PubMed] [Google Scholar]
- 52.Le S, Yao X, Lu S, Tan Y, Rao X, Li M, et al. (2014) Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci Rep 4: 4738 10.1038/srep04738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang H, Duan J, Sibley CD, Surette MG, Duan K (2011) Identification of mutants with altered phenazine production in Pseudomonas aeruginosa. Journal of medical microbiology 60: 22–34. 10.1099/jmm.0.022350-0 [DOI] [PubMed] [Google Scholar]
- 54.Han H, Iakovenko L, Wilson AC (2015) Loss of Homogentisate 1,2-Dioxygenase Activity in Bacillus anthracis Results in Accumulation of Protective Pigment. PloS one 10: e0128967 10.1371/journal.pone.0128967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hocquet D, Petitjean M, Rohmer L, Valot B, Kulasekara HD, Bedel E, et al. (2016) Pyomelanin-producing Pseudomonas aeruginosa selected during chronic infections have a large chromosomal deletion which confers resistance to pyocins. Environmental microbiology 18: 3482–3493. 10.1111/1462-2920.13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Lin B, Mostaghim A, Rubin RA, Glaser ER, Mittraparp-Arthorn P, et al. (2013) Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Frontiers in microbiology 4: 379 10.3389/fmicb.2013.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bierne H, Ehrlich SD, Michel B (1991) The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. The EMBO journal 10: 2699–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koskiniemi S, Sun S, Berg OG, Andersson DI (2012) Selection-driven gene loss in bacteria. PLoS genetics 8: e1002787 10.1371/journal.pgen.1002787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen M, Zhang H, Shen W, Zou Z, Lu S, Li G, et al. (2018) Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic acids research 46: 4505–4514. 10.1093/nar/gky160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen P, Huang HV (1986) Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112: 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehrlich SD, Bierne H, d'Alencon E, Vilette D, Petranovic M, Noirot P, et al. (1993) Mechanisms of illegitimate recombination. Gene 135: 161–166. [DOI] [PubMed] [Google Scholar]
- 62.Allgood ND, Silhavy TJ (1991) Escherichia coli xonA (sbcB) mutants enhance illegitimate recombination. Genetics 127: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Rojas A, Blazquez J (2009) The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. Journal of bacteriology 191: 844–850. 10.1128/JB.01081-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thrane SW, Taylor VL, Lund O, Lam JS, Jelsbak L (2016) Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. Journal of clinical microbiology 54: 1782–1788. 10.1128/JCM.00349-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan X, Cui X, Zhang F, He Y, Li L, Yang H (2016) Genetic Evidence for O-Specific Antigen as Receptor of Pseudomonas aeruginosa Phage K8 and Its Genomic Analysis. Front Microbiol 7: 252 10.3389/fmicb.2016.00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Islam ST, Lam JS (2014) Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60: 697–716. 10.1139/cjm-2014-0595 [DOI] [PubMed] [Google Scholar]
- 67.Broeker NK, Barbirz S (2017) Not a barrier but a key: How bacteriophages exploit host's O-antigen as an essential receptor to initiate infection. Molecular microbiology 105: 353–357. 10.1111/mmi.13729 [DOI] [PubMed] [Google Scholar]
- 68.Kiino DR, Rothman-Denes LB (1989) Genetic analysis of bacteriophage N4 adsorption. Journal of bacteriology 171: 4595–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiino DR, Licudine R, Wilt K, Yang DH, Rothman-Denes LB (1993) A cytoplasmic protein, NfrC, is required for bacteriophage N4 adsorption. Journal of bacteriology 175: 7074–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McPartland J, Rothman-Denes LB (2009) The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. Journal of bacteriology 191: 525–532. 10.1128/JB.01423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vecchietti D, Di Silvestre D, Miriani M, Bonomi F, Marengo M, Bragonzi A, et al. (2012) Analysis of Pseudomonas aeruginosa cell envelope proteome by capture of surface-exposed proteins on activated magnetic nanoparticles. PloS one 7: e51062 10.1371/journal.pone.0051062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi Y, Xiong X, Wang X, Duan C, Jia Y, Jiao J, et al. (2013) Proteome analysis and serological characterization of surface-exposed proteins of Rickettsia heilongjiangensis. PloS one 8: e70440 10.1371/journal.pone.0070440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imperi F, Ciccosanti F, Perdomo AB, Tiburzi F, Mancone C, Alonzi T, et al. (2009) Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics 9: 1901–1915. 10.1002/pmic.200800618 [DOI] [PubMed] [Google Scholar]
- 74.Moll I, Grill S, Grundling A, Blasi U (2002) Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Molecular microbiology 44: 1387–1396. [DOI] [PubMed] [Google Scholar]
- 75.Hershey AD (1946) Mutation of Bacteriophage with Respect to Type of Plaque. Genetics 31: 620–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran TA, Struck DK, Young R (2005) Periplasmic domains define holin-antiholin interactions in t4 lysis inhibition. Journal of bacteriology 187: 6631–6640. 10.1128/JB.187.19.6631-6640.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abedon ST, Culler RR (2007) Optimizing bacteriophage plaque fecundity. Journal of theoretical biology 249: 582–592. 10.1016/j.jtbi.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 78.Roychoudhury P, Shrestha N, Wiss VR, Krone SM (2014) Fitness benefits of low infectivity in a spatially structured population of bacteriophages. Proceedings Biological sciences 281: 20132563 10.1098/rspb.2013.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
PcyII-10-V03(1) PcyII-10-V10(4) and PcyII-10-V15(7) were transformed either by the vector pUCP24 (A) or by the recombinants (B). Colonies were observed on medium containing Gentamycin and the bacteria were tested for their susceptibility to four dilutions of phage Ab09 (1), Ab09-V03LP (2) and Ab27 (3).
(TIF)
(DOCX)
Data Availability Statement
The DNA sequence of the PcyII-10 strain has been deposited under accession number LT673656. The twelve archives of sequencing reads produced in the course of this study were deposited in the European Nucleotide Archive (ENA) under bioproject PRJEB18612 (https://www.ebi.ac.uk/ena/data/view/PRJEB18612; PcyII-10 resequenced colony POU32, variants V02 (POU35), V03 (POU33), V06 (POU43), V07 (POU44), V10 (POU45), V14 (POU34), V15 (POU36), V16 (POU29), V17 (POU23), V18 (POU27), V19 (POU25)).