Abstract
Recent advances in genetic analysis technologies such as next-generation sequencing (NGS) have considerably increased the incidental discovery of genetic abnormalities. Six heterozygous missense mutations of the human glucocorticoid receptor (GR; encoded by the NR3C1 gene) have been identified in the context of genetic screening of endocrine pathologies. GR, a nuclear receptor, hormone-induced transcription factor, is involved in many physiological processes. Nevertheless, the pathogenic significance of incidentally discovered mutations remains obscure. The aim of this work was to characterize these variants by evaluating their functional impact on GR signaling. Six original GR variants, located in exon 2, led to amino acid substitutions of the N-terminal domain of GR (F65V, M86V, A229T, A304E, N374S, and R386Q), excluding mainly the activation function tau core 1 domain, the potential site of functional interaction with transcriptional coregulators. Transient cotransfection in HEK293T cells of mutated GR-expressing vectors and a luciferase reporter established dose-response curves for dexamethasone. This excluded any major transactivation abnormality of the mutated GRs (ligand concentration leading to 50% maximal transactivation capacity ≈ 0.2 nM), with maximal transactivation capacity identical to that of the wild-type (WT) GR and without modification of the potentiation of transcriptional coactivator steroid receptor coactivator 2 except in N374S. Moreover, protein expression of mutated GRs and their cytonuclear translocation studied by immunocytochemistry were almost unchanged compared with WT GR. These results underline the silent nature of these missense GR variants and call for cautious interpretation of the discovery of genetic incidentalomas by NGS in the absence of detailed characterization in order to appropriately assess their functional impact on a particular signaling pathway.
Keywords: NR3C1, glucocorticoid signaling, transcription, genetic incidentalomas
Since the early 2000s, improvement in genetic technologies have allowed easier and faster sequencing with high fidelity. Next-generation sequencing (NGS), also referred to as high-throughput sequencing, has drastically changed classic genetic screening compared with earlier techniques, notably the time-consuming process of Sanger-based sequencing [1]. However, NGS technologies (e.g., Illumina®, Ion Torrent®) produce massive data that should be stocked and analyzed [2]. Clinical use of NGS has shortened the delay between blood sampling and results but has increased the number of both analyzable genes and patients, yet allowing earlier genetic diagnosis of patients [3, 4]. Thus, NGS has considerably changed genetic testing as a diagnostic tool—from select targeted genes to the association of known genes implicated in the pathogenicity of the disease [5, 6]. Some variants are of unknown significance according to the classifications proposed by the American College of Medical Genetics and Genomics in 2015 [7], and it is often difficult to appreciate whether they are pathogenic variants. Despite the use of predictive software examining the impact of genetic variants on protein functionality (i.e., PANTHER, MutationTaster, Human Splicing Finder, PhyloP, VarSome) [7] and because three-dimensional models may not be available for some proteins, it is clear that only detailed functional characterization of the protein variants can predict the deleterious impact of genetic variation.
Recently, using an NGS gene panel composed of 84 protein-coding genes to search for genetic alterations potentially associated with specific clinical endocrine phenotypes, we incidentally discovered six heterozygous missense variants of the nuclear receptor subfamily 3 group C member 1 (NR3C1) gene encoding the human glucocorticoid receptor (hGR) isoform α. This ligand-dependent transcription factor belongs to the nuclear receptor superfamily [8] and mediates most glucocorticoid (GC) actions in a large variety of tissues. hGRα is a modular 777‒amino acid protein of 97 kDa molecular weight composed of three distinct domains: the N-terminal domain (NTD), the DNA-binding domain, and the ligand-binding domain. The hinge region articulates the last two domains (Fig. 1). Fewer than 30 private loss-of-function mutations of hGRα have been reported to date, leading to GC resistance syndrome [9–12]. Up to now, two gain-of-function mutations located at the end of the NTD (D401H) and at the end of the 3′-UTR of the NR3C1 gene (G3134T), associated with GC hypersensitivity syndrome, have been described [13, 14]. No deleterious mutation has ever been reported in the NTD of the GR. Interestingly, all six heterozygous missense variants incidentally discovered by NGS are located in the NTD of hGRα.
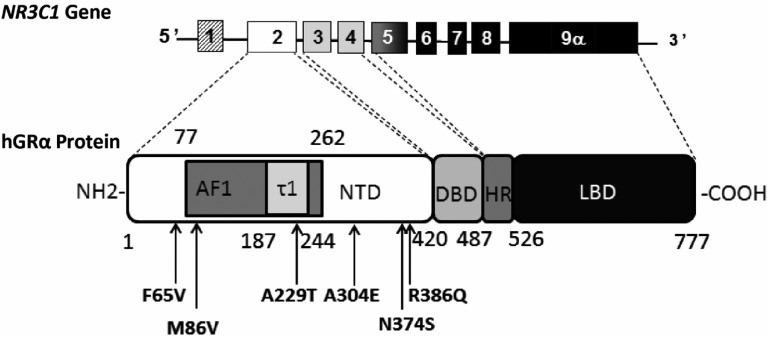
Figure 1.
Schematic representation of NR3C1 gene organization and localization of the six missense variants in the NTD of the GRα protein. The NR3C1 gene is located in chromosome 5 in humans and contains at least 10 exons (exon 9β is not illustrated). The first one is an untranslated exon. Exon 2, the first translated exon, encodes the entire NTD (aa 1–420) composed of the activation function 1 (AF1) domain (aa 77–262), which encompasses the tau core 1 (τ1) domain (aa 187–244). Exons 3 and 4 encode the two zinc fingers of the DNA-binding domain (DBD; aa 421–487), whereas exons 5 to 9 encode the hinge region (HR; aa 488–526) and the ligand-binding domain (LBD; aa 527–777) of the GRα protein. All six missense genetic variants (arrows) discovered by NGS are located in the NTD.
The NTD, composed of 420 amino acids, is the longest domain of hGRα and is entirely encoded by exon 2. NTD is the target for many posttranslational modifications (e.g., phosphorylation, ubiquitination, sumoylation), essential for optimal and efficient signal transduction. The tridimensional structure of the NTD is presently unsolved because of its intrinsic disorder state [15–18]. Amino acids 77 to 262 harbor a ligand-independent activation function of the hGR named activation function 1 [19–21], which contains a major activation domain also referred to as Tau1 core, a short 58-aa residue domain located between amino acids 187 and 244 (Fig. 1), essential for the activation function of hGRα. The tau core 1 is mostly composed of three α-helical prestructured motifs surrounded by repressive regions [16, 18, 19, 22]. The NTD is a privileged site for interaction and binding with regulatory molecules, namely transcriptional coregulators [steroid receptor coactivator (SRC) 2 or TIF2 and SRC1] [23–25].
The purpose of this study was to perform a functional characterization of the six heterozygous missense variants of hGRα, referred to as genetic incidentalomas, to clarify the potential pathogenic impact of these genetic variations and to improve our understanding of some aspects of GR-mediated signaling.
1. Methods
A. Genetic Studies
Two hundred patients in the Genetic Department of Bicêtre Hospital underwent genetic screening for gonadic diseases and steroidogenesis abnormalities. None of the patients presented with overt GC resistance syndrome. All patients gave their written consent for these genetic analyses. Genomic DNA was extracted from peripheral lymphocytes, and the entire coding exons and exon-intron junctions of the following genes were sequenced with the NGS technology Illumina®, as previously described [26]. All 84 genes are described in Table 1.
Table 1.
List of the 84 Genes Included in the NGS Panel
| Genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| AKR1C2 | CYP11A1 | ESR1 | GALT | LATS1 | MSH5 | NR3C2 | RSPO1 | WNT4 |
| AKR1C4 | CYP17A1 | ESR2 | GATA4 | LHB | NBN | NR5A1 | SOHLH2 | WT1 |
| AMH | CYP21A2 | FIGLA | GDF9 | LHCGR | NCOA1 | PAX6 | SOX3 | WWOX |
| AMHR2 | DHH | FMR1 | GJA4 | LHX8 | NCOA2 | PGR | SOX9 | XPNPEP2 |
| AR | DIAPH2 | FOXA1 | GPR3 | LHX9 | NCOA3 | PGRMC1 | SRD5A1 | |
| ATM | DMC1 | FOXA3 | HFM1 | MAMLD1 | NCOR1 | PITX2 | SRD5A2 | |
| ATRX | DMRT1 | FOXC1 | HSD17B3 | MAP3K1 | NCOR2 | POF1B | SRY | |
| BHLHB9 | DMRT2 | FOXL2 | HSD3B2 | MCM8 | NOBOX | POR | STAG3 | |
| BMP15 | EIF2S2 | FSHB | INHA | MCM9 | NR0B1 | RICTOR | STAR | |
| CBX2 | EMX2 | FSHR | INHBB | MID1 | NR3C1 | RPTOR | TSPYL1 | |
The NR3C1 gene was compared with National Center for Biotechnology Information references (NG 016367.1 RefSeqGene, NM 000176.2 Transcript, and NP 000167.1 Protein). Of 200 individuals, six patients were found to carry an NR3C1 variation corresponding to a heterozygous missense mutation of a single nucleotide. All variants were located in the NTD of the hGR. These patients had no other genetic variations in any of the 84 genes of the NGS panel.
B. Site-Directed Mutagenesis
pcDNA3hGRα (c.193T>G:p.F65V), pcDNA3hGRα (c.256A>G:p.M86V), pcDNA3hGRα (c.685G>A:p.A229T), pcDNA3hGRα (c.911C>A:p.A304E), pcDNA3hGRα (c.1121A>G:p.N374S), pcDNA3hGRα (c.1120A>G-1122C>A:p.N374E), and pcDNA3hGRα (c.1157G>A:p.R386Q) were obtained using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), with pcDNA3hGRα (NM 000176.2) as a template. Primers used are presented in Table 2.
Table 2.
Primer Sequences Used for Site-Directed Mutagenesis to Generate the Six Genetic GR Variants
| Variants | Primer Sequence |
|---|---|
| F65V | 5′-CGACGCAAGACTTTTGGTTGATGTTCCAAAAGGCTCA-3′ |
| M86V | 5′-TCTGTCAAAGCAGTTCCACTCTCAGTGGGACTGTATAT-3′ |
| A229T | 5′-TTTGCTTTCTCCTCTGACGGGAGAAGACGATTCA-3′ |
| A304E | 5′-CACAGTTTACTGTCAGGAAAGCTTTCCTGGAGCAA-3′ |
| N374S | 5′-GGATCTGGAGATGACAGCTTGACTCTCTGGGG-3′ |
| R386Q | 5′-CTCTGAACTTCCCTGGTCAAACAGTTTTTTCTAATGGC-3′ |
| N374E | 5′-GAGTCCCCAGAGAAGTCAATTCGTCATCTCCAGATCCTTGG-3′ |
The base in bold indicates the point mutation.
C. Cell Culture
Embryonic human kidney HEK293T and African green monkey kidney COS-7 cells were grown in DMEM (Life Technologies, Saint-Aubin, France) with 10% fetal calf serum (Biowest, Nuaille´, France) and penicillin/streptomycin (GE Healthcare, Vélizy-Villacoublay, France) and were incubated at 37°C in a humidified atmosphere containing 5% CO2 [27].
D. Transactivation Assays
GR-deficient HEK293T cells were transfected with wild-type (WT) or mutated GR-expressing plasmids (40 ng/well in 96-well plates), reporter luciferase plasmid pMMTV-luc (40 ng/well), and pMIR-β-gal encoding β-galactosidase as a normalizing vector (35 ng/well) using Lipofectamine 2000 (Thermo Fisher Scientific, Saint-Aubin, France). In addition, SRC2 or TIF2 or pcDNA3 (empty plasmid) (40 ng/well) was added in some transactivation assays together with the same amount of the three previously described plasmids. Six hours later, transfection medium was replaced with steroid-free DMEM medium. Twenty-four hours posttransfection, ethanol or increasing concentrations of dexamethasone (DXM) (0.1 to 1000 nM or 5 nM DXM for the cotransfection with SRC2) were added to the medium. Luciferase and β-galactosidase activities were measured in cell lysates as previously described [27, 28].
E. Western Blot Analysis
Twenty-four hours after transfection with the corresponding plasmids, HEK293T cells were lysed and 20 µg of the protein extracts were submitted to electrophoresis. After electroblotting onto C nitrocellulose membranes and incubation with blocking solution [5% fat-free dry milk in Tris-buffered saline containing 1% Tween 20], immunoblotting was performed overnight at 4°C using a mouse monoclonal anti-hGRα antibody directed against the NTD of the protein (SC393232, Santa Cruz Biotechnology ®) [29] at a final concentration of 1 μg/mL and with the anti‒β-actin antibody (Sigma, St Quentin-Falavier, France) [30] used as a loading control. Membranes were incubated with secondary fluorescent antibodies (Pierce; Dyelight 680 or 800) [31, 32]. Bands were visualized and quantified using Odyssey® Fc, Dual-Mode Western Imaging (Li-Cor, Lincoln, NE) [27].
F. Immunocytochemistry and Fluorescence Deconvolution Microscopy
GR-deficient COS-7 cells were transiently transfected with the corresponding plasmids and treated or not with 100 nM DXM for 1 hour and then processed for immunocytochemistry as previously described [33]. Briefly, cells were fixed, permeabilized, and incubated overnight at 4°C with a mouse anti-GR antibody [29], followed by 30 minutes of incubation with an anti-mouse Alexa 555‒coupled secondary antibody [34]. After 10 minutes of postfixation, nuclear counterstaining was performed with 0.5 μg/mL 4′,6-diamidino-2-phenylindole for 1 minute. Chambers were mounted using ProLong Gold mounting medium (Thermo Fisher Scientific). Fluorescence deconvolution microscopy was performed using Image Pro Plus AMS software (Media Cybernetics Inc., Marlow, UK) on a Mono Q Imaging Retiga 2000R Fast 1394 camera (Q Imaging Inc., Surrey, BC, Canada). Images were acquired with an automated upright BX61 microscope (Olympus, Rungis, France) equipped with an X60 objective (1.4 NA). A z-series of focal planes was digitally imaged and deconvolved with the three-dimensional blind iterative algorithm (Image Pro Plus AMS) to generate high-resolution images [27].
G. Automated High-Throughput Microscopy
Sequential images were acquired (20×/0.4 NA), analyzed, and quantified by the ArrayScan VTI imaging platform (Thermo Fisher Scientific). 4′,6-Diamidino-2-phenylindole and GR fluorescence was captured using sequential acquisition. The Molecular Translocation V4 Bioapplication algorithm (vHCS Scan, version 6.3.1, Build 6586) was used to quantify the relative subcellular distribution of immunodetected GR, essentially as described by [33, 35]. The translocation index measurement represents the average value ratio of the nuclear intensity of GR to the cytoplasmic intensity of GR calculated for each selected cell per well [27]. The higher this value, the more GR is translocated into the nuclear compartment.
H. Statistical Analysis
Results were expressed as mean ± SEM in the tables and figures. Statistical analyses were performed with Prism 7.0a software (Graph Pad Software, La Jolla, CA). Comparison of continuous variables among different groups was performed using ANOVA or nonparametric Kruskal-Wallis tests with Dunn posttests. When appropriate, nonparametric Mann-Whitney U tests were also performed. P values <0.05 were considered significant.
2. Results
A. Identification of the NR3C1 Genetic Variants
Among the six heterozygous genetic variants identified (Fig. 1; Table 3), three were private variants (A304E, N374S, and R386Q), indicating that they have been rarely if ever described in any databases (ExAC and 1000G), whereas the other three variants (F65V, M86V, and A229T) have already been reported. M86V has been described in nine patients, whereas the allelic frequency of A229T was at 0.0014 (165/120812) and that of F65V was at 0.0013 (153/121406) in the general population, although the latter variant is more frequently associated with the African population (0.014) (http://exac.broadinstitute.org/). All these variants except M86V were classified as having unknown significance according to Varsome, whereas the M86V variant was classified as likely pathogenic by Varsome. None of these variants has ever been functionally characterized. With respect to interspecies conservation, the University of California Santa Cruz Genome Browser website (https://genome.ucsc.edu/) indicates that all these amino acids except N374 were strictly conserved in mammals (humankind, Rhesus monkey, mouse, dog, and elephant) and in the chicken. Amino acid M86 and R386 residues were also conserved in the zebrafish, whereas M86 was also recovered in the Xenope Nr3c1 sequence. Although there is a potential pathogenicity of these variants and that no functional characterization has been performed, the MutationTaster website (www.mutationtaster.org/) predicted that all would be disease-causing mutations except for one hGR variant (i.e., N374S), which was predicted to be an NR3C1 polymorphism (Table 3). However, the association between genetic variation and disease is generally difficult to determine for rare variants in the absence of functional characterization.
Table 3.
Summary of the Genetic and Protein Characteristics of the Six Missense GR Variants
| DNA Modification | Protein Modification | Amino Acid Nature’s Modification | Predicted Pathogenicity (MutationTaster) | Allelic Frequency (ExAC) | Interspecies Conservation (UCSC Genome Browser) | |||
|---|---|---|---|---|---|---|---|---|
| Mammary | Chicken | Xenope | Zebrafish | |||||
| exon2: c.193T>G | F65V | 0 | Disease causing | 153/121406 | + | + | − | − |
| exon2: c.256A>G | M86V | 0 | Disease causing | 9/121218 | + | + | + | + |
| exon2: c.685G>A | A229T | Apolar → polar | Disease causing | 165/120812 | + | + | − | − |
| exon2: c.911C>A | A304E | Apolar → negative | Disease causing | 1/121284 | + | + | − | − |
| exon2: c.1121A>G | N374S | 0 | Polymorphism | 1/121308 | + | − | − | − |
| exon2: c.1157G>A | R386Q | Positive → polar | Disease causing | 0 | + | + | − | + |
Amino acid nature’s modification = conserved nature. Allelic frequency by ExAC and interspecies conservation by University of California Santa Cruz (UCSC) Genome Browser. Mammals correspond to human, Rhesus monkey, mouse, dog, and elephant (+ conservation and − variation of the amino acid between species), and predicted pathogenicity is by MutationTaster.
B. Functional Characterization of hGR Variants
Global functionality of the entire GR signaling pathway is well evaluated by reporter gene assays [36]. Indeed, transactivation studies indirectly reflect all GR-dependent steps, including ligand binding, transconformation, nuclear translocation of the GR-ligand complex, functional association with transcriptional coregulators, and finally induction of target gene expression. For this purpose, dose-response curves (from 0.1 to 1000 nM) of DXM, a GC agonist, were established by transiently transfecting the GRE-Luc reporter gene and the WT human GR expression vector or its genetic variants (F65V, M86V, A229T, A304E, N374S, and R386Q) into GR-deficient HEK293T cells.
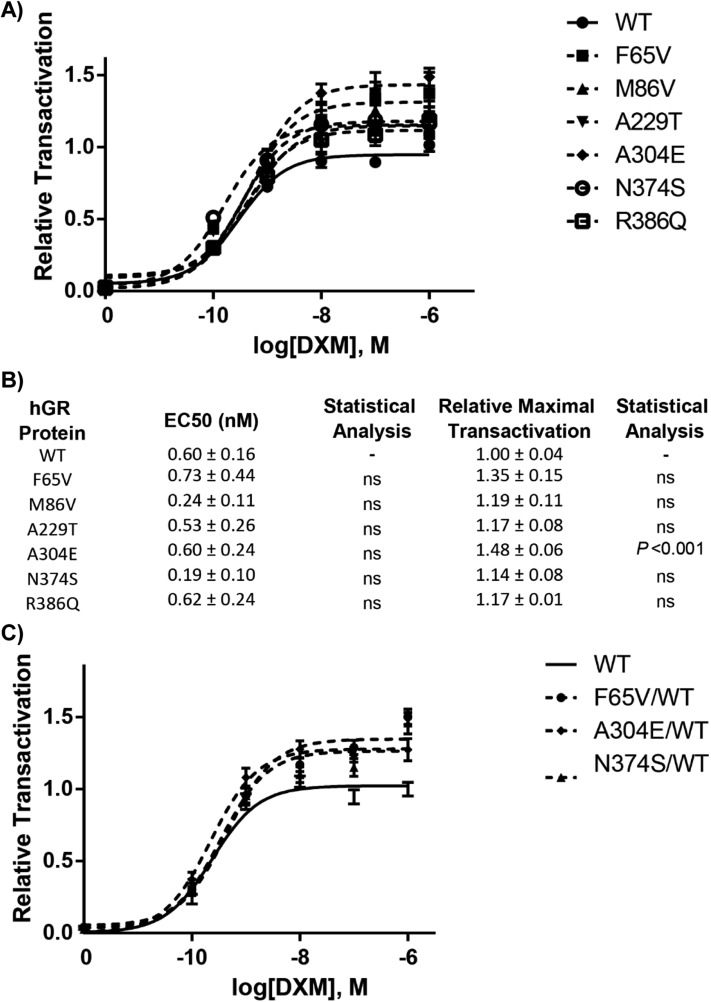
As illustrated in Fig. 2A, the transactivation capacity of WT reaches a plateau at 100 nM DXM concentration, with a calculated ligand concentration leading to 50% maximal transactivation activity, or Bmax (EC50) at 0.604 ± 0.160 nM DXM (mean ± SEM, n = 30 replicates from three independent experiments) (Fig. 2B). The Bmax of WT was arbitrarily set at 1.0. All genetic variants were able to transactivate the GRE-Luc reporter gene to the same extent as that driven by the WT (Fig. 2A). No difference in luciferase activity was observed between the WT and the F65V, M86V, and R386Q variants at any DXM concentration. Likewise, transcriptional activities of the A229T and N374S variants did not differ from that of the WT, except at the lower 0.1 nM DXM concentration, with a relative transactivation measured at 0.41 ± 0.02 (P < 0.01; Kruskal-Wallis) for A229T and 0.51 ± 0.03 (P < 0.001) for N374S, compared with 0.29 ± 0.02 for the WT. Lastly and unexpectedly, only the A304E variant had a higher transactivation capacity at any concentration of DXM tested except 1 nM (nonparametric statistical analysis of Kruskal-Wallis on 30 replicates from four independent experiences). However, the EC50 was not different from that of the WT, suggesting a conserved affinity of the ligand. In any case, no altered transcriptional activity was observed with all tested variants, excluding any major deleterious functional consequences of the substitutions.
Figure 2.
Dose-response curves of transcriptional activation of the GR variants compared with that of the WT receptor. (A) HEK293T cells plated in 96-well plates were cotransfected with 40 ng of plasmid encoding GR WT or GR variants (F65V, M86V, A229T, A304E, N374S, and R386Q), together with the reporter luciferase plasmid pMMTV-Luc, in which luciferase activity is under the control of GR response elements, and pMIR-β-gal encoding β-galactosidase. After 24-h stimulation by ethanol or increasing concentrations of DXM (10−10 to 10−6 M), enzymatic activities were measured. Transcriptional activities are expressed relative to the maximum transactivation of the WT (arbitrary set at 1). Each point represents the mean ± SEM of at least 30 replicates performed from at least three independent experiments. Representation was fitted with the “log(agonist) vs response three parameters” function of GraphPad software. (B) Comparison of EC50 (DXM concentration inducing half-maximum transactivation) and of maximum transcriptional activity (Bmax) of the six GR variants compared with the WT. Values are mean ± SEM, obtained from four independent experiments (nonparametric ANOVA Kruskal-Wallis tests followed by Dunn posttests). (C) The absence of dominant negative effects of the variants on the WT is shown. HEK293T cells plated in 96-well plates were cotransfected with 40 ng of plasmid encoding GR WT or 20 ng of plasmid encoding WT plus 20 ng encoding GR variants (F65V, A304E, and N374S), together with the reporter luciferase plasmid pMMTV-Luc, in which luciferase expression is under the control of GR response elements, and pMIR-β-gal coding β-galactosidase. After 24-h stimulation by ethanol or increasing concentrations of DXM (10−10 to 10−6 M), enzymatic activities were measured. Transcriptional activities are expressed relative to the maximum transactivation of the WT (arbitrary set at 1). Each point represents the mean ± SEM of at least 30 replicates performed from at least three independent experiments. Representation was fitted with the “log(agonist) vs response three parameters” function of GraphPad software. EC50, ligand concentration leading to 50% maximal transactivation capacity.
Figure 2B summarizes the main parameters (EC50 and Bmax) of all genetic GR variants. The EC50 (mean ± SEM) calculated from at least three independent experiments for each variant was in the nanomolar range, whereas the Bmax did not differ from that of the WT except for the A304E variant, as mentioned previously.
Owing to the heterozygous nature of the identified genetic variants, which supports biallelic expression, and given that some GR mutations have reportedly exerted a dominant negative effect [27], we next evaluated whether these variants exert adverse effects on WT GR signaling. Despite the normality of the intrinsic transactivating properties of the GR variants, dose-responses curves of DXM that were performed after cotransfection of equimolar amounts of WT and GR variant expression vector excluded any dominant negative effect of GR variants (Fig. 2C).
C. Protein Expression of the Receptor
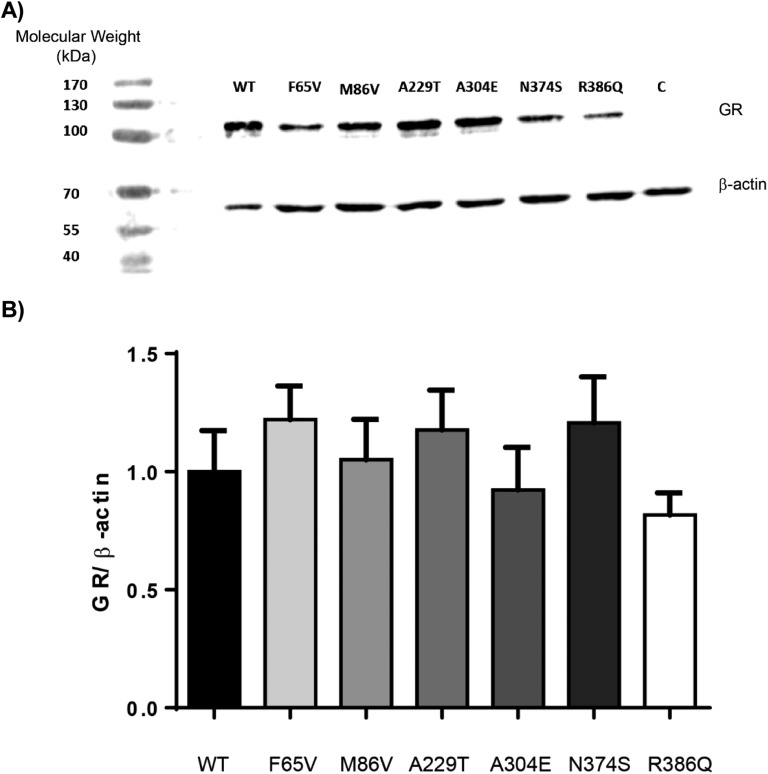
To examine whether slight modifications of GR variant transactivation capacity may be explained by different protein expression levels, western blot analyses were performed with whole cell extracts after transient transfections in HEK293T cells. As anticipated, WT and GR variants were detected with an anti-NTD GR antibody and were recovered as an ∼100-kDa band (Fig. 3A). Relative expression levels were identical for each GR variant and the WT as quantified by the hGR/β-actin ratio. Statistical analysis showed no significant difference between the GR WT and the six variants (P = 0.46 by Kruskal-Wallis test) (Fig. 3B). Given that western blot analyses gave only an overall estimation of the total GR cellular protein content (without exploring potential discrete modifications of their subcellular localization) and to further exclude subtle protein expression alterations, automated high-throughput microscopy—a more sensitive and quantitative approach—was used to evaluate the DXM-induced GR nuclear transfer, a major and critical step of transactivation.
Figure 3.
Protein expression of the WT and the six GR variants. (A) HEK293T cells were transiently transfected or not with a plasmid encoding the WT or the six GR variants. Twenty-four hours posttransfection, protein extracts (20 µg) were analyzed by western blot followed by immunostaining with an antibody recognizing the NTD of GR (29; dilution 1/500) and an anti‒β-actin antibody (30; dilution 1/1000). GRα protein expression was similar between the WT and the six variants but was below the detectable threshold in the untransfected cells used as control (i.e., C). Representative image of three independent experiments is shown. The molecular weight marker used was PageRulerTM (Thermo Fisher Scientific). (B) Quantification of the protein signal did not reveal any statistically significant difference between the WT and the GR variants. Results are expressed as means ± SEM.
D. Ligand-Induced Nuclear Translocation of GR WT and Its Variants
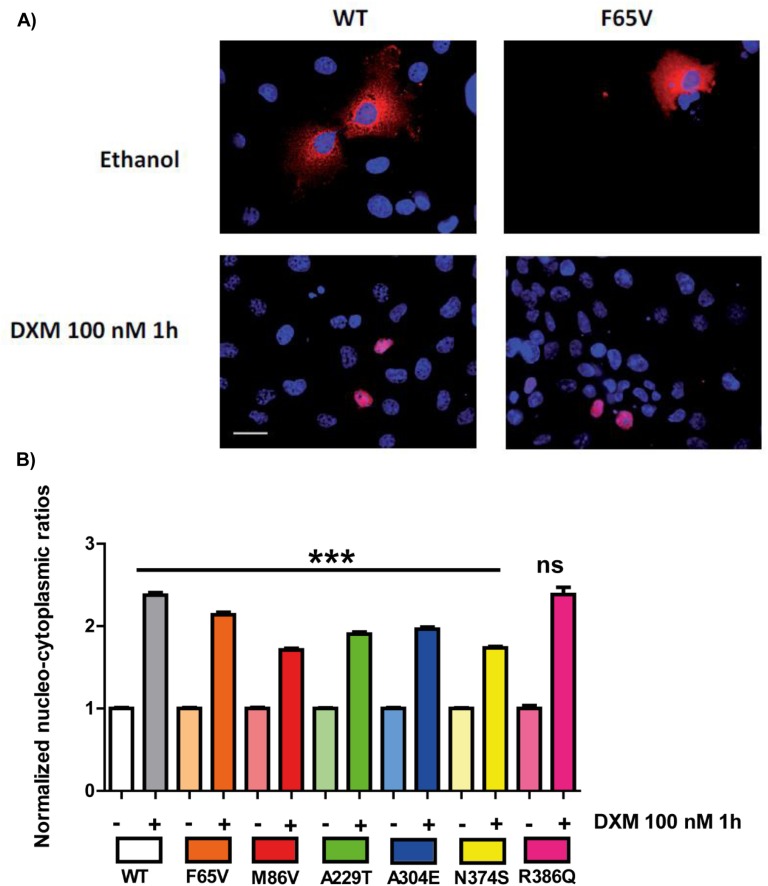
Concomitant analysis of the fluorescence from both cytoplasmic and nuclear compartments facilitated assessment of the hormone-induced nuclear translocation for each GR variant (F65V, M86V, A229T, A304E, N374S, and R386Q) after transient transfection into GR-deficient COS-7 cells. Figure 4A shows representative images of DXM-induced nuclear transfer for the WT and F65V GR variant. In the absence of ligand, most WT and F65V GR is in the cytoplasmic compartment, whereas with 1-hour DXM stimulation, a clear-cut transfer to the nucleus was observed for both receptors. All other variants behaved similarly.
Figure 4.
DXM-induced nuclear translocation of the WT and the six GR variants. COS-7 cells (grown in F6 or 100-mm petri dish flasks) were transiently transfected with 400 ng of plasmid encoding the WT or the six GR variants (F65V, M86V, A229T, A304E, N374S, and R386Q). After overnight recovery, cells were stimulated by ethanol (−) or 100 nM DXM (+) for 1 h and fixed and processed for immunofluorescence studies, followed by quantification analysis with high-throughput microscopy (see Methods section). (A) Representative images of cyto-nuclear translocation of the WT and F65V GR variant with ethanol (control) or with DXM for 1 h. Images obtained by deconvolution microscopy at 60× magnification and nuclear staining by 4′,6-diamidino-2-phenylindole (blue) and GR (red) using antibody [29] from Santa Cruz Biotechnology. White bar, 20 µm. (B) Comparison of the cyto-nuclear shuttling of the six variants compared with that of the WT after quantification by high-throughput microscopy on >1500 transfected cells per condition. Results are expressed as mean ± SEM. Nuclear-cytoplasmic ratios for each construction were normalized to the mean value arbitrarily set at 1 under basal conditions (i.e., in the absence of DXM stimulation). Statistical analysis was performed by an ANOVA test followed by Bonferroni posttests. ***P < 0.001. ns, not significant.
High-throughput microscopy analysis on >1500 transfected cells gave a precise quantification of subcellular GR shuttling (Fig. 4B). Under basal conditions (i.e., in the absence of DXM), the WT and all variants displayed identical cytoplasmic localization with similar intensities, as revealed by the nuclear/cytoplasmic (N/C) fluorescence ratio. After 100 nM DXM exposure for 1 hour, a strong increase in the N/C ratio was measured for these variants, indicating an unaltered DXM-induced translocation of GR, even though the N/C ratio values were slightly lower for F65V (2.14 ± 0.03), M86V (1.71 ± 0.02), A229T (1.90 ± 0.02), A304E (1.96 ± 0.02), and N374S (1.74 ± 0.02) than for WT (2.37 ± 0.03) and R386Q (2.38 ± 0.09).
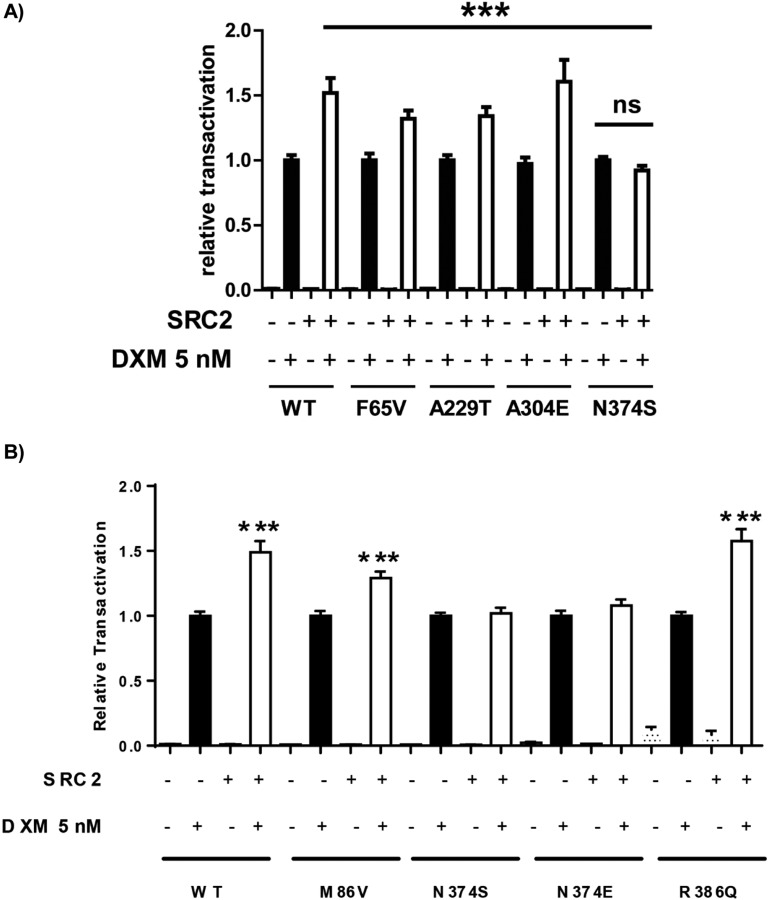
E. Transactivation Capacity in Association With a Coactivator SRC2 (or TIF2)
To more precisely explore the functional characteristics of GR variants, notably those exhibiting moderately higher transactivation capacity than the WT (i.e., F65V, A229T, A304E, and N374S; see Fig. 2), we addressed the question of their potential increased interaction with the coactivator (SRC2 or TIF2), which is known to interact with the NTD of GR [37]. Transient transactivation assays in the presence or absence of SRC2-expressing vector (Fig. 5A) were performed. The basal transcriptional activity of each variant was arbitrarily set at 1, similar to that of WT as previously demonstrated (see Fig. 2A). Upon DXM stimulation with a 5-nM concentration, which induced approximately half-maximum transactivation potency, the presence of SRC2 significantly increased luciferase activity driven by the WT and the F65V, A229T, and A304E variants, consistent with the coactivating property of SRC2. Surprisingly, this SRC2-induced stimulation was not observed with the N374S variant (Fig. 5A), suggesting a pivotal role of aa N374 residue for molecular interaction with SRC2. Because phosphorylations are important posttranslational modifications, occurring mostly in the NTD and known to greatly affect hGR transactivity [38, 39], we postulated that the serine 374 residue may constitute a new phosphorylation site impairing recruitment of the coactivator SRC2. To support this hypothesis, the phospho-mimetic variant N374E hGR was generated; interestingly, it was also shown to be insensitive to SRC2 coactivation (Fig. 5B).
Figure 5.
Effect of the coactivator SRC2 on GR transactivation. (A) HEK293T cells were plated on 96-well plates and cotransfected with 40 ng of plasmid encoding GR WT or the variants (F65V, M86V, A229T, A304E, and N374S), together with a plasmid encoding the coactivator SRC2 (40 ng/well) (+) as in the legend of Fig. 2. Cells were treated (+) or not (−) with 5 nM of DXM for 24 hours. Transcriptional activities were measured as in Fig. 2 and were expressed as mean ± SEM of 12 replicates from three independent experiments, normalized for each construct to the transactivation value obtained after stimulation by DXM in the absence of SRC2, arbitrarily set at 1. There was no difference between the induction of transactivation by SRC2 for the WT and all the variants, except for N374S, which showed no increase in transactivation following SRC2 cotransfection. ***P < 0.001, nonparametric ANOVA Kruskal-Wallis test followed by Dunn posttests. (B) HEK293T cells were plated on 96-well plates and cotransfected with 40 ng of plasmid encoding GR WT or the variants (M86V, N374E, and R386Q), together with a plasmid encoding the coactivator SRC2 (40 ng/well) (+) as in the legend of Fig. 2A. Cells were treated (+) or not (−) with 5 nM of DXM for 24 hours. Transcriptional activities were measured as in Fig. 2A and were expressed as mean ± SEM of eight to 12 replicates from three independent experiments, normalized for each construct to the transactivation value obtained after stimulation by DXM in the absence of SRC2, arbitrarily set at 1. SRC2 cotransfection stimulated transactivation of the WT as well as that of the M86V and R386Q variants but not that of N374S and N374E variants. ***P < 0.001, nonparametric Mann-Whitney U tests. ns, not significant.
3. Discussion
With the aim of characterizing genomic variation, we demonstrated that the six heterozygous genetic missense GR variants (F65V, M86V, A229T, A304E, N374S, and R386Q) incidentally discovered after NGS analysis exert a normal transactivation capacity compared with that of the WT, excluding any major impacts on the GC signaling pathway as well as their association with specific diseases.
The current study has two important facets. First, it offers improved insight into GC signaling related to NR3C1 variations and increases our understanding of certain critical steps of GR-mediated transcriptional activation. Second, it confidently assesses association information between a variant and the absence of disease, questioning the clinical decisions to be made following identification of such genetic alterations in the context of diagnostic uncertainty, and the potential pathogenicity of GR variants.
With respect to GC signaling, we ruled out any biologically relevant modifications in GR-mediated transcriptional activation of GR variants. Minor alterations were observed for some GR variants (F65V, A229T, A304E, and N374S); nevertheless, a potential cooperative activation of transcription through enhanced N/C interaction between the NTD and ligand-binding domain of the GR has not been explored. Although there are no absolute criteria to ascertain overactivity of GR variants [13, 40], higher endogenous GR-target gene expression in appropriate cell models, as well as stronger sensitivity of DXM suppression tests in patients, may be clearly indicative of higher GR activity for a given genetic variant.
Nonetheless, apart from few examples of genetic modifications that can positively impact the human GR pathway [i.e., the polymorphisms N363S, localized in the NTD (rs6195), and BclI (rs41423247), localized in the first intron of the NR3C1 gene] [8], the unique gain-of-function D401H mutation of the NTD [13] and a double mutant of the 3′-UTR of exon 9β, coupled with a polymorphism (A3669G/G3134T and BclI) alleviating the dominant negative effect of GRβ over GRα [14], we were able to unambiguously exclude any deleterious impacts of the GR variation identified in the current study.
Of note, our characterization of GR variants led us to an unexpected result that improved our understanding of GR-mediated signaling. Surprisingly, although residue 374 was located in the NTD, it did not directly reside in the activation function 1 domain, known to constitute the interacting site with SRC2 [37]. It was proposed that this binding structurally modifies the NTD structure from intrinsic disorder to an α helical structure, leading to an activating state of the GR [37]. We hypothesized that the asparagine to serine substitution at position 374 would alter the SRC2 binding site on the GR, as it was previously demonstrated that the 201 to 360 aa deletion of hGR led to a loss of SRC2 binding to hGR [40]. To test whether the novel serine 374 residue might constitute a phosphorylation site impairing recruitment of the coactivator SRC2, the phospho-mimetic variant N374E hGR was shown to be insensitive to SRC2 coactivation. This is reminiscent of the creation of the serine 363 phosphorylation site of the N363S polymorphism (rs6195), which created an environment leading to increased coactivating capacity of the coactivator SRC2 [41, 42], as opposed to the mirror image of the serine 374, which potentially reduced SRC2 interaction (present study). Further studies are required to validate this hypothesis.
Another important feature of this study is the clinical, biological, and bioethical issue of incidentally discovered genetic GR variants. The American College of Medical Genetics and Genomics published recommendations for managing incidental findings [5] as a function of high likelihood of pathogenicity with the use of standard terminology: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign according to the classification [7]. Two categories of genomic variation were established: One is referred to as an actionable gene responsible for irreversible diseases potentially prevented by specific therapeutics upon patient informed consent; the second class belongs to incidental findings, subsequently termed secondary findings [43], in which established and appropriate or future interventions may prevent or reduce mortality or morbidity of genetic disorders. Guidance on reporting secondary findings [44] as well as the need for functional characterization of such genetic variations remains very controversial. In this context, management of NGS data and of the discovery of multiple variants of unknown significance remains quite difficult [7, 45], especially after a recent study revealed that one-fourth of healthy volunteers carried a monogenic disease risk with uncertain clinical usefulness, as revealed by genome sequence analysis [46].
It is often necessary to avoid false variants and exclude false-positive and false-negative results [1]. The pathogenic potential of new variants, notably missense mutations, requires the careful use of shared international databases [6], examination of interspecies nucleotide conservation, and determination of variation frequency, as well as familial segregation of the variants [47]. Predicting the functionality of the translated peptide from the genetic variant may be possible through in silico analysis according to the localization of the mutated nucleotide, the nature of amino acid substitution in a specific domain, and the predicted modification of the secondary protein structure. In vitro and/or ex vivo analysis may help characterize the genetic variants [48]. However, some genes or a part of genes and their translated proteins may not be structured, and their modification cannot be predicted because of an intrinsic disorder state [16], as is the case for the NTD of GR.
In the current study, all predictions from MutationTaster and Varsome were somehow incorrect, pointing to the need for functional characterization to assess the pathogenicity or benignity of new variants. Thus, in vitro and/or ex vivo analysis of the newly identified variants may exclusively help to clarify their characterization. Although some variants may change classification [7, 47, 49–52], many authors nowadays strongly recommend functional characterization of new variants.
In sum, NGS has considerably improved genetic tests and access to an important number of genes in a large number of patients. However, a large amount of new data is generated, and identification of secondary findings requires functional characterization to adjust modeling and predict pathogenicity. Thus, this latter classification often leads to new concepts and novel mechanistic issues of well-known signaling pathways, as exemplified here for GR-mediated transcriptional activation.
Acknowledgments
The authors thank Isabelle Boucly and Chrislaine Saujot (CHU Bicêtre) for their excellent technical assistance in GR mutation screening.
Financial Support: This work was supported by grants from Institut National de la Santé et de la Recherche Médicale (Inserm) and Paris-Sud University. L.F. was the recipient of a master degree fellowship from the French Endocrine Society and HRA Pharma laboratories. G.V. is the recipient of a doctoral fellowship from the Fondation pour la Recherche Médicale. The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this study.
Author Contributions: M.L. contributed to the study concept and design. L.F. and G.V. performed experiments and statistical analysis. J.B. performed the NGS experiments and discovered the GR variants. L.A. contributed to the imaging analyses. L.F., G.V., and M.L. contributed to the drafting of the paper. M.L. took responsibility for submitting the manuscript for publication. All authors analyzed the data and corrected and approved the final version of the manuscript.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- aa
amino acid
- Bmax
maximal transcriptional activity
- DXM
dexamethasone
- EC50
ligand concentration leading to 50% maximal transactivation capacity
- GC
glucocorticoids
- GR
glucocorticoid receptor
- hGR
human glucocorticoid receptor
- N/C
nuclear/cytoplasmic ratio
- NGS
next-generation sequencing
- NR3C1
nuclear receptor subfamily 3 group C member 1
- NTD
N-terminal domain
- SRC
steroid receptor coactivator
- WT
wild-type
References and Notes
- 1. van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34(9):666–681. [DOI] [PubMed] [Google Scholar]
- 2. Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, Waterston RH. DNA sequencing at 40: past, present and future. Nature. 2017;550(7676):345–353. [DOI] [PubMed] [Google Scholar]
- 3. Tan TY, Dillon OJ, Stark Z, Schofield D, Alam K, Shrestha R, Chong B, Phelan D, Brett GR, Creed E, Jarmolowicz A, Yap P, Walsh M, Downie L, Amor DJ, Savarirayan R, McGillivray G, Yeung A, Peters H, Robertson SJ, Robinson AJ, Macciocca I, Sadedin S, Bell K, Oshlack A, Georgeson P, Thorne N, Gaff C, White SM. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171(9):855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams DR, Eng CM. Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med. 2018;379(14):1353–1362. [DOI] [PubMed] [Google Scholar]
- 5. Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roy S, Coldren C, Karunamurthy A, Kip NS, Klee EW, Lincoln SE, Leon A, Pullambhatla M, Temple-Smolkin RL, Voelkerding KV, Wang C, Carter AB. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn. 2018;20(1):4–27. [DOI] [PubMed] [Google Scholar]
- 7. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vitellius G, Trabado S, Bouligand J, Delemer B, Lombès M. Pathophysiology of glucocorticoid signaling. Ann Endocrinol (Paris). 2018;79(3):98–106. [DOI] [PubMed] [Google Scholar]
- 9. Vitellius G, Trabado S, Hoeffel C, Bouligand J, Bennet A, Castinetti F, Decoudier B, Guiochon-Mantel A, Lombes M, Delemer B; investigators of the MUTA-GR Study. Significant prevalence of NR3C1 mutations in incidentally discovered bilateral adrenal hyperplasia: results of the French MUTA-GR Study. Eur J Endocrinol. 2018;178(4):411–423. [DOI] [PubMed] [Google Scholar]
- 10. Nicolaides NC, Charmandari E. Chrousos syndrome: from molecular pathogenesis to therapeutic management. Eur J Clin Invest. 2015;45(5):504–514. [DOI] [PubMed] [Google Scholar]
- 11. Molnár Á, Patócs A, Likó I, Nyírő G, Rácz K, Tóth M, Sármán B. An unexpected, mild phenotype of glucocorticoid resistance associated with glucocorticoid receptor gene mutation case report and review of the literature. BMC Med Genet. 2018;19(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Al Argan R, Saskin A, Yang JW, D’Agostino MD, Rivera J. Glucocorticoid resistance syndrome caused by a novel NR3C1 point mutation. Endocr J. 2018;65(11):1139–1146. [DOI] [PubMed] [Google Scholar]
- 13. Charmandari E, Ichijo T, Jubiz W, Baid S, Zachman K, Chrousos GP, Kino T. A novel point mutation in the amino terminal domain of the human glucocorticoid receptor (hGR) gene enhancing hGR-mediated gene expression. J Clin Endocrinol Metab. 2008;93(12):4963–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santen RJ, Jewell CM, Yue W, Heitjan DF, Raff H, Katen KS, Cidlowski JA. Glucocorticoid receptor mutations and hypersensitivity to endogenous and exogenous glucocorticoids. J Clin Endocrinol Metab. 2018;103(10):3630–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar R, Thompson EB. Folding of the glucocorticoid receptor N-terminal transactivation function: dynamics and regulation. Mol Cell Endocrinol. 2012;348(2):450–456. [DOI] [PubMed] [Google Scholar]
- 16. Motlagh HN, Anderson JA, Li J, Hilser VJ. Disordered allostery: lessons from glucocorticoid receptor. Biophys Rev. 2015;7(2):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D-H, Wright A, Han K-H. An NMR study on the intrinsically disordered core transactivation domain of human glucocorticoid receptor. BMB Rep. 2017;50(10):522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salamanova E, Costeira-Paulo J, Han K-H, Kim D-H, Nilsson L, Wright APH. A subset of functional adaptation mutations alter propensity for α-helical conformation in the intrinsically disordered glucocorticoid receptor tau1core activation domain. Biochim Biophys Acta, Gen Subj. 2018;1862(6):1452–1461. [DOI] [PubMed] [Google Scholar]
- 19. Dahlman-Wright K, Almlöf T, McEwan IJ, Gustafsson JA, Wright AP. Delineation of a small region within the major transactivation domain of the human glucocorticoid receptor that mediates transactivation of gene expression. Proc Natl Acad Sci USA. 1994;91(5):1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391(3):449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giguère V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of the human glucocorticoid receptor. Cell. 1986;46(5):645–652. [DOI] [PubMed] [Google Scholar]
- 22. Dahlman-Wright K, Baumann H, McEwan IJ, Almlöf T, Wright AP, Gustafsson JA, Härd T. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1995;92(5):1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wallberg AE, Neely KE, Hassan AH, Gustafsson JA, Workman JL, Wright AP. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol Cell Biol. 2000;20(6):2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bohen SP, Yamamoto KR. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90(23):11424–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kakar M, Davis JR, Kern SE, Lim CS. Optimizing the protein switch: altering nuclear import and export signals, and ligand binding domain. J Control Release. 2007;120(3):220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bouali N, Francou B, Bouligand J, Imanci D, Dimassi S, Tosca L, Zaouali M, Mougou S, Young J, Saad A, Guiochon-Mantel A. New MCM8 mutation associated with premature ovarian insufficiency and chromosomal instability in a highly consanguineous Tunisian family. Fertil Steril. 2017;108(4):694–702. [DOI] [PubMed] [Google Scholar]
- 27. Vitellius G, Fagart J, Delemer B, Amazit L, Ramos N, Bouligand J, Le Billan F, Castinetti F, Guiochon-Mantel A, Trabado S, Lombès M. Three novel heterozygous point mutations of NR3C1 causing glucocorticoid resistance. Hum Mutat. 2016;37(8):794–803. [DOI] [PubMed] [Google Scholar]
- 28. Bouligand J, Delemer B, Hecart A-C, Meduri G, Viengchareun S, Amazit L, Trabado S, Fève B, Guiochon-Mantel A, Young J, Lombès M. Familial glucocorticoid receptor haploinsufficiency by non-sense mediated mRNA decay, adrenal hyperplasia and apparent mineralocorticoid excess. PLoS ONE. 2010;5(10):e13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RRID:AB_2687823, https://scicrunch.org/resolver/AB_2687823.
- 30.RRID:AB_476693, https://scicrunch.org/resolver/AB_476693.
- 31.RRID:AB_614946, https://scicrunch.org/resolver/AB_614946.
- 32.RRID:AB_2556797, https://scicrunch.org/resolver/AB_2556797.
- 33. Amazit L, Le Billan F, Kolkhof P, Lamribet K, Viengchareun S, Fay MR, Khan JA, Hillisch A, Lombès M, Rafestin-Oblin ME, Fagart J. Finerenone impedes aldosterone-dependent nuclear import of the mineralocorticoid receptor and prevents genomic recruitment of steroid receptor coactivator-1. J Biol Chem. 2015;290(36):21876–21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.RRID:AB_2633276, https://scicrunch.org/resolver/AB_2633276.
- 35. Williams RG, Kandasamy R, Nickischer D, Trask OJ Jr, Laethem C, Johnston PA, Johnston PA. Generation and characterization of a stable MK2-EGFP cell line and subsequent development of a high-content imaging assay on the Cellomics ArrayScan platform to screen for p38 mitogen-activated protein kinase inhibitors. Methods Enzymol. 2006;414:364–389. [DOI] [PubMed] [Google Scholar]
- 36. Raivio T, Palvimo JJ, Kannisto S, Voutilainen R, Jänne OA. Transactivation assay for determination of glucocorticoid bioactivity in human serum. J Clin Endocrinol Metab. 2002;87(8):3740–3744. [DOI] [PubMed] [Google Scholar]
- 37. Khan SH, Awasthi S, Guo C, Goswami D, Ling J, Griffin PR, Simons SS Jr, Kumar R. Binding of the N-terminal region of coactivator TIF2 to the intrinsically disordered AF1 domain of the glucocorticoid receptor is accompanied by conformational reorganizations. J Biol Chem. 2012;287(53):44546–44560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carruthers CW, Suh JH, Gustafsson J-A, Webb P. Phosphorylation of glucocorticoid receptor tau1c transactivation domain enhances binding to CREB binding protein (CBP) TAZ2. Biochem Biophys Res Commun. 2015;457(1):119–123. [DOI] [PubMed] [Google Scholar]
- 39. Khan SH, McLaughlin WA, Kumar R. Site-specific phosphorylation regulates the structure and function of an intrinsically disordered domain of the glucocorticoid receptor. Sci Rep. 2017;7(1):15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang D, Wang Q, Awasthi S, Simons SS Jr. Amino-terminal domain of TIF2 is involved in competing for corepressor binding to glucocorticoid and progesterone receptors. Biochemistry. 2007;46(27):8036–8049. [DOI] [PubMed] [Google Scholar]
- 41. Russcher H, Smit P, van den Akker ELT, van Rossum EF, Brinkmann AO, de Jong FH, Lamberts SW, Koper JW. Two polymorphisms in the glucocorticoid receptor gene directly affect glucocorticoid-regulated gene expression. J Clin Endocrinol Metab. 2005;90(10):5804–5810. [DOI] [PubMed] [Google Scholar]
- 42. Feng J, Zheng J, Bennett WP, Heston LL, Jones IR, Craddock N, Sommer SS. Five missense variants in the amino-terminal domain of the glucocorticoid receptor: no association with puerperal psychosis or schizophrenia. Am J Med Genet. 2000;96(3):412–417. [DOI] [PubMed] [Google Scholar]
- 43. Scheuner MT, Peredo J, Benkendorf J, Bowdish B, Feldman G, Fleisher L, Mulvihill JJ, Watson M, Herman GE, Evans J. Reporting genomic secondary findings: ACMG members weigh in. Genet Med. 2015;17(1):27–35. [DOI] [PubMed] [Google Scholar]
- 44. Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR, McKelvey KD, Ormond KE, Richards CS, Vlangos CN, Watson M, Martin CL, Miller DT. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics [published correction appears in Genet Med. 2017;19(4):484] Genet Med. 2017;19(2):249–255. [DOI] [PubMed] [Google Scholar]
- 45. Kuhlenbäumer G, Hullmann J, Appenzeller S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat. 2011;32(2):144–151. [DOI] [PubMed] [Google Scholar]
- 46. Vassy JL, Christensen KD, Schonman EF, Blout CL, Robinson JO, Krier JB, Diamond PM, Lebo M, Machini K, Azzariti DR, Dukhovny D, Bates DW, MacRae CA, Murray MF, Rehm HL, McGuire AL, Green RC; MedSeq Project. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med. 2017;167(3):159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karbassi I, Maston GA, Love A, DiVincenzo C, Braastad CD, Elzinga CD, Bright AR, Previte D, Zhang K, Rowland CM, McCarthy M, Lapierre JL, Dubois F, Medeiros KA, Batish SD, Jones J, Liaquat K, Hoffman CA, Jaremko M, Wang Z, Sun W, Buller-Burckle A, Strom CM, Keiles SB, Higgins JJ. A standardized DNA variant scoring system for pathogenicity assessments in Mendelian disorders. Hum Mutat. 2016;37(1):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guillermin Y, Lopez J, Chabane K, Hayette S, Bardel C, Salles G, Sujobert P, Huet S. What does this mutation mean? The tools and pitfalls of variant iInterpretation in lymphoid malignancies. Int J Mol Sci. 2018;19(4):1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3(65):65ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tabor HK, Auer PL, Jamal SM, Chong JX, Yu JH, Gordon AS, Graubert TA, O’Donnell CJ, Rich SS, Nickerson DA, Bamshad MJ; NHLBI Exome Sequencing Project. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet. 2014;95(2):183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue W-Q, He Y-Q, Zhu J-H, Ma J-Q, He J, Jia W-H. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci Rep. 2014;4(1):6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shearer AE, Kolbe DL, Azaiez H, Sloan CM, Frees KL, Weaver AE, Clark ET, Nishimura CJ, Black-Ziegelbein EA, Smith RJ. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6(5):37. [DOI] [PMC free article] [PubMed] [Google Scholar]