Abstract
Integration of environmental and endogenous cues at plant shoot meristems determines the timing of flowering and reproductive development. The MADS box transcription factor FLOWERING LOCUS C (FLC) of Arabidopsis thaliana is an important repressor of floral transition, which blocks flowering until plants are exposed to winter cold. However, the target genes of FLC have not been thoroughly described, and our understanding of the mechanisms by which FLC represses transcription of these targets and how this repression is overcome during floral transition is still fragmentary. Here, we identify and characterize TARGET OF FLC AND SVP1 (TFS1), a novel target gene of FLC and its interacting protein SHORT VEGETATIVE PHASE (SVP). TFS1 encodes a B3-type transcription factor, and we show that tfs1 mutants are later flowering than wild-type, particularly under short days. FLC and SVP repress TFS1 transcription leading to deposition of trimethylation of Iysine 27 of histone 3 (H3K27me3) by the Polycomb Repressive Complex 2 at the TFS1 locus. During floral transition, after downregulation of FLC by cold, TFS1 transcription is promoted by SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), a MADS box protein encoded by another target of FLC/SVP. SOC1 opposes PRC function at TFS1 through recruitment of the histone demethylase RELATIVE OF EARLY FLOWERING 6 (REF6) and the SWI/SNF chromatin remodeler ATPase BRAHMA (BRM). This recruitment of BRM is also strictly required for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) binding at TFS1 to coordinate RNAPII recruitment through the Mediator complex. Thus, we show that antagonistic chromatin modifications mediated by different MADS box transcription factor complexes play a crucial role in defining the temporal and spatial patterns of transcription of genes within a network of interactions downstream of FLC/SVP during floral transition.
Author summary
The initiation of flowering in plants is exquisitely sensitive to environmental signals, ensuring that reproduction occurs at the appropriate time of year. The sensitivity of these responses depends upon strong repression of flowering under inappropriate conditions. FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) are related transcription factors that act in concert to strongly inhibit flowering in crucifer plants through repressing transcription of their target genes. Many direct FLC/ SVP targets have been identified in genome-wide studies, however few of these genes have been characterized for their roles in regulating flowering time or other aspects of reproductive development. Here, we characterize TARGET OF FLC AND SVP1 (TFS1) as a novel target of FLC and SVP, and demonstrate that TFS1 contributes to proper flowering-time control. Moreover, we provide a detailed mechanistic view of how TFS1 transcription is controlled during reproductive development through the repressive activity of FLC/SVP being overcome by the transcriptional activator SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1. Thus we further elucidate the network of genes repressed by FLC/SVP to block flowering and determine mechanisms by which their repressive activity is overcome during the initiation of flowering.
Introduction
The transition from vegetative to reproductive development in plants is controlled by a complex transcriptional network that responds both to environmental cues and endogenous hormonal signals [1, 2]. In Arabidopsis thaliana, the MADS-box transcription factor FLOWERING LOCUS C (FLC) plays a major role in this network as an inhibitor of floral transition [3, 4]. Transcription of FLC is repressed by extended exposure to cold that mimics winter conditions (vernalization) so that flowering can proceed when plants are subsequently exposed to warm in spring. The repression of FLC transcription by accumulation of histone modifications in response to cold has been extensively studied [5] and FLC target genes have been described by whole genome chromatin immunoprecipitation (ChIPseq) [6–8]. Nevertheless, our understanding of how FLC influences the transcriptional network that controls floral transition and how it represses expression of its target genes is still fragmentary. Here, we utilize data derived from genome-wide binding studies of FLC and its partner MADS box transcription factor SHORT VEGETATIVE PHASE (SVP) [7, 9–12] to identify a common target gene that we named TARGET of FLC and SVP 1 (TFS1). We show that this gene acts in the network downstream of FLC and other floral regulators, and has an important role on the flanks of the meristem during the early stages of floral development.
FLC binds to several hundred target genes, but only a small subset of these are conserved between species [8]. Genes involved in flowering control are enriched among the conserved targets, and FLC represses transcription of several of these, including FLOWERING LOCUS T (FT), SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15 (SPL15), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and SEPALLATA 3 (SEP3). MADS box transcription factors are proposed to bind DNA as dimers and tetramers [13, 14], and FLC interacts with and binds to a subset of its targets in complexes with the related protein SHORT VEGETATIVE PHASE (SVP) [7, 10]. Mutants at SVP are early flowering and exhibit increased levels of SOC1 and FT mRNAs [9, 10, 15, 16]. How FLC represses transcription of its targets is not completely clear, but appears to involve modification of histones. FLC and its homologue FLOWERING LOCUS M (FLM) recruit EMBRYONIC FLOWER 1 (EMF1), a plant-specific Polycomb Repressive Complex 1 (PRC1) component [17, 18], to FLOWERING LOCUS T (FT) in leaf veins to repress its transcription [19]. Moreover, cooperativity between FLC or FLM and PRC1 contributes to maintenance of PRC2-induced trimethylation of lysine-27 at histone H3 (H3K27me3) at FT chromatin, most probably through the activity of the histone methyltransferase CURLY LEAF (CLF) and the H3K27me3-associated protein LIKE HETEROCHROMATIN PROTEIN1 (LHP1) [19]. Additionally, the JmjC domain-containing trimethyl histone H3 lysine-4 demethylase JUMONJI14/PKDM7B (JMJ14/PKDM7B) also associates with PRC1 to further antagonize the active chromatin state at FT [20, 21].
Other targets of FLC and SVP that are involved in floral induction encode the MADS-box transcription factor SOC1 and the plant-specific transcription factor SPL15 [22–27]. Both SOC1 and SPL15 are expressed in the shoot apical meristem where they cooperate at the promoters of target genes such as FRUITFULL (FUL) and MIR172b to activate the basal floral promotion pathway under non-inductive environmental conditions [12, 22, 28]. Interestingly, SOC1 coordinates the recruitment of the histone demethylase RELATIVE OF EARLY FLOWERING 6 (REF6) to the promoter of FUL and MIR172b to orchestrate the removal of the H3K27me3 mark and activate transcription [22]. Furthermore, SPL15 activity is repressed post-transcriptionally by miR156 and post-translationally through its physical interaction with the gibberellin (GA)-labile DELLA protein REPRESSOR OF GA1-3 (RGA) [22]. By contrast, SPL9, a paralogue of SPL15 that is expressed after floral induction at the periphery of the SAM, requires interaction with DELLA proteins to potentiate its trans-activation activity and contribute to the induction of expression of the floral meristem identity gene APETALA1 (AP1) in the low gibberellin context present in the cells that give rise to the floral primordium [29].
Here, we show that TFS1, which encodes a B3-type transcription factor that is a member of the REPRODUCTIVE MERISTEM (REM) family [30, 31], constitutes a target of FLC and SVP and that its transcription is repressed through cooperation with PRC complexes during vegetative growth. After floral induction, TFS1 expression is induced at the periphery of the SAM by SOC1 and the age-regulated transcription factor SPL9 through coordinated recruitment of the histone demethylase REF6 and the chromatin remodeler BRAHMA (BRM) [32–34]. This analysis deepens our understanding of the mechanism by which FLC represses the floral network and indicates the importance of antagonistic histone modifications mediated by different MADS box complexes on common target genes.
Results
TFS1 is a target of the floral repressors FLC and SVP
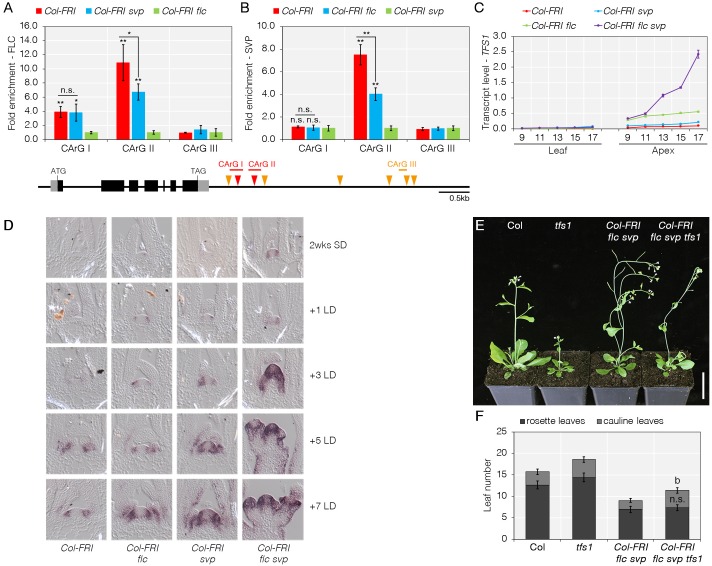
To further define the regulatory network controlling floral transition at the shoot apex, recently published ChIP-Seq and tissue-specific RNA-Seq datasets were examined to identify genes that are expressed specifically at the shoot apex and are bound by the floral repressor transcription factors FLC and SVP [6–8]. Cross-referencing these datasets identified the gene encoding the B3-type transcription factor TARGET OF FLC AND SVP1 (TFS1), which was formerly annotated as REPRODUCTIVE MERISTEM 17 (REM17) [30, 31]. A phylogenetic analysis revealed that TFS1 is part of a gene family including REDUCED VERNALIZATION RESPONSE1 (VRN1), VERDANDI (VDD) and VALKYRIE (VAL), and that REM18 and REM19 are the closest homologs of TFS1 within a sub-branch of the phylogenetic tree (S1A Fig). To verify the binding of FLC and SVP at TFS1, chromatin-immunoprecipitation (ChIP) analysis was performed using chromatin extracted from the aerial parts of 15-day old plants grown under inductive long days (LDs) and using antibodies that were raised against FLC [8] and SVP (S1B Fig, S3 and S4 Tables, Methods). In agreement with the ChIP-seq data, specific enrichment of a fragment that encompasses the putative CArG-boxes, designated as CArG-box II, located at the 3’ end of TFS1 was detected after ChIP of FLC or SVP (Fig 1A and 1B). Moreover, the ChIP-qPCR analyses also demonstrated a mutual co-operation between these floral repressors at TFS1, because binding of FLC and SVP was enhanced in the presence of the other protein (Fig 1A and 1B), as reported previously for several other targets of these transcription factors [7, 10].
Fig 1. TFS1 is directly regulated by FLC and SVP during floral induction.
A and B) Analysis of FLC (A) and SVP (B) binding to TFS1 by ChIP-qPCR. Grey and black boxes depict UTR and exons, respectively. Orange and red triangles indicate CArG-boxes with CCW(6)GG and CCW(6)RG consensus, respectively. Plant material used and replicates performed are described in Methods. Values were scaled to set value of CArG-box I in Col-FRI flc svp to 1. Statistical significance was calculated using Student’s t-test against Col-FRI flc (A) and Col-FRI svp (B). Also for CArG II the significance of the difference in enrichment between Col-FRI and Col-FRI svp was tested. *P < 0.05, **P < 0.01, n.s. P > 0.05. C) Increased expression of TFS1 in apices of Col-FRI flc svp plants by RT-qPCR. Numbers on X-axis indicate number of long-days (LD) for which plants were grown prior to RNA extraction. D) Spatial pattern of expression of TFS1 assessed by in situ hybridization during floral transition. E and F) Representative photo of plants of the illustrated genotypes 28 days after germination under LDs (E) and their respective flowering time (F). Statistical significance was calculated using Student’s t-test; P < 0.01.
To further characterize the regulation of TFS1 by FLC and SVP, the abundance of TFS1 mRNA in leaves and apices was tested by RT-qPCR in Col-FRI flc-3, Col-FRI svp-41 and Col-FRI flc-3 svp-41 mutants as well as Col-FRI wild-type. TFS1 mRNA was exclusively detected at the shoot apex and its abundance was increased throughout a developmental time course in the single mutants compared to wild-type and was strongly increased in the double mutant (Fig 1C, S1C Fig). Furthermore, TFS1 expression was increased at the shoot apex, but not in leaves, after Col-FRI plants were exposed to vernalisation and returned to normal growth temperature (S1D Fig), consistent with the repression of TFS1 transcription by FLC. To determine whether the spatial pattern of expression of TFS1 differs in Col-FRI flc-3 and Col-FRI svp-41 mutants compared to Col-FRI, in-situ hybridisation analysis of the shoot apex was performed during floral transition after transfer of plants from short days (SDs) to inductive LDs (Fig1D, S1E Fig). The Col-FRI and Col-FRI flc-3 plants grown for 2 wks under SD were still vegetative (Fig 1D), but TFS1 expression level was clearly elevated in Col-FRI flc-3 (S1C Fig), indicating that FLC represses TFS1 expression even in the vegetative stage. After transfer to LDs, TFS1 mRNA appeared at the periphery of the SAM in the Col-FRI wild-type. This spatial pattern was not changed in the Col-FRI svp-41, Col-FRI flc-3 or Col-FRI flc-3 svp-41 mutants, but the mRNA appeared more rapidly after transfer to LDs in the mutants than in Col-FRI wild-type (Fig 1D, S1C Fig). Therefore, TFS1 transcription is induced during floral transition, while the timing and amplitude of its expression are modulated by SVP and FLC.
The temporal expression pattern of TFS1 coincided with the transition to flowering, so the flowering time of tfs1 mutants was determined under inductive and non-inductive conditions. Interestingly, tfs1-1 mutants flowered significantly later than wild-type plants under both conditions, suggesting that TFS1 is involved in promoting floral transition (Fig 1E and 1F, S2A–S2F Fig). The role of TFS1 was confirmed by transgenic complementation of tfs1-1 (see later), and by showing that a second allele (tfs1-2) caused a similar late-flowering phenotype under LDs (S2E and S2F Fig). The tfs1-1 mutation also delayed flowering in the Col-FRI flc-3 svp-41 background, supporting the idea that TFS1 acts downstream of FLC and SVP to promote flowering (Fig 1E and 1F). In addition, the Col-FRI flc-3 svp-41 tfs1-1 plants showed impaired flower development, suggesting redundancy between these genes in flower and inflorescence development (S2G and S2H Fig). Overall, these results demonstrate that TFS1 is a direct target of FLC and SVP and is specifically expressed at the periphery of the SAM during floral transition to promote flowering and floral development.
Histone modification of TFS1 through the activity of POLYCOMB REPRESSIVE COMPLEX 2 is dependent on FLC/SVP
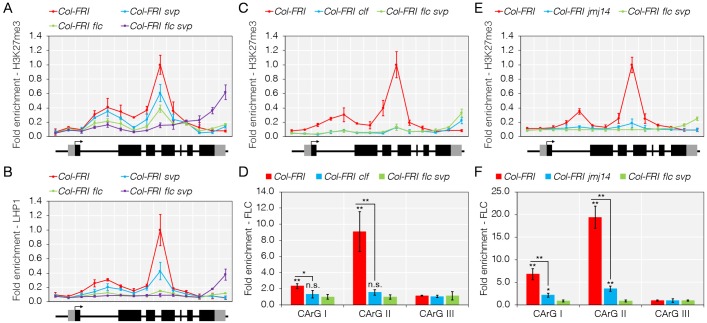
Transcriptional repression by FLC and SVP has been linked to activity of Polycomb Repressive Complex (PRC) 2 (PRC2) [10, 19]. PRC2 catalyzes the methylation of histone 3 (H3) at lysine 27 (H3K27me3) and is associated with the repression of transcription [35]. Therefore, whether TFS1 is subjected to PRC-mediated regulation in a FLC and SVP dependent manner was tested by performing ChIP-qPCR using chromatin extracted from 15-day old plants grown under LDs and antibodies directed against H3K27me3 or H3K4me3 (S3 Table). H3K27me3 was detected in the gene body of TFS1 in Col-FRI plants at much higher levels than in Col-FRI flc-3, Col-FRI svp-41 and Col-FRI flc-3 svp-41 mutants (Fig 2A). However, in Col-FRI flc-3 svp-41 plants an additional peak in H3K27me3 levels was detected close to the transcriptional termination site of TFS1 (Fig 2A, S3A Fig). Furthermore, a commercially available antibody (S3 Table) was used for ChIP-qPCR of LHP1, which is frequently found associated with H3K27me3 marked chromatin [36, 37], and the protein was detected at TFS1 in a similar pattern to H3K27me3 (Fig 2B). Therefore, H3K27me3 and LHP1 are present in the gene body of TFS1 in a FLC and SVP dependent manner correlating with reduced transcription of TFS1.
Fig 2. Binding of FLC and SVP to TFS1 is associated with accumulation of H3K27me3.
A) Accumulation of H3K27me3 at TFS1 is reduced in Col-FRI flc svp plants. B) LHP1 is associated with a region of TFS1 that heavily accumulates H3K27me3. C) Accumulation of H3K27me3 at TFS1 is reduced in Col-FRI clf mutants. D) Binding of FLC to TFS1 is reduced in Col-FRI clf plants. E) Accumulation of H3K27me3 at TFS1 is reduced in Col-FRI jmj14 mutants. F) Binding of FLC to TFS1 is reduced in Col-FRI jmj14 plants. A to C and E, values were scaled to set value of first primers in Col-FRI to 1. D and F, values were scaled to set value of CArG-box I in Col-FRI flc svp to 1. Statistical significance was calculated against Col-FRI flc svp using Student’s t-test as described in Fig 1A and 1B; *P < 0.05, **P < 0.01, n.s. P > 0.05.
In contrast to H3K27me3, K4-trimethylated H3 (H3K4me3) is present at genes that are actively transcribed [38]. Enrichment of H3K4me3 was detected close to the transcriptional start site (TSS) of TFS1 in the Col-FRI flc-3, Col-FRI svp-41 and Col-FRI flc-3 svp-41 mutants, whereas it was not present at the gene in Col-FRI plants (S3B Fig). These data are in agreement with previous reports of the dynamic and antagonistic relationship between H3K27me3 and H3K4me3 during development and the floral transition [35, 39]. Therefore, under these conditions the dynamic change in these chromatin marks at TFS1 correlates with the repression of FLC transcription, the induction of TFS1 and the transition to flowering.
The PRC2 mutation curly leaf (clf), which impairs the activity of an enzyme that catalyses H3K27me3 deposition [40], partially suppresses the late-flowering phenotype of Col-FRI plants [41]. Strikingly, early flowering Col-FRI clf-2 mutant plants expressed highly elevated levels of TFS1 mRNA in the apex, while still expressing high levels of FLC and SVP mRNA (S3D–S3F Fig) [41, 42]. These observations supported the idea that PRC2 might contribute to transcriptional repression of FLC target genes such as TFS1. To examine this possibility, ChIP analyses were performed to test for H3K27me3 enrichment across the coding region of TFS1 in Col-FRI clf-2 and Col-FRI flc-3 svp-41 mutant plants. In agreement with the functional role of CLF in the deposition of H3K27me3, a strong reduction in this mark at TFS1 was detected in Col-FRI clf-2 and this was similar to the reduction observed in Col-FRI flc-3 svp-41 mutant plants when compared to wild-type Col-FRI (Fig 2C). In contrast, ChIP analyses for H3K4me3 at TFS1 showed higher enrichment patterns in Col-FRI clf-2 as well as in Col-FRI flc-3, Col-FRI svp-41 and Col-FRI flc-3 svp-41 mutants than in wild-type Col-FRI (S3B and S3C Fig). These data indicate that reduction in H3K27me3 at TFS1 in Col-FRI clf-2 mutants correlates with an increase in H3K4me3, consistent with the antagonistic role of these marks during floral transition. Finally, to determine whether the reduction in H3K27me3 levels at TFS1 in Col-FRI clf-2 plants correlated with reduced FLC binding, ChIP analyses were performed for FLC. FLC binding was strongly compromised in Col-FRI clf-2, indicating that FLC binding requires and is sustained by PRC2 function (Fig 2D).
SVP and FLC interact respectively with LHP1 and the Polycomb Repressive Complex 1 protein EMBRYONIC FLOWER (EMF1) [18, 19, 43, 44], suggesting a link between PRC function and the activity of these floral repressors. Furthermore, a complex including LHP1, EMF1 and the H3K4me3 demethylase JMJ14/PKDM7B, has been described to play roles related to PRC1, including delaying flowering in non-inductive photoperiods [19–21]. These observations, together with the result described above that H3K4me3 levels are lower at TFS1 in the presence of SVP and FLC (S3B Fig), suggested that the H3K4me3 demethylase activity of JMJ14 might be required for FLC and PcG mediated repression of TFS1. To test this idea, ChIP-qPCR analyses of H3K27me3 were performed on TFS1 in Col-FRI jmj14-2 mutants. In these plants the enrichment levels of the repressive mark H3K27me3 were strongly reduced compared to Col-FRI wild-type (Fig 2E). By contrast, the active chromatin mark H3K4me3 was increased at TFS1 in Col-FRI jmj14-2 compared to Col-FRI and was present at a similar level as in Col-FRI flc-3 svp-41 mutants (S3C Fig). Consistent with the increased levels of H3K4me3, Col-FRI jmj14-2 also showed higher mRNA levels of TFS1 in apices, but did not affect the expression of SVP or FLC (S3G–S3I Fig). In support of the notion that FLC binding to TFS1 requires PRC2 activity and higher levels of H3K27me3 (Fig 2D), reduced binding of FLC to CArG-box II at TFS1 was also detected in Col-FRI jmj14-2 plants, although FLC mRNA level was unaffected (Fig 2F, S3H Fig). Collectively, these data suggest that for FLC mediated transcriptional repression of TFS1, the recruitment of PRC2 and deposition of H3K27me3 are required, and that these can also be inhibited by increasing the levels of H3K4me3 through mutation of the JMJ14 demethylase.
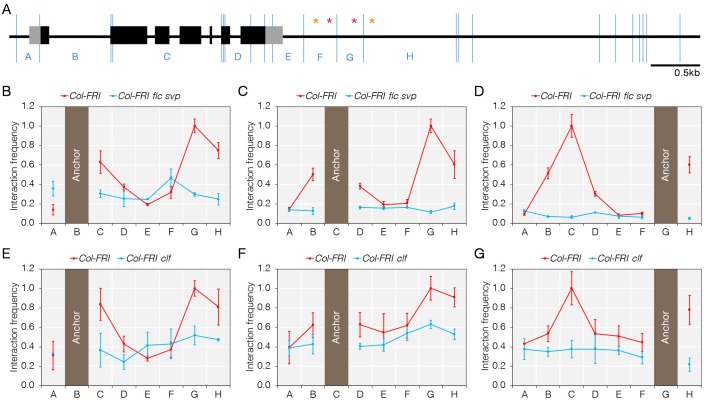
FLC and SVP mediated repression of TFS1 is coupled to a gene loop
The observations that FLC and SVP bind 3’ of the TFS1 stop codon (Figs 1A, 1B, 2D and 2F) and that they interact with a PRC complex and LHP1 [19, 43] that are associated with the gene body of TFS1 (Fig 2B), suggested that a chromosomal loop might form between the 3’ distal region and the gene body. To test for the presence of such a loop, a chromosome conformation capture (3C) assay was performed. Indeed, in Col-FRI plants fragments B, C and D (middle region) were found to interact with the 3’ end (G and H) of TFS1, suggesting that a ‘locked’ DNA loop was formed (Fig 3). The presence of this loop was then tested in different mutants to determine whether it required FLC/SVP and PcG function. In Col-FRI flc-3 svp-41 mutants, the interaction between the middle region and the 3’ end was strongly reduced compared to Col-FRI, indicating that FLC and SVP are required for the formation of the ‘locked’ DNA loop (Fig 3A–3D). To define the contribution of PRC2 in the formation of this loop at TFS1, Col-FRI clf-2 plants were examined. In this genotype, the loop appeared significantly weaker than in Col-FRI plants, indicating that CLF activity is required to support the formation of the ‘locked’ DNA loop at TFS1 (Fig 3E–3G). These data indicate therefore that transcriptional repression of TFS1 by FLC and SVP is associated with the formation of a chromatin loop that requires FLC and SVP binding at the 3’ end of the gene and high levels of H3K27me3 within the gene body.
Fig 3. FLC and SVP are associated with looping between the 3’ end of TFS1 and gene body.
A) High resolution digestion map of the TFS1 locus by the enzyme Sau3AI. Primers were designed on 8 restriction fragments as indicated (A to H). Restriction sites are highlighted with blue vertical lines. CArG-boxes are presented as red and orange asterisks. Exons are indicated as black boxes and 5’ and 3’ UTR are indicated as grey boxes. B to D) DNA looping between coding region and 3’ end of TFS1 is reduced in Col-FRI flc svp plants, but not in Col-FRI. E to G) DNA looping between coding region and 3’ end of TFS1 is reduced in Col-FRI clf plants, but not in Col-FRI. Plants were collected for 3C experiments after 14 LD at ZT4. Values were scaled to set the highest interaction value per fixed primer to 1. The position of the fixed primers is indicated by a filled column designated as ‘Anchor’.
Activation of TFS1 transcription by SOC1
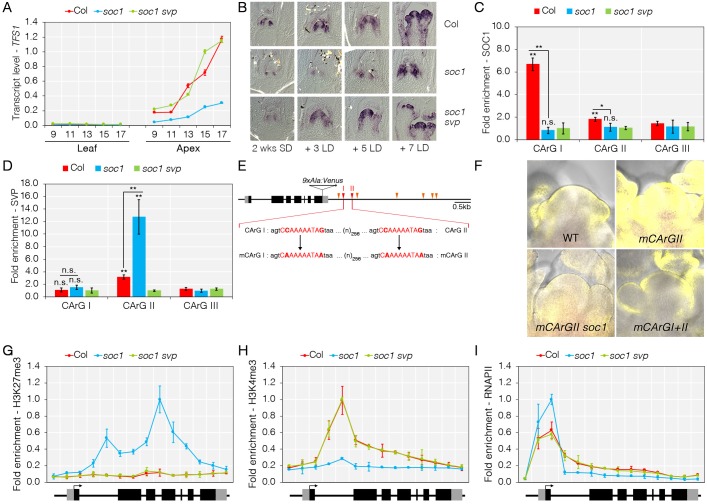
In a genome-wide study of binding sites of the MADS-box protein SOC1, a site at the 3’ end of TFS1 was detected [28]. To determine whether TFS1 is regulated by SOC1, TFS1 transcript abundance was tested by RT-qPCR using RNA extracted from leaves and apices in soc1-2, soc1-2 svp-41 and Col genotypes. TFS1 mRNA levels were much lower in apices of soc1-2 mutants than Col, but were largely restored to Col levels in soc1-2 svp-41 double mutants (Fig 4A). To determine the spatial pattern of TFS1 expression in soc1-2 and soc1-2 svp-41, in-situ hybridisations were performed on apices during floral transition. The overall spatial expression pattern was similar in Col and soc1-2 svp-41, however, a significant delay in TFS1 expression at the periphery of the SAM was observed in soc1-2 (Fig 4B).
Fig 4. TFS1 is activated during floral transition by direct binding of SOC1.
A) TFS1 transcript is reduced in apices of soc1 mutants during floral transition. Numbers on X-axis indicate number of long-days (LD) for which plants were grown prior to RNA harvest. B) Spatial pattern of expression of TFS1 in soc1 and soc1 svp mutants. C) SOC1 binds to the 3’ end of TFS1 as detected by ChIP-qPCR. Statistical significance was calculated against soc1 svp. D) SVP binding to the 3’ end of TFS1 is reduced in the presence of SOC1. Statistical significance was calculated against soc1 svp. E) Model of TFS1 genomic region and the mutations introduced into CArG-boxes. CArG-box with consensus sequence CW6RG (red triangle) and CW6GG (orange triangle) are shown. F) SOC1 activates TFS1 through CArG box I at the 3’ end of TFS1. Images were taken from 17 LD-grown seedlings. G) H3K27me3 levels at TFS1 are increased in soc1 mutants compared to Col. H) H3K4me3 levels are reduced at TFS1 in soc1 mutants compared to Col. I) RNA PolII stalls at the 5’ end of TFS1 in soc1 mutants. G to I, values were scaled to set value of the first primers in Col to 1. Statistical significance was calculated using Student’s t-test; *P < 0.05, **P < 0.01, n.s. P > 0.05.
ChIP analyses were then performed with antibodies that were directed against endogenous SOC1 and SVP (S1B Fig, S3 Table, [22]). To verify the results of the previous report on the genome-wide study for SOC1, ChIP-qPCR was performed and detected SOC1 binding in the region of CArG-box I (CArGI), which is located at the 3’ end of TFS1 (Figs 1A and 4C). The ChIP-qPCR experiment with SVP generated a specific enrichment in the region of CArG-box II (CArGII) in Col, and this was enhanced in the soc1-2 mutant, suggesting that SOC1 reduces SVP recruitment to TFS1 (Fig 4D). Similarly, Dexamethasone (DEX)-induced translocation of SOC1:GR into the nucleus in 35S::SOC1:GR plants caused higher TFS1 transcription and increased binding of SOC1 to CArGI as well as reduced binding of SVP to CArGII (S4A–S4C Fig). Therefore, binding of SOC1 to the 3’ end of TFS1 occurs during floral transition, whereas SVP binds during vegetative development, and is in agreement with their observed roles in the transcriptional regulation of TFS1.
To test in vivo whether the CArG-boxes identified within the ChIP-qPCR amplicons are responsible for the regulation of TFS1 by SOC1 and SVP, a TFS1::TFS1:9xAla-Venus (TFS1::TFS1:9AV) gene fusion was constructed that contained the entire intergenic region flanking TFS1 on the 5’ and 3’ sides (Fig 4E). This gene fusion complemented the tfs1-1 mutant phenotype (S5A and S5B Fig) and in the transgenic plants VENUS signal was detected at the periphery of the SAM in a similar pattern as observed by in situ hybridization of TFS1 mRNA (Figs 1D and 4F, S4E Fig). Also, the confocal imaging of the TFS1:9xAla-Venus fusion protein indicated that it predominately localized in the cytosol of slowly dividing meristematic cells (S6A and S6B Fig) while treatment with leptomycin B (LMB), which impairs the activity of nuclear exportin [45], suggested that this was due to active export from the nucleus (S6C Fig). However, in actively dividing cells in young sepals the VENUS signal appeared to be nuclear (S6D to S6G Fig), suggesting the nuclear accumulation and presumably the activity of TFS1 may be closely related to cell division.
The CArG-boxes identified in the ChIP amplicons were then mutated in this gene fusion construct. Two mutant plasmids were generated in which CArGII or both CArGI and CArGII were mutated (Fig 4E). Several independent transformants carrying each construct were analysed (Fig 4F, S4E–S4H Fig). Transformants harbouring the mCArGII construct displayed a stronger and broader VENUS signal than those carrying the wild-type construct, whereas the mCArGI+II construct conferred a VENUS signal that was similar to the wild-type construct (Fig 4F, S4E–S4G Fig). The relative strength of these mutant constructs was supported by RT-qPCR analysis performed on RNA extracted from apices of the transgenic plants (S4H Fig). Furthermore, the strong signal of the mCArGII construct was greatly reduced when it was introduced into the soc1-2 mutant by crossing (Fig 4F). The low level of TFS1:9AV expression detected in the soc1 TFS1::TFS1:9AV mCArGII and TFS1::TFS1:9AV mCArGI+II plants was consistent with their delayed flowering time compared to TFS1::TFS1:9AV plants (S5A–S5H Fig). Taken together, these observations were consistent with the transcriptional profile of TFS1 in Col and soc1-2 svp-41 (Fig 4A), and supported the proposal that SOC1 activates and SVP represses transcription of TFS1 at least partly through binding to CArG-box I and CArG-box II, respectively.
Consistent with the role of SOC1 in promoting TFS1 transcription, increased levels of the repressive mark H3K27me3 were detected in aerial parts of 15-day old soc1-2 mutants across the TFS1 gene body (Fig 4G). Furthermore, in DEX-induced 35S::SOC1:GR plants, H3K27me3 levels were reduced following binding of SOC1 (S4D Fig). Additionally, the presence of the active chromatin mark H3K4me3 and of RNA polymerase II (RNAPII) were monitored along the transcribed region of TFS1 in soc1-2 mutants. ChIP-qPCR analysis revealed that H3K4me3 levels were strongly reduced in soc1-2 mutants, similar to those observed in Col-FRI wild-type (Figs 2A and 4H). Also, the ChIP profile of RNAPII demonstrated enrichment throughout the transcribed region of TFS1 in Col and soc1-2 svp-41. By contrast, in soc1-2 mutants the loss of RNAPII enrichment was most apparent in regions of the gene body (Fig 4I), which is reminiscent of inactive genes that display a poised RNAPII machinery at promoter regions.
Overall, these experiments demonstrate that SOC1-induced activation of TFS1 transcription is invoked by eviction of SVP and reduction in H3K27me3 as well as by releasing RNAPII to transcribe the gene.
SOC1 recruits REF6 and BRM to TFS1 to activate transcription
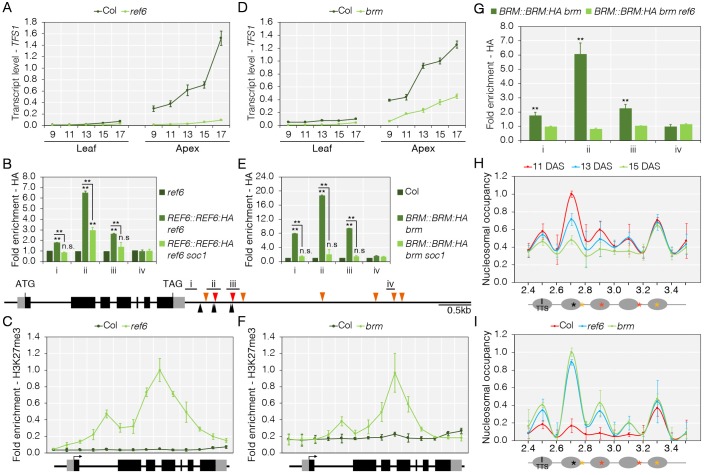
The histone demethylase REF6 and the chromatin remodeler BRM physically interact to antagonize PcG proteins at target loci, and both proteins were detected at the 3’ end of TFS1 in a genome-wide study [33, 34, 46]. Furthermore, REF6 and SOC1 co-purified in the same complex, which was required to facilitate transcriptional activation of target genes through the removal of the repressive histone mark H3K27me3 [22, 47]. Thus, to understand the effects of REF6 and BRM on TFS1 transcription, TFS1 transcript abundance was monitored by RT-qPCR in ref6-1 and brm-1 mutants grown for 9 to 17 days in LDs. Throughout the time-course, the transcript profile of TFS1 was not changed in leaves of ref6-1 or brm-1 mutants compared to Col, but a dramatic reduction in TFS1 transcript abundance was detected in apices of both mutants (Fig 5A and 5D). Therefore, REF6 and BRM are required for the activation of TFS1 in apices.
Fig 5. Activation of TFS1 transcription requires direct recruitment of REF6 demethylase and BRAHMA chromatin remodelling ATPase.
A) Reduced TFS1 mRNA levels in ref6 mutants during floral transition compared to Col. B) Top: REF6 recruitment to TFS1 depends on SOC1. Bottom: TFS1 gene model highlighting putative REF6 binding sites. CArG-box are indicated in red and orange triangles and black triangles show putative binding sites for REF6. C) REF6 is required to remove H3K27me3 at TFS1. D) Reduced TFS1 mRNA levels in brm during floral transition compared to Col. E) BRM recruitment to TFS1 depends on SOC1. F) BRM is required to remove H3K27me3 at TFS1. Values were scaled to set the value of the first primer set in Col to 1. G) BRM binding to 3’ end of TFS1 requires REF6. Values were scaled to set peak value of ii in BRM::BRM:HA brm to 1. Statistical significance was calculated against BRM::BRM:HA brm ref6. H) Nucleosome positioning at 3’end of TFS1. MNase digestion followed by tiled oligo qPCR (MNase-qPCR) to monitor nucleosome positioning at 3’ end of TFS1 in 11-, 13- and 15-day-old LD-grown Col plants. Values were scaled to set highest peak value to 1. CArG-boxes are indicated in red and orange asterisks and black asterisk shows putative SPL-binding sites. TTS, transcriptional start site. I) Nucleosome positioning at 3’end of TFS1 in Col, ref6 and brm plants that were grown for 15LD. Values were scaled to set highest peak value to 1. In A and D, Numbers on x-axis indicate number of long-days (LD) for which plants grown prior to harvest. In B and E, Values were scaled to set value of amplicon i to 1 in ref6 and Col, respectively. Statistical significance was calculated against ref6 (B) and Col (E) using Student’s t-test; **P < 0.01, n.s. P > 0.05.
To validate the previously reported binding of REF6 and BRM to TFS1 [46], ChIP-qPCR analyses were performed using REF6::REF6:HA ref6-1 and BRM::BRM:HA brm-1 transgenic lines [47, 48]. Binding of REF6:HA and BRM:HA to sites located at the 3’ end of TFS1 was detected and these sites flank CArGI, to which SOC1 binds (Fig 5B and 5E). To understand whether association of REF6 and BRM with chromatin is dependent on SOC1 (Fig 4C), the soc1-2 mutation was introduced into the REF6::REF6:HA ref6-1 and BRM:BRM:HA brm-1 transgenic lines by genetic crossing. A strong reduction in binding of REF6:HA and BRM:HA was detected by ChIP-qPCR in soc1-2 mutants, indicating that SOC1 supports REF6 and BRM binding to the 3’-end of TFS1 (Fig 5B and 5E). Additionally, the BRM:BRM:HA brm-1 transgene was introduced into the ref6-1 mutant to study binding behaviour of BRM:HA to the 3’ end of TFS1. Chromatin association of BRM:HA was strongly reduced in ref6-1 compared to Col (Fig 5G), which further corroborated the idea that REF6 is required for BRM recruitment.
REF6 is a H3K27me3 demethylase [47] and BRM and REF6 appeared to act as direct activators of TFS1 transcription, so H3K27me3 levels were tested at TFS1 in the respective mutants. Compared to Col, increased H3K27me3 levels were detected by ChIP-qPCR along the TFS1 genomic locus in ref6-1 and brm-1 mutants. The pattern of increase of H3K27me3 was identical in both mutants and consistent with the observed increase in soc1-2 mutants (Figs 4G, 5C and 5F). In summary, these findings suggest that the histone demethylase REF6 and the chromatin remodeler BRM are required to activate transcription of TFS1 in association with SOC1.
To test whether BRM recruitment leads to activation of TFS1 through changes in chromatin accessibility, limited Micrococcal nuclease (MNase) digestion followed by tiled oligo qPCR was employed to identify well-positioned nucleosomes near the SOC1, REF6 and BRM bound site at the 3’ end of TFS1. The MNase-qPCR analysis identified in ref6-1 and brm-1 mutants a nucleosome at a position that encompasses the binding site for SOC1 and this nucleosome was destabilized in Col (Fig 5H and 5I). Therefore, SOC1 appears to increase chromatin accessibility and transcription of TFS1 through recruitment of REF6 and BRM.
Co-operativity between SOC1 and SPL9 transcription factors in the activation of TFS1
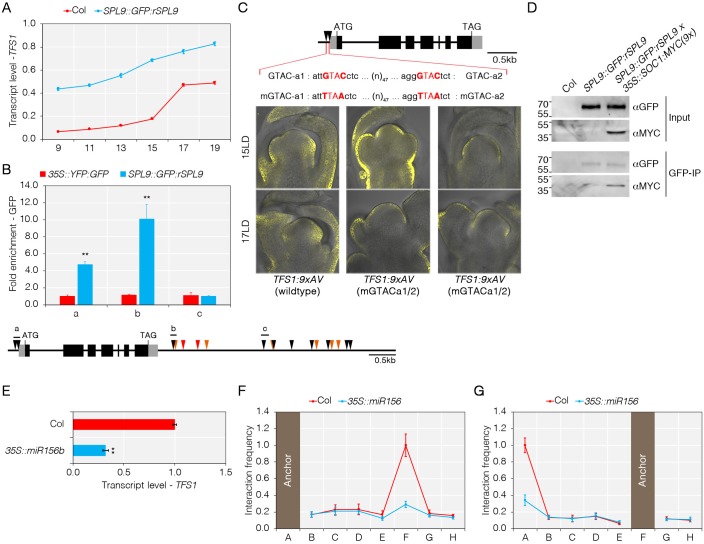
The spatial and temporal expression patterns of TFS1 appeared similar to those of SPL9 [22, 49, 50], which encodes a transcription factor that binds to regulatory sequences in the promoter of the floral meristem-identity gene APETALA1 (AP1) to regulate floral fate [29, 50]. Moreover, SPL15 and SOC1 co-operate to regulate floral commitment under non-inductive conditions [22]. Taken together, the data suggested that SPL9 and SOC1 might cooperate to activate TFS1. To test for the molecular effect of SPL9, RNA was extracted from apices of SPL9::GFP:rSPL9 to monitor TFS1 transcript abundance by RT-qPCR. TFS1 transcript levels were strongly increased in SPL9::GFP:rSPL9 plants compared to wild-type (Fig 6A). Next, ChIP-qPCR analysis was employed to test binding of GFP:rSPL9 at the 5’ and 3’ ends of TFS1 (Fig 6B, S7A and S7B Fig). Consistent with direct activation of TFS1 by SPL9, fragments at the 5’ and 3’ ends of the gene were enriched after immunoprecipitation of GFP:rSPL9 (Fig 6B, S7A and S7B Fig).
Fig 6. Cooperativity between SPL9 and SOC1 in the activation of TFS1.
A) Increased expression of TFS1 is enhanced by rSPL9. Numbers on X-axis indicate number of long-days (LD) for which plants were grown prior to harvest. B) Top: SPL9 binds to the 5’ and 3’ end of TFS1. Bottom: Cartoon of potential SPL9 binding sites and amplicons used for ChIP-PCR. Binding sites for SPL9 are depicted in black and CArG-boxes in red and orange triangles. Values were scaled to set value of amplicon a in 35S::YFP:GFP to 1. Statistical significance was calculated using Student’s t-test; **P < 0.01. C) Top: Model of TFS1 genomic region and the mutations introduced into SPL-binding sites. Bottom: Mutation of two putative SPL binding sites reduces TFS1 expression during floral transition. D) SOC1 co-immunoprecipitates with SPL9. Values on the left of the Western-blots indicated molecular weight (kDa). E) Overexpression of miR156b reduces TFS1 mRNA levels. Statistical significance was calculated using Student’s t-test; **P < 0.01. F and G) Overexpression of miR156b reduces loop formation between 5’ and 3’ end of TFS1.
The presence of markers for transcriptional activity at TFS1, particularly the Mediator head-module component Med18, RNAPII and H3K4me3, was scored by ChIP-qPCR in different genotypes. Higher enrichment levels of these markers were detected at TFS1 in SPL9::GFP:rSPL9 than in wild-type, supporting that SPL9 activates TFS1 (S7C–S7F Fig). In contrast, reduced TFS1 transcript abundance was detected in spl9-1, spl15-1 and spl9-1 spl15-1 mutants (S8A–S8E Fig) and this was accompanied with a reduction in H3K4me3 and an increase in H3K27me3 at the TFS1 locus (S8F–8H Fig).
Whether the identified binding sites for SPL9 are responsible for in vivo regulation of TFS1 was then examined. To this end, a reporter gene cassette was constructed, mGTACa1/2, in which the two GTAC motifs overlapping with the ChIP-qPCR peak of GFP:rSPL9 at the 5’ end of TFS1 were mutated (Fig 6C). Transformants harbouring the mGTACa1/2 mutated form were generated and compared by confocal microscopy with plants harbouring a wild-type construct. VENUS fluorescent signal detected at the periphery of the SAM in wild-type was missing in the mGTACa1/2 plants, supporting the idea that SPL9 binds to these sites to activate transcription (Fig 6C, S7G Fig). Surprisingly, however, VENUS fluorescent signal was retained in the epidermis of mGTACa1/2 plants, indicating that expression in these cells likely takes place independently of SPL9 and other SPL transcription factors (Fig 6C, S7G Fig). Additionally, in-situ hybridisation indicated that TFS1 mRNA appeared more rapidly on the flanks of the meristem after transferring SPL9::GFP:rSPL9 plants from SDs to LDs than after transferring Col wild-type (S7F Fig). These studies are consistent with SPL9 binding to the 5’ end of TFS1 to activate transcription at the periphery of the SAM.
In support of the notion of functional cooperativity between SPL9 and SOC1, co-immunoprecipitation of GFP:rSPL9 and SOC1:MYC(9x) was detected in protein extracts from shoot apical tissue of SPL9::GFP:rSPL9 35S::SOC1:MYC(9x) transgenic lines (Fig 6D). The cooperativity between SPL9 binding at the 5’ end of TFS1 and SOC1 binding at the 3’ end suggested that DNA loop formation might occur between their binding regions. Therefore, chromosome conformation capture (3C) was employed to test for DNA loop formation in Col and 35S::miR156b, in which several redundant SPL transcription factors are reduced in expression [51] leading to reduced transcription of TFS1 (Fig 6E). The 3C analyses suggested that interaction between SPL-binding sites located at the 5’ end and the CArG-box predicted to bind SOC1 that is located at the 3’ end of TFS1 occurred in a SPL9 dependent manner (Figs 4C, 6F and 6G). Together with results described above, the data suggest that SPL9 cooperates with SOC1 to form an ‘active’ DNA-loop that is required for active TFS1 transcription.
BRAHMA recruitment by SOC1 is required for SPL9 binding
The chromatin remodeler BRM is recruited to TFS1 in a SOC1 dependent-manner to increase chromatin accessibility and TFS1 transcription (Fig 5E and 5I). In addition, SOC1 is required for increased H3K4me3 at TFS1 (Fig 4H). This chromatin mark is also supported by the COMPASS-like (Complex Proteins Associated with Set1) histone H3 lysine-4 methyltransferase complex component WD40 REPEAT HOMOLOG 5 (WDR5), which associates with the active elongating RNAPII [52]. Therefore, ChIP analyses were performed using commercial antibody (S3 Table) that recognises WDR5a and WDR5b [53] to test WDR5 enrichment at TFS1 in different genotypes. Consistently, the presence of WDR5 at TFS1 was decreased across the gene body in soc1-2, brm-1 as well as spl9-1, spl15-1 and spl9-1 spl15-1 mutants (S9A–S9C Fig). Additionally, using commercial antibody (S3 Table) ChIP analyses for the histone variant H2A.Z, which marks both transcriptionally active and inactive genes [54, 55], detected colocalization of H2A.Z with WDR5 at TFS1 in Col and decreased enrichment in soc1-2 and brm-1 (S9D–S9F Fig). In contrast, no difference in H2A.Z enrichment was detected between spl mutants and Col, further corroborating the idea that SPL functions to orchestrate transcriptional machinery rather than influencing nucleosomal composition (S9D–S9F Fig).
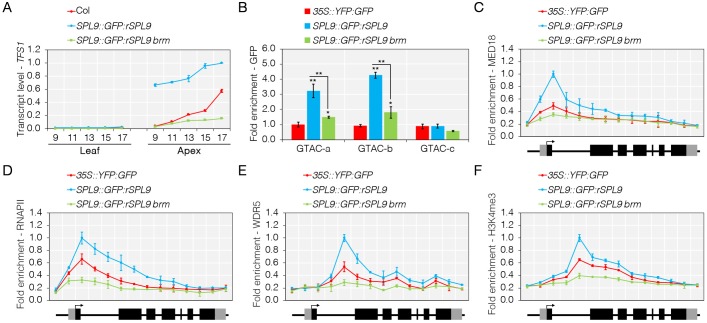
The data described so far suggested that SOC1 mediated recruitment of BRM might enable association of SPL9 to chromatin. To test this idea, SPL9::GFP:rSPL9 was introduced into brm-1 mutants by genetic crossing. Unexpectedly, in SPL9::GFP:rSPL9 brm-1 most of the floral structures were converted into carpelloid structures at the primary inflorescence, a more severe phenotype than either parental line (S10A and S10B Fig). To further characterize the molecular effect, TFS1 transcript abundance was examined by RT-qPCR using RNA extracted from leaves and apices. The enhanced apex specific expression of TFS1 in SPL9::GFP:rSPL9 was strongly suppressed by brm-1, supporting the idea that BRM is required to support SPL9 activity (Fig 7A). Similarly, expression of other floral marker genes such as SOC1, FUL, LEAFY (LFY), AP1 and SEPALLATA3 (SEP3) was also reduced in this genotype, although SPL9 protein level was not affected (S10C–S10E Fig).
Fig 7. BRM is required for SPL9 dependent activation of TFS1.
A) Elevated TFS1 transcript levels in rSPL9 plants suppressed by brm mutation. Numbers at x-axis indicate number of long-days (LD) for which plants were grown prior to harvest. B) Binding of SPL9 at TFS1 is reduced in brm mutants. Values were scaled to the value of amplicon GTAC-a in 35S::YFP:GFP. Statistical significance was calculated against 35S::YFP::GFP using Student’s t-test; *P < 0.05, **P < 0.01. C to F) SPL9-dependent recruitment of MED18 (C), RNAPII (D), WDR5 (E) and H3K4me3 (F) across gene body of TFS1 is reduced in SPL9::GFP:rSPL9 brm.
Consistent with the idea that BRM alters nucleosomal positioning leading to changes in the exposure of a critical SPL-binding site located at the 3’ end of TFS1 (Fig 5I), reduced binding of GFP:rSPL9 was detected to the 5’ and 3’ end of TFS1 in brm-1 (Fig 7B). This result suggested that BRM facilitates binding of SPL9 to its cognate binding sites. SPL15 recruits RNAPII through Mediator [22], and consistent with SPL9 having a similar role at TFS1, a strong reduction in the recruitment of MED18, RNAPII and markers of active transcription such as WDR5 and H3K4me3 was detected in brm-1 (Fig 7C–7F). Taken together, these data indicate that SOC1-dependent recruitment of BRM is required to allow SPL9 to bind to TFS1 and that Mediator conveys regulatory information from SPL9 to the basal RNAPII transcriptional machinery that is coupled with the COMPASS-like complex to activate TFS1 transcription.
Discussion
The MADS box transcription factors FLC and SVP are well-established negative regulators of floral induction in Arabidopsis, however only fragmentary information is available on the roles of their direct targets in floral transition and the architecture of the regulatory network they control. Here, we addressed these issues by characterizing TFS1, an immediate target gene of FLC/SVP that encodes a B3-type transcription factor, which is expressed specifically on the flanks of the shoot apical meristem and promotes floral transition under LDs and SDs. TFS1 transcription is repressed by FLC/SVP and promoted by SOC1, another MADS box transcription factor that is also encoded by a primary target of FLC/SVP. We show that FLC/SVP and SOC1 have opposing effects on transcription through mediating antagonistic histone modifications at TFS1. These data provide insight into the complexity of the regulatory network controlling floral transition downstream of FLC/SVP and define mechanisms by which MADS box transcription factors antagonistically regulate transcription of their direct targets.
B3-Type transcription factors and plant reproductive development
TFS1 is a member of the B3-type transcription factor superfamily that is specific to the Viridiplantae [56]. Within this superfamily, TFS1 falls in the REM family, several of which have established or proposed roles in reproduction of Arabidopsis [30, 31]. Loss of function alleles of two members of this family, VERNALIZATION 1 (VRN1) and VERDANDI (VDD), provided genetic support for roles in reproductive development [57, 58]. VRN1 is required for stable transcriptional repression of FLC during induction of flowering by vernalization [57, 59], and appears to bind DNA non-specifically [57], while VDD is involved in ovule development [58]. In addition, several other members of this family are specifically expressed in the inflorescence meristem or developing flowers [31, 60–62]. REM transcription factors and MADS box proteins, another family of transcription factors with multiple roles in reproductive development, appear to often regulate one another’s expression. For example, VRN1 regulates FLC, VDD transcription is controlled by SEEDSTICK, TFS1 is repressed by FLC/SVP and genome-wide studies of binding sites of MADS box factors AG, AP3, PI and AGL15 identified several REM genes as direct targets [31]. Both families of transcription factors are amplified in higher plants [30, 63], and they may have co-evolved to act in common pathways during the evolution of reproductive development.
The mechanism of action of REM proteins is not known, although they are believed to bind DNA via their B3 domains. A GFP-tagged form of VRN1 was found to associate widely with Arabidopsis chromosomes, and this association persisted through mitosis but was lost at meiosis [59]. Interestingly, TFS1 was also previously identified in a targeted proteomics approach as interacting with PCNA, a component of the DNA replication complex [64]. Also, our confocal imaging suggested that the nuclear localization and activity of TFS1 is closely related to cell division. The molecular functions of REM proteins such as TFS1 and how they are related to chromatin structure and cell division are interesting areas for future experimentation.
Antagonistic effects of MADS box transcription factors on target gene expression
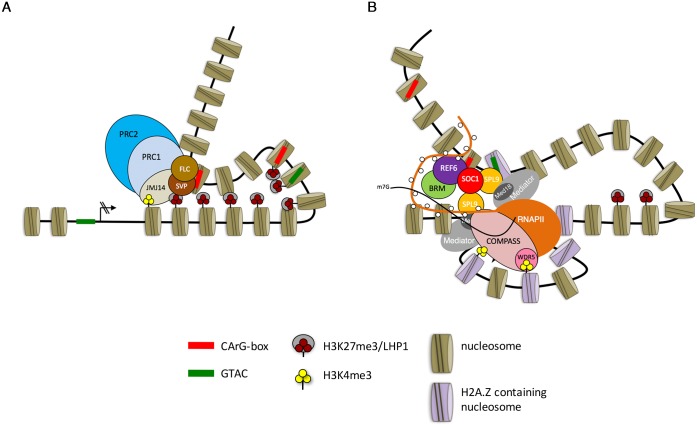
Genome-wide studies demonstrated that binding of FLC and SVP is predominately associated with transcriptional repression of target genes [6–8]. One of these targets is the flowering-time gene FT, whose expression in the vascular tissue of leaves is repressed by FLC and SVP [16, 65]. The capacity of FLC-like transcription factors to repress FT transcription has been reported to be associated with their ability to recruit PRC components and maintain H3K27me3 levels at the gene [19]. We also found that transcriptional repression of TFS1 by FLC at the shoot meristem is associated with H3K27me3 accumulation, and that this involves formation of a chromatin loop between the 3’-end and intragenic regions of TFS1 that requires PRC complexes. The JMJ14 H3K4 demethylase also associates with EMF1 and LHP1 [19–21], and we found that in jmj14 mutants H3K27me3 levels as well as binding of both FLC and SVP were significantly reduced at TFS1, although the expression levels of FLC and SVP were not compromised. Collectively, these data suggest a model whereby PRC complexes involving EMF1, LHP1 and JMJ14 are recruited by FLC and SVP to TFS1 to sustain H3K27me3 levels and binding of these transcription factors, thereby stably repressing TFS1 transcription (Fig 8A). In this model, how the PRC1-like complexes reinforce binding of FLC and SVP and whether binding of these transcription factors is a prerequisite for PcG recruitment and PcG-mediated gene silencing remain to be resolved. Another possibility is that a co-factor for FLC binding, perhaps another MADS box transcription factor such as AGL16 [66], is reduced in expression in circumstances in which H3K27me3 levels are reduced. In this case, reduction in H3K27me3 would indirectly lower FLC binding.
Fig 8. Proposed model for TFS1 regulation and function by different flowering pathways.
A) FLC and SVP mediated repression of TFS1 requires PRC activity and a locked chromatin conformation. B) Activation of TFS1 requires loop formation between 5’ and 3’ end and is mediated by the cooperativity between SOC1 and SPL9.
In contrast to FLC/SVP, the MADS box factor SOC1 activates TFS1 transcription during floral transition. Furthermore, SOC1 binds directly to TFS1 as defined in genome-wide [12, 28] and targeted ChIP-qPCR experiments performed here. Induction of SOC1:GR was sufficient to activate TFS1 transcription in the presence of SVP, demonstrating that SOC1 activation is epistatic to the repression mediated by SVP and that after SOC1:GR activation SVP binding to TFS1 was strongly reduced. This reduction of SVP at TFS1 could be due to its displacement by SOC1 binding to an adjacent CArG box or to the transcriptional repression of SVP by SOC1 [12, 15, 28]. Furthermore, our ChIP data indicate that the repressive state imposed by FLC/SVP is overcome by SOC1 through recruitment of the H3K27me3 demethylase REF6 and the chromatin remodeler BRM to the TFS1 locus. Similarly, REF6 was recently shown to be recruited to targets by other MADS box transcription factors [67]. Our observations suggest that SOC1 displays characteristics associated with pioneer transcription factors, as it resolves condensed chromatin structures and opens chromatin through the combinatorial activity of REF6 and BRM. Similarly, a recent report in Caenorhabditis elegans demonstrated that the pioneer factor PHA-4 binds to promoters required for foregut development to recruit RNAPII and promote chromatin opening [68]. PHA-4 was proposed to facilitate chromatin opening by depositing RNAPII at target gene promoters. Similarly, the Drosophila maternal pioneer factor ZELDA (Zld) recruits poised RNAPII to Dorsal (Dl) target genes, facilitating chromatin accessibility for Dl which then mediates their zygotic activation [69, 70]. Accordingly, we found that the SOC1-REF6-BRM complex relaxes and opens chromatin at TFS1 to facilitate binding of SPL9 and to activate poised RNAPII, resulting in a reduction in H3K27me3 levels across the TFS1 genomic locus. Many genes directly repressed by FLC or SVP to maintain vegetative development are likely to be subsequently bound and activated by other MADS box transcription factors during reproductive development. Thus the mechanisms defined here by which SOC1 antagonises the repression of TFS1 transcription imposed by FLC/SVP are likely to be more broadly relevant during the transition to flowering.
Activating chromatin loop formed at TFS1 through functional cooperation between SPL9 and SOC1
SOC1 functionally co-operates with SPL15 to form a chromatin loop associated with activation of FUL transcription [22]. Similarly, we showed by co-immunoprecipitation a physical interaction between SPL9 and SOC1 at TFS1. Similarly, we detected looping at the TFS1 locus between the SPL9-binding region close to the TSS and the SOC1 binding region at the 3’-end of TFS1 that might enable a higher turn-over rate of RNAPII to yield higher transcriptional activity. These observations suggest that the formation of an active chromatin loop could enable SOC1 and SPL9 to recruit respectively REF6 and RNAPII to the TSS, and then the active elongating RNAPII could cause the gene body of TFS1 to change its position relative to the stable SOC1-SPL9 complex enabling REF6 to track along the gene with RNAPII progressively removing H3K27me3 (Fig 8B). This model predicts that the SPL9-SOC1 interaction induces dynamic chromatin folding that facilitates movement of the RNAPII along the gene body, rather than that RNAPII separates from the pre-initiation complex and tracks along the TFS1 gene body. It will be interesting to determine in a genome-wide context whether other targets of SPL9 and SOC1 display similar features.
TFS1 contributes to an inter-related network of interactions downstream of FLC
The analysis presented here incorporates TFS1 into a network of interactions among FLC target genes. At the shoot meristem, FLC directly binds to and represses transcription of SOC1 and TFS1 [6, 7, 27, 43]. Furthermore, SOC1 directly activates the transcription of TFS1. Thus repression of TFS1 by FLC involves both direct repression of expression of its positive activator SOC1 as well as direct repression of TFS1, a relationship characterized as a coherent feed forward loop type II [71]. The temporal and spatial patterns of TFS1 expression on the flanks of the inflorescence meristem are overlapping with and partially conferred by SPL9, and may indicate an important role for TFS1 in modulating the expression of genes in cells that will give rise to floral primordia. This suggestion is strengthened by the observation that the Col-FRI flc-3 svp-41 tfs1-1 triple mutant shows a floral morphology defect not shown by any of the single mutants. Previously, the soc1-2 agl24-1 svp-41 combination was also demonstrated to have a synergistic effect on floral development due to redundancy among these transcription factors in the repression of genes involved in floral organ development [43]. Our data suggest that there may also be redundancy among FLC, SVP and TFS1 in the regulation of downstream genes, which could be characterized in a future analysis of TFS1 targets. More generally, our work emphasises that defining the network of genes negatively regulated by FLC/SVP, and understanding how these then interact during the progression to flowering when FLC expression is repressed or lost by mutation, is proving to be a productive approach in defining critical mechanisms controlling floral transition.
Materials and methods
Plant material and growth conditions
All seed stocks are in the Columbia-0 (Col-0) genetic background and were obtained from the Nottingham Arabidopsis Stock Centre (NASC; S1 Table) except for 35S::SOC1:GR soc1-1 (Hyun et al., 2016), which is in a Landsberg erecta (Ler-0) genetic background. Seeds were sown on soil or on full-strength Murashige and Skoog (MS) medium containing 1% sucrose, stratified for 3 days at 4°C, and grown at 22°C under either long-days (16hrs light/8hrs dark; 150μmol.m-1.s-1) or short-days (8hrs light/16hrs dark; 150μmol.m-1.s-1). Plant age was measured when seeds were transferred from stratifying to ambient growth conditions.
Plasmid construction and plant transformation
Full-length TFS1 genomic region was cloned by PCR with Phusion Enzyme (New England Biolabs) according to the manufacturer’s recommendations and used to generate TFS1::TFS1::9xAla-Venus. To introduce 9xAla-Venus coding sequence, we employed Polymerase Incomplete Primer Extension (PIPE) cloning method [72] and plasmids were then introduced into Agrobacterium to transform Col plants by floral dip [73]. The sequences of the primers used for PIPE cloning are listed in S2 Table.
RNA-extraction and real-time quantitative PCR
Total RNA of indicated genotypes at different days after sowing from leaves and apices was isolated with NucleoSpin RNA plant kit (Macherey-Nagel). DNA was removed by an on-column treatment with rDNase and 2 μg RNA was reverse transcribed with an oligo(dT) primer, RNAseOUT Recombinant Ribonuclease Inhibitor (Thermo Fisher Scientific) and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). The cDNA equivalent of 20ng of total RNA was used in a 12 μL qPCR reaction on a Roche Light Cycler 480 instrument (Roche) with either iQ SYBR Green Supermix (BioRad) or GoTaq qPCR Master Mix (Promega) and quantified using the UBC21 (AT5G25760) as a reference gene to which data was normalized [74]. The mean of three biological replicates with standard deviation is shown and list of primers used for expression analyses can be found in S2 Table.
Chromatin-immunoprecipitation (ChIP) assays
ChIP was performed as previously described with minor modifications [22]. In brief, above-ground tissue of 15LD-grown plants was collected at ZT8 and fixed in PBS solution containing 1.5% formaldehyde. ChIP-assays in which indirect binding of the protein of interest to chromatin was studied, Di(N-succinimidyl) glutarate (DSG; Synchem) at a final concentration of 1 μM was used to introduce protein-protein crosslinks prior to formaldehyde-assisted protein-chromatin crosslinking. To determine fold enrichment levels, ChIP-DNA was quantified on a Roche Light Cycler 480 instrument (Roche) with iQ SYBR Green Supermix (BioRad) and normalized against ACT8 (AT1G49240). In ChIP assays in which histone modifications were tested, the values of the tested histone marks were normalized against histone H3. The average of three biological replicates is shown and list of primers used for fold enrichment analyses can be found in S2 Table.
Chromatin conformation capture (3C) assay
3C assay was performed as described previously with minor modifications. A total of 2g of above-ground tissue of 15 day-old LD-grown plants was used for 3C study. Chromatin DNA was digested for 16 hrs at 37°C with 400U Sau3AI (New England Biolabs, S3 Table) while agitating at 900 r.p.m. For intramolecular ligation, digested nuclei were incubated for 5 hrs at 16°C in 500 U T4 DNA Ligase (Promega, S3 Table). In parallel, the cloned TFS1:9xAla-Venus construct was digested and ligated. The 3C DNA ligation products were quantified by RT-qPCR and normalised to the TFS1:9xAla-Venus control using the delta-delta Ct method. The sequences of the primers used in the 3C-assay are listed in S2 Table.
Micrococcal nuclease-assay (MNase-assay)
Micrococcal nuclease-assay was performed as described previously with minor modifications [75]. For nuclear extraction, above-ground tissue of 15 day-old LD-grown plants was harvested, ground in liquid nitrogen and resuspended in lysis buffer (LB) [50 mM HEPES pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol, 5 mM ß-mercaptoethanol and protease inhibitor cocktail (Roche)]. After 1 hr of lysis, the lysis mixture was filtered twice through 2 layers of Miracloth (Calbiochem) and protocol was followed as previously described. For MNase treatment, nuclei were treated with 5U MNase (Thermo Fisher Scientific) for 15 min and digest was stopped by adding 16 μL 250 mM EDTA, and then treated with RNase A and Proteinase K (Sigma Aldrich), each for 1 hr.
Protein analyses
Total protein extraction and in vivo co-immunoprecipitation were performed as described previously with minor modifications [22, 76]. For SVP Western-analysis, roughly 50 apices of 15 day-old LD-grown plants were harvested. Protein concentration was determined by Bradford series and a total of 50μg for crude extract and 1mg for immunoprecipitation was used. The amino acid sequences of the epitopes for generating SVP antibody are presented in S4 Table.
In-situ hybridisation and imaging
In-situ hybridisation was performed according to the method described previously [77]. The sequences of the primers used for the in-situ hybridisation experiments are listed in S2 Table. For confocal microscopy, shoot apices at different developmental stages were collected and fixed with 4% paraformaldehyde (PFA) prepared in phosphate-buffered saline (PBS) at pH7.0. Samples were then vacuum infiltrated for 20 min on ice, transferred to fresh 4% PFA, and stored at 4°C overnight. The fixed samples were washed twice for 1 min in PBS, then cleared with ClearSee [10% (w/v) xylitol, 15% (w/v) sodium deoxycholate and 25% (w/v) urea][78] for 3 to 8 days at room temperature. After clearing, the shoot meristems were imaged by confocal laser scanning microscopy (Zeiss LSM780), as described previously [79].
Image processing and figure construction
All image processing and figure construction was performed with Photoshop (www.adobe.com).
Accession numbers
Mutant and transgenic lines used in this study, including references for their origin and description in literature, and their respective AGI identifiers are listed in S1 Table.
Supporting information
A) Phylogenetic tree for B3 type gene family based on ClustalW alignment with a bootstrap value of 1.000 replicates. B) Immunoblot to test for cross-reactivity of SVP. Triangle indicates specific whereas asterisks indicate non-specific bands. Values on the left of the Western-blots indicated molecular weight (kDa). C) Transcript level of TFS1. Plants were grown for 2 weeks in short-days and then transferred to long-day. Values were scaled to set value of apices of 2 weeks short-day grown Col-FRI plants to 1. D) Increased expression of TFS1 in apices of Col-FRI plants when transferred to ambient growth condition after vernalisation. NV, non-vernalised; V, weeks vernalised in short-days (SD). E) Spatial pattern of expression of TFS1 assessed by in-situ hybridization after floral transition. DAS, days after sowing.
(TIF)
A to D) Comparison of tfs1 mutant plants and Columbia grown in either LDs (A) or in SDs (C). Leaf number of tfs1 plants compared to Columbia grown in LDs (B) and in SDs (D). Statistical significance was calculated using Student’s t-test; **P < 0.01. E) Representative gene model for TFS1 and T-DNA insertion used in this study. F) Total leaf number of tfs1-1 and tfs1-2 plants compared to Col. Statistical significance was calculated using Student’s t-test; *P < 0.05. G) Inflorescence of Col-FRI flc svp tfs1 plants. H) Floral structures of Col-FR flc svp tfs1 plants. Particular features are marked: blue arrow, misplaced floral organs; white arrow, ectopic ovules; yellow arrow, stigmatic papillae on leaf-like structures; red arrow, leaf-like anthers.
(TIF)
A) H3K27me3 levels are increased at TTS of TFS1 in Col-FRI flc svp. B and C) H3K4me3 levels are increased in Col-FRI flc svp (B) and Col-FRI clf and Col-FRI jmj14 (C) plants. Values were scaled to set highest value in Col-FRI flc svp to 1. D) Reduced TFS1 transcript levels in Col-FRI plants suppressed by clf mutation. E and F) Transcript levels of FLC (E) and SVP (F) in leaves and apices of Col-FRI clf plants. Numbers at x-axis indicate number of long-days (LD) for which plants were grown prior to harvest. G) Reduced TFS1 transcript levels in Col-FRI plants suppressed by jmj14 mutation. H and I) Transcript levels of FLC (H) and SVP (I) in leaves and apices of Col-FRI jmj14 plants. Numbers at x-axis indicate number of long-days (LD) for which plants were grown prior to harvest. Statistical significance was calculated using Student’s t-test; *P < 0.05, **P < 0.01, n.s. P > 0.05.
(TIF)
A) SOC1:GR-induced transcriptional activation of TFS1. B and C) Top: ChIP-qPCR analysis of SOC1 (B) and SVP (C) on CArG-boxes at TFS1 in 35S::SOC1:GR soc1 plants after DEX treatment. Bottom: Diagram of TFS1 locus with CArG-boxes and amplicons used for ChIP-qPCR. Red and orange triangles indicate CArG-boxes in TFS1. Green and red lines represent amplicons used for ChIP-qPCR study. D) ChIP-qPCR analysis of H3K27me3 at TFS1 in 35S::SOC1:GR soc1 plants after DEX treatment. B to D, values were scaled to set value of first primers at 20min after DEX treatment in 35S::SOC1:GR soc1 to 1. GB, indicates Gene Body amplicon used for ChIP-qPCR. Statistical significance was calculated against mock treatment using Student’s t-test; *P < 0.01, n.s. P > 0.05. Representative confocal images of independent transformants of E) TFS1:9AV, F) TFS1:9AV mCArGII and G) TFS1:9AV mCArGI+II. H) Abundance of TFS1 mRNA in indicated transgenic plants as well as in Col and tfs1 controls.
(TIF)
A and B) Representative photograph (A) and total leaf number (B) of TFS1::TFS1:9AV in tfs1. C and D) Representative photograph (C) and total leaf number (D) of TFS1::TFS1:9AV mCArGII in tfs1. E and F) Representative photograph (E) and total leaf number (F) of TFS1::TFS1:9AV mCArGII in soc1. G and H) Representative photograph (G) and total leaf number (H) of TFS1::TFS1:9AV mCArGI+II in tfs1. In each case, transformants are shown in the same order in the representative photograph as in the leaf number plot. Statistical significance was calculated using Student’s t-test; *P < 0.1, **P < 0.05, ***P < 0.01, n.s. P > 0.05.
(TIF)
A) Top view of TFS1:9xAla-Venus (yellow) expression at the apical meristem. DAPI (blue) serves to highlight individual cells. B) Side view of TFS1:9xAla-Venus (yellow) localisation at the shoot apical meristem. C to G) TFS1:9xAla-Venus localises to the nucleus of sepal cells (C), in young flowers and at the base of pedicels (D) and in flowers of stage stage 3 (E), stage 2 (F) and stage 4 (G) after LMB treatment.
(TIF)
A) ChIP-qPCR for rSPL9 binding to putative SPL-binding sites at TFS1 locus. Statistical significance was calculated against the respective amplicon of 35S::YFP:GFP. B) ChIP-qPCR for temporal binding of SPL9:GR at TFS1 after DEX treatment. Statistical significance was calculated against the 20min DEX treatment. C to E) ChIP-qPCR for MED18 (C), RNAPII (D) and H3K4me3 (E) at TFS1 locus. Plants were grown for 15 LD and harvested at ZT8. F) Spatial pattern of expression of TFS1 assessed by in-situ hybridization during floral transition in Col and GFP:rSPL9. Plants were grown for 2 weeks in SD and then transferred to permissive LD. G) Confocal images of TFS1:9AV mGTACa1/a2 mutant transgenic lines. Statistical significance was calculated using Student’s t-test; *P < 0.05, **P < 0.01, n.s. P > 0.05.
(TIF)
A and B) TFS1 transcript abundance in short-days is increased by SPL and SOC1 (A) and REF6 (B). Data are shown for 3–8 weeks after germination and values were scaled to set value of 3 weeks SD-grown Col plants to 1. C) Reduced expression of TFS1 in ga1 under LD conditions. D) Spatio-temporal expression pattern of TFS1 assessed by in situ hybridization in LD. E) TFS1 mRNA levels are reduced in apices of spl mutants during floral transition. Values were scaled to set value of 7 LD of Col to 1. F to H) Reduced RNAPII (F) and H3K4me3 (G) and increased H3K27me3 (H) levels at TFS1 in spl mutants. Values were scaled to highest value in Col (F and G) and spl9 spl15 (H) to 1. In A to H, Plants were harvested at ZT8.
(TIF)
A to C) WDR5 levels at TFS1 depend on SOC1 (A), BRM (B) and SPL (C). D to F) H2A.Z incorporation at TFS1 depends on SOC1 (D) and BRM (E) but not on SPL (F). ChIP-PCR was performed on 15 LD grown plants that were harvested at ZT8 (A to F).
(TIF)
A and B) 5-weeks old long-day grown GFP:rSPL9 brm plants display altered plant architecture (A) and partial transformation of flowers into carpelloid structures (B). C) GFP Western analysis to detect GFP:SPL9 in Col and brm. Values on the left of the Western-blots indicated molecular weight (kDa). D and E) Comparative expression analysis of marker genes expressed in leaves (D) and apices (E) of Col, GFP:rSPL9 and GFP:rSPL9 brm plants. X-axis indicates number of long-days (LD) for which plants were grown prior to harvest.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Franziska Turck and Fernando Andrés for critical reading of the manuscript. We thank Yeonhee Choi for Col-FRI clf-2, Xiaofeng Cao for REF6::REF6:HA ref6-1 and 35S::REF6:YFP-HA lines, Doris Wagner for BRM::BRM::HA brm-1 line, Detlef Weigel and Jia-Wei Wang for SPL9::GFP:rSPL9 and SPL9::rSPL9:GR lines and Hao Yu for 35S::SOC1:myc(9x) line.
Data Availability
The underlying numerical data for the figures has been submitted to the Dryad database. (DOI: 10.5061/dryad.b0j2td6)
Funding Statement
This work was funded by the following grants to G.C., N° 339113 HyLife European Research Council https://erc.europa.eu/, SPP1530 Deutsche Forschungsgemeinschaft http://www.dfg.de/en/ and a Core Grant from the Max Planck Society https://www.mpg.de/en. Also, AK and JLM were funded by post-doctoral fellowships from the Alexander von Humboldt Foundation https://www.humboldt-foundation.de/web/home.html. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–39. 10.1038/nrg3291 . [DOI] [PubMed] [Google Scholar]
- 2.Bouche F, Lobet G, Tocquin P, Perilleux C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016;44(D1):D1167–D71. 10.1093/nar/gkv1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, et al. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11(3):445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whittaker C, Dean C. The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu Rev Cell Dev Bi. 2017;33:555–75. [DOI] [PubMed] [Google Scholar]
- 6.Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(16):6680–5. 10.1073/pnas.1103175108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateos JL, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, et al. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015;16:31 10.1186/s13059-015-0597-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, et al. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. P Natl Acad Sci USA. 2017;114(51):E11037–E46. 10.1073/pnas.1618075114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant Journal. 2000;21(4):351–60. [DOI] [PubMed] [Google Scholar]
- 10.Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008;15(1):110–20. 10.1016/j.devcel.2008.05.002 . [DOI] [PubMed] [Google Scholar]
- 11.Gregis V, Andres F, Sessa A, Guerra RF, Simonini S, Mateos JL, et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013;14(6):R56 10.1186/gb-2013-14-6-r56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao Z, Shen LS, Liu C, Liu L, Yan YY, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant Journal. 2012;70(4):549–61. 10.1111/j.1365-313X.2012.04919.x [DOI] [PubMed] [Google Scholar]
- 13.Theissen G, Saedler H. Plant biology—Floral quartets. Nature. 2001;409(6819):469–71. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- 14.de Folter S, Immink RGH, Kieffer M, Parenicova L, Henz SR, Weigel D, et al. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell. 2005;17(5):1424–33. 10.1105/tpc.105.031831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009;60(4):614–25. 10.1111/j.1365-313X.2009.03986.x . [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21(4):397–402. 10.1101/gad.1518407 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calonje M, Sanchez R, Chen LJ, Sung ZR. EMBRYONIC FLOWER1 participates in Polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell. 2008;20(2):277–91. 10.1105/tpc.106.049957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calonje M. PRC1 Marks the Difference in Plant PcG Repression. Mol Plant. 2014;7(3):459–71. 10.1093/mp/sst150 [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Gu X, Yuan W, Schmitz RJ, He Y. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell. 2014;28(6):727–36. 10.1016/j.devcel.2014.01.029 . [DOI] [PubMed] [Google Scholar]
- 20.Lu FL, Cui X, Zhang SB, Liu CY, Cao XF. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 2010;20(3):387–90. 10.1038/cr.2010.27 [DOI] [PubMed] [Google Scholar]
- 21.Yang WN, Jiang DH, Jiang JF, He YH. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant Journal. 2010;62(4):663–73. 10.1111/j.1365-313X.2010.04182.x [DOI] [PubMed] [Google Scholar]
- 22.Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. Multi-layered Regulation of SPL15 and Cooperation with SOC1 Integrate Endogenous Flowering Pathways at the Arabidopsis Shoot Meristem. Dev Cell. 2016;37(3):254–66. 10.1016/j.devcel.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 23.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67(1–2):183–95. 10.1007/s11103-008-9310-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–6. . [DOI] [PubMed] [Google Scholar]
- 25.Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, et al. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24(5):591–9. . [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14(18):2366–76. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002;21(16):4327–37. 10.1093/emboj/cdf432 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Immink RGH, Pose D, Ferrario S, Ott F, Kaufmann K, Valentim FL, et al. Characterization of SOC1's Central Role in Flowering by the Identification of Its Upstream and Downstream Regulators. Plant Physiology. 2012;160(1):433–49. 10.1104/pp.112.202614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, et al. Gibberellin Acts Positively Then Negatively to Control Onset of Flower Formation in Arabidopsis. Science. 2014;344(6184):638–41. 10.1126/science.1250498 [DOI] [PubMed] [Google Scholar]
- 30.Romanel EA, Schrago CG, Counago RM, Russo CA, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One. 2009;4(6):e5791 10.1371/journal.pone.0005791 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantegazza O, Gregis V, Mendes MA, Morandini P, Alves-Ferreira M, Patreze CM, et al. Analysis of the arabidopsis REM gene family predicts functions during flower development. Ann Bot. 2014;114(7):1507–15. 10.1093/aob/mcu124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131(20):4965–75. 10.1242/dev.01363 . [DOI] [PubMed] [Google Scholar]
- 33.Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li ZT, et al. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. P Natl Acad Sci USA. 2012;109(9):3576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li CL, Gu LF, Gao L, Chen C, Wei CQ, Qiu Q, et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nature Genetics. 2016;48(6):687–+. 10.1038/ng.3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez-Venegas R. Regulation by polycomb and trithorax group proteins in Arabidopsis. Arabidopsis Book. 2010;8:e0128 10.1199/tab.0128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3(6):e86 10.1371/journal.pgen.0030086 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14(9):869–71. 10.1038/nsmb1283 . [DOI] [PubMed] [Google Scholar]
- 38.Fromm M, Avramova Z. ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr Opin Plant Biol. 2014;21:75–82. 10.1016/j.pbi.2014.07.004 . [DOI] [PubMed] [Google Scholar]
- 39.Engelhorn J, Blanvillain R, Kröner C, Parrinello H, Rohmer M, Pose Δ, et al. Dynamics of H3K4me3 Chromatin Marks Prevails over H3K27me3 for Gene Regulation during Flower Morphogenesis in Arabidopsis thaliana. Epigenomes. 2017;1:8–40. [Google Scholar]
- 40.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386(6620):44–51. 10.1038/386044a0 [DOI] [PubMed] [Google Scholar]
- 41.Doyle MR, Amasino RM. A Single Amino Acid Change in the Enhancer of Zeste Ortholog CURLY LEAF Results in Vernalization-Independent, Rapid Flowering in Arabidopsis. Plant Physiology. 2009;151(3):1688–97. 10.1104/pp.109.145581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Vernaza M, Yang SX, Muller R, Thorpe F, de Leau E, Goodrich J. Antagonistic Roles of SEPALLATA3, FT and FLC Genes as Targets of the Polycomb Group Gene CURLY LEAF. Plos One. 2012;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Xi WY, Shen LS, Tan CP, Yu H. Regulation of Floral Patterning by Flowering Time Genes. Dev Cell. 2009;16(5):711–22. 10.1016/j.devcel.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 44.Sung ZR, Belachew A, Shunong B, Bertrandgarcia R. Emf, an Arabidopsis Gene Required for Vegetative Shoot Development. Science. 1992;258(5088):1645–7. 10.1126/science.258.5088.1645 [DOI] [PubMed] [Google Scholar]
- 45.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A. 1999;96(16):9112–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui X, Lu FL, Qiu Q, Zhou B, Gu LF, Zhang SB, et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nature Genetics. 2016;48(6):694–+. 10.1038/ng.3556 [DOI] [PubMed] [Google Scholar]
- 47.Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nature Genetics. 2011;43(7):715–U144. 10.1038/ng.854 [DOI] [PubMed] [Google Scholar]
- 48.Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, et al. Regulation of Leaf Maturation by Chromatin-Mediated Modulation of Cytokinin Responses. Dev Cell. 2013;24(4):438–45. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell. 2009;138(4):750–9. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang JW, Czech B, Weigel D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–49. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 51.Xu ML, Hu TQ, Zhao JF, Park MY, Earley KW, Wu G, et al. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. Plos Genetics. 2016;12(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang DH, Kong NC, Gu XF, Li ZC, He YH. Arabidopsis COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development. Plos Genetics. 2011;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang DH, Gu XF, He YH. Establishment of the Winter-Annual Growth Habit via FRIGIDA-Mediated Histone Methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2009;21(6):1733–46. 10.1105/tpc.109.067967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman-Derr D, Zilberman D. Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. Plos Genetics. 2012;8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.To TK, Kim JM. Epigenetic regulation of gene responsiveness in Arabidopsis. Front Plant Sci. 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehti-Shiu MD, Panchy N, Wang PP, Uygun S, Shiu SH. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Bba-Gene Regul Mech. 2017;1860(1):3–20. 10.1016/j.bbagrm.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 57.Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297(5579):243–6. 10.1126/science.1072147 [DOI] [PubMed] [Google Scholar]
- 58.Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, et al. VERDANDI Is a Direct Target of the MADS Domain Ovule Identity Complex and Affects Embryo Sac Differentiation in Arabidopsis. Plant Cell. 2010;22(6):1702–15. 10.1105/tpc.109.068627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. P Natl Acad Sci USA. 2006;103(13):5012–7. 10.1073/pnas.0507427103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romanel E, Das P, Amasino RM, Traas J, Meyerowitz E, Alves-Ferreira M. Reproductive Meristem22 is a unique marker for the early stages of stamen development. Int J Dev Biol. 2011;55(6):657–64. 10.1387/ijdb.113340er [DOI] [PubMed] [Google Scholar]
- 61.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. Plos Genetics. 2006;2(7):1012–24. 10.1371/journal.pgen.0020117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franco-Zorrilla JM, Cubas P, Jarillo JA, Fernandez-Calvin B, Salinas J, Martinez-Zapater JM. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiology. 2002;128(2):418–27. 10.1104/pp.010323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139(17):3081–98. 10.1242/dev.074674 [DOI] [PubMed] [Google Scholar]
- 64.Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van de Slijke E, Stals H, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6 10.1038/msb.2010.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Searle I, He YH, Turck F, Vincent C, Fornara F, Krober S, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Gene Dev. 2006;20(7):898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, et al. miR824-Regulated AGAMOUS-LIKE16 Contributes to Flowering Time Repression in Arabidopsis. Plant Cell. 2014;26(5):2024–37. 10.1105/tpc.114.124685 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan WH, Chen DJ, Smaczniak C, Engelhorn J, Liu HY, Yang WJ, et al. Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat Plants. 2018;4(9):681–9. 10.1038/s41477-018-0219-5 [DOI] [PubMed] [Google Scholar]
- 68.Hsu HT, Chen HM, Yang Z, Wang J, Lee NK, Burger A, et al. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science. 2015;348(6241):1372–6. 10.1126/science.aab1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boija A, Mannervik M. Initiation of diverse epigenetic states during nuclear programming of the Drosophila body plan. Proc Natl Acad Sci U S A. 2016;113(31):8735–40. 10.1073/pnas.1516450113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foo SM, Sun YJ, Lim BY, Ziukaite R, O’Brien K, Nien CY, et al. Zelda Potentiates Morphogen Activity by Increasing Chromatin Accessibility. Current Biology. 2014;24(12):1341–6. 10.1016/j.cub.2014.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–61. 10.1038/nrg2102 . [DOI] [PubMed] [Google Scholar]
- 72.Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. 10.1007/978-1-59745-196-3_6 . [DOI] [PubMed] [Google Scholar]
- 73.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. . [DOI] [PubMed] [Google Scholar]
- 74.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li G, Liu S, Wang J, He J, Huang H, Zhang Y, et al. ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. 2014;78(4):706–14. 10.1111/tpj.12499 . [DOI] [PubMed] [Google Scholar]
- 76.Richter R, Behringer C, Muller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010;24(18):2093–104. 10.1101/gad.594910 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torti S, Fornara F, Vincent C, Andres F, Nordstrom K, Gobel U, et al. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell. 2012;24(2):444–62. 10.1105/tpc.111.092791 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142(23):4168–79. 10.1242/dev.127613 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andres F, Romera-Branchat M, Martinez-Gallegos R, Patel V, Schneeberger K, Jang S, et al. Floral Induction in Arabidopsis by FLOWERING LOCUS T Requires Direct Repression of BLADE-ON-PETIOLE Genes by the Homeodomain Protein PENNYWISE. Plant Physiol. 2015;169(3):2187–99. 10.1104/pp.15.00960 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Phylogenetic tree for B3 type gene family based on ClustalW alignment with a bootstrap value of 1.000 replicates. B) Immunoblot to test for cross-reactivity of SVP. Triangle indicates specific whereas asterisks indicate non-specific bands. Values on the left of the Western-blots indicated molecular weight (kDa). C) Transcript level of TFS1. Plants were grown for 2 weeks in short-days and then transferred to long-day. Values were scaled to set value of apices of 2 weeks short-day grown Col-FRI plants to 1. D) Increased expression of TFS1 in apices of Col-FRI plants when transferred to ambient growth condition after vernalisation. NV, non-vernalised; V, weeks vernalised in short-days (SD). E) Spatial pattern of expression of TFS1 assessed by in-situ hybridization after floral transition. DAS, days after sowing.
(TIF)
A to D) Comparison of tfs1 mutant plants and Columbia grown in either LDs (A) or in SDs (C). Leaf number of tfs1 plants compared to Columbia grown in LDs (B) and in SDs (D). Statistical significance was calculated using Student’s t-test; **P < 0.01. E) Representative gene model for TFS1 and T-DNA insertion used in this study. F) Total leaf number of tfs1-1 and tfs1-2 plants compared to Col. Statistical significance was calculated using Student’s t-test; *P < 0.05. G) Inflorescence of Col-FRI flc svp tfs1 plants. H) Floral structures of Col-FR flc svp tfs1 plants. Particular features are marked: blue arrow, misplaced floral organs; white arrow, ectopic ovules; yellow arrow, stigmatic papillae on leaf-like structures; red arrow, leaf-like anthers.
(TIF)
A) H3K27me3 levels are increased at TTS of TFS1 in Col-FRI flc svp. B and C) H3K4me3 levels are increased in Col-FRI flc svp (B) and Col-FRI clf and Col-FRI jmj14 (C) plants. Values were scaled to set highest value in Col-FRI flc svp to 1. D) Reduced TFS1 transcript levels in Col-FRI plants suppressed by clf mutation. E and F) Transcript levels of FLC (E) and SVP (F) in leaves and apices of Col-FRI clf plants. Numbers at x-axis indicate number of long-days (LD) for which plants were grown prior to harvest. G) Reduced TFS1 transcript levels in Col-FRI plants suppressed by jmj14 mutation. H and I) Transcript levels of FLC (H) and SVP (I) in leaves and apices of Col-FRI jmj14 plants. Numbers at x-axis indicate number of long-days (LD) for which plants were grown prior to harvest. Statistical significance was calculated using Student’s t-test; *P < 0.05, **P < 0.01, n.s. P > 0.05.
(TIF)
A) SOC1:GR-induced transcriptional activation of TFS1. B and C) Top: ChIP-qPCR analysis of SOC1 (B) and SVP (C) on CArG-boxes at TFS1 in 35S::SOC1:GR soc1 plants after DEX treatment. Bottom: Diagram of TFS1 locus with CArG-boxes and amplicons used for ChIP-qPCR. Red and orange triangles indicate CArG-boxes in TFS1. Green and red lines represent amplicons used for ChIP-qPCR study. D) ChIP-qPCR analysis of H3K27me3 at TFS1 in 35S::SOC1:GR soc1 plants after DEX treatment. B to D, values were scaled to set value of first primers at 20min after DEX treatment in 35S::SOC1:GR soc1 to 1. GB, indicates Gene Body amplicon used for ChIP-qPCR. Statistical significance was calculated against mock treatment using Student’s t-test; *P < 0.01, n.s. P > 0.05. Representative confocal images of independent transformants of E) TFS1:9AV, F) TFS1:9AV mCArGII and G) TFS1:9AV mCArGI+II. H) Abundance of TFS1 mRNA in indicated transgenic plants as well as in Col and tfs1 controls.
(TIF)
A and B) Representative photograph (A) and total leaf number (B) of TFS1::TFS1:9AV in tfs1. C and D) Representative photograph (C) and total leaf number (D) of TFS1::TFS1:9AV mCArGII in tfs1. E and F) Representative photograph (E) and total leaf number (F) of TFS1::TFS1:9AV mCArGII in soc1. G and H) Representative photograph (G) and total leaf number (H) of TFS1::TFS1:9AV mCArGI+II in tfs1. In each case, transformants are shown in the same order in the representative photograph as in the leaf number plot. Statistical significance was calculated using Student’s t-test; *P < 0.1, **P < 0.05, ***P < 0.01, n.s. P > 0.05.
(TIF)
A) Top view of TFS1:9xAla-Venus (yellow) expression at the apical meristem. DAPI (blue) serves to highlight individual cells. B) Side view of TFS1:9xAla-Venus (yellow) localisation at the shoot apical meristem. C to G) TFS1:9xAla-Venus localises to the nucleus of sepal cells (C), in young flowers and at the base of pedicels (D) and in flowers of stage stage 3 (E), stage 2 (F) and stage 4 (G) after LMB treatment.
(TIF)
A) ChIP-qPCR for rSPL9 binding to putative SPL-binding sites at TFS1 locus. Statistical significance was calculated against the respective amplicon of 35S::YFP:GFP. B) ChIP-qPCR for temporal binding of SPL9:GR at TFS1 after DEX treatment. Statistical significance was calculated against the 20min DEX treatment. C to E) ChIP-qPCR for MED18 (C), RNAPII (D) and H3K4me3 (E) at TFS1 locus. Plants were grown for 15 LD and harvested at ZT8. F) Spatial pattern of expression of TFS1 assessed by in-situ hybridization during floral transition in Col and GFP:rSPL9. Plants were grown for 2 weeks in SD and then transferred to permissive LD. G) Confocal images of TFS1:9AV mGTACa1/a2 mutant transgenic lines. Statistical significance was calculated using Student’s t-test; *P < 0.05, **P < 0.01, n.s. P > 0.05.
(TIF)
A and B) TFS1 transcript abundance in short-days is increased by SPL and SOC1 (A) and REF6 (B). Data are shown for 3–8 weeks after germination and values were scaled to set value of 3 weeks SD-grown Col plants to 1. C) Reduced expression of TFS1 in ga1 under LD conditions. D) Spatio-temporal expression pattern of TFS1 assessed by in situ hybridization in LD. E) TFS1 mRNA levels are reduced in apices of spl mutants during floral transition. Values were scaled to set value of 7 LD of Col to 1. F to H) Reduced RNAPII (F) and H3K4me3 (G) and increased H3K27me3 (H) levels at TFS1 in spl mutants. Values were scaled to highest value in Col (F and G) and spl9 spl15 (H) to 1. In A to H, Plants were harvested at ZT8.
(TIF)
A to C) WDR5 levels at TFS1 depend on SOC1 (A), BRM (B) and SPL (C). D to F) H2A.Z incorporation at TFS1 depends on SOC1 (D) and BRM (E) but not on SPL (F). ChIP-PCR was performed on 15 LD grown plants that were harvested at ZT8 (A to F).
(TIF)
A and B) 5-weeks old long-day grown GFP:rSPL9 brm plants display altered plant architecture (A) and partial transformation of flowers into carpelloid structures (B). C) GFP Western analysis to detect GFP:SPL9 in Col and brm. Values on the left of the Western-blots indicated molecular weight (kDa). D and E) Comparative expression analysis of marker genes expressed in leaves (D) and apices (E) of Col, GFP:rSPL9 and GFP:rSPL9 brm plants. X-axis indicates number of long-days (LD) for which plants were grown prior to harvest.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The underlying numerical data for the figures has been submitted to the Dryad database. (DOI: 10.5061/dryad.b0j2td6)