Abstract
Early ripening apples are usually used for fresh marketing because of short storage life, although they are with high acid and low sugar contents. Understanding the malate metabolism in fleshy fruit and underpinning process during ripening is crucial for particular crop improvement where acidity is a concern for direct consumption or further processing. In this research, a traditional Chinese apple cultivar ‘Hongyu’, which belongs to early ripening apple cultivar, were freshly harvested at commercial maturity stage (120 Days after full bloom) and used for different storage temperature (4°C, 20°C) and UV-C treatment (following storage at 20°C after treatment). Simple sugars (glucose, sucrose, and fructose) and organic acids (malic, and oxalic) were assessed after 14 d of storage. Compared to fruits stored at 20°C, the malate content in fruits stored at 4°C significantly higher, while it was decreased significantly in UV-C treated fruits stored at 20°C after 14 d of storage. The sugar content was almost similar throughout the UV-C-treated fruits and fruits stored at different temperature. The higher ratios of total sugars to total organic acids in UV-C treated fruits after 14 d suggest that UV-C treatment has the potential to improve the taste of early ripening apple cultivars. Considering the significant difference in malate the samples at 14 d of storage were subjected for RNA-seq analysis. Transcriptome analysis revealed that the phenomena underlying this change were governed by metabolism of malate by the regulation of NADP-malic enzyme (NADP-ME) and phosphoenolpyruvate carboxylase kinase (PEPCK) in apple during postharvest storage. This transcriptome profiling results have specified the transcript regulation of malate metabolism and lead to possible taste improvement without affecting the other fruit quality attributes.
Introduction
Apple (Malus domestica Borkh.) is an economically important and popularly grown fruit crop all over the world due to high nutritive value, good taste and availability throughout the year. The postharvest storage period varied among different cultivars and different harvesting time [1]. In general, early harvested apple cultivars such as ‘Gala’, ‘Royal Gala’ tended to be relatively less suitable for the long-term cold storage, comparing the late harvested apple cultivars such as ‘Fuji’, ‘Red Delicious’, ‘Golden Delicious’ [2,3,4]. Therefore, summer apple cultivars or early harvested apple cultivars have been in sale for fresh market but not for long-term storage [1]. ‘Granny Smith’ apples, which were belonging to the late ripening cultivar, can be stored for up to seven months under the cold storage condition, while most of the early ripening apples can be stored for about one and a half months under cold storage [5].
Occurrence of definite organic acids determine the organoleptic properties and overall acceptability of fruits either by promoting or reducing the fruit taste [6]. Therefore, acidity has been considered as an important factor of fruit harvesting due to its direct influence on fruit quality particularly where acidity is crucial for further processing. Although fruit flavor largely depends on a multifaceted interaction of several metabolites, the ratio of sugars and acids are considered as a key determinant factor of fruit taste and sensory evaluation [7,8]. When at the expected level, this parameter will lead to repeated purchases and consumer loyalty [9]. In apple fruit, the primary soluble sugars are glucose, fructose, and sucrose, and the predominant organic acid is malate [10]. The early ripening apples are usually of high acidity and low sugar contents, which results in a low ratio of total sugars to total organic acids and reduce the market preference for fresh consumption. Thus, improvement of fruit taste for better consumption of early ripening apple is needed. Storage temperature is one of the most important factors affecting fruit storability [11] by regulating the rate of associated physiological and biochemical process. Many studies elucidated a prominent influence on the variation of organic acids and sugars in post-harvest fruit quality and shelf life [12–14].
Ultraviolet-C treatment (at hormetic dose) is a non-thermal, non-ionizing and safest practice inducing beneficial responses in fruits and vegetables and considered as an effective measure for maintaining postharvest quality [15]. Low-temperature storage and controlled atmosphere storage of apple fruit are common practices in recent years. However, UV-C treatment on apple has been rarely practiced, although it has promising effects on the extension of storage life of fresh horticultural crops and vegetables [16]. Previous studies stated the effectiveness of UV-C treatment on postharvest produce mainly by delaying ripening and senescence, maintaining relatively higher fruit firmness, flavonoid biosynthesis (e.g., anthocyanin) and increasing antioxidant ability and defense molecules [15,17]. Exposure to different doses of UV-C irradiations has reduced the titratable acidity of kiwifruits during cold-storage [18]. Furthermore, UV-C treatment lowered the amount of reducing sugar accumulation during low-temperature storage in potato tubers [19]. The accumulation and degradation of particular organic acid in mesocarp cell are modulated by both genetic and environmental factor. Several transcriptomics [20–22], metabolomics [22,23], proteomics [23,24], and quantitative trait loci (QTLs) [25,26] studies have been conducted recent years which described some of the mechanisms that control fruit acidity at the cellular level. As stated above, UV-C treatment has eminent effects on several quality aspects in fresh produce, it must regulate the induction of primary metabolites and interactive compounds. However, the underlying regulation of UV-C treatment on the malate metabolism in early ripening apple fruit during postharvest storage is still indistinct.
Considering ‘Hongyu’ apple is one of the traditional Chinese variety with high acidity and also an early ripening cultivar with shorter ripening period, it could be a good model to study apple fruit acidity modulation during post-harvest storage and find out the possible alternative treatment and storage condition which promote the fruit taste by regulating the fruit acidity of early ripening apple.
Materials and methods
Fruit materials and treatments
The ‘Hongyu’ apple fruits (Malus domestica Borkh.) were randomly harvested from different trees and different plant rows at commercial maturity stage (120 Dayes After Full Bloom) from an orchard located in Changping district, Beijing, P.R. China. The maturity stage was considered as the predefined date by the appropriate orchard management. The growing conditions of apple trees and the management practices during fruits development and ripening were controlled and maintained by the corresponding orchard management. Fruits of uniform size, appearance, and without any physical injuries or infections were selected and randomly divided into three groups (at least 15 fruits in each group). The fruits of different groups were subjected to the following postharvest treatments: (1) Cold storage: the fruits were stored at 4°C, 85–95% relative humidity (RH) throughout. (2) Room temperature storage: the fruits were stored at 20°C, 85–95% RH throughout. (3) UV-C treatment (storage at 20°C after treatment): A low-pressure mercury (LPM) UV light reactor was assembled using a UV tube (Ruisente Ltd., Tianjin, China); it delivered UV photons at 254 nm. The UV-C dose rate was determined by a digital ILT1700 radiometer (10 Technology Drive, Peabody, MA, USA). The different UV-C illumination doses were obtained by the exposure at a fixed distance. The apples were placed 20 cm below the lamps, and irradiated for 9.0 kJ m-2, and stored at 20°C after treatment, with a successive relative humidity of 85–95% throughout the storage period. The fruits were sampled at 14 d of storage. Fifteen fruits were sampled for individual treatment which has been divided into three biological replicates. Fruits were transported shortly to the laboratory, flesh sample was taken and immediately frozen by using liquid nitrogen and stored at -80°C until futher analysis.
Organic acids and sugars measurement
The contents of organic acids and soluble carbohydrates were measured by following a modified method previously described by Lin et al. [19]. Briefly, a total of 1 g sample powder which was obtained by using high throughput tissue lapping apparatus (QM100, Wuzhou Ding Chuang technology, Beijing, China) homogenized with 4 mL of 80% ethanol. The mixture was extracted with ultra-sonication for 30 min at room temperature and centrifuged at 10,000 g for 15 min. Aliquots of 1 mL of the upper phase were dried under pure nitrogen. The residue was dissolved in 1 mL ddH2O and filtered through a membrane with 0.22 μm pore size. For sugar analysis, a volume of 10 μL for each sample was injected into the ion chromatograph (ICS-3000, Dionex, Sunnyvale, CA, USA) fitted with Carbo PacTMPA20 column (3 mm × 150 mm). The column temperature was 35°C, and the flow rate was 0.5 mL min-1. The gradient elution buffer was used as follows: A, ddH2O; B, 250 mmol L-1 NaOH; equal gradient of 92.5% A and 7.5% B were used for elution. A pulsed amperometric detector with gold electrode was used. For organic acid analysis, a volume of 25 μL for each sample was injected into the ion chromatograph (ICS-3000, Dionex, Sunnyvale, CA, USA) fitted with IonPac AS11-HC column (4 mm × 250 mm) (Dionex, Sunnyvale, CA, USA). The column temperature was 30°C, and the flow rate was 1 mL min-1. The gradient elution procedure was 0.8 mmol L-1 KOH, 0–12 min; 0.8–34 mmol L-1 KOH, 12–40 min; 34 mmol L-1 KOH, 40–50 min. The electrical conductivity detector was used for detection. Identification of the substances was according to the retention time (RT) of standard compounds. The contents of glucose, fructose, sucrose, malate, and oxalate were calculated by comparison with standard curves.
Total RNA extraction
Total RNA was extracted using a modified CTAB method [27]. Quality of RNA was assesed by using gel electrophoresis. Purity of RNA detected by Nanodrop (OD260/280 ratio) spectrophotometer (IMPLEN, CA, USA). Qubit 2.0 Flurometer (Life Technologies, CA, USA) was used for accurate quantification of RNA concentration. RNA integrity was assessed by Agilent 2100 (Agilent Technologies, CA, USA).
Library construction and RNA-Seq analysis
The RNA-Seq analysis was performed by following previously used methods described elaborately by onik et al. [28]. Total RNA from 4°C, 20°C post-harvest storage and UV-C treated fruits were pooled before library preparation and an equimolar quantity of total RNA from each sample was considered as one pool. Prior to cDNA library construction, poly-T oligo-attached magnetic beads were used to purify the mRNA, which was added to a fragmentation buffer to break mRNA into short segments of approximately 150–200 bp followed by cDNA synthesis using random hexamers and reverse transcriptase. Then buffer, dNTPs, RNase H and DNA polymerase I were added to synthesize the second strand cDNA. Both ends of sequencing adapters were ligated after terminal repairing of double-stranded cDNA fragments. The AMPure XP system (Beckman Coulter, Beverly, MA, USA) was used to purify the final cDNA library and selectively enriched by PCR enrichment and library quality was assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) on an Illumina HiSeq PE150 platform. Three replications were used in each group for the precision of results. To obtain high-quality clean reads, raw reads were further filtered by removing adapter sequences, poly-N reads, and low-quality sequencesand all the clean reads were mapped to the Apple reference genome (https://www.rosaceae.org/organism/Malus/x-domestica v1.0) using TopHat [29]. Differentially expressed genes (DEGs) among the treatment groups werw identified using previously reported method [30], and P-values were adjusted to eliminate the false discovery rate (FDR) via multiple testing [31]. DESeq package (v1.10.1) was used and a corrected P-value threshold of 0.05 (using the Benjamini and Hochberg method) and |log2foldchang|>1 were considered for filtering the DEGs value. For gene expression quantification, the read numbers mapped to each gene were counted using HTSeq v0.6.1 and then normalized to FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) [30]. For gene ontology (GO) enrichment analysis of differentially expressed genes, GOseq (v2.12) was used [32]. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed using the KOBAS (v 2.0) software (Center for Bioinformatics, Peking University) [33].
Quantitative real-time PCR (qRT-PCR)
First strand cDNA was synthesized from 1 μg RNA, after digestion of genomic DNA, using Bio-Rad iScript cDNA Synthesis Kit. qRT-PCR was performed using Power SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA, USA) on an ABI 7500 instrument (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). The PCR reactetion protocol initiated by 5 min at 95°C, then followed by 45 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 15 s, and completed with a melting curve analysis program. No-template controls and melting curve analyses were included in every reaction. The β-Actin was used as an internal control. The comparative CT method (2-ΔΔCT method) was used to analyze the relative expression levels of the different genes [34]. Primers used in this analysis were listed in S1 Table.
Statistical analysis
Figures were drawn using Origin 8.6 (Microcal Software Inc., Northampton, MA, USA). The least significant difference (LSD) at 0.05 level was calculated by DPS 7.05 (Zhejiang University, Hangzhou, China).
Results
Variation of organic acids and sugars contents in response to temperature and UV-C treatment in apple
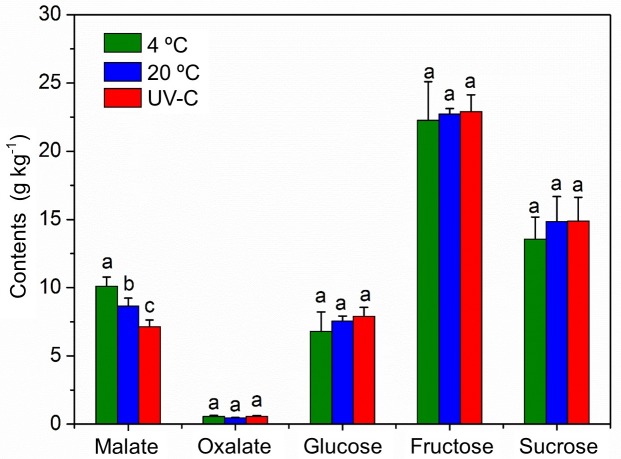
Among the primary organic acids and sugars present in apple fruit, malate, oxalate, glucose, sucrose and fructose content in different treatment group (4°C, 20°C, and UV-C treatment following 20°C storage after treatment) were measured. Results indicated that malate and fructose was the most prominent in content. The oxalate contents were below 0.5 g kg-1, while the malate contents varied among 7 to 12 g kg-1. Compared to the fruits stored at 4°C, the malate content in fruits stored at 20°C was significantly decreased at 14 d of storage. According to the results, the content of glucose, fructose and sucrose were not significantly varied among the different treatments. However, glucose and sucrose were slightly higher in 20°C stored fruit and UV-C treated fruit comparing 4°C temperature stored fruit. Considering the significant variation of malic acid content, all the samples of different temperature storage and UV-C treated fruits at 14 d of storage was considered for RNA-seq analysis. The variation of acids and sugars content was shown in Fig 1.
Fig 1. Variation of sugars (glucose, fructose and sucrose) and organic acids (oxalate and malate) in apple fruit under different temperature storage and postharvest UV-C treatment.
The error bar represents the standard error of the mean value of three biological replicates. Same letters indicated the mean values are not statistically different according to the least significant difference test (p < 0.05).
RNA-Seq libraries and the differently expressed genes (DEGs) analysis
Three replications of each treatment were used for RNA-Seq analysis to investigate the molecular mechanism underlying malate metabolism in apple fruit during postharvest storage. After removing adapter reads, low-quality regions, and possible contamination, more than six clean gigabases with a GC percentage above 46.84% and a Q20 percentage above 95.81% were obtained. The proportion of clean reads in the apple transcriptome libraries that mapped to the apple reference genome ranged from 87.87% to 93.39%, while the adjusted proportion ranged from 73.35% to 78.85% (Table 1).
Table 1. Quality evaluation of RNA-Seq data.
| Groups | Replicates | Clean bases (G) | Error rate (%) | Q20 (%) | GC content (%) | Total mapped (%) | Uniquely mapped (%) |
|---|---|---|---|---|---|---|---|
| 4°C | A | 7.80 | 0.02 | 97.42 | 46.84 | 94.31 | 78.85 |
| B | 7.33 | 0.02 | 96.5 | 47.1 | 93.39 | 77.65 | |
| C | 9.19 | 0.02 | 96.55 | 47.12 | 93.29 | 77.51 | |
| 20°C | A | 8.47 | 0.02 | 95.81 | 48.20 | 90.05 | 75.19 |
| B | 7.31 | 0.02 | 95.43 | 47.74 | 87.87 | 73.35 | |
| C | 7.37 | 0.02 | 97.30 | 47.20 | 92.36 | 76.89 | |
| UV-C | A | 7.79 | 0.02 | 97.19 | 47.63 | 92.91 | 77.48 |
| B | 7.42 | 0.02 | 96.53 | 47.91 | 90.92 | 75.53 | |
| C | 6.55 | 0.02 | 96.87 | 47.85 | 89.39 | 74.39 |
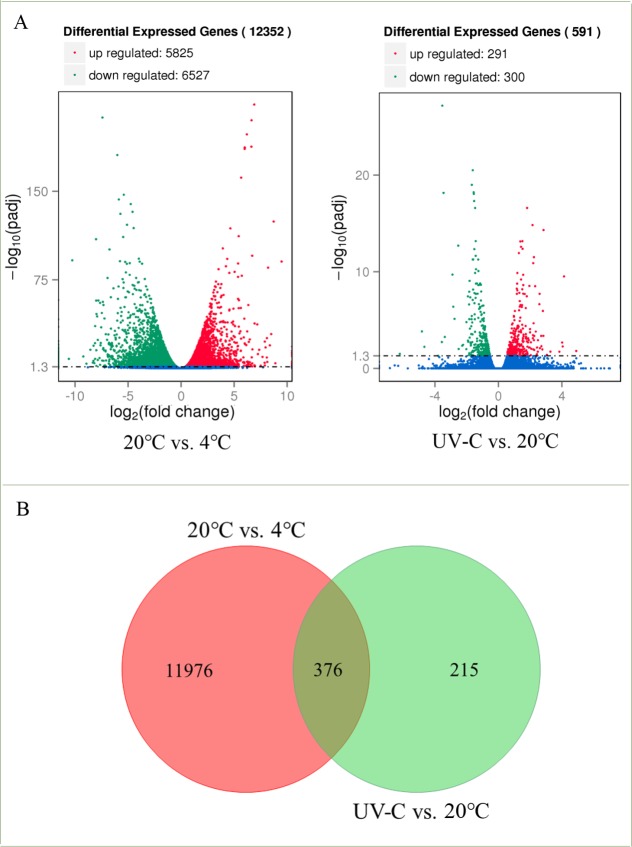
A total of 70,163 unique genes had perfect matches with apple genes, including 6,648 new predicted genes (S2 Table). We compared the transcript levels of each gene among different samples, 12,352 and 591 DEGs were found in the clusters of 20°C vs. 4°C and UV-C vs. 20°C, respectively (Fig 2A). 5,825 up-regulated and 6,527 down-regulated DEGs were observed in 20°C vs. 4°C cluster, while 291 up-regulated and 300 down-regulated DEGs were found in UV-C vs. 20°C cluster. Venn diagram showed that 376 DEGs were overlapped between clusters of 20°C vs. 4 ºC and UV-C vs. 20°C (Fig 2B). 11,976 and 215 DEGs were exclusively shown in 20°C vs. 4°C cluster and UV-C vs. 20°C cluster, respectively.
Fig 2. Volcano plots and venn diagrams of DEGs.
DEGs of 20°C vs. 4°C and UV-C vs. 20°C displayed by volcano plots (A). The horizontal and vertical coordinates indicate the fold change differences and adjusted p-values; respectively for differential expression of genes. The up-regulated genes are represented by red dots, and the down-regulated genes are represented by green dots. Genes without significant differences are indicated by blue dots. Venn diagrams showing the overlap in DEGs between 20°C vs. 4°C and UV-C vs. 20°C groups (B).
GO classification of the DEGs in apple during postharvest storage
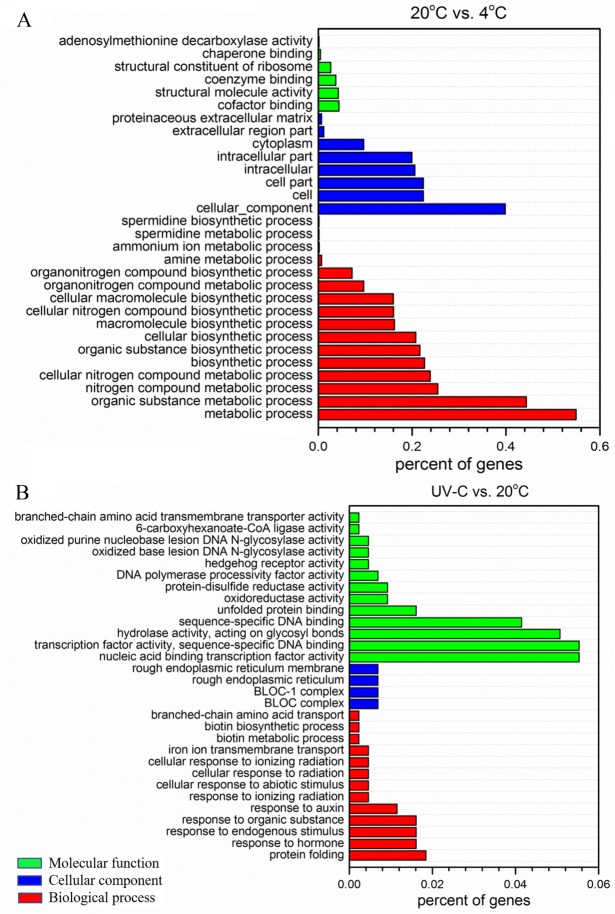
We further assigned gene ontology (GO) terms to annotated DEGs in apple during postharvest storage (S3 Table). In 20°C vs. 4°C cluster, the largest terms within the molecular function category were ‘cofactor binding’, ‘structural molecule activity’, and ‘coenzyme binding’. Within the cellular component category, ‘cell’, ‘cell part’ and ‘intracellular part’ were the three most abundant terms. Within the biological process category, the most highly represented terms were ‘metabolic process’, ‘organic substance metabolic process’, and ‘nitrogen compound metabolic process’ (Fig 3A). In UV-C vs. 20°C cluster, the largest terms within the molecular function category were ‘nucleic acid binding transcription factor activity’, ‘transcription factor activity, sequence-specific DNA binding’, and ‘hydrolase activity, acting on glycosyl bonds’. Within the cellular component category, ‘rough endoplasmic reticulum membrane’ and ‘BLOC-1 complex’ were the two most abundant terms. Within the biological process category, the most highly represented terms were ‘protein folding’, ‘response to hormone’, ‘response to endogenous stimulus’, and ‘response to organic substance’ (Fig 3B).
Fig 3.
Classification of GO terms concerning differently expressed genes in apple under different temperature (A) and UV-C treatment (B) during postharvest storage. The GO terms were classified into three categories, including molecular function, cellular component, and biological process. Top 30 enriched GO terms were exhibited in each cluster.
KEGG pathway enrichment of the DEGs in apple during postharvest storage
To identify the biological pathways activated in selected apple fruits, we mapped the annotated sequences to the reference pathways in the KEGG database (S4 Table) [35]. The top 20 most enriched pathways between the two groups were shown in Fig 4. In 20°C vs. 4°C cluster, the ‘carbon metabolism’, ‘fatty acid biosynthesis’, ‘pyruvate metabolism’, ‘glycolysis’ and ‘biosynthesis of amino acids’ were the five most enriched pathways (Fig 4A). In UV-C vs. 20°C cluster, the ‘plant hormone signal transduction’, ‘protein processing in endoplasmic reticulum’, ‘Glycosaminoglycan degradation’, ‘Circadian rhythm’ and ‘starch and sucrose metabolism’ were the five most enriched pathways (Fig 4B).
Fig 4.
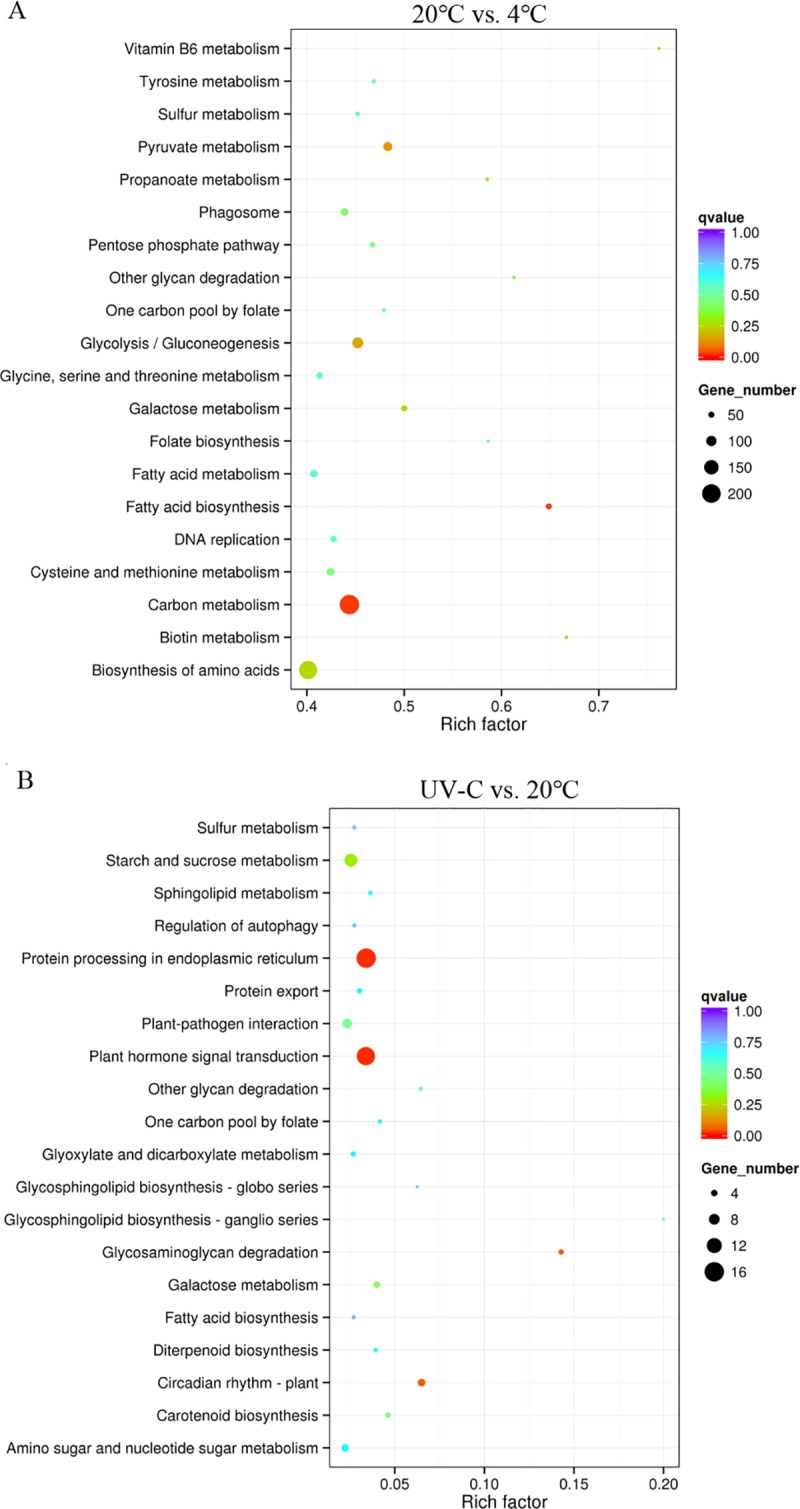
KEGG pathway enrichment concerning differently expressed genes in apple under different temperature (A) and UV-C treatment (B) during postharvest storage. Top 20 enriched KEGG pathways were exhibited in each cluster. Rich factor is the ratio of DEGs counts to this pathway in the annotated genes counts. The more the q-value is close to zero, the more significant is the enrichment.
Malate metabolism in response to temperature and UV-C treatment in apple during postharvest storage
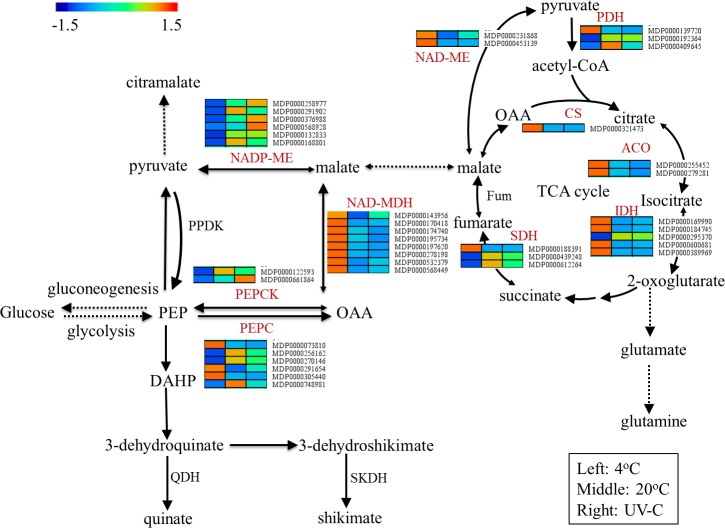
According to our results, the malate content was significantly influenced by cold-storage and UV-C treatment. Compared to the fruits stored at room temperature, the malate content was significantly higher in the fruits stored at cold-storage. The expression of tricarboxylic acid cycle (TCA) related genes, including citrate synthase (CS), aconitase (ACO), isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), and the malate synthesis-related genes, including phosphoenolpyruvate carboxylase (PEPC) and NAD-malate dehydrogenase (NAD-MDH) were up-regulated, whereas a significant down-regulation was showed by malate degradation related genes, NADP-malic enzyme (NADP-ME) and phosphoenolpyruvate carboxylase kinase (PEPCK). On the other hand, a significant declination of malate content in UV-C treated fruits was observed in contrast to the fruits stored at room temperature. Consequently, no significant difference was observed in the expression of TCA cycle and malate synthesis-related genes. The expression of NADP-ME and PEPCK were significantly induced by UV-C treatment, which might be the key genes regulating malate degradation in UV-C treated apples during postharvest storage (Table 2).
Table 2. Differently expressed genes related to malate metabolism in apple under different temperature and UV-C treatment during postharvest storage.
| Enzymes | Gene ID | FPKM ratio (4°C/20°C) |
FPKM ratio (UV-C/20°C) |
|---|---|---|---|
| NADP-malic enzyme (NADP-ME) | MDP0000258977 | 0.52 | 1.31 |
| MDP0000291902 | 0.40 | 0.75 | |
| MDP0000376988 | 0.45 | 1.42 | |
| MDP0000568928 | 0.45 | 2.33 | |
| MDP0000132833 | 0.66 | 1.01 | |
| MDP0000168801 | 0.43 | 0.80 | |
| NAD-malate dehydrogenase (NAD-MDH) | MDP0000143956 | 2.46 | 1.75 |
| MDP0000170418 | 2.29 | 1.11 | |
| MDP0000174740 | 1.62 | 0.89 | |
| MDP0000195734 | 1.97 | 0.77 | |
| MDP0000197620 | 1.86 | 0.93 | |
| MDP0000278198 | 5.56 | 0.76 | |
| MDP0000532379 | 3.54 | 1.48 | |
| MDP0000568449 | 2.85 | 1.24 | |
| Phosphoenolpyruvate carboxylase (PEPC) | MDP0000073810 | 5.16 | 0.74 |
| MDP0000256162 | 0.47 | 0.80 | |
| MDP0000270146 | 0.34 | 0.81 | |
| MDP0000291654 | 2.56 | 1.60 | |
| MDP0000305440 | 5.65 | 0.72 | |
| MDP0000748981 | 0.71 | 0.80 | |
| Phosphoenolpyruvate carboxylase kinase (PEPCK) | MDP0000122593 | 0.08 | 0.70 |
| MDP0000661864 | 0.16 | 2.44 | |
| NAD-malic enzyme (NAD-ME) | MDP0000231868 | 2.13 | 1.45 |
| MDP0000453139 | 2.02 | 1.04 | |
| Pyruvate dehydrogenase (PDH) | MDP0000139720 | 2.12 | 0.90 |
| MDP0000192364 | 0.56 | 0.98 | |
| MDP0000409645 | 0.46 | 0.65 | |
| Citrate synthase (CS) | MDP0000321473 | 2.45 | 0.98 |
| Aconitase (ACO) | MDP0000255452 | 2.17 | 0.83 |
| MDP0000279281 | 2.87 | 0.69 | |
| Isocitrate dehydrogenase (IDH) | MDP0000169990 | 5.79 | 1.29 |
| MDP0000184745 | 4.52 | 1.25 | |
| MDP0000295370 | 0.59 | 0.97 | |
| MDP0000600681 | 5.08 | 1.25 | |
| MDP0000389969 | 2.65 | 0.98 | |
| Succinate dehydrogenase (SDH) | MDP0000188391 | 2.54 | 1.05 |
| MDP0000439248 | 0.44 | 0.86 | |
| MDP0000612264 | 0.43 | 0.80 |
Briefly, higher expression of TCA cycle and malate synthesis-related genes in the cold stored fruit (4°C) resulting in promotion of malate synthesis. On the other hand, higher degradation of malate was taken place by up-regulation of NADP-ME and PEPCK in UV-C treated fruits. The interactive expressions of the genes in primary pathway of malate metabolism regarding different storage temperature and postharvest UV-C treatment were shown in Fig 5.
Fig 5. The regulation of malate metabolism pathway in apple under different temperature and UV-C treatments during postharvest storage.
The rectangles from left to right represent 4°C, 20°C and UV-C treated fruits. ACO, Aconitase; CS, Citrate synthesis; IDH, Isocitrate dehydrogenase; PEPC, Phosphoenolpyruvate carboxylase; PEPCK, Phosphoenolpyruvate carboxylase kinase; PDH, Pyruvate dehydrogenase; NAD-MDH, NAD-malate dehydrogenase; NADP-ME, NADP- malic enzyme; PPDK, Pyruvate orthophosphate dikinase; SDH, Succinate dehydrogenase; SKDH, Shikimate dehydrogenase.
Quantitative Real-Time PCR (qRT-PCR) validation of DEGs
To validate the credibility of the gene expression profiles obtained from RNA-seq analysis, we selected the DEGs involved in the malate metabolism pathway for qRT-PCR using specific primers to confirm the gene expression changes detected in the transcriptome analysis. The exact fold change of DEGs at several data points varied between RNA-seq and qPCR methods. However, the overall expression trends were strongly consistent (Pearson correlation coefficients R2 = 0.9753) (Fig 6), confirming the reliability of the RNA-seq results.
Fig 6. Correlation analysis of relative expression levels determined by RNA-seq and qPCR on selected genes NADP-ME (MDP0000568928), NAD-MDH (MDP0000174740), PEPCK (MDP0000661864) and PEPC (MDP0000073810).
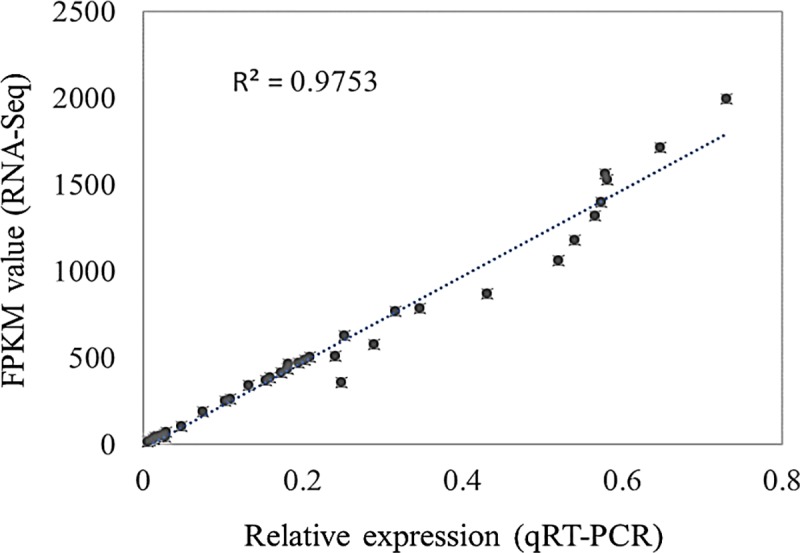
All qPCR reactions were carried out by using three independent biological replication and repeated three times. Relative expression levels were calculated from CT values according to the 2-ΔΔCT method. Actin was used as the reference gene for these qPCR experiments.
Discussion
Considering the health benefits associated with apple progressively understood through consumers, demand is increasing for cultivars with preferred quality features, such as size, texture, flavor and nutritional value [36]. For an apple cultivar, fruit quality characteristics are underlain by metabolite composition and contents at maturity and during postharvest storage. The content of predominant organic acid and metabolism also plays an important role in food processing [37] and quality [38]. The early ripening apples are mostly used for fresh marketing because of short storage life, but they are usually with high acidity and low sugar contents [5,39].
UV-C treatment has already been established as a positive regulator for postharvest preservation of several fruits and vegetables, such as tomato [40], strawberry [41], apple [42], mango [43] and peach [44] by inducing plant defense response, inhibition of particular microbes as bacteria, disease resistance gene regulation and modulating antioxidant capacity. It has been reported that, exposure to short wavelength UV-C (200–280 nm) irradiation at low (hormetic) doses, resulted in the enhancement of antioxidant capacity, anthocyanin content, and phenolic content in mangoes, edible mushrooms, strawberries, blueberries and grape [45]. UV-C irradiation in the range of 0.125–9 kJ m−2 showed beneficial effects including increased resistance to pathogens, delayed ripening and senescence, and increased the content of beneficial phytochemicals [15,46]. In general, our results showed significant differences between the UV-C treated fruits over low temperature stored fruits and room temperature stored fruits but the sugar contents were not significantly diverged between different storage conditions. The similar trend was observed in a previous study on the action of UV-C on ‘Trust’ tomato fruit at mature-green stage where the fruit acidity (pH and titratable acidity) was apparently more sensitive to the treatment, with a significantly lower acidity in UV-C treated fruit [47]. However, another study showed higher malic acid in UV-C treated tomato fruits at breaker stage which was reduced in UV-treated vine-ripe tomato after 15d of storage [48]. In addition, titratable acid content was comparatively higher in UV-C treated ‘Fuji’ apple than those of control fruit stored at 20°C [49]. Following different effects observed in different horticultural produce, important things must be taken into consideration to achieve successful implementation such as plant parts used for treatment, dose, cultivar specificity, developmental stages, harvesting date, pre-harvest factors and postharvest storage conditions [49].
The final malate content of fruit is generally determined by the net balance of acid synthesis, degradation, and compartmentation [50,51]. Malate biosynthesis occurs primarily in the cytosol and is catalyzed by PEPC and NAD-MDH [52]. Loss of acidity signifies decarboxylation associated with carboxylates which can occur with the conversion of tricarboxylates into dicarboxylates, but additionally through decarboxylation of the malate and oxaloacetic acid (OAA), leading to the actual degradation involving organic acids. Decarboxylation connected with OAA as well as malate enables the production of phosphoenolpyruvic acid (PEP), which may additionally originate from the activity with PEPCK, directed to the gluconeogenesis process [6]. Those findings supported by the study indicating the involvement of PEPCK in the dissimilation of malate of several fleshy fruits [24]. The results also revealed a potential association between PEPCK enzyme activity, and malate loss in raspberry, blueberry, and red currant fruits. Involvement of PEPCK was observed in the dissimilation of malate and possibly in the lack of malate in low acid apple cultivars [53]. Another study demonstrated continuous revenues of PEPCK in maize protoplasts through the ubiquitin protease pathway, may act for rapid reversal of PEPC phosphorylation, thus considered as another level of control for malate metabolism [54]. The instigation of PEP can also be taken place by the conversion of pyruvate through pyruvate orthophosphate dikinase (PPDK) activity and the required pyruvate for PPDK may be supplied through the carboxylation of malate by a cytosolic NADP-dependent malic enzyme (NADP-cytME) [6]. Involvement of NADP-cytME in the decrease in malate content during the ripening has been demonstrated in several fruit species [6,55]. NADP-cytME appears to be involved in the lack of malate in the ripe pulp of low acid apple cultivars [56]. Increasing the activity of NADP-ME enzymes has also been observed during grape berry ripening, indicating the functions of this enzyme in malate degradation [57]. Thus, the rapid decrease in malate content during fruit ripening has usually been attributed to its degradation by cytosolic NADP-ME [55].
The NADP-ME gene promoter is activated by different effectors, such as fungal elicitors, UV irradiation and wounding, and agents producing redox perturbations in bean [58,59]. NADP-ME activity was also increased by UV-B radiation in bean [60] and in maize [61,62]. Our results showed that the increased expression of NADP-ME played a significant role in malate metabolism during postharvest storage in UV-C treatment of apples, which provides further evidence that NADP-ME has a role in response to UV-C stress. As a primary contributor in fruit flavor development, maintaining the sugars and acids contents is of greatest interest to select a postharvest treatment. In general, the postharvest condition or treatment that promotes TSS: TA would have a positive effect on flavor development. Therefore, the significantly higher sugar: acid ratios in the UV-C treated fruit of this cultivar under the described conditions, UV-C treatment positively affected fruit quality. Although there are some homology and as well as deviation of our results with previous studies, the following trend of malate degradation through the UV-C treatment during postharvest storage at room temperature and stimulated response of the genes involved in the degradation of malate in fruit would lead to a better fruit quality of particular cultivar. The potential effects of UV-C hormesis should therefore be considered as a way to enhance quality without negatively impacting quality attributes of early ripening apple fruit. It may also be possible that UV treatment stimulates a class of compounds that could counteract the positive sugar acid ratio attribute. However, for successful implementation of UV-C treatment on early harvested apple fruit, the deviation of the current study must be taken into consideration for further study by adjusting the dose, storage parameters and cultivars of apple fruit.
Conclusions
Low temperature maintained significantly higher malate contents by up-regulating the expression of genes (NAD-MDH and PEPC) related to the TCA cycle and citrate synthesis and down-regulating the expression of NADP-ME and PEPCK. UV-C treatment significantly increased the ratio of total sugars to total acids compared with room temperature stored fruits, which was carried out by the up-regulation expressions of NADP-ME and PEPCK. This finding suggested UV-C as an effective measure to maintain and promote the fruit taste during post-harvest storage of early ripening apple cultivar at room temperature.
Supporting information
(PDF)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files. Raw sequencing datasets of the current study have been deposited in the NCBI Sequence Read Archive repository, with sample accession numbers SAMN10963045, SAMN10963046, SAMN10963047, SAMN10963072, SAMN10963073, SAMN10963074, SAMN10963323, SAMN10963324, SAMN10963325 under the following project (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA522961).
Funding Statement
This research was funded by National Key Research and Development Program of China (2016YFD0400101) and the Young Scientist’s Fund of National Natural Science Foundation of China (31601527).
References
- 1.Musacchi S, Serra S. Apple Fruit Quality: Overview on Pre-Harvest Factors. Sci Hortic (Amsterdam). 2018;234: 409–430. [Google Scholar]
- 2.Jan I, Rab A, Sajid M. Storage Performance of Apple Cultivars Harvested at Different Stages of Maturity. J Anim Plant Sci. 2012;22 (2): 438–447. [Google Scholar]
- 3.Ahmadi-Afzadi M, Tahir I, Nybom H. Impact of Harvesting Time and Fruit Firmness on the Tolerance to Fungal Storage Diseases in an Apple Germplasm Collection. Postharvest Biol Technol. 2013;82: 51–58. [Google Scholar]
- 4.Neri F, Gualanduzzi S, Brigati S. Effects of Harvest Maturity on Quality, Physiological and Pathological Disorders during Storage of “Gala” Apples. In Acta Horticulturae. 2005;682(682): 2069–2076. [Google Scholar]
- 5.Hajnajari H, Peyghambari A, Koochaki MF. Investigation on Cold Storage Capacity of Early and Mid Ripening Apple Cultivars of Iran. Acta Hort. 2010;877: 905–910. [Google Scholar]
- 6.Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of Malate Metabolism in Grape Berry and Other Developing Fruits. Phytochemistry. 2009;70: 1329–1344. 10.1016/j.phytochem.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 7.Kader AA. Flavor Quality of Fruits and Vegetables. J. Sci. Food Agric. 2008;88(11): 1863–1868. [Google Scholar]
- 8.Beckles DM. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biology and Technology. 2012;63: 129–140. [Google Scholar]
- 9.Casals J, Pascual L, Cañizares J, Cebolla-Cornejo J., Casañas F, Nuez F. The risks of success in quality vegetable markets: possible genetic erosion in Marmande tomatoes (Solanum lycopersicum L.) and consumer dissatisfaction.Scientia Horticulturae. 2011;130: 78–84. [Google Scholar]
- 10.Jing C, Ma C, Zhang J, Jing S, Jiang X, Yang Y, Zhao Z. Effect of Debagging Time on Pigment Patterns in the Peel and Sugar and Organic Acid Contents in the Pulp of “Golden Delicious” and “Qinguan” Apple Fruit at Mid and Late Stages of Development. PLoS One. 2016;11(10): e0165050 10.1371/journal.pone.0165050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, Kader AA. Preharvest and Postharvest Factors Influencing Vitamin C Content of Horticultural Crops. Postharvest Biol Technol. 2000;20(3): 207–220. [Google Scholar]
- 12.Chen M, Jiang Q, Yin XR, Lin Q, Chen JY, Allan AC, Xu CJ, Chen KS. Effect of Hot Air Treatment on Organic Acid- and Sugar-Metabolism in Ponkan (Citrus Reticulata) Fruit. Sci Hortic (Amsterdam). 2012;147:118–125. [Google Scholar]
- 13.Lin Q, Qian J, Zhao C, Wang D, Liu C, Wang Z, Sun C, Chen K. Low Temperature Induced Changes in Citrate Metabolism in Ponkan (Citrus Reticulata Blanco Cv. Ponkan) Fruit during Maturation. PLoS One. 2016;11(6): e0156703 10.1371/journal.pone.0156703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcilla A, Zarzo M, del Rio MA. Effect of Storage Temperature on the Flavour of Citrus Fruit. Spanish J Agric Res. 2006;4(4): 336–344. [Google Scholar]
- 15.Charles MT, Arul J. UV Treatment of Fresh Fruits and Vegetables for Improved Quality: A Status Report. Stewart Postharvest Rev. 2007;3(3): 1–8. [Google Scholar]
- 16.Ben-Yehoshua S. Effects of Postharvest Heat and UV Applications on Decay, Chilling Injury and Resistance against Pathogens of Citrus and Other Fruits and Vegetables. In Acta Horticulturae. 2003;599: 159–173. [Google Scholar]
- 17.Artés F, Gómez P, Aguayo E, Escalona V, Artés-Hernández F. Sustainable Sanitation Techniques for Keeping Quality and Safety of Fresh-Cut Plant Commodities. Postharvest Biol Technol. 2009;51(3): 287–296. [Google Scholar]
- 18.Bal E, Kok D. Effects of UV-C Treatment on Kiwifruit Quality during the Storage Period. J Cent Eur Agric. 2009;10(4): 375–382. [Google Scholar]
- 19.Lin Q, Xie Y, Liu W, Zhang J, Cheng S, Xie X, Guan W, Wang Z. UV-C Treatment on Physiological Response of Potato (Solanum Tuberosum L.) during Low Temperature Storage. J Food Sci Technol. 2017;54(1): 55–61. 10.1007/s13197-016-2433-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etienne C, Moing A, Dirlewanger E, Raymond P, Monet R, Rothan C. Isolation and Characterization of Six Peach CDNAs Encoding Key Proteins in Organic Acid Metabolism and Solute Accumulation: Involvement in Regulating Peach Fruit Acidity. Physiol Plant. 2002;114(2): 259–270. [DOI] [PubMed] [Google Scholar]
- 21.Cercós M, Soler G, Iglesias DJ, Gadea J, Forment J, Talón M. Global Analysis of Gene Expression during Development and Ripening of Citrus Fruit Flesh. A Proposed Mechanism for Citric Acid Utilization. Plant Mol Biol. 2006;62(4–5): 513–527. 10.1007/s11103-006-9037-7 [DOI] [PubMed] [Google Scholar]
- 22.Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. Transcriptomic and Metabolite Analyses of Cabernet Sauvignon Grape Berry Development. BMC Genomics 2007;8: 429 10.1186/1471-2164-8-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz E, Boo KH, Kim HY, Eigenheer RA, Phinney BS, Shulaev V, Negre-Zakharov F, Sadka A, Blumwald E. Label-Free Shotgun Proteomics and Metabolite Analysis Reveal a Significant Metabolic Shift during Citrus Fruit Development. J Exp Bot. 2011;62(15): 5367–5384. 10.1093/jxb/err197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Famiani F, Cultrera NGM, Battistelli A, Casulli V, Proietti P, Standardi A, Chen ZH, Leegood RC, Walker RP. Phosphoenolpyruvate Carboxykinase and Its Potential Role in the Catabolism of Organic Acids in the Flesh of Soft Fruit during Ripening. J Exp Bot. 2005;56(421): 2959–2969. 10.1093/jxb/eri293 [DOI] [PubMed] [Google Scholar]
- 25.Famiani F, Cultrera NGM, Battistelli A, Casulli V, Proietti P, Standardi A, Chen ZH, Leegood RC, Walker RP. Genetic Dissection of Fruit Quality Traits in the Octoploid Cultivated Strawberry Highlights the Role of Homoeo-QTL in Their Control. Theor Appl Genet. 2012;124(6): 1059–1077. 10.1007/s00122-011-1769-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, Wang A, Brown S. Genetic Characterization of the Ma Locus with PH and Titratable Acidity in Apple. Mol Breed. 2012;30(2): 899–912. [Google Scholar]
- 27.Zong X, Wang W, Wang J, Wei H, Yan X, Liu Q. The application of SYBR Green I real-time quantitative RT-PCR in quantitative analysis of sweet cherry viruses in different tissues. Acta Phytophylacica Sin. 2012;39: 497–502. [Google Scholar]
- 28.Onik JC, Hu X, Lin Q, Wang Z. Comparative Transcriptomic Profiling to Understand Pre- and Post-Ripening Hormonal Regulations and Anthocyanin Biosynthesis in Early Ripening Apple Fruit. Molecules. 2018;23: 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics 2009;25(9): 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat Biotechnol. 2010;28(5): 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Yekutieli D. THE CONTROL OF THE FALSE DISCOVERY RATE IN MULTIPLE TESTING UNDER DEPENDENCY By Yoav Benjamini 1 and Daniel Yekutieli 2. Ann Stat. 2001;29(4): 1165–1188. [Google Scholar]
- 32.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010;11(2): R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao X, Cai T, Olyarchuk JG, Wei L. Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 2005;21(19): 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the the 2-ΔΔ CT Method. Methods. 2001;25(4): 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2008;36: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan MA, Olsen KM, Sovero V, Kushad MM, Korban SS. Fruit Quality Traits Have Played Critical Roles in Domestication of the Apple. Plant Genome 2014;7(3): 1–18. [Google Scholar]
- 37.Silva BM, Andrade PB, Gonçalves AC, Seabra RM, Oliveira MB, Ferreira MA. Influence of Jam Processing upon the Contents of Phenolics, Organic Acids and Free Amino Acids in Quince Fruit (Cydonia Oblonga Miller). Eur. Food Res Technol. 2004;218(4): 385–389. [Google Scholar]
- 38.Tang M, Bie Z, Wu M, Yi H, Feng J. Changes in Organic Acids and Acid Metabolism Enzymes in Melon Fruit during Development. Sci Hortic (Amsterdam). 2010;123(3): 360–365. [Google Scholar]
- 39.Liu Y, Che F, Wang L, Meng R, Zhang X, Zhao Z. Fruit Coloration and Anthocyanin Biosynthesis after Bag Removal in Non-Red and Red Apples (Malus × Domestica Borkh.). Molecules 2013;18(2): 1549–1563. 10.3390/molecules18021549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiecher A, de Paula LA, Chaves FC, Rombaldi CV. UV-C Effect on Ethylene, Polyamines and the Regulation of Tomato Fruit Ripening. Postharvest Biol Technol. 2013;86: 230–239. [Google Scholar]
- 41.Kim JY, Kim HJ, Lim GO, Jang SA, Song KB. The Effects of Aqueous Chlorine Dioxide or Fumaric Acid Treatment Combined with UV-C on Postharvest Quality of “Maehyang” Strawberries. Postharvest Biol Technol. 2010;56(3): 254–256. [Google Scholar]
- 42.Kondo S, Fiebig A, Okawa K, Ohara H, Kowitcharoen L, Nimitkeatkai H, Kittikorn M, Kim M. Jasmonic Acid, Polyamine, and Antioxidant Levels in Apple Seedlings as Affected by Ultraviolet-C Irradiation. Plant Growth Regul. 2011;64(1): 83–89. [Google Scholar]
- 43.Gonzalez-Aguilar GA, Wang CY, Buta GJ, Krizek DT. Use of UV-C Irradiation to Prevent Decay and Maintain Postharvest Quality of Ripe “Tommy Atkins” Mangoes. Int J Food Sci Technol. 2001;36(7): 767–773. [Google Scholar]
- 44.Gonzalez-Aguilar G, Wang CY, Buta GJ. UV-C Irradiation Reduces Breakdown and Chilling Injury of Peaches during Cold Storage. J. Sci. Food Agric. 2004;84(5): 415–422. [Google Scholar]
- 45.Ribeiro C, Canada J, Alvarenga B. Prospects of UV Radiation for Application in Postharvest Technology. Emirates Journal of Food and Agriculture. 2012; 24(6):586–597. [Google Scholar]
- 46.Shama G, Alderson P. UV Hormesis in Fruits: A Concept Ripe for Commercialisation. Trends in Food Science and Technology. 2005;128–136. [Google Scholar]
- 47.Charles MT, Kalantari S, Corcuff R, Arul J. Postharvest quality and sensory evaluation of UV-treated tomato fruit. Acta Horticulturae. 2005;682: 537e542. [Google Scholar]
- 48.Charles MT, Arul J, Charlebois D, Yaganza ES, Rolland D, Roussel D, et al. Postharvest UV-C treatment of tomato fruits: Changes in simple sugars and organic acids contents during storage. LWT—Food Sci Technol. 2016;65: 557–564 [Google Scholar]
- 49.Yujuan LA., Yin M, Ren X. Effects of UV-C radiation on storability and quality in harvested apple. Food Science. 2015;36: 14. [Google Scholar]
- 50.Laval-Martin D, Farineau J, Diamond J. Light versus Dark Carbon Metabolism in Cherry Tomato Fruits. Plant Physiol. 1977;60(6): 872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaki S. Isolation of Vacuoles from Immature Apple Fruit Flesh and Compartmentation of Sugars, Organic Acids, Phenolic Compounds and Amino Acids. Plant Cell Physiol. 1984;25(1): 151–166. [Google Scholar]
- 52.Moing A, Rothan C, Svanella L, Just D, Diakou P, Raymond P, Gaudillere JP, Monet R. Role of Phosphoenolpyruvate Carboxylase in Organic Acid Accumulation during Peach Fruit Development. Physiol Plant. 2000; 108(1): 1–10. [Google Scholar]
- 53.Berüter J. Carbohydrate Metabolism in Two Apple Genotypes That Differ in Malate Accumulation. J Plant Physiol. 2004;161(9): 1011–1029. 10.1016/j.jplph.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 54.Agetsuma M, Furumoto T, Yanagisawa S, Izui K. The Ubiquitin-Proteasome Pathway Is Involved in Rapid Degradation of Phosphoenolpyruvate Carboxylase Kinase for C4 Photosynthesis. Plant Cell Physiol. 2005;46(3): 389–398. 10.1093/pcp/pci043 [DOI] [PubMed] [Google Scholar]
- 55.Chen FX, Liu XH, Chen LS. Developmental Changes in Pulp Organic Acid Concentration and Activities of Acid-Metabolising Enzymes during the Fruit Development of Two Loquat (Eriobotrya Japonica Lindl.) Cultivars Differing in Fruit Acidity. Food Chem. 2009;114(2): 657–664. [Google Scholar]
- 56.Yao YX, Li M, Liu Z, You CX, Wang DM, Zhai H, Hao YJ. Molecular Cloning of Three Malic Acid Related Genes MdPEPC, MdVHA-A, MdcyME and Their Expression Analysis in Apple Fruits. Sci Hortic (Amsterdam). 2009;122(3): 404–408. [Google Scholar]
- 57.Ruffner HP, Possner D, Brem S, Rast DM. The Physiological Role of Malic Enzyme in Grape Ripening. Planta. 1984;160(5): 444–448. 10.1007/BF00429761 [DOI] [PubMed] [Google Scholar]
- 58.Schaaf J, Walter MH, Hess D. Primary Metabolism in Plant Defense (Regulation of a Bean Malic Enzyme Gene Promoter in Transgenic Tobacco by Developmental and Environmental Cues). Plant Physiol. 1995;108(3): 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter MH, Grima‐Pettenati J, Feuillet C. Characterization of a Bean (Phaseolus Vulgaris L.) Malic‐enzyme Gene. Eur J Biochem. 1994;224(3): 999–1009. [DOI] [PubMed] [Google Scholar]
- 60.Casati P, Andreo CS. UV-B and UV-C Induction of NADP-Malic Enzyme in Tissues of Different Cultivars of Phaseolus Vulgaris (Bean). Plant, Cell Environ. 2001;24(6): 621–630. [Google Scholar]
- 61.Casati P, Drincovich MF, Andreo CS, Donahue R, Edwards GE. UV-B, Red and Far-Red Light Regulate Induction of the C4isoform of NADP-Malic Enzyme in Etiolated Maize Seedlings. Aust J Plant Physiol. 1998;25(6): 701–708. [Google Scholar]
- 62.Drincovich MF, Casati P, Andreo CS, Donahue R, Edwards GE. UV-B Induction of NADP-Malic Enzyme in Etiolated and Green Maize Seedlings. Plant Cell Environ. 1998;21(1): 63–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. Raw sequencing datasets of the current study have been deposited in the NCBI Sequence Read Archive repository, with sample accession numbers SAMN10963045, SAMN10963046, SAMN10963047, SAMN10963072, SAMN10963073, SAMN10963074, SAMN10963323, SAMN10963324, SAMN10963325 under the following project (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA522961).