ABSTRACT
The generation of neuronal diversity is essential for circuit formation and behavior. Morphological differences in sequentially born neurons could be due to intrinsic molecular identity specified by temporal transcription factors (henceforth called intrinsic temporal identity) or due to changing extrinsic cues. Here, we have used the Drosophila NB7-1 lineage to address this issue. NB7-1 generates the U1-U5 motor neurons sequentially; each has a distinct intrinsic temporal identity due to inheritance of different temporal transcription factors at its time of birth. We show that the U1-U5 neurons project axons sequentially, followed by sequential dendrite extension. We misexpressed the earliest temporal transcription factor, Hunchback, to create ‘ectopic’ U1 neurons with an early intrinsic temporal identity but later birth-order. These ectopic U1 neurons have axon muscle targeting and dendrite neuropil targeting that are consistent with U1 intrinsic temporal identity, rather than with their time of birth or differentiation. We conclude that intrinsic temporal identity plays a major role in establishing both motor axon muscle targeting and dendritic arbor targeting, which are required for proper motor circuit development.
KEY WORDS: Temporal identity, Heterochronic, Temporal transcription factors, Hunchback, Motor neuron, Dendrite morphology, Neural circuits, Motor circuits, Larval locomotion
Summary: Hunchback misexpression in the Drosophila NB7-1 lineage reveals that intrinsic temporal identity is important for neuronal connectivity.
INTRODUCTION
Axon and dendrite targeting is an essential step in neural circuit formation, and may even be sufficient for proper connectivity in some cases, as postulated in Peters’ rule (Peters and Feldman, 1976; Rees et al., 2017; Stepanyants and Chklovskii, 2005). In both Drosophila and mammals, individual progenitors generate a series of neurons that differ in axon and dendrite targeting (Doe, 2017; Kohwi and Doe, 2013; Li et al., 2013a; Pearson and Doe, 2004; Rossi et al., 2016). In these examples, neurons born at different times have intrinsic molecular differences due to temporal transcription factors (TTFs) present at their time of birth (reviewed by Kohwi and Doe, 2013), which could specify neuronal morphology. Conversely, there are likely to be changing extrinsic cues present at the time of neuronal differentiation that could also influence neuronal morphology, such as modulation of global pathfinding cues or addition of axon and dendrite processes throughout neurogenesis. Teasing out the relative contributions of intrinsic or extrinsic factors requires heterochronic experiments where either intrinsic or extrinsic cues are altered to create a mismatch, and the effects on axon and dendrite targeting are assessed.
Several experiments highlight the importance of extrinsic cues present at the time of neuronal differentiation in establishing axon or dendrite targeting. For example, transplantation of rat fetal occipital cortical tissue into the rostral cortex of a more developmentally mature newborn host results in axonal projections characteristic of the host site (O'Leary and Stanfield, 1989; Schlaggar and O'Leary, 1991; Stanfield and O'Leary, 1985). Similarly, transplantation of embryonic day 15 fetal occipital tissue into newborn occipital cortex reveals that the transplanted tissue receives thalamic projections typical of the host site and developmental stage (Chang et al., 1986). More recent work in zebrafish shows that vagus motor neurons extend axons sequentially to form a topographic map, and that the time of axon outgrowth directs axon target selection (Barsh et al., 2017). Similarly, recent work in Drosophila has shown that sequential axon outgrowth of R7/R8 photoreceptor neurons, coupled with a temporal gradient in the levels of the transcription factor Sequoia, is essential for axon spacing during retinotopic map formation within the medulla neuropil (Kulkarni et al., 2016). In all of these systems, the relative importance of neuron intrinsic factors and changing environmental cues remain unknown.
In contrast, heterochronic experiments where donor neurons maintain donor identity are more consistent with intrinsic temporal identity specifying neuronal axon and dendrite targeting. For example, heterochronic experiments in ferrets show that late cortical progenitors transplanted into younger hosts generate neurons with late-born deep layer position and subcortical axonal projections (McConnell, 1988). Transplantation of older post-natal cerebellum into embryonic host mice results in the neurons maintaining donor ‘late-born’ identity based on molecular markers and neuronal morphology (Jankovski et al., 1996). Similarly, experiments carried out in grasshopper embryos show that delaying the birth of the first-born aCC motor neuron in the NB1-1 lineage leads to defects in the initial axon trajectory (extending anterior instead of posterior) but the temporally delayed aCC invariably finds and exits through the proper nerve root in the adjacent anterior segment (Doe et al., 1986). In all of these heterochronic experiments, it is likely that intrinsic temporal identity is unaltered and helps maintain donor neuron identity despite their altered time of differentiation. However, none of these experiments shows that intrinsic temporal identity is unchanged in the transplanted neurons, and none of these experiments manipulates intrinsic temporal identity to directly assess its role in establishing proper axon or dendrite targeting.
We sought to test the relative contribution of neuronal time of differentiation versus neuronal intrinsic temporal identity in establishing motor neuron axon and dendrite targeting. Our model system is the NB7-1 lineage in the Drosophila ventral nerve cord (VNC), a segmentally repeated structure analogous to the mammalian spinal cord. The VNC offers several benefits for the study of neurogenesis due to its individually identifiable neuroblasts (NBs), which produce a stereotyped sequence of distinct neuronal cell types whose identities are determined by a well-characterized temporal transcription factor (TTF) cascade (reviewed by Doe, 2017; Kao and Lee, 2010; Kohwi and Doe, 2013; Rossi et al., 2016; Skeath and Thor, 2003) (Fig. 1; Fig. S1). For example, NB7-1 sequentially expresses the four TTFs: Hunchback (Hb), Kruppel, Pdm and Castor. During each NB TTF expression window, a different motor neuron is born: U1 and U2 during the Hb window; U3, U4 and U5 during the later three TTF windows (Isshiki et al., 2001; Kanai et al., 2005; Kohwi et al., 2013; Pearson and Doe, 2003). Importantly, the two Hb+ U1-U2 motor neurons have a morphology, neuropil targeting and connectivity that is clearly different from the later-born U3-U5 motor neurons (Fig. 1; Fig. S1). The ability to individually identify the U1-U5 neurons, and to cleanly change their intrinsic temporal identity in an otherwise normal CNS, make the NB7-1 lineage an ideal system to study the relative contribution of time of differentiation and intrinsic temporal identity in establishing neuron morphology, targeting and connectivity.
Fig. 1.
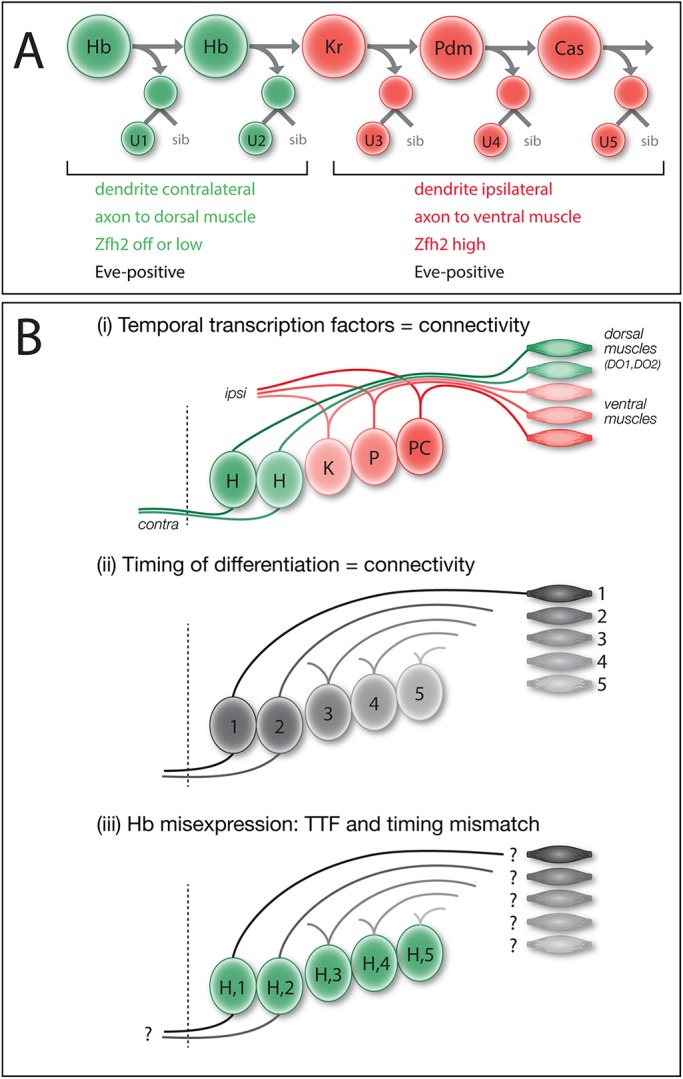
Models: intrinsic temporal identity or time of differentiation determines U1-U5 motor neuron morphology. (A) NB7-1 (top ) sequentially expresses the temporal transcription factors Hb, Kr, Pdm and Cas. The U1-U2 neurons (bottom) born during the Hb window have an ‘early-born’ identity (green) characterized by contralateral dendrites, an axon projection to dorsal body wall muscles DO1 and DO2, and little or no nuclear Zfh2. The U3-U5 neurons born after the Hb window have a ‘late-born’ identity (red) characterized by ipsilateral dendrites, an axon projection to more ventral muscles DA3/LL1 and high nuclear Zfh2. All U1-U5 neurons have nuclear Eve. (B) Models for specification of U1-U5 axon and dendrite targeting. (i) Intrinsic temporal identity could determine axon and dendrite targeting. (ii) Neuronal time of differentiation could determine axon and dendrite targeting. (iii) Misexpression of Hb can generate late-differentiating neurons with an early intrinsic temporal identity; this mismatch reveals which mechanism is more important for axon and dendrite targeting.
Previously, we have shown that misexpression of Hb throughout the NB7-1 lineage results in an extended series of ‘ectopic U1’ motor neurons based on molecular markers and axon projections to dorsal body wall muscles (Isshiki et al., 2001; Pearson and Doe, 2003); however, the ectopic U1 motor neurons were not assayed for their specific muscle targets, nor was dendrite morphology and targeting assessed, nor was it known whether U1-U5 motor neurons extended axons or dendrites synchronously or sequentially. Here, we focus on the difference between early-born Hb+ U1-U2 neurons and later-born U3-U5 neurons. U1-U2 are bipolar, have contralateral dendrites and innervate dorsal body wall muscles; in contrast, U3-U5 neurons are monopolar, have ipsilateral dendrites and innervate more ventral body wall muscles. Although there are molecular differences between U1-U2 and between U3-U5 (Isshiki et al., 2001), in this article we focus on the major morphological differences between these two groups of neurons. We show for the first time that the U1-U5 neurons extend axons sequentially, and subsequently extend dendrites sequentially. We test whether U1-U5 motor neurons project to their normal CNS and muscle targets due to their intrinsic temporal identity (Fig. 1Bi) or due to their time of differentiation (Fig. 1Bii) – two mechanisms that are normally tightly correlated. To break this correlation, we misexpress the early TTF Hb specifically in the NB7-1 lineage to create ‘ectopic U1’ motor neurons with an early intrinsic temporal identity but late time of differentiation (Fig. 1Biii). Moreover, we show that the heterochronic placement of an ‘ectopic U1’ into the later developmental environment does not affect the ability of the ‘ectopic U1’ to project dendrites to the proper neuropil domain or axons to the proper body wall muscle. Our results show that intrinsic temporal identity is an important determinant of neuronal morphology, and axon and dendrite targeting.
RESULTS
U1-U5 motor neurons extend axons and dendrites sequentially
To determine whether U1-U5 motor neuron axon target selection is correlated with intrinsic temporal identity or time of differentiation, we first needed to investigate the timing of U1-U5 motor neuron axon outgrowth. If the U1-U5 motor neurons have synchronous axon outgrowth, despite being born sequentially, we can rule out time of axon outgrowth as a mechanism for specifying their differential axon target selection. Conversely, if the U1-U5 motor neurons extend their axons sequentially, then both models remain possible.
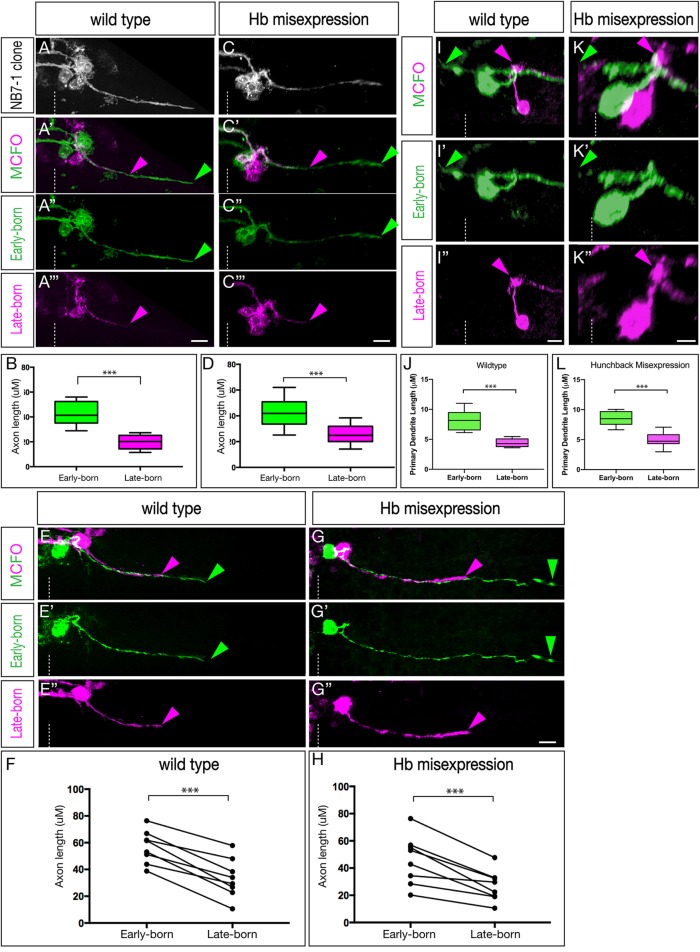
To determine the time of U1-U5 axon outgrowth, we used MultiColorFlpOut (MCFO) (Nern et al., 2015) which produces randomized multi-color labeling of neurons within the expression domain of any Gal4 line. We restricted labeling to the NB7-1 lineage using a new split-gal4 killer zipper line (NB7-1-Gal4KZ). This new line is based on our published NB7-1-Gal4 line (ac-VP16 gsb-DBD) (Kohwi and Doe, 2013) but also includes an R25A05-KillerZipper construct to block expression in NB6-1, which was commonly observed in the previously described NB7-1 split Gal4 line (Kohwi and Doe, 2013; see Materials and Methods for quantification). This new NB7-1-Gal4KZ line was used for all MCFO or Hb misexpression experiments. Within the NB7-1 lineage, early-born neurons are located close to the midline and later-born neurons are located more laterally (Pearson and Doe, 2003) (Fig. S1A). As expected, MCFO labeling of the entire wild-type NB7-1 lineage shows neurons spread from medial to lateral within the CNS, with ipsilateral motor projections and contralateral dendrite projections (Fig. 2A); we call these dendrites because they have a large number of post-synaptic densities but no pre-synaptic sites when analyzed by electron microscopy (Fig. S2). We analyzed embryos where MCFO differentially labeled early-born and late-born neurons in the NB7-1 lineage at embryonic stages 12-15 (staging according to Hartenstein, 1993). In all cases, the medial early-born neurons invariably extended axons further than the lateral later-born neurons (Fig. 2A,B; n=10, P<0.0001, two-tailed unpaired t-test). This observation remained consistent at all tested embryonic stages and independent of the position at which the lineage was subdivided along the medial-lateral axis. In all cases, the U neurons showed axon projections that are staggered until they reach their muscle targets; they never stall and become synchronized prior to innervating their target muscle. Furthermore, in every case where MCFO differentially labeled only a pair of neurons, we always found the medial (early-born) neuron had a longer axon projection than the lateral (later-born) neuron (Fig. 2E,F, n=8, P<0.0001, two-tailed paired t-test). We conclude that during wild-type embryonic development, the U1-U5 motor neurons project axons sequentially out the nerve root.
Fig. 2.
The U1-U5 motor neurons extend axons and dendrites sequentially. (A-H) Axon outgrowth timing in early- and late-born neurons of the NB7-1 lineage in stage 13-15 embryos. (A-B) Wild-type multicellular MCFO labeling (NB7-1-Gal4KZ UAS-MCFO). Analysis was restricted to lineages in which early-born medially located neurons were stochastically labeled in one MCFO color, and in which the later-born laterally located neurons were stochastically labeled in a different MCFO color. (A) All labeled cells. (A′-A‴) Early-born medial neurons (green) project out of the CNS ahead of later-born lateral neurons (magenta). (B) Quantification of axon length as a representation of timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (***P<0.001). (C-D) Hb misexpression (NB7-1-Gal4KZ UAS-hb UAS-MCFO) multicellular MCFO labeling. (C) All labeled cells. (C′-C‴) Early-born medial neurons (green) project out of the CNS ahead of later-born lateral neurons (magenta). (D) Quantification of axon length as a representation of timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (***P<0.001). (E-F) Wild-type single-neuron MCFO labeling (NB7-1-Gal4KZ UAS-MCFO). (E) A single early-born medial neuron (green) always projects out of the CNS ahead of a single later-born lateral neuron (magenta). (F) Quantification of axon length as a representation of timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (***P<0.001). (G-H) Hb misexpression (NB7-1-Gal4KZ UAS-hb UAS-MCFO) single-neuron MCFO. (G-G″) A single early-born medial neuron (green) always projects out of the CNS ahead of a single later-born lateral neuron (magenta). Quantification of axon length as a representation of timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (***P<0.001). (I-L) Dendrite outgrowth timing in early- and late-born neurons of the NB7-1 lineage in stage 13-15 embryos. (I-I″) Wild-type single-neuron MCFO labeling (NB7-1-Gal4KZ UAS-MCFO). A single early-born medial neuron (green) extends a dendrite before a single later-born lateral neuron (magenta). (J) Quantification of axon length as a representation of the timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (***P<0.001). (K-K″) Hb misexpression (NB7-1-Gal4KZ UAS-hb UAS-MCFO) single neuron MCFO labeling. A single early-born medial neuron (green) extends a dendrite before a single later-born lateral neuron (magenta). (L) Quantification of axon length as a representation of timing of axon outgrowth; early-born neurons project further (earlier) than late-born neurons (P<0.001). Arrowheads indicate the early-born axon (green) and late-born axon (magenta); dashed line indicates the midline. Scale bars: 5 μm.
We next wanted to determine whether misexpression of Hb throughout the NB7-1 lineage, known to produce many ectopic U1 motor neurons (Isshiki et al., 2001; Kohwi and Doe, 2013; Pearson and Doe, 2003), would alter the timing of motor axon outgrowth. We misexpressed Hb in the NB7-1 lineage, and used MCFO to differentially label early-born and late-born neurons. MCFO marking the entire NB7-1 lineage did not change the gross distribution of neurons (Fig. 2C). Importantly, in every case where MCFO differentially labeled early-born and late-born neurons, we found that early-born neurons projected axons out of the CNS before later-born neurons (Fig. 2C-D; n=10, P<0.0001, two-tailed unpaired t-test). As in the wild type, in every case where MCFO differentially labeled just a pair of neurons, we always found the more medial (early-born) neuron had a longer axon projection than the lateral (later-born) neuron (Fig. 2G-H, n=8, P<0.001, two-tailed paired t-test). Moreover, the axon length differential between early-born and late-born neurons was indistinguishable in wild-type and Hb-misexpression lineages (n=17, P=0.41, two-tailed unpaired t-test, data not shown).
We next examined the time course of dendrite extension. In wild type, we observed that earlier-born cells elaborated their dendritic processes before their later-born counterparts (Fig. 2I,J; n=7, P<0.003, two-tailed unpaired t-test); the same was observed in Hb misexpression animals (Fig. 2K,L; n=7, P<0.001, two-tailed unpaired t-test). We conclude that sequentially born motor neurons project axons and dendrites sequentially, in both wild type and following Hb misexpression. This raises the question: is intrinsic temporal identity or time of differentiation more important for U1-U5 axon or dendrite target selection?
Late-born neurons with early intrinsic temporal identity have ‘early’ contralateral dendrite targeting
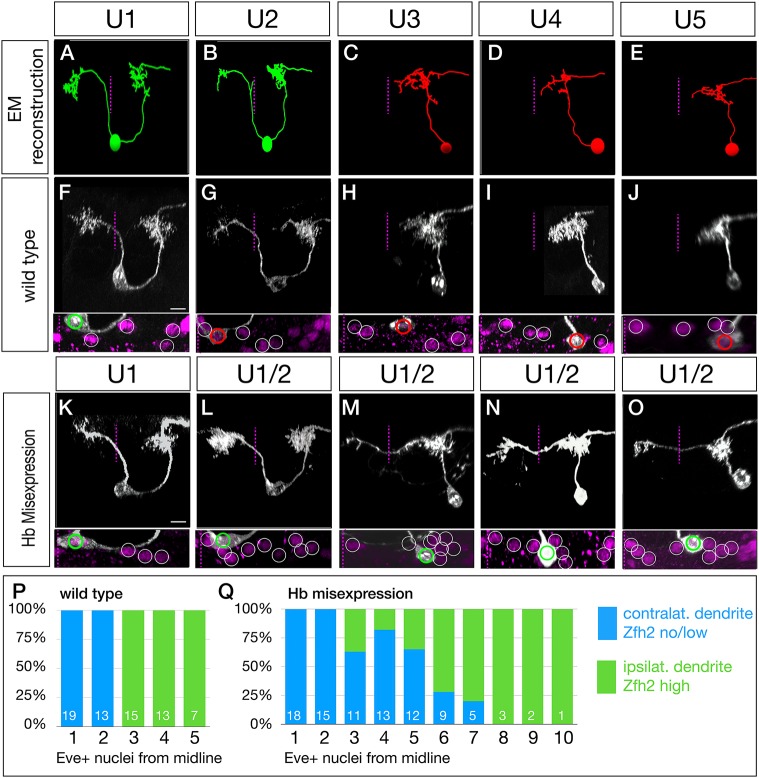
To determine whether neuronal morphology was correlated with intrinsic temporal identity or time of differentiation, we first needed to define the morphology of the U1-U5 motor neurons. Previous work has mapped generic U neuron axonal projections (Landgraf et al., 1997), but did not identify muscle targets for specific U1-U5 motor neurons. To precisely define U1-U5 motor neuron identity, we used the serial section transmission electron microscopy (EM) (Ohyama et al., 2015) to reconstruct U1-U5 morphology (Fig. 3A-E; Fig. S2). We detected two striking differences in morphology between early-born U1-U2 neurons and later-born U3-U5 neurons: U1-U2 have bipolar projections, whereas U3-U5 have monopolar projections; and U1-U2 have contralateral dendrites, whereas U3-U5 have ipsilateral dendrites (Fig. 3A-E).
Fig. 3.
Late-born neurons with early intrinsic temporal identity have ‘early’ dendrite morphology. (A-E) U1-U5 neuronal morphology determined by EM reconstruction in the first instar larval CNS. Early-born U1-U2 neurons (green) have a bipolar morphology with a contralateral dendrite arbor (left of dashed midline), whereas later-born U3-U5 neurons (red) have a monopolar morphology and ipsilateral dendritic arbors. Neuronal birth-order is determined by mediolateral position (U1 most medial/earliest, U5 most lateral/latest). (F-J) Wild-type U1-U5 single neuronal morphology by MCFO in L1 larvae (NB7-1-Gal4KZ UAS-MCFO). Neurons are shown from left to right based on birth-order, determined by their position within the five Eve+ neurons (inset). Scale bar: 5 μm. (K-O) Hb misexpression U1-U5 single neuronal morphology by MCFO in L1 larvae (NB7-1-Gal4KZ UAS-MCFO). Neurons are shown from left to right based on birth-order, determined by their position within the Eve+ neurons (inset). The later-born neurons (‘ectopic U1’) have acquired a contralateral dendrite, more consistent with their early intrinsic temporal identity than their late time of differentiation. Scale bar: 5 μm. (P,Q) Quantification. In wild type, early-born U1-U2 neurons have low/no nuclear Zfh2, a marker for their early intrinsic temporal identity, and contralateral dendrites; later-born neurons have high Zfh2 and no contralateral projection. In Hb misexpression embryos, all neurons with low/no Zfh2 have a contralateral dendrite, even when they have a late-born time of differentiation (>3 Eve+ nuclei from the midline). The number of neurons scored is shown within each bar.
To determine whether the U1-U5 morphology seen in the larval EM reconstruction are reproducible and present in the larval VNC, we generated MCFO labeling of single U1-U5 motor neurons in early L1 larvae (Fig. 3F-J). Previous work showed that U1-U5 are positive for the Even-skipped (Eve) transcription factor, and are arranged from medial (U1) to lateral (U5) (Isshiki et al., 2001; Kohwi and Doe, 2013; Pearson and Doe, 2003), which we confirm here (Fig. 3F-J, bottom panels, and quantified in Fig. 3P). We observed that the larval U1-U5 motor neurons had a morphology closely matching the larval U1-U5 motor neurons in the EM reconstruction (compare Fig. 3A-E with F-J). We conclude that the early-born U1-U2 neurons and the late-born U3-U5 neurons have distinctive, stereotyped neuronal morphologies.
In wild type, intrinsic temporal identity and time of axon outgrowth are tightly linked; neurons with early intrinsic temporal identity extend axons first, neurons with late intrinsic temporal identity extend axons later. We sought to break this correlation by misexpressing Hb in the NB7-1 lineage so that both early-differentiating and late-differentiating neurons have an early U1 intrinsic temporal identity (Isshiki et al., 2001; Kohwi and Doe, 2013; Pearson and Doe, 2003). To perform this experiment, we needed to monitor neuronal birth order (a proxy for time of differentiation), intrinsic temporal identity and neuronal morphology. Birth order was determined by the neuron position in the medio-lateral series of Eve+ nuclei (medial, early-differentiating; lateral, late-differentiating); intrinsic temporal identity was determined by molecular markers (U1 is Eve+ Zfh2−, whereas neurons with later temporal identities are Eve+ Zfh2+; and neuronal morphology was determined by MCFO (Fig. 3K-O). As expected, misexpression of Hb had no effect on the morphology of the endogenous Hb+ U1 or U2 neurons (Fig. 3K,L; U1 n=13). In contrast, all late-differentiating neurons with an ectopic U1 intrinsic temporal identity (Eve+ Zfh2−) had a morphology similar the endogenous U1 neurons: both producing a dorsal contralateral dendritic arbor (Fig. 3M-O, arrowheads). The penetrance of the transformation declined in neurons with progressively later birthdates (quantified in Fig. 3Q). The failure to project a contralateral dendrite was perfectly correlated with the failure to repress Zfh2 (Fig. 3Q; Fig. S3), leading us to conclude that these Zfh2+ late-born neurons are simply not being transformed to a U1 identity, and thus fail to project contralaterally. Interestingly, even the transformed ectopic U1 neurons (Eve+ Zfh2−) had their contralateral process emerging from a dorsal location, rather than from the cell body, as observed for endogenous U1 neurons, indicating that the morphological transformation was not complete (Fig. 3M-O, Movies 1 and 2). Nevertheless, the ectopic U1 neurons are more similar to the endogenous early-born U1-U2 neurons than to the later-born U3-U5 neurons. We conclude that neuronal morphology is more tightly linked to intrinsic temporal identity than to neuronal birth order.
Late-born neurons with early intrinsic temporal identity target their dendrites to the early-born U1 dendritic domain
The experiments described above show that late-differentiating neurons with early intrinsic temporal identity have gross morphological features matching their intrinsic temporal identity, rather than their time of differentiation. In this section and the next, we investigate whether these ectopic U1 neurons target their axons to the normal U1 muscle target (the dorsal DO1 and DO2 muscles) and target their dendrites to the normal U1 neuropil target (a contralateral, dorsal volume of neuropil). In this section we assay dendritic projections; in the following section we assay axonal projections.
In wild type, the endogenous U1 neurons have ipsilateral and contralateral dendrites that are colocalized in the same region of dorsal neuropil, as seen by EM reconstruction (Fig. 4A) or dual color MCFO labeling (Fig. 4B). To map dendrite targeting of ‘heterochronic’ ectopic U1 motor neurons, we misexpressed Hb in the NB7-1 lineage and then screened for MCFO labeling in which one hemisegment had the endogenous U1 motor neuron labeled (identified by its medial position and ‘U’ shaped neuronal morphology), and the opposite hemisegment had an ectopic U1 neuron labeled (identified by its lateral cell body position and dorsal contralateral dendrite process). In every case, we found the ‘heterochronic’ ectopic U1 neuron dendrite precisely targeted to the normal dorsal neuropil target of the U1 neuron, with both ectopic and endogenous U1 dendritic arbors tightly intermingled (Fig. 4B,B‴; n=10). For each dendrite assessed, correct neuropil localization was confirmed through quantification of the distance of the dendrite from the midline, the anteroposterior distance of the dendrite from the directly anterior hemisegment in relation to the labelled cell, and the position of the dendrite in the dorsoventral axis (Fig. 4C). We observed no significant differences between the dendritic localization of the wild-type U1 neurons and our ectopic early-born cells across any of our positional metrics (Fig. 4C, two-way ANOVA, P=0.71). We conclude that intrinsic temporal identity, not time of dendrite outgrowth, generates precise dendrite targeting to the appropriate region of the neuropil.
Fig. 4.
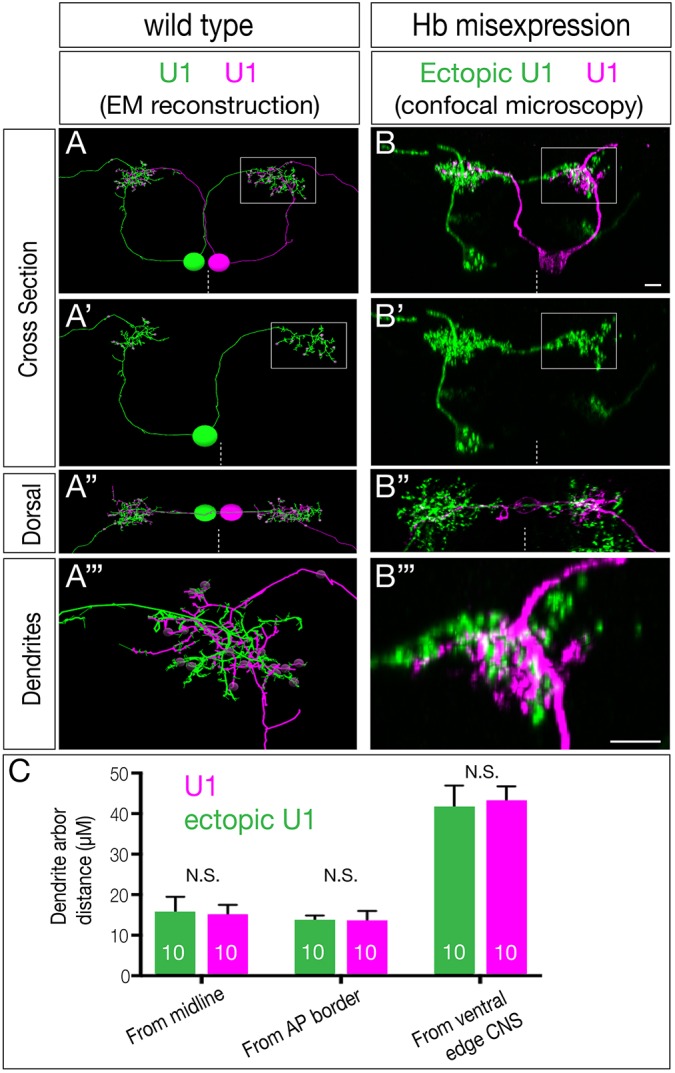
Ectopic U1 dendrites target the normal U1 neuropil domain. (A-A‴) Wild-type bilateral U1 neurons (green, magenta) assayed in the EM reconstruction of the L1 larval CNS. A U1 neuron (magenta) targets its contralateral dendrite to the same neuropil volume as the ipsilateral dendrite of the contralateral U1 neuron (green; boxed region in A,A′, shown enlarged in A″). (B-B‴) Hb misexpression (NB7-1-Gal4KZ UAS-hb UAS-MCFO) assayed by MCFO labeling in L1 larvae, showing an endogenous U1 (magenta; defined by its medial cell body position, bipolar morphology and contralateral projection) and an ectopic U1 neuron (green; defined by its lateral cell body position, monopolar morphology and contralateral projection). The endogenous and ectopic U1 neurons target the same dorsal neuropil domain (boxed in B,B′ shown enlarged in B″). Dashed line indicates the midline; all views are cross-sections; dorsal is upwards except A″ and B″ (dorsal views, anterior upwards). Scale bars: 5 μm. (C) Quantification. Endogenous and ectopic U1 dendrites are the same distance from the midline, anterior-posterior (AP) border and ventral edge of the CNS. n=10 for U1 and ectopic U1.
Ectopic U1 axons project to dorsal body wall muscles and lack ventral muscle targets
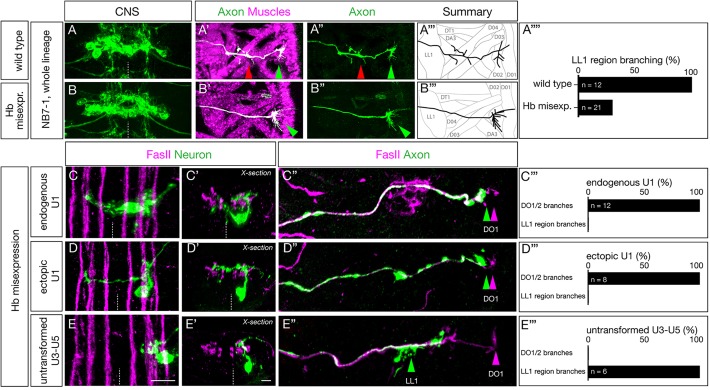
Here, we determine whether late-born neurons with early intrinsic temporal identity project their axons to dorsal muscles normally targeted by neurons with early temporal identity or to more ventral muscles normally targeted by late-born neurons. We focus our analysis on L1 larvae, where neuromuscular junctions have formed and are functional for locomotion. In wild type, we find that the U1-U2 motor neurons innervate the dorsal-most oblique muscles DO1and DO2, and the U3-U5 motor neurons innervate more ventral muscles in the area of DA3 and DO4 (Fig. 5A-A‴; quantified in Fig. 5A‴). All motor neurons make varicosities, indicating presynaptic differentiation at their muscle targets, but here we do not assay functional synaptic connectivity, simply axon targeting. In contrast, Hb misexpression results in a complete loss of the more ventral axon varicosities, while still exhibiting varicosities at the site of the DO1 and DO2 dorsal muscles (Fig. 5B-B‴; quantified in Fig. 5A‴). These results suggest that later-born motor neurons in the lineage have been transformed into an early intrinsic temporal identity and thereby target the normal early U1-U2 muscle targets.
Fig. 5.
Ectopic U1 axons project to dorsal muscles and lack ventral muscle targets. (A-B‴) Wild-type (NB7-1-Gal4KZ UAS-GFP) and Hb-misexpression (NB7-1-Gal4KZ UAS-hb UAS-GFP) L1 larvae stained for U motor neurons (green) and muscles (magenta). (A-A‴) Wild type: U motor neurons project axons to dorsal muscles (DO1/DO3) and more ventral muscles in the LL1/DA3 region. (B-B‴) Hb misexpression: U motor neurons project only to dorsal muscle targets (magenta, shown in B′) consistent with ectopic U1-U2 identity at the expense of U3-U5 neuronal identity. Arrowheads indicate U1-U2 muscle targets (green) and U3-U5 muscle targets (red). (A″″) Quantification. (C-C″,D-D″,E-E″) Hb misexpression L1 larvae (NB7-1-Gal4KZ UAS-hb UAS-MCFO) showing MCFO-labeled single neurons. (C-C″) The endogenous U1 motor neuron (green; closest to midline). (C) Dorsal view showing medial cell body position, contralateral dendrites and ipsilateral axon (Zfh2 negative; not shown). (C′) Cross-sectional view of the same U1 neuron. (C″) Dorsal view of the body wall showing the U1 axon (green arrowhead) projecting to the most dorsal extent of the FasII+ motor neurons (magenta arrowhead). (C‴) Quantification. (D-D″) An ectopic U1 motor neuron (green). (D) Dorsal view showing lateral cell body position, contralateral dendrites and ipsilateral axon (Zfh2 negative; not shown). (D′) Cross-sectional view of the same neuron; the contralateral dendrite has a dorsal origin and there is a lack of bipolar morphology. (D″) Dorsal view of body wall showing the ectopic U1 axon (green arrowhead) projecting to the most dorsal extent of the FasII+ motor neurons (magenta arrowhead). (D‴) Quantification. (E-E″) A late-born laterally positioned U3, U4 or U5 motor neuron that was not transformed (based on being Zfh2+; not shown). (E) Dorsal view showing far lateral cell body position, ipsilateral dendrites and ipsilateral axon. (E′) Cross-sectional view of the same neuron. (E″) Dorsal view of the body wall showing the U3-U5 axon (green arrowhead) projecting to a more ventral region along the FasII+ motor neurons (magenta arrowhead). (E‴) Quantification.
To examine individual motor neurons, we used MCFO following Hb misexpression throughout the NB7-1 lineage. We observed endogenous early-born U1 motor neurons, identified by their medial position and bipolar morphology (Fig. 5C-C′), that project to DO1/DO2 dorsal muscles (Fig. 5C″; quantified in Fig. 5C‴). We also observed ‘ectopic U1’ motor neurons identified by their displacement from the midline, their monopolar morphology and their dorsal originating contralateral dendrite projection (Fig. 5D,D′); these neurons project to the same DO1/DO2 dorsal muscles as the endogenous U1 motor neuron (Fig. 5D″; quantified in Fig. 5D‴). These heterochronic ‘ectopic U1’ motor neurons are clearly different from the normal late-born U3-U5 motor neurons, identified by their lateral position and lack of contralateral dendrites (Fig. 5E,E′), that project to the region of the more ventral muscle LL1 (Fig. 5E″; quantified in Fig. 5E‴). We conclude that intrinsic temporal identity, not time of axon outgrowth, generates precise axon targeting to the appropriate body wall muscles.
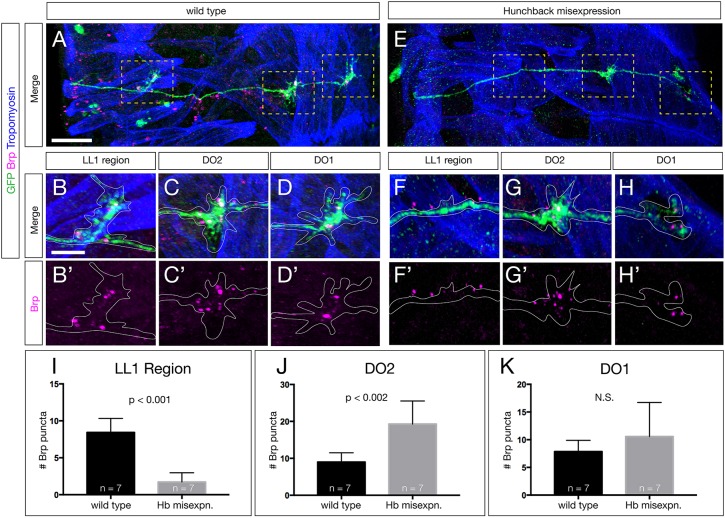
To examine the ability of these ‘ectopic U1’ motor neurons to create putative synaptic inputs onto the DO1/DO2 dorsal muscles, we quantified the numbers of pre-synaptic Bruchpilot (Brp) puncta formed by U neurons on their dorsal longitudinal muscle targets. Brp is an active zone marker that is a good measure of pre-synapse location. In wild type, U1-U2 neurons form Brp+ synapses on the most dorsal longitudinal muscles DO1/DO2, and the later-born U3-U5 neurons form synapses with the slightly more ventral muscles in the LL1 region (Fig. 6A-D; quantified in Fig. 6I-K). In contrast, the ‘ectopic U1’ motor neurons have a significant shift towards more dorsal muscle targets (Fig. 6E-H). There is a significant loss of pre-synaptic Brp+ puncta in the LL1 region (Fig. 6F, quantified in Fig. 6I), while simultaneously increasing their amount of synaptic input onto the dorsal muscle DO2 (Fig. 6G,H, quantified in Fig. 6J). We saw an insignificant difference in synaptic input onto DO1 between wild-type and Hb-misexpression samples (Fig. 6H quantified in Fig. 6K). We conclude that intrinsic temporal identity, not time of axon outgrowth, determines the position of Brp+ presynaptic puncta on the dorsal longitudinal muscle targets.
Fig. 6.
Ectopic U1 axons shift synaptic input from ventral to dorsal muscle targets. (A-D′) Wild-type L1 larva stained for all U motor neurons in the NB7-1 lineage (GFP, green), Brp+ puncta (magenta) and body wall muscles (Tropomyosin, blue). The U motor neurons have Brp+ puncta contacting muscles around LL1 (B,B′), the DO2 muscle (C,C′) and the DO1 muscle (D,D′). (E-H′) Hb-misexpression L1 larva (NB7-1-Gal4KZ UAS-hb) stained for all U motor neurons in the NB7-1 lineage (GFP, green), Brp+ puncta (magenta) and body wall muscles (Tropomyosin, blue). There are reduced Brp+ puncta around LL1 (F,F′), increased Brp+ puncta on the DO2 muscle (G,G′) and a similar number of Brp+ puncta on the DO1 muscle (H,H′). Areas outlined in A and E are shown at higher magnification in B-D′ and F-H′, respectively. (I-K) Quantification. Scale bars: 15 μm in A,E; 5 μm in B-H′.
DISCUSSION
During neurogenesis, intrinsic temporal identity and time of differentiation are typically tightly correlated. For example, the Drosophila NB7-1 sequentially generates the U1-U5 motor neurons, which have distinct intrinsic temporal identities and distinct times of differentiation. Our work shows that intrinsic temporal identity is more important than the time of neuronal differentiation for establishing proper axon and dendrite targeting. We generated ectopic motor neurons with an early-born U1 intrinsic temporal identity in a later extracellular environment, breaking the correlation between intrinsic temporal identity and time of differentiation. These late-born ectopic U1 neurons sent their axons to the DO1/2 muscles (together with endogenous U1 neurons), and their dendrites to a dorsal contralateral neuropil domain (together with endogenous U1-U2 neurons). Furthermore, ectopic U1 neurons are also born in a much more lateral location in the CNS, and yet are able to find their correct axon and dendrite targets. Overexpression of Hunchback in other neuroblast lineages generates early-born neuronal identity based on molecular marker expression (Isshiki et al., 2001; Moris-Sanz et al., 2015; Novotny et al., 2002; Tran and Doe, 2008), but here we characterize the pre- and post-synaptic targeting of these ‘heterochronic’ neurons. Our data show that intrinsic temporal identity is an important determinant of neuronal axon and dendrite targeting.
Temporal transcription factors (TTFs) are known to regulate neuronal cell fate in multiple neuroblast lineages in Drosophila (Doe, 2017). In mushroom body neuroblasts, TTFs are known to specify the molecular and morphological features of the Kenyon cells (Kao and Lee, 2010; Liu et al., 2015; Zhu et al., 2006). Similarly, in type II neuroblast intermediate neural progenitor (INP) lineages, recent work has shown that the late TTF Eyeless specifies the molecular identity and axon/dendrite targeting of several classes of central complex neurons (Sullivan et al., 2019). In contrast, the INP parental type II neuroblasts express a different set of TTFs (Syed et al., 2017), but nothing is yet known about their role in axon/dendrite targeting (Ren et al., 2017; Syed et al., 2017). Recent work has shown that optic lobe neuroblasts express TTFs that specify the molecular identity and axon targeting of visual system neuronal subtypes, but it is unknown whether sequentially born neurons project axons sequentially or synchronously (Bertet et al., 2014; Erclik et al., 2017; Li et al., 2013b). The antero-dorsal larval brain neuroblast expresses TTFs that govern the identity of olfactory projection neurons, as well as regulating the dendritic targeting to specific antennal lobe glomeruli, but it is unknown whether the projection neuron dendrites project sequentially or synchronously (Jefferis et al., 2001; Liu et al., 2015). Taken together, abundant data suggest that TTFs control neuronal molecular identity, with a growing number of studies showing that TTFs also regulate axon/dendrite targeting to specific neuropil domains or muscles.
Previous work has demonstrated that motor neurons in different lineages project axons at different times, e.g. aCC prior to RP2 (Sanchez-Soriano and Prokop, 2005). Similarly, we have found that the U1-U5 motor neurons extend axons sequentially, and independently of their intrinsic temporal identity. This suggests that the initial timing of axon extension is regulated by an internal clock mechanism in each cell, likely beginning upon its terminal cell division. In C. elegans, the HSN motor neurons require expression of lin-18 mRNA to initiate axon extension (Olsson-Carter and Slack, 2010); whether a similar mechanism is used by U1-U5 motor neurons is unknown. We also show that dendrite elaboration occurs much later than axon extension in the U motor neurons. The observation that axon outgrowth precedes dendrite outgrowth has been widely reported (Gerhard et al., 2017; Mason, 1983; Mumm et al., 2006; Ramon y Cajal, 1909), although the mechanism that sets the time of axon or dendrite outgrowth is poorly understood.
Hb misexpression robustly transformed later-born U motor neurons into ectopic U1 neurons, yet there were two limitations. First, ectopic U1 neurons do not have a bipolar cell body: they branch off dendrites from the dorsal axon, rather than from the cell body. Nevertheless, despite their novel dorsal outgrowth, ectopic U1 dendrites targeted a contralateral neuropil volume indistinguishable from the endogenous U1 neurons. The failure of the ectopic U1 neurons to generate a bipolar somata may be due to: (1) incomplete transformation of neuronal identity; (2) an abnormal lateral cell body position; (3) changing extrinsic cues; or (4) intrinsic changes in the neuronal cytoskeletal that are not under Hb regulation. A second limitation is the decline in Hb potency as the NB7-1 lineage progresses. We find that, following Hb misexpression, there are always some laterally positioned Eve+ motor neurons that fail to repress Zfh2 and fail to extend contralateral dendrites (Fig. S3); we conclude these neurons are simply untransformed. The inability of Hb to fully transform late-born neurons has been well documented (Kohwi et al., 2013; Pearson and Doe, 2003). The striking correlation between Zfh2 expression and ipsilateral dendrite projection raises the possibility that Zhf2 levels regulate dendrite midline crossing. For example, Zfh2 might activate Robo expression in late-born neurons to keep them ipsilateral, whereas early-born neurons lacking Zfh2 would lack Robo expression, allowing midline crossing. Testing this hypothesis would require generating UAS-zfh2 transgenics for NB7-1-specific overexpression, and an FRT zfh2 mutant fourth chromosome to make zfh2 mutant clones in NB7-1.
The NB7-1 cell cycle is ∼50 min (Hartenstein et al., 1987), which means that ectopic U1 motor neurons can be born up to six divisions or 300 min later than normal and yet still find their normal axon and dendrite targets. This suggests that the guidance cues used for endogenous U1 pathfinding are still present many hours later. Consistent with this, the major pathfinding ligands regulating sensory axon targeting and motor dendrite targeting in the CNS – NetrinA/B, Slit, Semaphorin1/2 and Wnt5 (Mauss et al., 2009; Wu et al., 2011; Yoshikawa et al., 2016; Zlatic et al., 2003, 2009) – all maintain their graded expression patterns during this window of neurogenesis (Fradkin et al., 2004; Harris et al., 1996; Mitchell et al., 1996; Rothberg et al., 1988; Yoshikawa et al., 2003; Zlatic et al., 2009). Although we cannot exclude the possibility of the endogenous and ectopic U1 neurons using different cues to find their proper targets, e.g. later-born neurons may project along ‘pioneer neuron’ processes formed earlier in neurogenesis, it is more likely that both early-born endogenous U1 neurons and later-born ectopic U1 neurons use the same guidance cues for axon and dendrite targeting.
In the future, it will be important to understand the mechanism by which the Hb transcription factor confers U1 neuron axon and dendrite targeting. As mentioned above, it is likely that the endogenous and ectopic U1 motor dendrites target the proper neuropil domain by responding to the known Netrin, Slit, Semaphorin and Wnt5 ligand gradients (Mauss et al., 2009; Wu et al., 2011; Yoshikawa et al., 2016; Zlatic et al., 2003, 2009). Thus, we hypothesize that Hb induces expression of distinct receptor combinations that allow the endogenous and ectopic U1 axon and dendrite to respond to these persistent pathfinding ligand gradients. Hb may directly regulate receptor gene expression, or it may act via an intermediate tier of transcription factors, similar to the ‘morphology transcription factors’ that act downstream of temporal transcription factors in establishing adult leg motor neuron axon and dendrite targeting (Enriquez et al., 2015). Understanding how Hb directs axon and dendrite targeting will require characterization of receptor expression in endogenous and ectopic U1 neurons, and/or single cell RNA-seq to characterize the endogenous and ectopic U1 neuron transcriptomes.
MATERIALS AND METHODS
Fly stocks
Male and female Drosophila melanogaster were used. The chromosomes and insertion sites of transgenes (if known) are shown in parentheses next to genotypes. Previously published Gal4 lines, mutants and reporters used were: hs-FLPG5;;MCFO (I and III; FBst0064086), UAS-hunchback/CyO (II) (Isshiki et al., 2001) and 10XUAS-IVS-mCD8::GFP (III, FBst0032185).
New NB7-1-Gal4KZ line
We generated a new NB7-1-Gal4KZ line that uses an enhancer killer zipper construct to eliminate the NB6-1 off-target expression seen in the published NB7-1 split-Gal4 line (Kohwi et al., 2013). The previous NB7-1-Gal4 line showed NB6-1 expression in 65% of hemisegments (n=20); the NB7-1-Gal4KZ line shows NB6-1 expression in just 25% of hemisegments (n=20). The new split gal4 genotype is ac-VP16 gsb-DBD, R25A05-KillerZipper/CyO (II; attP40), where R25A05 is an enhancer expressed in NB6-1. The full stock was created as follows. The Syn21-KZip(+)-P10 fragment was PCR amplified from CCAP-IVS-Syn21-KZip(+)-P10 (Dolan et al., 2017) (a gift from Benjamin White, National Institutes of Health) and fused via Gibson assembly with NheI/HindIII-digested pBPGal80Uw-6 (Pfeiffer et al., 2008) to create pBP-Syn21-KZip. The Janelia Research Campus enhancer R25A05 (FBst0000162964) was introduced into pBP-Syn21-KZip by gateway cloning (Pfeiffer et al., 2008) to generate R25A05-KZip, which was then integrated into attP40 site by standard injection (Bestgene).
Immunofluorescence staining
Primary antibodies were: rabbit anti-Hunchback #5-25 (1:200) (Tran and Doe, 2008), rabbit anti-Eve #2472 (1:100, Doe Lab), chicken anti-GFP (1:1000, Abcam, 13970), rat anti-Zfh2 (1:250) (Tran et al., 2010), mouse anti-HA-Alexa Fluor 488 conjugate (1:200, Cell Signaling, 2350S), rat anti-HA (1:100, Sigma, 11867423001), chicken anti-V5 (1:1000, Bethyl, A190-218A), rat anti-FLAG (1:400, Novus, NBP1-06712) and mouse anti-FasII (1:60, DSHB, 1D4). Secondary antibodies were from Jackson ImmunoResearch and were used at 1:350: anti-Rb 405 (Dylight 405, 711-475-152), anti-Rat 647 (AlexaFluor 647, 712-605-153), anti-Ck 488 (AlexaFluor 488, 703-545-155), anti-Mouse 647 (AlexaFluor 647, 715-605-151).
Embryos were blocked overnight in 0.3% PBST (1X PBS with 0.3% Triton X-100) with 5% normal goat serum and 5% donkey serum (PDGS), followed by incubation in primary antibody overnight at 4°C. Next, embryos underwent three 30 min washes in PBST, followed by an overnight secondary antibody incubation at 4°C. Embryos were then dehydrated in a glycerol series (10%, 50%, 90%) for 20 min each followed by 90% glycerol with 4% n-propyl Gallate overnight before imaging.
Whole L1 larvae were washed for 2 h in methanol, blocked overnight in 0.3% PBST (1×PBS with 0.3% Triton X-100) with 5% normal goat serum and 5% donkey serum (PDGS), followed by incubation in primary antibody for 2 nights at 4°C. Next, larvae were washed overnight in PBST, followed by secondary antibody incubation for 2 nights at 4°C. Embryos were dehydrated in a glycerol series (10%, 50% and 90%) for 20 min each followed by 90% glycerol with 4% n-propyl Gallate overnight before imaging. Larval brains were dissected in 0.3% PBST, fixed in 4% paraformaldehyde in PBST, rinsed and blocked in PDGS with 0.3% Triton X-100. Staining was carried out as above for embryos, but after the secondary antibody incubation brains were mounted in Vectashield (Vector Laboratories).
MCFO labeling
MCFO labeling in wild type used ac-gsb-Gal4, R25A05-KillerZipper (II) x hsFLPG5;;UAS-MCFO (I and III) and in Hb misexpression used ac-gsb-Gal4, R25A05-KillerZipper (II) x hsFLPG5;UAS-Hunchback;UAS-MCFO (I, II and III). Embryos were collected for 2 h at 25°C, aged for 4 h and heat shocked at 37°C (15-20 min for dense labeling, 8-10 min for sparse labelling), then left to develop until desired stages.
Imaging
Images were captured with a Zeiss LSM 710 or Zeiss LSM 800 confocal microscope with a z-resolution of 0.35 µm. Images were processed using the open-source software FIJI (https://fiji.sc) and Photoshop (Adobe). Figures were assembled in Illustrator CS5 (Adobe). Three-dimensional reconstructions, morphometrics and level adjustments were generated using Imaris (Bitplane). Any level adjustment was applied to the entire image.
Statistical analysis
Statistical significance is indicated by asterisks: ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; n.s., not significant. The following statistical tests were performed: two-tailed unpaired t-test (Fig. 2B,D,J,I,H,I,J); two-tailed paired t-test (Fig. 2F,H); and two-way ANOVA (Fig. 4). All analyses were performed using Prism8 (GraphPad). The results are stated as mean±s.d., unless otherwise noted.
Serial section electron microscopy
We accessed a previously published serial section transmission electron microscopic volume of the newly hatched larval CNS using CATMAID software (Ohyama et al., 2015) to describe the U1-U5 motor neurons in the first abdominal segment. U1-U5 motor neurons were identified based on their published unique dendritic morphology (Landgraf et al., 1997).
Note added in proof
While this manuscript was in review, a paper with similar findings was published (Heckscher et al., 2019 preprint).
Supplementary Material
Acknowledgements
We thank Luis Sullivan and Judith Eisen for comments on the manuscript; Sen-Lin Lai for making the new NB7-1-Gal4KZ line; Aref Zarin for annotating U3-U5 neurons in the EM reconstruction; Brandon Mark for providing helpful advice through the work; and Cooper Doe for assistance with sample preparation and imaging. Transgenic lines were made by BestGene. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.Q.D., A.Q.S.; Methodology: A.Q.S.; Formal analysis: A.Q.S.; Investigation: C.Q.D., A.Q.S.; Data curation: A.Q.S.; Writing - original draft: C.Q.D., A.Q.S.; Writing - review & editing: C.Q.D., A.Q.S.; Visualization: A.Q.S.; Supervision: C.Q.D.; Project administration: C.Q.D.; Funding acquisition: C.Q.D.
Funding
Funding was provided by the Howard Hughes Medical Institute (A.Q.S. and C.Q.D.; where C.Q.D. is an HHMI Investigator) and the National Institutes of Health (HD27056 to C.Q.D. and T32HD007348-24 to A.Q.S.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.175570.supplemental
References
- Barsh G. R., Isabella A. J. and Moens C. B. (2017). Vagus motor neuron topographic map determined by parallel mechanisms of hox5 expression and time of axon initiation. Curr. Biol. 27, 3812-3825.e3813. 10.1016/j.cub.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C., Li X., Erclik T., Cavey M., Wells B. and Desplan C. (2014). Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell 158, 1173-1186. 10.1016/j.cell.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F.-L. F., Steedman J. G. and Lund R. D. (1986). The lamination and connectivity of embryonic cerebral cortex transplanted into newborn rat cortex. J. Comp. Neurol. 244, 401-411. 10.1002/cne.902440311 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219-240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Bastiani M. J. and Goodman C. S. (1986). Guidance of neuronal growth cones in the grasshopper embryo. IV. Temporal delay experiments. J. Neurosci. 6, 3552-3563. 10.1523/JNEUROSCI.06-12-03552.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M.-J., Luan H., Shropshire W. C., Sutcliffe B., Cocanougher B., Scott R. L., Frechter S., Zlatic M., Jefferis G. and White B. H. (2017). Facilitating neuron-specific genetic manipulations in Drosophila melanogaster Using a split GAL4 repressor. Genetics 206, 775-784. 10.1534/genetics.116.199687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J., Venkatasubramanian L., Baek M., Peterson M., Aghayeva U. and Mann R. S. (2015). Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron 86, 955-970. 10.1016/j.neuron.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erclik T., Li X., Courgeon M., Bertet C., Chen Z., Baumert R., Ng J., Koo C., Arain U., Behnia R. et al. (2017). Integration of temporal and spatial patterning generates neural diversity. Nature 541, 365-370. 10.1038/nature20794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin L. G., van Schie M., Wouda R. R., de Jong A., Kamphorst J. T., Radjkoemar-Bansraj M. and Noordermeer J. N. (2004). The Drosophila Wnt5 protein mediates selective axon fasciculation in the embryonic central nervous system. Dev. Biol. 272, 362-375. 10.1016/j.ydbio.2004.04.034 [DOI] [PubMed] [Google Scholar]
- Gerhard S., Andrade I., Fetter R. D., Cardona A. and Schneider-Mizell C. M. (2017). Conserved neural circuit structure across Drosophila larval development revealed by comparative connectomics. eLife 6, e29089 10.7554/eLife.29089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R., Sabatelli L. M. and Seeger M. A. (1996). Guidance cues at the Drosophila CNS midline: identification and characterization of two Drosophila Netrin/UNC-6 homologs. Neuron 17, 217-228. 10.1016/S0896-6273(00)80154-3 [DOI] [PubMed] [Google Scholar]
- Hartenstein V. (1993). Atlas of Drosophila Development, pp. 57 Cold Spring HArbor Laboratory Press. [Google Scholar]
- Hartenstein V., Rudloff E. and Campos-Ortega J. A. (1987). The pattern of proliferation of the neuroblasts in the wild-type embryo of Drosophila melanogaster. Roux's Arch. Dev. Biol. 196, 473-485. 10.1007/BF00399871 [DOI] [PubMed] [Google Scholar]
- Heckscher E., Meng J. L., Marshall Z. D. and Lobb-Rabe M. (2019). How prolonged expression of Hunchback, a temporal transcription factor, re-wires locomotor circuits. bioRxiv. doi:10.1101/556209. 10.1101/556209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S. and Doe C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511-521. 10.1016/S0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Jankovski A., Rossi F. and Sotelo C. (1996). Neuronal precursors in the postnatal mouse cerebellum are fully committed cells: evidence from heterochronic transplantations. Eur. J. Neurosci. 8, 2308-2319. 10.1111/j.1460-9568.1996.tb01194.x [DOI] [PubMed] [Google Scholar]
- Jefferis G. S. X. E., Marin E. C., Stocker R. F. and Luo L. (2001). Target neuron prespecification in the olfactory map of Drosophila. Nature 414, 204-208. 10.1038/35102574 [DOI] [PubMed] [Google Scholar]
- Kanai M. I., Okabe M. and Hiromi Y. (2005). seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev. Cell 8, 203-213. 10.1016/j.devcel.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Kao C. F. and Lee T. (2010). Birth time/order-dependent neuron type specification. Curr. Opin. Neurobiol. 20, 14-21. 10.1016/j.conb.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M. and Doe C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823-838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M., Lupton J. R., Lai S. L., Miller M. R. and Doe C. Q. (2013). Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152, 97-108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A., Ertekin D., Lee C.-H. and Hummel T. (2016). Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. Elife 5, e13715 10.7554/eLife.13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M., Bossing T., Technau G. M. and Bate M. (1997). The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. 17, 9642-9655. 10.1523/JNEUROSCI.17-24-09642.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chen Z. and Desplan C. (2013a). Temporal patterning of neural progenitors in Drosophila. Curr. Top. Dev. Biol. 105, 69-96. 10.1016/B978-0-12-396968-2.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z., Voutev R., Venkatesh S., Morante J., Celik A. and Desplan C. (2013b). Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456-462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yang C.-P., Sugino K., Fu C.-C., Liu L.-Y., Yao X., Lee L. P. and Lee T. (2015). Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 350, 317-320. 10.1126/science.aad1886 [DOI] [PubMed] [Google Scholar]
- Mason C. A. (1983). Postnatal maturation of neurons in the cat's lateral geniculate nucleus. J. Comp. Neurol. 217, 458-469. 10.1002/cne.902170410 [DOI] [PubMed] [Google Scholar]
- Mauss A., Tripodi M., Evers J. F. and Landgraf M. (2009). Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 7, e1000200 10.1371/journal.pbio.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S. K. (1988). Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J. Neurosci. 8, 945-974. 10.1523/JNEUROSCI.08-03-00945.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K. J., Doyle J. L., Serafini T., Kennedy T. E., Tessier-Lavigne M., Goodman C. S. and Dickson B. J. (1996). Genetic analysis of Netrin genes in Drosophila: Netrins guide CNS commissural axons and peripheral motor axons. Neuron 17, 203-215. 10.1016/S0896-6273(00)80153-1 [DOI] [PubMed] [Google Scholar]
- Moris-Sanz M., Estacio-Gomez A., Sanchez-Herrero E. and Diaz-Benjumea F. J. (2015). The study of the Bithorax-complex genes in patterning CCAP neurons reveals a temporal control of neuronal differentiation by Abd-B. Biol. Open 4, 1132-1142. 10.1242/bio.012872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J. S., Williams P. R., Godinho L., Koerber A., Pittman A. J., Roeser T., Chien C. B., Baier H. and Wong R. O. (2006). In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron 52, 609-621. 10.1016/j.neuron.2006.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D. and Rubin G. M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967-E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T., Eiselt R. and Urban J. (2002). Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development 129, 1027-1036. [DOI] [PubMed] [Google Scholar]
- O'Leary D. D. and Stanfield B. B. (1989). Selective elimination of axons extended by developing cortical neurons is dependent on regional locale: experiments utilizing fetal cortical transplants. J. Neurosci. 9, 2230-2246. 10.1523/JNEUROSCI.09-07-02230.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T., Schneider-Mizell C. M., Fetter R. D., Aleman J. V., Franconville R., Rivera-Alba M., Mensh B. D., Branson K. M., Simpson J. H., Truman J. W. et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633-639. 10.1038/nature14297 [DOI] [PubMed] [Google Scholar]
- Olsson-Carter K. and Slack F. J. (2010). A developmental timing switch promotes axon outgrowth independent of known guidance receptors. PLoS Genet. 6, e1001054 10.1371/journal.pgen.1001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J. and Doe C. Q. (2003). Regulation of neuroblast competence in Drosophila. Nature 425, 624-628. 10.1038/nature01910 [DOI] [PubMed] [Google Scholar]
- Pearson B. J. and Doe C. Q. (2004). Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20, 619-647. 10.1146/annurev.cellbio.19.111301.115142 [DOI] [PubMed] [Google Scholar]
- Peters A. and Feldman M. L. (1976). The projection of the lateral geniculate nucleus to area 17 of the rat cerebral cortex. I. General description. J. Neurocytol. 5, 63-84. 10.1007/BF01176183 [DOI] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H., Laverty T. R. et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S. (1909). Histology of the Nervous System of Man and Vertebrates. Paris, France: Maloine. [Google Scholar]
- Rees C. L., Moradi K. and Ascoli G. A. (2017). Weighing the evidence in peters’ rule: does neuronal morphology predict connectivity? Trends Neurosci. 40, 63-71. 10.1016/j.tins.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Yang C.-P., Liu Z., Sugino K., Mok K., He Y., Ito M., Nern A., Otsuna H. and Lee T. (2017). Stem cell-intrinsic, seven-up-triggered temporal factor gradients diversify intermediate neural progenitors. Curr. Biol. 27, 1303-1313. 10.1016/j.cub.2017.03.047 [DOI] [PubMed] [Google Scholar]
- Rossi A. M., Fernandes V. M. and Desplan C. (2016). Timing temporal transitions during brain development. Curr. Opin. Neurobiol. 42, 84-92. 10.1016/j.conb.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg J. M., Hartley D. A., Walther Z. and Artavanis-Tsakonas S. (1988). slit: an EGF-homologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell 55, 1047-1059. 10.1016/0092-8674(88)90249-8 [DOI] [PubMed] [Google Scholar]
- Sanchez-Soriano N. and Prokop A. (2005). The influence of pioneer neurons on a growing motor nerve in Drosophila requires the neural cell adhesion molecule homolog FasciclinII. J. Neurosci. 25, 78-87. 10.1523/JNEUROSCI.2377-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaggar B. L. and O'Leary D. D. (1991). Potential of visual cortex to develop an array of functional units unique to somatosensory cortex. Science 252, 1556-1560. 10.1126/science.2047863 [DOI] [PubMed] [Google Scholar]
- Skeath J. B. and Thor S. (2003). Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13, 8-15. 10.1016/S0959-4388(03)00007-2 [DOI] [PubMed] [Google Scholar]
- Stanfield B. B. and O'Leary D. D. (1985). Fetal occipital cortical neurones transplanted to the rostral cortex can extend and maintain a pyramidal tract axon. Nature 313, 135-137. 10.1038/313135a0 [DOI] [PubMed] [Google Scholar]
- Stepanyants A. and Chklovskii D. B. (2005). Neurogeometry and potential synaptic connectivity. Trends Neurosci. 28, 387-394. 10.1016/j.tins.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Sullivan L. F., Warren T. L. and Doe C. Q. (2019). Temporal identity establishes columnar neuron morphology, connectivity, and function in a Drosophila navigation circuit. Elife 8, e43482 10.7554/eLife.43482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed M. H., Mark B. and Doe C. Q. (2017). Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. Elife 6, 26287 10.7554/eLife.26287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K. D. and Doe C. Q. (2008). Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development 135, 3491-3499. 10.1242/dev.024349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K. D., Miller M. R. and Doe C. Q. (2010). Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development 137, 1421-1430. 10.1242/dev.048678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Sweeney L. B., Ayoob J. C., Chak K., Andreone B. J., Ohyama T., Kerr R., Luo L., Zlatic M. and Kolodkin A. L. (2011). A combinatorial semaphorin code instructs the initial steps of sensory circuit assembly in the Drosophila CNS. Neuron 70, 281-298. 10.1016/j.neuron.2011.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S., Long H. and Thomas J. B. (2016). A subset of interneurons required for Drosophila larval locomotion. Mol. Cell. Neurosci. 70, 22-29. 10.1016/j.mcn.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S., McKinnon R. D., Kokel M. and Thomas J. B. (2003). Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature 422, 583-588. 10.1038/nature01522 [DOI] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C. F., Awasaki T., Chiang A. S. and Lee T. (2006). Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127, 409-422. 10.1016/j.cell.2006.08.045 [DOI] [PubMed] [Google Scholar]
- Zlatic M., Landgraf M. and Bate M. (2003). Genetic specification of axonal arbors: atonal regulates robo3 to position terminal branches in the Drosophila nervous system. Neuron 37, 41-51. 10.1016/S0896-6273(02)01131-5 [DOI] [PubMed] [Google Scholar]
- Zlatic M., Li F., Strigini M., Grueber W. and Bate M. (2009). Positional cues in the Drosophila nerve cord: semaphorins pattern the dorso-ventral axis. PLoS Biol. 7, e1000135 10.1371/journal.pbio.1000135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.