Abstract
The recently developed 3D bioprinting technology has greatly improved the ability to generate biomimetic tissues that are structurally and functionally relevant to their human counterparts. The selection of proper biomaterials as the bioinks is a key step toward successful bioprinting. For example, viscosity of a bioink is an important rheological parameter to determine the flexibility in deposition of free-standing structures and the maintenance of architectural integrity following bioprinting. This requirement, however, has greatly limited the selection of bioinks, especially for those naturally derived due to their commonly low mechanical properties. Here the generalization of a mechanism for extrusion bioprinting of bio-macromolecular components, mainly focusing on collagen and its derivatives including gelatin and gelatin methacryloyl, is reported. Specifically, a templating strategy is adopted using a composite bioink containing both the desired bio-macromolecular component and a polysaccharide alginate. The physically crosslinkable alginate component serves as the temporal structural support to stabilize the shape of the construct during bioprinting; upon subsequent chemical or physical crosslinking of the bio-macromolecular component, alginate can be selectively removed to leave only the desired biomacromolecule. It is anticipated that this strategy is general, and can be readily expanded for use of a wide variety of other bio-macromolecular bioinks.
Keywords: alginate, bioink, bio-macromolecule, extrusion bioprinting, microfluidic bioprinting
Tissue engineering has emerged as a promising solution to the unmet demand of tissues and organs for regenerative medicine. Tissue engineering uses a combination of cells, biomaterials, bioactive molecules, and engineering technologies to fabricate biological constructs that mimic and improve the functions of their counterparts in human body.[1–13] The field has significantly advanced during the past decades with the advent of numerous biofabrication strategies.[14,15] However, most of these conventional approaches do not possess the capacity to produce sophisticated architectures consisting of compositional complexity, and have therefore been insufficient in reconstituting structure and functionality of most biological tissues.[16]
The recently emerged three-dimensional (3D) bioprinting technology offers improved versatility and capability to deliver both cells and biomaterials with accurate control over spatial distributions, and is therefore capable of recreating tissue constructs and their models with desired architecture and function of target tissues.[15,17–20] While a wide range of synthetic biomaterials have been successfully utilized as the bioink, rarely have naturally derived biomacromolecules been adopted with only a few reports to date.[21,22] However, bio-macromolecular components are strongly desired for use as the bioink, which provide well-matched microenvironments in addition to architectural fidelity between the bioprinted biological structures and the native tissues.[18] The limitation is largely attributed to the relatively weak mechanical properties of the extracellular matrix (ECM)-like bio-macro molecules that do not allow for suitable printability to sustain the architectural integrity of the bioprinted constructs, especially in the case of extrusion bioprinting.[17,18,23,24]
Here, we have generalized a mechanism for extrusion bioprinting of bio-macromolecular bioinks. Specifically, we adopted a templating strategy using a composite bioink containing both the desired bio-macromolecular component and a nonhuman polysaccharide (i.e., alginate), generalized from our previously reported microfluidic bioprinting method.[25,26] As such, the physically crosslinkable alginate component, achieved by co-delivered calcium chloride (CaCl2) solution in the sheath of the printhead, serves as the temporal structural support to stabilize the shape of the construct during the bioprinting procedure. Subsequently, the bio-macromolecular component is crosslinked in a secondary step, through either photo-crosslinking (e.g., for gelatin methacryloyl (GelMA)), chemical crosslinking (e.g., for gelatin), or physical crosslinking (e.g., for collagen type I). Finally, the inert alginate component is selectively removed by immersing the bioprinted construct in a bath containing Ca2+-chelator to leave only the desired bio-macromolecule, supporting the function of the encapsulated cells.
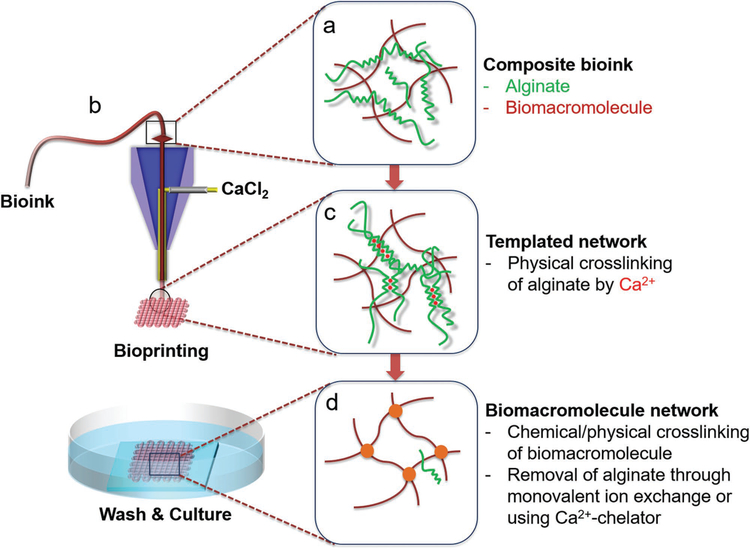
Figure 1 shows the general schematics of the microfluidic bioprinting-based templating method, from both macroscopic and microscopic views, for extrusion of the composite bioink containing an ECM-derived bio-macromolecule and the polysaccharide alginate (Figure 1a). To perform the microfluidic bioprinting, a custom nozzle made of two concentric needles in a core–sheath structure was fabricated and fitted onto the printhead of a bioprinter (Figure 1b).[25–27] In a typical process, the bioink is first extruded from the inner needle of the nozzle to produce a microfibrous structure of any desired shape and architecture, where the alginate component of this composite bioink is immediately physically crosslinked by the sheath CaCl2 solution co-delivered from the outer needle, until a desired scaffold is bioprinted (Figure 1c). Upon initial temporal fixation of the structure during the bioprinting process, the construct is further crosslinked chemically or physically for the ECM bio-macromolecular component; finally, the residual CaCl2 is removed by extensive washing with phosphate-buffered saline (PBS), and alginate can be selectively removed using a Ca2+-chelating agent, such as ethylenediaminetetraacetic acid (EDTA), to leave only the ECM bio-macromolecule in the bioprinted construct (Figure 1d). It should be noted that this strategy is general, and can be possibly applied for bioprinting of a wide variety of bio-macromolecular bioinks. Specifically, we first reproduced our previous reports in using such a strategy for bioprinting microfibrous GelMA-based constructs[25–28] to illustrate the entire microfluidic bioprintingenabled alginate-templating process, and then extended it to the use of gelatin and collagen as the bio-macromolecular bioinks demonstrated in this work.
Figure 1.
Schematics showing the bioprinting process: a) preparation of the composite bioink containing alginate and the desired bio-macromolecular bioink; b) construction of a core–sheath coaxial nozzle, where the composite bioink is delivered through the core flow and the CaCl2 solution is co-delivered through the sheath flow; c) physical crosslinking of the alginate component by Ca2+ during the bioprinting process, to ensure shape fidelity of the bioprinted microfibrous structures; and d) formation of the bio-macromolecular network through a secondary chemical/physical crosslinking, followed by gradual removal of alginate through monovalent ion exchange during culture in the medium or using a bath of Ca2+ chelator.
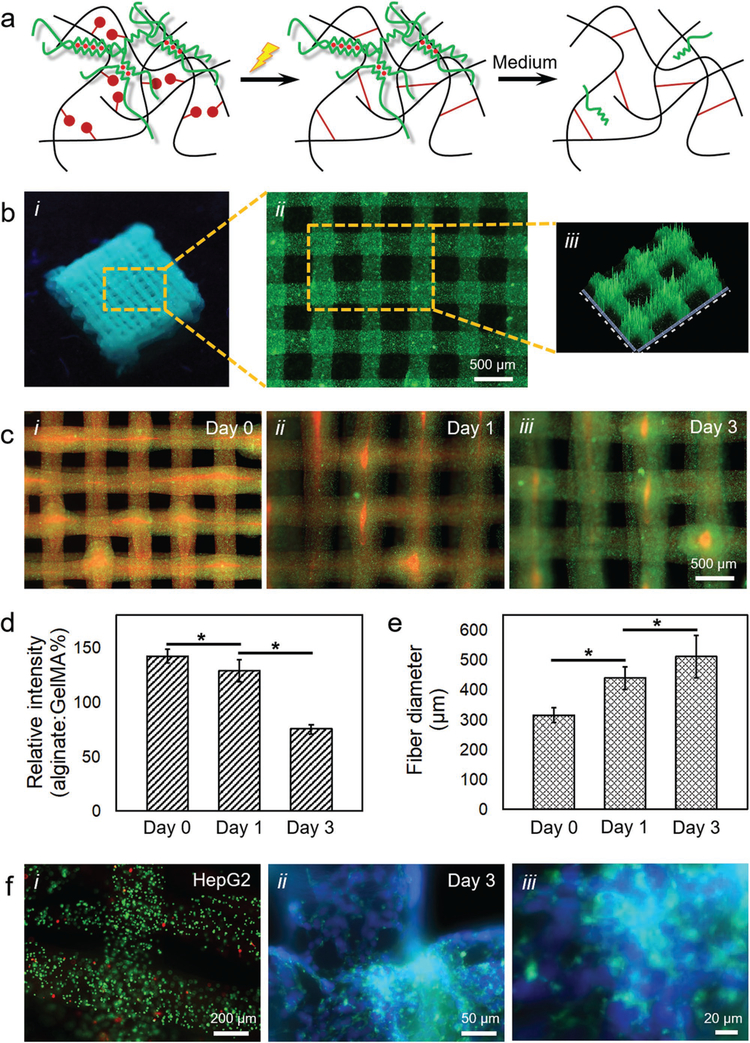
Figure 2a shows the schematics of alginate–GelMA bioprinting. After Ca2+-induced physical crosslinking of the alginate component, the methacryloyl groups in the GelMA component are subsequently photo-crosslinked by UV light illumination in the presence of the photoinitiator. Eventually, the bioprinted construct is immersed in a buffer of medium to dissociate the Ca2+-crosslinked alginate molecules, leading to the left-over of a hydrogel scaffold containing almost pure GelMA with minimum presence of alginate.
Figure 2.
Templated microfluidic bioprinting of the alginate–GelMA bioink. a) Schematics showing the dual-step crosslinking process and subsequent removal of alginate. b-i) Photograph showing a bioprinted five-layer construct; b-ii) fluorescence microscopic image showing the top view of the bioprinted microfibrous structure, where green fluorescent microbeads were incorporated into the bioink to assist visualization; and b-iii) pseudo-3D view showing the uniformity of the bioprinted microfibers. c) Fluorescence microscopic images showing the release of alginate from an individual bioprinted microfibrous construct at c-i) day 0, c-ii) day 1, and c-iii) day 3 post-bioprinting. The alginate was labeled in red by RITC while GelMA in green by FITC. d) Quantification of relative intensity (alginate-to-GelMA ratio) at different time points. e) Quantification of microfiber diameter at different time points. f) Fluorescence microscopic images showing f-i) live/dead staining (live cells in green and dead in red) at day 0, as well as f-ii,f-iii) F-actin/nuclei staining (F-actin in green and nuclei in blue) at day 3 of the encapsulated HepG2/C3A cells in the bioprinted constructs.
As an example, we bioprinted a five-layer microfibrous construct using a bioink containing 2% (w/v) alginate and 7% (w/v) GelMA, delivered through the interior flow at a flow rate of 10 μL min−1, and a 0.3 m CaCl2 solution co-delivered through the exterior flow at a flow rate of 10 μL min−1, while the nozzle movement speed was maintained at 4 mm s−1. The bioink formulation was stable in rheology over the period tested that matched the time needed for a typical microfluidic bioprinting process (<5 min; Figure S1a, Supporting Information). The bioink also exhibited a shear-thinning property with overall low viscosities (Figure S1b, Supporting Information). It was clear that, however, the bioprinted multilayer construct was structurally robust after photo-crosslinking at a power of 800 mW cm−2 for 45 s (Figure 2b-i), featuring evenly spaced microfibers with an average diameter of 313 ± 25 μm (Figure 2b-ii). Pseudo-3D reconstruction image further revealed the uniform sizes of the microfibers bioprinted in the two adjacent layers (Figure 2b-iii).
We next analyzed the ability to remove the alginate component from the bioprinted construct post-photo-crosslinking of GelMA. Alginate is a brown algae–derived polysaccharide composed of α-l-guluronic acid (G) and β-d-mannuronic acid (M) residues.[29] The G-blocks within the polymer of alginate can rapidly gel in the presence of certain divalent cations such as Ca2+ or Ba2+, resulting in the formation of an “eggbox” configuration where chain pairs pack and lock the ions in between.[30] This gelation process is, nevertheless, unstable and reversible.[20,31] When immersed in a solution containing monovalent ions, these ions can replace the divalent ions leading to the breakdown of the “egg-box” configuration and dissociation of the physically crosslinked alginate hydrogel.[32] Indeed, when our bioprinted alginate/GelMA microfibrous construct was incubated in a standard Dulbecco’s modified Eagle’s medium (DMEM) containing ≈154.6 × 10−3 m Na+ and ≈5.4 × 10−3 m K+, we were able to observe a gradual leach-out of the dissociated alginate (labeled in red with rhodamine isothiocyanate (RITC)) from the GelMA matrix (labeled in green with fluorescein isothiocyanate (FITC)) over a 3 day duration (Figure 2c). The relative intensities of the alginate component, expressed by the percentage intensity ratios of alginate to GelMA, decreased from 142 ± 6 at day 0 (immediate following bioprinting/photo-crosslinking) to 129 ± 10 at day 1 and 75 ± 4 at day 3, indicating a >50% drop in alginate intensity during the period (Figure 2d). This dissociation of physically crosslinked alginate may also be accelerated when a chelator for the divalent ions, such as EDTA, is added to the surrounding medium of the alginate hydrogel.[25,27,33,34]
Meanwhile, as the crosslinked alginate was gradually dissociated and leached out of the bioprinted construct, the diameter of the remaining GelMA/alginate composite hydrogel microfibers increased due to swelling (Figure 2e). The diameters of the microfibers increased from 313 ± 25 μm at day 0 (immediately following bioprinting/photo-crosslinking) to 437 ± 38 μm at day 1 and 509 ± 70 μm at day 3, consistent with the literature.[25,26] Accompanying the swelling, the removal of the alginate could also induce the enlargement of the pores of the remaining hydrogel microfibers, potentially leading to improved behaviors of the encapsulated cells, as we previously reported.[34]
Of note, this microfluidic bioprinting procedure has been demonstrated benign to the encapsulated cells.[25–27,34] As revealed in Figure 2f-i, the viability of the HepG2/C3A cells was maintained at a level of >85% post-bioprinting analyzed from semiquantitative calculations based on live/dead staining, suggesting the biocompatibility of the CaCl2 crosslinker at the selected concentration, as well as the minimal damage exerted by the shear stress during the extrusion of the cell-laden bioink. F-actin/nuclei staining results of the HepG2/C3A cells at day 3 in culture further proved the proliferation of the cells in the bioprinted microfibrous structures, evidenced by formation of aggregates (Figure 2f-ii,f-iii) as opposed to the uniform single-cell distributions at day 0 (Figure 2f-i).
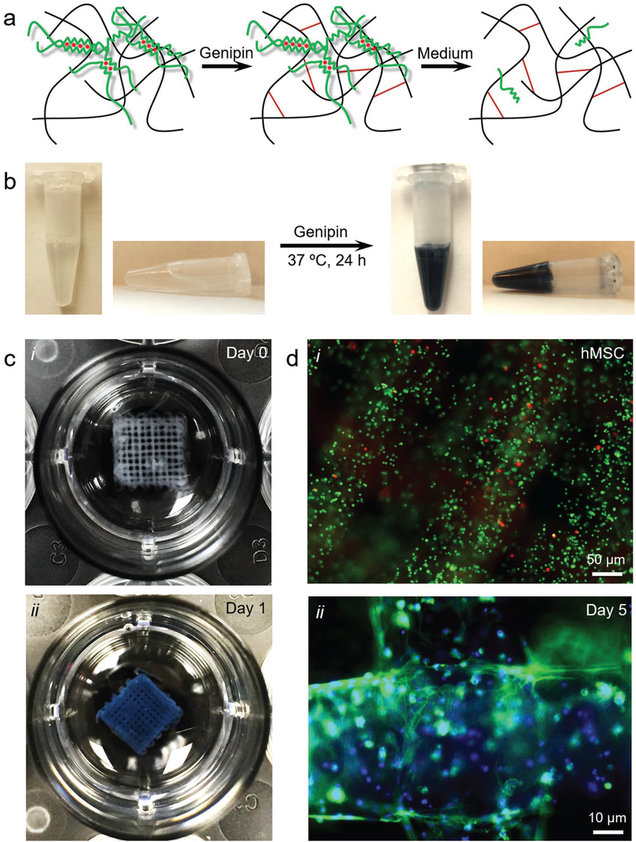
We then extended this strategy to the bioprinting of other variations of bio-macromolecular bioinks besides GelMA. Figure 3a shows the schematics of alginate–gelatin bioprinting. After physical crosslinking of the alginate component by the co-delivered sheath CaCl2 solution during the bioprinting process, the gelatin component is subsequently chemically crosslinked by genipin treatment. Finally, the physically crosslinked alginate is removed to leave a hydrogel scaffold containing mostly gelatin.
Figure 3.
Templated microfluidic bioprinting of the alginate–gelatin bioink. a) Schematics showing the dual-step crosslinking process and subsequent removal of alginate. b) Photographs showing the crosslinking process of gelatin by genipin. c) Photographs showing a bioprinted five-layer construct at c-i) day 0 and c-ii) day 1 post-bioprinting. d) Fluorescence microscopic images showing d-i) live/dead staining (live cells in green and dead in red) at day 0, as well as d-ii) F-actin/nuclei staining (F-actin in green and nuclei in blue) at day 5 of the encapsulated hMSCs in the bioprinted constructs.
It should be noted that we chose genipin as a crosslinking agent due to its remarkably lower cytotoxicity when compared to other crosslinkers based on aldehydes,[35,36] allowing for reactions to occur in the presence of living cells and thus the use of cell-laden bioinks. Genipin is a derivative of geniposide isolated from the fruits of Genipa americana and Gardenia jasminoides Ellis, obtained through enzymatic hydrolysis.[37,38] Two possible reactions are involved in the crosslinking process of gelatin by genipin: i) the faster reaction relying on a nucleophilic attack on genipin by a primary amine group in gelatin leading to the formation of a heterocyclic compound of genipin linked to the basic residues also in gelatin; and ii) the slower reaction relying on the nucleophilic substitution of the ester group possessed by genipin resulting in the formation of a secondary amide link with gelatin.[37] Interestingly, additional complex reactions, mainly the oxygen radical–induced polymerization of genipin and dehydrogenation of intermediate compounds, generate blue pigments,[37–40] enabling the crosslinking reaction to be visually monitored (Figure 3b).
Photograph of a bioprinted five-layer alginate/gelatin microfibrous construct is shown in Figure 3c-i. Similar to the case of alginate/GelMA scaffold, the construct had evenly spaced and uniformly sized microfibers across the entire area, at the gelatin concentration of 10% (w/v) and the alginate concentration of 2% (w/v). The chemical crosslinking of gelatin is completed in roughly a day’s time during its incubation at 37 °C, indicated by the prominent color change from milky white to deep blue (Figure 3c-ii). This timeline is well balanced with the time needed for the dissociation of physically crosslinked alginate from the composite matrix, which had possibly contributed to the maintenance of the 3D microfibrous structure with the initial presence of alginate chains holding the gelatin bio-macromolecules in place before they formed a robust interconnected network through chemical crosslinking. The bioprinting process and the genipin crosslinking step were benign, resulting in a high viability of encapsulated human mesenchymal stem cells (hMSCs) at >90% calculated from live/dead staining (Figure 3d-i). At day 5 of culture, most of the encapsulated hMSCs could stretch to assume elongated morphologies, consistent with our previous observations for similar bioprinted structures post-release of alginate from composite matrices (Figure 3d-ii).[34] It should be noted, however, while the bioink formulation was also stable in rheology over a 5 min period (Figure S1c, Supporting Information), the bioink exhibited fairly high viscosities with a shear-thinning behavior due to the gelation behavior of gelatin at the room temperature range (Figure S1d, Supporting Information). This fact indicated the flexibility of our microfluidic bioprinting technology in supporting the extrusion of bioinks with a wide viscosity range.
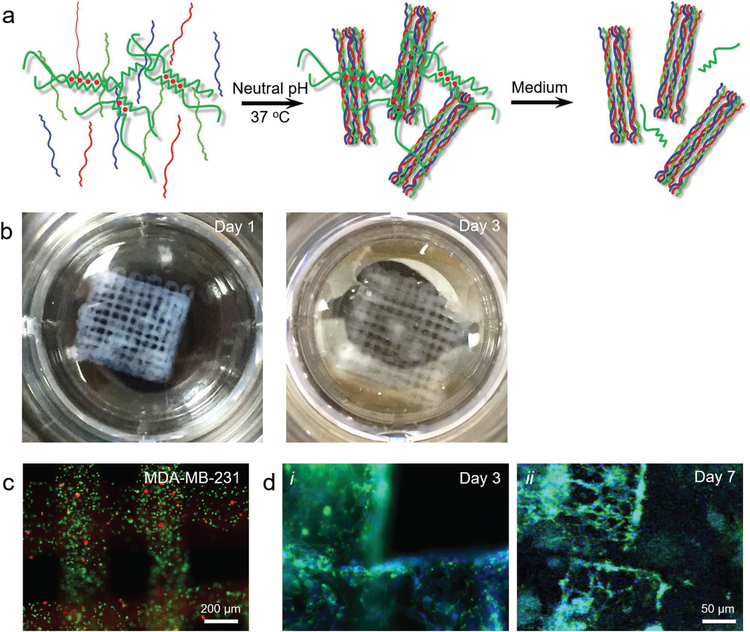
Besides gelatin-based bio-macromolecules, we finally demonstrated the use of the strategy for extrusion bioprinting of collagen type I, which is the most abundant type of collagen in the human body. It is expressed almost unanimously in all connective tissues and functions as the key structural protein of several tissue such as the bone, tendon, skin, and cornea.[41] It also exists in large quantities in many tumor microenvironments.[42,43] Figure 4a shows the schematics of alginate–collagen I bioprinting. Prior to bioprinting, the composite bioink is adjusted to neutral pH, and the bioprinting is undertaken at room temperature to reduce the chain coiling/packing rate of the collagen molecules. Following bioprinting and Ca2+ crosslinking of alginate, the physical crosslinking of the collagen I component is expedited by incubating the construct at 37 °C for 30 min. Alginate is then released from the matrix leaving the main composition as collagen I. Indeed, it was found that at a collagen concentration of 1 mg mL−1 and an alginate concentration of 2% (w/v), the extrusion was smooth leading to generation of multilayered microfibrous constructs, which possessed good stability showing slight swelling when immersed in PBS for 3 days (Figure 4b).
Figure 4.
Templated microfluidic bioprinting of the alginate–collagen type I bioink. a) Schematics showing the dual-step crosslinking process and subsequent removal of alginate. b) Photographs showing bioprinted microfibrous structures at days 1 and 3. c) Fluorescence microscopic images showing live/dead staining (live cells in green and dead in red) of the encapsulated hMSCs in the bioprinted constructs at day 0. d) Fluorescence microscopic images showing F-actin/nuclei staining (F-actin in green and nuclei in blue) of the encapsulated MDA-MB-231 breast cancer cells in the bioprinted constructs at i) day 3 and ii) day 5.
The compatibility of this strategy with bioprinting was subsequently evaluated. When MDA-MB-231 breast cancer cells were encapsulated in the bioink, they could uniformly distribute within the entire volume of the bioprinted microfibers, exhibiting a relative high viability of >90% (Figure 4b). Over a culture period of 7 days, the MDA-MB-231 cells were observed to stretch and proliferate, forming an interconnected network within the boundaries of the microfibers (Figure 4c), indicating the bioactive microenvironments of the bioprinted collagen scaffold in supporting the growth of the encapsulated cells. Analogous to the GelMA–alginate bioink, the collagen–alginate bioink had overall low viscosities with a shear-thinning behavior, and maintained stable rheological properties over a 5 min test period (Figure S1e,f, Supporting Information).
It should be noted that direct extrusion bioprinting of bio-macromolecular ECM-derived bioinks has been historically challenging due to their usually weak mechanical properties. While gelatin-based bio-macromolecules, including gelatin and GelMA, may be extrudable by taking advantages of their thermoresponsive characteristics (i.e., formation of gels at lower temperature),[44–47] the deposition of collagens can be difficult and unstable. Direct extrusion bioprinting of pure collagen type I at extremely high concentrations of 10–20 mg mL−1 possessing strong viscosities was reported, which, however, had poor resolutions.[48] Alternatively, a viscous mixture of collagen, agarose, and alginate may be directly extruded to form microfibrous scaffolds with higher resolution.[49] These approaches, nevertheless, can potentially exert unwanted shear stresses on the encapsulated cells during extrusion, compared to situations when microfluidic bioprinting is employed.
In summary, we have generalized a strategy for extrusion bioprinting of bio-macromolecular components. A templating mechanism was adopted, where a combination of the desired bio-macromolecule and alginate both at relatively low concentrations was formulated as a composite bioink. With the use of a core–sheath coaxial microfluidic printhead, the alginate component of the bioink delivered through the core flow was immediately physically crosslinked by the Ca2+ present in the outer sheath flow, ensuring the shape stabilization of the bioprinted microfibrous structure. Subsequently, further crosslinking of the bio-macromolecular component was achieved, whether photochemically (e.g., for GelMA), chemically (e.g., for gelatin), or physically (e.g., for collagen). Finally, over the culture period, alginate was selectively removed through dissociation, leaving only the desired bio-macromolecule. Specifically, we demonstrated the application of such a strategy in extrusion bioprinting of several common ECM-derived bio-macromolecules, including GelMA, gelatin, and collagen type I. We also proved the biocompatibility of the strategy with multiple cell types of HepG2/C3A hepatocytes, hMSCs, and MDA-MB-231 breast cancer cells.
As bio-macromolecules making up ECM-like hydrogels, GelMA, gelatin, and collagen hold great promise in their applications in tissue engineering. However, certain limitations such as poor printability arising from their relatively low mechanics remain challenging for them to be used as bioinks, especially in the case of extrusion bioprinting. The alginate-based templating strategy in combination with microfluidic bioprinting reported in this current work presents a reliable approach for extrusion of ECM-like bio-macromolecular bioinks, allowing widespread utility of these favorable naturally derived biomaterials in generation of biomimetic tissues through 3D bioprinting. While we have demonstrated three independent categories of crosslinking mechanisms, namely photo-crosslinking, chemical crosslinking, and physical crosslinking, we anticipate many other crosslinking mechanisms to be compatible with the bioprinting strategy leading to its generalization to most bio-macromolecules (e.g., hyaluronic acid, laminin, fibrin, and their derivatives) to target various tissue types to be fabricated.
Experimental Section
Materials:
Alginic acid sodium salt from brown algae (low-viscosity, 100–300 cP), CaCl2, gelatin (type A from porcine skin, 300 Bloom), collagen (type I), methacrylic anhydride, and 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (photoinitiator, PI) were purchased from Sigma-Aldrich (St. Louis, MO, USA). PBS, fetal bovine serum (FBS), penicillin–streptomycin, amphotericin, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), DMEM, LIVE/DEAD Viability/Cytotoxicity Kit, and PrestoBlue Kit were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
GelMA (substituting degree ≈80%) was synthesized according to the previously reported protocol.[25,28,34,47] Described in short, gelatin was dissolved at 60 °C in PBS. With continuous stirring and maintaining temperature, methacrylic anhydride was added dropwise to the gelatin solution. The ratio of gelatin to methacrylic anhydride was 1:0.6. Unreacted methacrylic anhydride was removed through dialysis against deionized water, with a 12–14 kDa molecular weight cut-off range, at 50 °C for 7 days. Finally, the solution was freeze-dried for 5–7 days and stored at room temperature until use.
Cell Culture:
HepG2/C3A hepatocytes, hMSCs, and MDA-MB-231 breast cancer cells were all purchased from American Type Culture Collection (ATCC, Manassas, MD, USA). All cells were cultured in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, and 0.1% amphotericin at 37 °C and 5% CO2 in an incubator with a humidified atmosphere. The cells were passaged every 2–3 days at 70% confluency.
Bioink Preparation:
GelMA (14%, w/v), gelatin (20%, w/v), or collagen (2 mg mL−1) was dissolved in PBS at 37 °C. To obtain the cell-laden bioink, an alginate solution (4%, w/v) at an equal volume was mixed with the desired bio-macromolecule solution to constitute the final composite bioink. Afterward, cells were mixed with the composite bioink and mildly pipetted for 10 times to obtain the cell-laden bioink.
Microfluidic Bioprinting:
The coaxial nozzle, customized as previously reported,[25,26,28] was installed on an NovoGen MMX Bioprinter (Organovo, San Diego, CA, USA) to perform the continuous 3D deposition of computer-designed models. The internal nozzle (i.d. = 200 μm) was connected with a plastic tubing to a 1 mL syringe loaded with the composite bioink, while the external nozzle (i.d. = 600 μm) was fed with the CaCl2 solution (0.3 m). Extrusion was controlled by syringe pumps (Harvard Apparatus, Holliston, MA, USA). The first crosslinking procedure was designed to start when both flows reached the tip of the coaxial nozzle. It has been well-reported that the temperature, the distance of the nozzle to the substrate, the moving speed of the nozzle, and the extrusion rate of the bioink are main factors affecting the fidelity of the bioprinted structures. These parameters were varied to be in the ranges of 0.5–3.0 mm, 1–10 mm s−1, and 1–20 μL min−1, respectively. To bioprint a continuous microfibrous construct, it was found that the optimized parameters of bioprinting were 1.0 mm of nozzle distance, 4–6 mm s−1 of nozzle moving speed, and 5–10 μL min−1 of extrusion rate.
At the end of the bioprinting, the GelMA–alginate constructs were exposed to UV light (800 mW cm−2) for 30 s to undergo the second photo-crosslinking. The gelatin–alginate constructs were chemically crosslinked by incubation with a genipin-containing medium (0.01% w/v). Collagen–alginate were adjusted to neutral pH value and mixed with cells. After bioprinting, constructs were physically crosslinked at 37 °C. After the completion of the secondary crosslinking step of the bioprinted constructs, they were put into culture to gradually leach out the alginate component. Alternatively, the constructs could also be immersed in an EDTA solution for 1 min to quickly remove the alginate molecules.[25,26] The tensile moduli of the constructs generated from the concentrations of GelMA (7% w/v), gelatin (10% w/v), and collagen (1 mg mL−1) were measured to be 44.7 ± 0.8, 67.0 ± 1.3, and 41.7 ± 0.5 kPa, respectively (Figure S2, Supporting Information).
Cellular Characterizations:
The cell viability in the bioprinted constructs was measured by live/dead assay based on the manufacturer’s protocol. Ethidium homodimer-1 when bound to the nuclei of the dead cells fluoresces red and calcein acetoxymethyl can hydrolize in the cytoplasm of live cells to fluoresce green. After washing the samples, 300 μL solution of the two dyes (2 and 0.5 μL mL−1, respectively) was added into each sample and incubated at 37 °C for 30 min. Images were taken with a fluorescence microscope (Axio Observer D1, Carl Zeiss, Thornwood, NY, USA).
For observing the cytoskeleton, the samples were fixed for 30 min in 4% paraformaldehyde (Sigma-Aldrich) PBS solution at room temperature. After fixation, the cells were permeabilized with 0.15% Triton X-100 (Sigma-Aldrich) for 30 min, and subsequently stained with F-actin. The nuclei were counterstained with 4′6-diamidino-2-phenylindole (DAPI, 1:1000 dilution, Thermo Fisher Scientific) in deionized water. The stained samples were then imaged with a fluorescence microscope.
Supplementary Material
Acknowledgements
K.Z. and N.C. contributed equally to this work. The authors acknowledge funding from the National Cancer Institute of the National Institutes of Health (K99CA201603) and Science and Technology Commission of Shanghai Municipality (STCSM, 17JC 1400200). K.Z. acknowledges funding from Shanghai Pujiang Program (Program No. 17PJ1401500), National Natural Science Foundation of China (Grant No. 81771971), and the “Chen Guang” Project Supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (Program No. 14CG06).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Kai Zhu, Department of Cardiac Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, P. R. China; Shanghai Institute of Cardiovascular Disease, Shanghai 200032, P. R. China.
Nan Chen, Department of Cardiac Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, P. R. China; Shanghai Institute of Cardiovascular Disease, Shanghai 200032, P. R. China.
Prof. Xiao Liu, Beijing Advanced Innovation Center for Biomedical Engineering, Key Laboratory for Biomechanics and Mechanobiology of the Ministry of Education, School of Biological Science and Medical Engineering, Beihang University, Beijing 100083, P. R. China
Xuan Mu, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
Prof. Weijia Zhang, Department of Chemistry and Institute of Biomedical Science, Fudan University, Shanghai 200433, P. R. China
Prof. Chunsheng Wang, Department of Cardiac Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, P. R. China Shanghai Institute of Cardiovascular Disease, Shanghai 200032, P. R. China.
Yu Shrike Zhang, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
References
- [1].Langer R, Vacanti JP, Science 1993, 260, 920. [DOI] [PubMed] [Google Scholar]
- [2].Langer R, Vacanti JP, Vacanti CA, Atala A, Freed LE, Vunjak-Novakovic G, Tissue Eng. 1995, 1, 151. [DOI] [PubMed] [Google Scholar]
- [3].Langer R, e-Biomed 2000, 1, 5. [Google Scholar]
- [4].Khademhosseini A, Vacanti JP, Langer R, Sci. Am 2009, 300, 64. [DOI] [PubMed] [Google Scholar]
- [5].Rice JJ, Martino MM, De Laporte L, Tortelli F, Briquez PS, Hubbell JA, Adv. Healthcare Mater 2013, 2, 57. [DOI] [PubMed] [Google Scholar]
- [6].Zhang YS, Xia Y, Nanomedicine 2015, 10, 689. [DOI] [PubMed] [Google Scholar]
- [7].Mao AS, Mooney DJ, Proc. Natl. Acad. Sci. USA 2015, 112, 14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berthiaume F, Maguire TJ, Yarmush ML, Annu. Rev. Chem. Biomol. Eng 2011, 2, 403. [DOI] [PubMed] [Google Scholar]
- [9].Choi S-W, Zhang Y, Xia Y, Langmuir 2010, 26, 19001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi S-W, Zhang Y, Yeh Y-C, Lake Wooten A, Xia Y, J. Mater. Chem 2012, 22, 11442. [Google Scholar]
- [11].Zhang Y, Choi S-W, Xia Y, Macromol. Rapid Commun 2012, 33, 296. [DOI] [PubMed] [Google Scholar]
- [12].Zhang YS, Choi S-W, Xia Y, Soft Matter 2013, 9, 9747. [Google Scholar]
- [13].Zhang YS, Regan KP, Xia Y, Macromol. Rapid Commun 2013, 34, 485. [DOI] [PubMed] [Google Scholar]
- [14].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [15].Zhang YS, Yue K, Aleman J, Moghaddam K, Bakht SM, Dell’Erba V, Assawes P, Shin SR, Dokmeci MR, Oklu R, Khademhosseini A, Ann. Biomed. Eng 2017, 45, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Leijten J, Rouwkema J, Zhang YS, Nasajpour A, Dokmeci MR, Khademhosseini A, Small 2016, 10, 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Malda J, Visser J, Melchels FP, Jungst T, Hennink WE, Dhert WJ, Groll J, Hutmacher DW, Adv. Mater 2013, 25, 5011. [DOI] [PubMed] [Google Scholar]
- [18].Skardal A, Atala A, Ann. Biomed. Eng 2015, 43, 730. [DOI] [PubMed] [Google Scholar]
- [19].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [20].Zhang YS, Khademhosseini A, Science 2017, 356, eaaf3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pati F, Jang J, Ha D-H, Kim SW, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Nat. Commun 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jang J, Kim TG, Kim BS, Kim S-W, Kwon S-M, Cho D-W, Acta Biomater 2016, 33, 88. [DOI] [PubMed] [Google Scholar]
- [23].Chung JHY, Naficy S, Yue Z, Kapsa R, Quigley A, Moulton SE, Wallace GG, Biomater. Sci 2013, 1, 763. [DOI] [PubMed] [Google Scholar]
- [24].Murphy SV, Skardal A, Atala A, J. Biomed. Mater. Res., Part A 2013, 101, 272. [DOI] [PubMed] [Google Scholar]
- [25].Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Malekabadi ZG, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Biomaterials 2016, 110, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang YS, Pi Q, van Genderen AM, Visualized Exp J 2017, 126, e55957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A, Adv. Mater. 2016, 28, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhu K, Shin SR, van Kempen T, Li YC, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J, Adv. Funct. Mater 2017, 27, 1605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rowley JA, Madlambayan G, Mooney DJ, Biomaterials 1999, 20, 45. [DOI] [PubMed] [Google Scholar]
- [30].Braccini I, Pérez S, Biomacromolecules 2001, 2, 1089. [DOI] [PubMed] [Google Scholar]
- [31].Augst AD, Kong HJ, Mooney DJ, Macromol. Biosci 2006, 6, 623. [DOI] [PubMed] [Google Scholar]
- [32].Moe ST, Skjaak-Braek G, Elgsaeter A, Smidsroed O, Macromolecules 1993, 26, 3589. [Google Scholar]
- [33].Zhang Y, Xia Y, Adv. Funct. Mater 2012, 22, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sung H-W, Huang R-N, Huang LLH, Tsai C-C, J. Biomater. Sci., Polym. Ed 1999, 10, 63. [DOI] [PubMed] [Google Scholar]
- [36].Mi F-L, Tan Y-C, Liang H-C, Huang R-N, Sung H-W, J. Biomater. Sci., Polym. Ed 2001, 12, 835. [DOI] [PubMed] [Google Scholar]
- [37].Butler MF, Ng Y-F, Pudney PDA, J. Polym. Sci., Part A: Polym. Chem 2003, 41, 3941. [Google Scholar]
- [38].Park J-E, Lee J-Y, Kim H-G, Hahn T-R, Paik Y-S, Agric J Food Chem. 2002, 50, 6511. [DOI] [PubMed] [Google Scholar]
- [39].Touyama R, Takeda Y, Inoue K, Kawamura I, Yatsuzuka M, Ikumoto T, Shingu T, Yokoi T, Inouye H, Chem. Pharm. Bull 1994, 42, 668. [Google Scholar]
- [40].Touyama R, Inoue K, Takeda Y, Yatsuzuka M, Ikumoto T, Moritome N, Shingu T, Yokoi T, Inouye H, Chem. Pharm. Bull 1994, 42, 1571. [Google Scholar]
- [41].Henriksen K, Karsdal MA, in Biochemistry of Collagens, Laminins and Elastin (Ed: Karsdal MA), Academic Press, San Diego, CA: 2016, p. 1. [Google Scholar]
- [42].Zhang YS, Duchamp M, Oklu R, Ellisen LW, Langer R, Khademhosseini A, ACS Biomater. Sci. Eng 2016, 2, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pickup MW, Mouw JK, Weaver VM, EMBO Rep. 2014, 15, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schuurman W, Levett PA, Pot MW, van Weeren PR, Dhert WJA, Hutmacher DW, Melchels FPW, Klein TJ, Malda J, Macromol. Biosci 2013, 13, 551. [DOI] [PubMed] [Google Scholar]
- [45].Luiz EB, Juliana CC, Vijayan M, Ana LC, Nupura SB, Wesleyan AA, Pinar Z, Nihal EV, Amir MG, Mehmet RD, Ali K, Biofabrication 2014, 6, 024105.24695367 [Google Scholar]
- [46].Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P, Biomaterials 2014, 35, 49. [DOI] [PubMed] [Google Scholar]
- [47].Liu W, Heinrich MA, Zhou Y, Akpek A, Hu N, Liu X, Guan X, Zhong Z, Jin X, Khademhosseini A, Zhang YS, Adv. Healthcare Mater 2017, 6, 1601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rhee S, Puetzer JL, Mason BN, Reinhart-King CA, Bonassar LJ, ACS Biomater. Sci. Eng 2016, 2, 1800. [DOI] [PubMed] [Google Scholar]
- [49].Yang X, Lu Z, Wu H, Li W, Zheng L, Zhao J, Mater. Sci. Eng., C 2018, 83, 195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.