Abstract
Despite advances in the bioprinting technology, biofabrication of circumferentially multilayered tubular tissues or organs with cellular heterogeneity, such as blood vessels, trachea, intestine, colon, ureter, and urethra, remains a challenge. Herein, a promising multichannel coaxial extrusion system (MCCES) for microfluidic bioprinting of circumferentially multilayered tubular tissues in a single step, using customized bioinks constituting gelatin methacryloyl, alginate, and eight-arm poly(ethylene glycol) acrylate with a tripentaerythritol core, is presented. These perfusable cannular constructs can be continuously tuned up from monolayer to triple layers at regular intervals across the length of a bioprinted tube. Using customized bioink and MCCES, bioprinting of several tubular tissue constructs using relevant cell types with adequate biofunctionality including cell viability, proliferation, and differentiation is demonstrated. Specifically, cannular urothelial tissue constructs are bioprinted, using human urothelial cells and human bladder smooth muscle cells, as well as vascular tissue constructs, using human umbilical vein endothelial cells and human smooth muscle cells. These bioprinted cannular tissues can be actively perfused with fluids and nutrients to promote growth and proliferation of the embedded cell types. The fabrication of such tunable and perfusable circumferentially multilayered tissues represents a fundamental step toward creating human cannular tissues.
Keywords: bioinks, cannular tissues, coaxial extrusion systems, microfluidic bioprinting, perfusion
The 3D bioprinting technology provides an excellent platform to fabricate biomimetic complex tissue constructs.[1,2] An important advantage offered by this technology is the ability to deposit biomaterials or cell-loaded biomaterials individually or in tandem, producing desired 3D tissue-like structures.[3] In the past two decades, biofabrication has been explored extensively for generating 3D tubular tissues, particularly vasculature, by simultaneous deposition of biocompatible materials and cells, through layer-by-layer assembly of 2D patterns to form 3D well-organized structures[4] or by 3D sacrificial bioprinting.[5–7] However, most of these available approaches generate cell-laden hydrogels with embedded microchannels that require the delicate process of precise stacking of individually fabricated layers. In addition to slow and multistep processes, these approaches lack ability to perfuse fluids directly through the tissue constructs.
More recently, extrusion bioprinting of biomaterials and blends using coaxial nozzles has been shown to produce perfusable hollow fibers,[5,8] which is the basic requirement for generating functional tubular tissues. Several studies have reported the fabrication of hollow fibers using various 3D bioprinting approaches. For example, Zhang et al. employed a coaxial-nozzle extrusion method that enabled direct bioprinting of cellular microfluidic channels in the form of hollow tubes.[9,10] Similarly, Nishiyama et al.,[11] Christensen et al.,[12] and Blaeser et al.[13] separately demonstrated layer-by-layer fabrication of vascular-like cellular structures using an inkjet-based approach. Notably, Xiong et al. demonstrated the bioprinting of 3D cellular tubes using laser-induced forward transfer,[14] whereas Ouyang et al. showed the 3D bioprinting of heterogeneous and hollow filaments allowing fabrication of complex engineered cell-laden constructs, using a coaxial extrusion printing system.[15] Moreover, some groups employed layer-by-layer fabrication of hydrogel mixtures containing two different cell types to produce porous tissues with vertical channels,[16] and others bioprinted multiple cell types along with sacrificial Pluronic F127 to generate hollow channels.[17] However, 3D bioprinting of tunable circumferentially multilayered hollow structures that would mimic tissues such as gastrointestinal tract, trachea, urinary bladder, ureter, or urethra with structural and functional integrity, still remains a challenging task in tissue engineering.[18]
In the present study, we introduce a digitally coded coaxial extrusion device that can directly bioprint 3D complex tubular hollow fibers with multiple circumferential layers in a single step, without the need of pre-/postprocessing. The hollow filaments are directly extruded by a pressure-assisted bioprinting system using gelatin methacryloyl (GelMA)-based hydrogels as bioinks. This system enables continuous fabrication of perfusable and tunable circumferentially multilayered tubular fibers that can be switched between monolayer and double layers, at regular intervals, or even triple layers, across the length of the tube. As a proof-of-concept study, to mimic the native cannular tissues, we demonstrated bioprinting of several tubular tissues using relevant cell types. These include muscle and endothelial layers of vascular tissues with bioinks containing human smooth muscle cells (hSMCs) and human umbilical vein endothelial cells (HUVECs), respectively, representing the cellular heterogeneity of different layers and cellular phenotypes of the blood vessel-like tissues. Similarly, we were also able to fabricate tubular urothelial tissues by bioprinting inner human urothelial cells (HUCs) and outer human bladder smooth muscle cells (HBdSMCs), simultaneously. These bioprinted tubular tissues can be actively perfused with fluids and nutrients to promote the growth and proliferation of cells in different layers of the hollow fibers. The bioprinted constructs showed characteristic features of human tubular tissues allowing continuous perfusion of fluids for the growth and interaction of different cell types within the bioprinted layers exhibiting sustained viability for up to 2 weeks in vitro. Our current work expands the bioprinting platform to create multitude of tubular tissue architectures for tissue regeneration and modeling.
GelMA hydrogel is a biocompatible biomaterial that has been extensively used in tissue engineering,[2,19–23] and bioprinting.[24–26] Alginate is also commonly combined with GelMA to achieve proper viscosity for bioprinting. In our previous reports, we developed GelMA/alginate-based bioinks for the encapsulation of cells.[5,27] We used alginate in our customized bioink to improve printability and mechanical strength of the hollow tubes. However, alginate is a bioinert material that can inhibit the adhesion of cells. Since the stability of alginate depends on the stability of calcium complexes within the hydrogel, the chelating agents such as ethylenediaminetetraacetic acid (EDTA) that strongly bind calcium can quickly solubilize the alginate. Therefore, to achieve adhesion, spreading, and proliferation of cells in the bioprinted construct by minimizing the alginate concentration, we immersed the bioprinted hollow constructs in an EDTA solution for 5 min. Our previous reports have shown that most alginate can be removed within this time frame.[5]
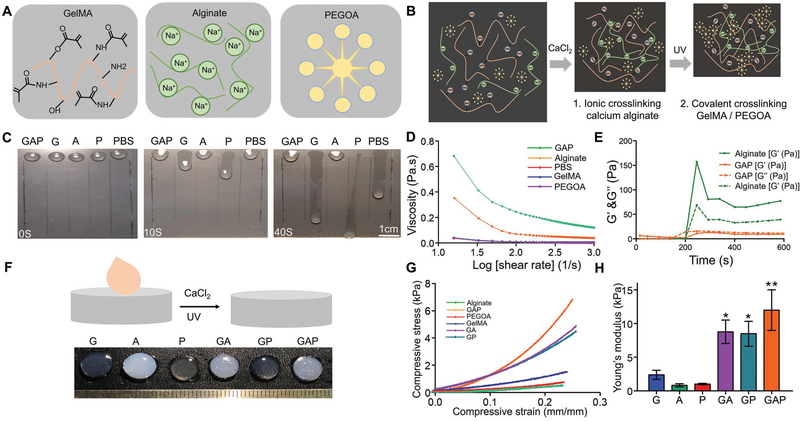
In this study, we further customized a blend bioink formulation by mixing eight-arm poly(ethylene glycol) (PEG) acrylate with tripentaerythritol core (PEGOA) in the GelMA/alginate hydrogel to enhance the mechanical strength and stability of the crosslinked matrix. Previously, it has been demonstrated that increasing the branching of PEG molecules (e.g., using poly(ethylene glycol) tetraacrylate, PEGTA) not only improved the mechanical strength of the hydrogels but also promoted growth and proliferation of the bioprinted cells as compared to the linear PEG derivatives.[5,28] Thus, the purpose of replacing PEGTA with PEGOA in our customized bioink in this work was to further promote growth and proliferation of the encapsulated cells while maintaining printability and mechanical strength of the 3D-bioprinted tubular constructs, which are prerequisites for the generation of functional 3D cannular tissues. To obtain the optimum printable bioink, we evaluated the printability of different formulations of customized bioinks containing 5 and 7% (w/v) GelMA, 2 and 3% (w/v) alginate, and 1 and 2% (w/v) PEGOA. As shown in Figure 1, the optimum bioprintability was obtained at 7% GelMA, 2% alginate, and 2% PEGOA, which altogether was termed as the GAP bioink. Thus, our customized bioink, GAP (Figure 1A) and the two-step crosslinking strategy (Figure 1B) could potentially provide favorable and sustained physicochemical environment for the proliferation and maturation of different cell types in the bioprinted constructs. The viscosity of GAP increased significantly as compared to its individual components as shown by a slope test (Figure 1C). Both GelMA and PEGOA could flow down the slope rapidly, exhibiting very low viscosities, whereas GAP showed higher viscosity with no movement even at the perpendicular surface. These results were consistent with the rheological studies. A logarithmic plot of the viscosity as a function of shear rate for individual components and GAP showed a decrease in viscosity with increasing shear rate for all materials. In addition, GAP exhibited a higher viscosity as compared to pure alginate, GelMA, or PEGOA (Figure 1D). In addition, when calcium chloride (CaCl2) was added, both pure alginate and GAP indicated obvious changes in storage (G′) and loss (G″) moduli as a function of time representing stiffness of the hydrogel (Figure 1E). After crosslinking with CaCl2 and/or ultraviolet (UV) light, GelMA, PEGOA, and GelMA/PEGOA exhibited transparent morphologies, whereas alginate, GelMA/alginate, and GAP showed translucent appearances due to the presence of crosslinked alginate (Figure 1F). Similarly, the stress–strain curves plotted for all materials showed that the mechanical strength of GAP was significantly higher as compared to those of other groups (Figure 1G,H).
Figure 1.
Optimization of the bioink. A) Schematic structures of GelMA, alginate, and PEGOA. B) Schematic depiction of formation of the GAP hydrogel via crosslinking with CaCl2 and UV exposure. C) Illustration of viscosity of hydrogels on a slope surface (scale bar, 1 cm). D) Influence of the shear rate on the rheological behavior of the customized bioink and the different components of the bioinks. E) G′ and G″ as a function of time of the bioink and the different components of the bioink. F) Visual inspection of the customized bioink and the different components of the bioink after CaCl2 and UV-light exposure. G) Compressive stress–strain curves for the customized bioink and the different components of the bioink. H) Young’s modulus of the hydrogels determined from the stress–strain curves. G: 7% GelMA; A: 2% alginate; P: 2% PEGOA; GA: 7% GelMA and 2% alginate; GP: 7% GelMA and 2% PEGOA; GAP: 7% GelMA, 2% alginate, and 2% PEGOA.
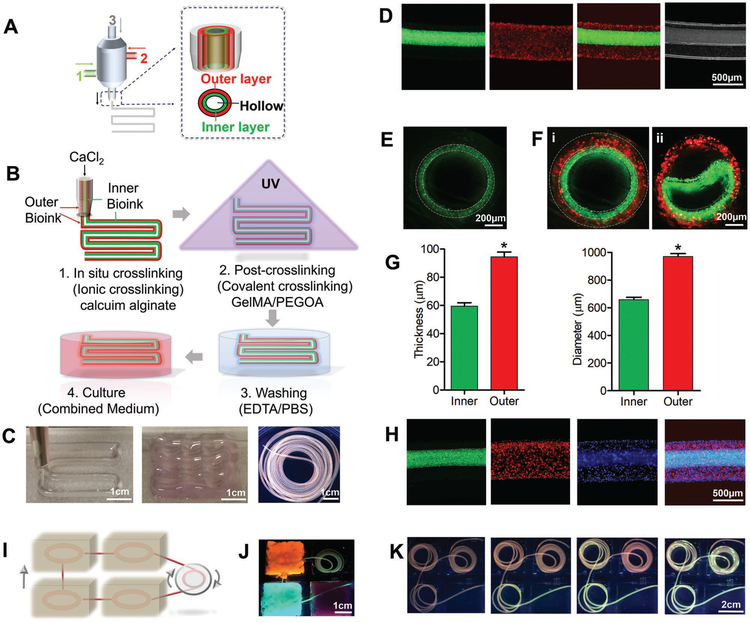
Figure 2A,B represents the schematics showing the components of the multichannel coaxial extrusion system (MCCES) and the process of bioprinting using the blend bioink composed of GelMA, alginate, and PEGOA, respectively. Figure S1A (Supporting Information) shows the actual bioprinting setup to generate a complex and highly organized 3D hollow tube in a single step using the MCCES. The MCCES, consisting of three concentrically assembled needles, enabled direct extrusion of multicircumferential and perfusable hollow tubes with varying diameters. With optimized flow rate of bioink in each channel during the bioprinting process (Figure S1B, Supporting Information), different cell-laden bioinks were delivered through the second and third layers of the nozzle, while the CaCl2 solution was simultaneously delivered through the core channel.
Figure 2.
3D bioprinting of circumferentially multilayered cannular tissues using the GAP bioink. A,B) Schematic illustration of the MCCES and the process of bioprinting of a multilayered hollow tube. C) Images showing bioprinted perfusable tubes with various shapes. D) Representative longitudinal fluorescence microscopy images of double-layered hollow fibers. E,F) Representative cross-sectional view fluorescent microscopy images of i) monolayered and ii) double-layered hollow fibers with fluorescent beads. G) Diameter and thickness of inner and outer layers of the bioprinted tube. H) Fluorescent microscopy images showing bioprinted tri-layered hollow tubes, where green fluorescent beads were embedded into the innermost layer, red fluorescent beads were embedded into the middle layer, and blue fluorescent beads were embedded into the outermost layer during the bioprinting process. I) Schematic of perfusion among tissues. J,K) Images showing the simulation of perfusable multiple tissues.
The alginate component in the bioink was first crosslinked with the calcium ion (Ca2+) in the solution and the GelMA component was subsequently photocrosslinked by exposing the bioprinted constructs under UV light, resulting in generation of organized and stable hollow tubes (Figures 1B and 2B). Although many reports suggest that high concentration and persistent existence of alginate might adversely affect the cell proliferation and migration,[29] the bioinert alginate can be removed by using Ca2+-chelating agents, such as ethylenediaminetetraacetic acid (EDTA), when necessary, to facilitate cell spreading, migration, and proliferation in the bioprinted constructs (Figure 2B).[5]
The unique feature of our MCCES is its ability to fabricate multilayered hollow tubes, consisting of continuously altering shapes and sizes, without the need of changing the nozzles. To observe the different layers in the bioprinted multilayered hollow tube, we used different fluorescent microbeads, at the concentration of 20 μL/3 mL bioink, with distinct fluorescence in each channel during bioprinting, and we confirmed that a tubular structure with multiple layers could be constructed simultaneously in a single step (Figure 2C,D, Figure S2A,B, and Movies S1 and S2, Supporting Information). Figure 2E,F represents the cross-sectional views of the hollow structures, distinctly showing walls of a singlelayered tube and a double-layered tube, respectively, each colored fluorescence representing a layer (Figure 2Fi). Furthermore, we intentionally delaminate the layers of a bioprinted double-layered hollow tube to show the two distinct layers (Figure 2Fii). We obtained hollow tubes with varying layers either by turning on/off the bioink flow in the desired channel or by changing the flow rates for the bioink extrusion. On average, the diameters of the inner lumen and the outer lumen were measured to be 663 ± 52 and 977 ± 63 μm, respectively, while the thicknesses of the inner wall and the outer wall were 62 ± 10 and 94 ± 10 μm, respectively (Figure 2G and Figure S2A, Supporting Information).
Using our designed MCCES, we were able to develop advanced tubular structures with multiple channels demonstrating a platform to create complex cannular tissue architectures. Figure 2H shows fluorescence microscopy images of a bioprinted tri-layered hollow tube with the innermost green layer, the middle red layer, and the outermost blue layer. To test the perfusability of the bioprinted hollow tubes, we successfully bioprinted long hollow tubes and traced the active perfusion in the tubes with fluorescent microbeads along the entire length. Figure 2I–K represents schematic and experimental demonstrations of the perfusable tubular networks mimicking multiple tissues to simulate the supply of nutrients among them. During perfusion, the structural integrity of the continuous hollow fiber remained intact. Thus, we elucidated that the bioprinted multilayered tubes with varying diameters and layers maintain their perfusable hollow structures throughout the entire length.
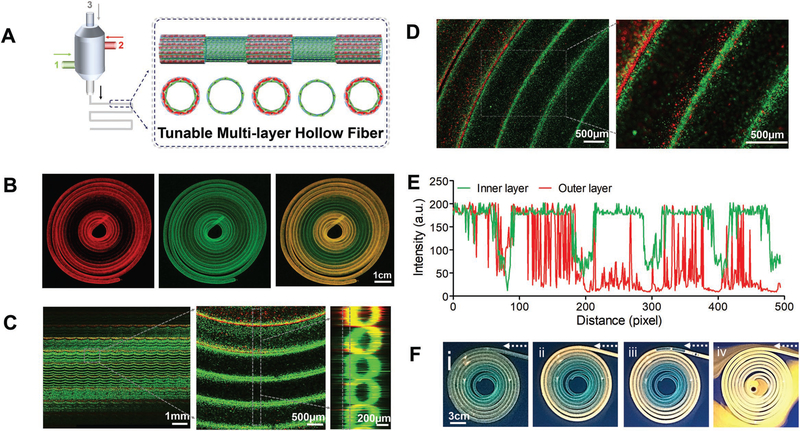
Most importantly, we further demonstrated precise control over the architecture of the bioprinted tubes constructing a wide range of multilayered hollow tubes with desired diameters and numbers of circumferential layers at the same time, in a continuous manner, by switching the coaxial channels on or off at desired intervals (Figure 3A). To construct a tube with such features, we initially bioprinted a double-layered hollow tube, switched smoothly to a single-layered tube by turning off the flow of bioink in the outmost channel, and then switched again to the double-layered tube, all in a single continuous process (Figure 3B). We labeled the outer and inner layers of the tube with red and green fluorescent microbeads, respectively, and observed the dynamic conversion between the single and double-layered hollow tubes by confocal laser scanning microscopy. The confocal images clearly demonstrated the outer red and inner green fluorescent microbeads representing the double-layered region while the green microbeads represent the inner single-layered region of the tunable hollow tube (Figure 3C). Figure 3D further represents the close-up confocal images of the junction regions where the double-layered tube (possessing both outer red and inner green fluorescence signals) gradually transitioned into a single-layered tube (possessing only green signal). Indeed, intensity analyses of the signals revealed the persistent existence of the green signal throughout the tube, representing the inner green layer. In contrast, the red fluorescence signal appeared at regular intervals, present at high intensity in the outer layer of the tube which gradually disappeared, indicating the conversion of the double-layered lumen into a single-layered tube (Figure 3E). These results were consistent with results shown in Figure 3C. Finally, perfusability of a bioprinted hollow tube with continuous single- and double-layered walls at regular intervals was tested. The perfusion of fluorescent microbeads throughout the length of tube is a clear indication of perfusability of the bioprinted tunable tube (Figure 3F).
Figure 3.
3D bioprinting of tunable and perfusable double-layered constructs with MCCES. A) Schematic illustration of tuning of layers of a hollow fiber (between single and double layers at regular intervals) with MCCES during a 3D bioprinting process. B) Fluorescence confocal microscopy images showing dynamic conversion between single and double-layered hollow tubes during the bioprinting process (scale bar: 1 cm). C,D) Simulation of 150 scanned layers showing dynamic conversion from double-layered to single-layered hollow tube with clear and gradual demarcation of single-and double-layered regions of the hollow tube. E) Dynamic changing of intensity of green and red signals at demarcation of single and double-layered regions of the hollow tube. F) Perfusion of the bioprinted hollow fiber with continuous single and double-layered walls at regular intervals.
We further interrogated the biocompatibility of the GAP bioink on several cell lines and subsequent perfusability of the double-layered hollow tissue construct. First, we encapsulated C2C12 skeletal myocytes in GAP, crosslinked and cultured for up to 3 weeks. The F-actin/nuclei staining of encapsulated cells showed that there was prominent proliferation and spreading of C2C12 cells within the GAP bioink indicating muscle-like tissue formation (Figure S3A, Supporting Information). Afterward, we labeled NIH/3T3 fibroblasts and C2C12 with green and red cell trackers, respectively, and mixed separately with the GAP bioink. We then bioprinted these cell-laden bioinks using the MCCES, resulting in a double-layered hollow construct and allowed the cells to grow under standard cell culture conditions. As shown in Figure S3B (Supporting Information), the bioprinted hollow construct possessed two distinct layers, with the outer layer containing C2C12 cells and the inner layer, NIH/3T3 cells. Finally, we obtained a bioprinted double-layered hollow tissue with smooth perfusion (Figure S3C, Supporting Information), analogous to those without cells (Figure 2D). During the entire process of perfusion, the bioprinted lumen maintained good structural integrity without fluid leakage.
We next explored our MCCES along with the customized bioink for bioprinting of biomimetic urothelial tissue construct. The urethra is a fibromuscular tube through which urine is discharged from the bladder to the exterior of the body. The native urethra consists of three cell layers including the urothelial epithelium, fibroblasts, and smooth muscle cells. Urothelial epithelial lining and smooth muscle cell layer form the most important layers of the urethra.[30] Various urothelial pathologies such as inflammatory lesions, urothelial strictures, congenital anomalies, and malignancy often necessitate the replacement of urothelial tissue. Currently, urethroplasty remains the most effective treatment option for urothelial defects.[31] The most commonly used urethroplasty are anastomotic urethroplasty and widening the urothelial lumen using flaps or grafts of tissues from different origin such as skin, bladder, or buccal mucosa.[32] However, in many circumstances, shortage of local tissues, immunosuppression, and complications arising from the use of non-native tissues have urged to develop bioengineered urothelial tissues.[30] Conventional tissue engineering approaches for in vitro urethra reconstruction involves formation of porous scaffolds such as collagen-based matrices, synthetic polyesters, and protein-derived biological scaffolds, which are manually seeded with urothelial cells and smooth muscle cells or unseeded. Although these approaches have been successful in several animal and clinical studies, they show several limitations associated with homogenous seeding of cells throughout the entire scaffolds, distribution of multiple cell types, and control of the scaffold microarchitecture.[32,33]
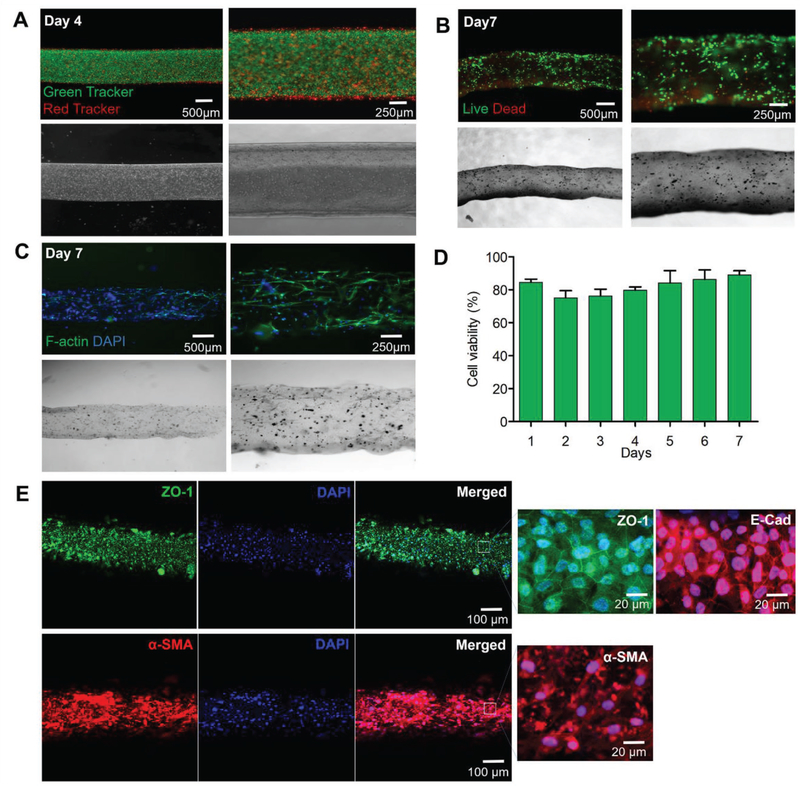
Hence, to address these issues, we bioprinted a biomimetic urothelial tissue construct using HUCs and HBdSMCs prelabeled with green and red trackers, respectively. These cells were encapsulated in the GAP individually and bioprinted using MCCES, resulting in a double-layered urothelial construct constituting the inner urothelial layer (green) and the outer HBdSMCs (red) (Figure 4A). The viability of the cells was 85 ± 2% on day 1 and remained as high as 89 ± 3% on day 7 (Figure 4B,D). Likewise, in a separate experiment, the bioprinted HUCs and HBdSMCs were stained for F-actin, a major component of the cytoskeleton and nuclei on day 7 to observe the cell proliferation and cellular phenotypes within the bioprinted tissue constructs. Both HUCs and HBdSMCs were homogenously distributed within their respective layers without losing the structural integrity of the tubular tissue construct (Figure 4C). These results demonstrated that our customized bioink created a favorable microenvironment for encapsulated cells and MCCES facilitated the formation of perfusable urothelial tissue-like tubular structures.
Figure 4.
3D bioprinting of urethelial tissue constructs. A) Fluorescence images of the bioprinted inner human urothelial cells labeled with green cell tracker and the outer human bladder smooth muscle cells labeled with red cell tracker on day 14. B) Live/dead assay of human urothelial cells within different layers of the bioprinted urothelial tissue construct on day 7. C) F-actin and nuclei staining images of human urothelial cells within the bioprinted urothelial tissue construct on day 7. D) Viability of human urethra cells within the bioprinted urothelial tissue construct on day 7. E) Confocal microscopy images of the immunostained urothelial tubes after 14 d showing the expressions of urethral cell-specific biomarkers, ZO-1 (green) and E-cadherin (red) by HUCs, and α-SMA (red) by hSMCs.
Prior to confirming the biofunctionality of the bioprinted urothelial tissue constructs, we first examined the behaviors of HUCs and HBdSMCs in the GAP bioink by immunocytochemical analysis. The cells were encapsulated in the GAP bioink separately and crosslinked under UV without bioprinting, followed by incubation at standard cell culture conditions for 7 d. At day 7, the cells in the GAP hydrogel were fixed and immunocytochemical staining of HUCs with E-cadherin and zonula occludens-1 (ZO-1, tight junction markers), and HBdSMCs with α-smooth muscle actin (α-SMA) was performed. Figure S4 (Supporting Information) shows the high viability and proliferation of both HUCs and HBdSMCs in GAP, with HUCs expressing ZO-1, tight junction protein essential for normal function of urothelium,[34] and E-cadherin, cell–cell adhesion molecule that plays important roles in epithelial cell behavior and tissue formation,[35] while HBdSMCs expressed α-SMA. The biofunctionality of the bioprinted urothelial tissue constructs was further analyzed via immunocytochemical staining at day 14 after bioprinting. Confocal microscopy images of the immunostained urothelial tubes after 14 d showed the formation of mature monolayer of spreading HUCs, with the expression of both ZO-1 and E-cadherin, confirming the presence of critical tight junction protein-1 and cell–cell adhesion molecules necessary for proper urothelial function. In addition, expression of α-SMA by HBdSMCs[36] was also observed (Figure 4E).
Similarly, diseases affecting blood vessels such as arteritis, atherosclerosis, and thrombosis are a major health concern worldwide and have a great financial impact.[37] Hence, it is an urgent necessity to engineer blood vessel substitutes for the replacement of damaged or diseased vascular tissues. Typically, the walls of blood vessels are composed of the innermost endothelial cell layer, the middle smooth muscle cell layer, and the outer layer of fibroblasts along with their associated matrices. Among them, the endothelial cells and smooth muscle cells play crucial roles in maintaining homeostasis and mechanical properties of blood vessels, respectively.[38] Thus, both smooth muscle cells and endothelial cells are required to be incorporated into the tubular scaffold to build a blood vessel-like structure.[39] In addition, due to the limited proliferation ability and loss of contractility of smooth muscle cells, creating blood vessel substitutes remains a challenge in tissue engineering.
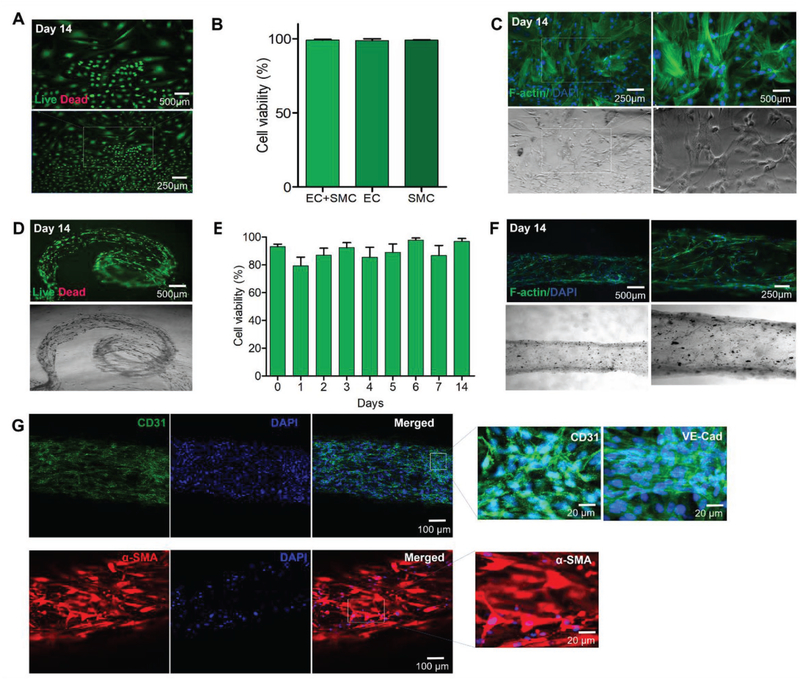
To address these issues, we encapsulated hSMCs and HUVECs in the GAP bioink separately and bioprinted directly through the different channels of the MCCES, resulting in a biologically relevant hollow vascular construct that represented the cellular heterogeneity of different layers as seen in normal blood vessels. We further incubated the bioprinted vascular constructs at 37 °C and 5% CO2 in a common medium consisting of 1:1 volume ratio of the endothelial growth medium (EGM-2) and the smooth muscle growth medium-2 (SmGM-2) for up to 14 d. Prior to bioprinting, we studied the survival and proliferation of HUVECs and hSMCs in the GAP as monocultures and coculture systems using this common medium for 14–21 d. We observed the viability, confluency, and proliferation of the encapsulated cells in GAP under fluorescence microscope after live/dead assay and F-actin/nuclei staining. As shown in Figure 5, both HUVECs and hSMCs could proliferate and spread in GAP demonstrating their capability to form networks (Figure 5A,C). The viability of HUVECs and hSMCs in GAP was similar in monocultures in their respective media and cocultures in the common medium (HUVECs + hSMCs 99 ± 1%; HUVECs 99 ± 1%; hSMCs 99 ± 1%, Figure 5B). In addition, we also confirmed the biofunctionality of vascular cells in the GAP bioink by immunocytochemical analysis. The cells were encapsulated in the GAP bioink separately and crosslinked under UV without bioprinting, followed by incubation under similar cell culture conditions for 7 d. Figure S5 (Supporting Information) shows the high viability and proliferation of both cell types in GAP, with strong expressions of VE-cadherin and CD31 for HUVECs and expression of α-SMA for hSMCs. CD31 and VE-cadherin are endothelial-specific adhesion molecules expressed on the membranes of endothelial cells and at junctions between endothelial cells, respectively.[40–42]
Figure 5.
3D bioprinting of blood vessel tissue constructs. A) Fluorescence images of GFP-HUVECs within GAP on day 21. B) Comparison of viability of cocultured HUVECs and hSMCs in combined media on day 14 with respective individual cell types and media. C) F-actin and nuclei staining images of cocultured HUVECs and hSMCs in combined media on day 14. D) Live/dead assay of HUVECs and hSMCs within different layers of the bioprinted blood vessel on day 14. E) Viability of the bioprinted vascular cells on day 14. F) F-actin and DAPI images of vascular cells within the bioprinted blood vessel construct on day 14. G) Confocal microscopy images of the immunostained vascular tubes after 14 d showing the expression of vascular cell-specific biomarkers, CD31 (green) and VE-cadherin (green) by HUVECs, and α-SMA (red) by hSMCs.
Similar to the bioprinted urothelial tissues, we monitored the viability of the cells in the bioprinted double-layered hollow vascular tissues using the live/dead assay on days 0–7 and day 14. The viability of vascular cells in the bioprinted tissue was found to be 93 ± 2% immediately after bioprinting (Figure 5D,E). Thus, the bioprinting of vascular tissues using GAP and other parameters optimized in this study had minimal effects on vascular cells during the extrusion process. Overall the viability of the vascular cells, both HUVECs and hSMCs, was found to be in between 85 and 97% with prolonged culture time (Figure 5D,E). However, slight fluctuation in cell viability was found due to the fact that there existed shear stress during cell extrusion from channels, which affected the cells in the bioprinted constructs. The damaged cells or the cells under stress gradually died with time. As a result, the viability of cells in days 2–4 was found to reduce. However, the healthy cells could then proliferate leading to increased viability reaching more than 90% within 14 d. Our results of viability of the vascular cells are consistent with previous reports.[43,44]
Prior to immunocytochemical studies with vascular cell-specific markers, the bioprinted cells were stained with F-actin/nuclei to characterize the cell proliferation and vascular cell phenotypes. We observed the proper spreading and proliferation of the cells throughout the bioprinted vascular tissue constructs (Figure 5F). We then performed immunofluorescence staining of VE-cadherin and CD31 for the vascular tissue constructs at day 14 after bioprinting to show the biofunctionality of bioprinted vascular tissue constructs. In addition, immunofluorescent staining of α-SMA was performed for hSMCs. Confocal microscopy images of the immunostained vascular tubes after 14 d showed the formation of mature monolayer of endothelium with the expression of both CD31 and VE-cadherin by the spreading HUVECs, confirming the presence of critical junctions necessary for proper vascular endothelial function, and expression of α-SMA by hSMCs, an actin isoform predominantly present in vascular smooth muscle cells that plays an important role in fibroblast contractility (Figure 5G).[45,46]
In summary, we have reported a digitally tunable multilayer coaxial nozzle that enables the bioprinting of complex and perfusable hollow tubes with multiple circumferential layers in a single step. This generalized bioprinting method can be extended for the fabrication of many different types of tubular tissues possessing hierarchically layered structures, using a single, digitally controlled extruder system. In our previous studies, we demonstrated the bioprinting of only single-layered hollow tubes using GelMA-based bioinks. Based on our previous work, we further studied the possibility to produce the more complicated hollow structure with 3D bioprinting technology. In this study, we successfully bioprinted circumferentially multilayered hollow tissue constructs using the customized GelMA-based bioinks. We used PEGOA that enhanced the mechanical strength and stability of the crosslinked hydrogel. Our customized blend bioink, based on a mixture of GelMA, alginate, and PEGOA with optimized concentrations, represents a favorable physicochemical matrix for proliferation and early maturation of a wide range of cell types. However, there is still plenty of room for improvement of this 3D bioprinting system and optimization of the blend bioink for enhanced mechanical strength and stability of the bioprinted constructs to achieve proper organization and eventually function of the tissue constructs. The bioprinting of circumferentially multilayered hollow tubes is the basis for reconstruction of complex hollow tissues or organs and the fabrication of such perfusable circumferentially multilayered tissue constructs represents a fundamental step toward creating human cannular tissues. We believe that our system has the potential to expand current bioprinting platforms to create multitude of tubular tissue architectures for tissue regeneration and modeling.
Supplementary Material
Acknowledgements
Q.P., S.M., X.Y., and X.L. contributed equally to this work. The authors acknowledge funding from the National Institutes of Health (AR057837, DE021468, AR068258, AR066193, EB022403, EB021148, HL137193, EB021857, and EB024403) and the Presidential Early Career Award for Scientists and Engineers (PECASE). Q.P. gratefully acknowledges funding by China Scholarship Council (No. 201406235036). S.M. acknowledges New England Anti-Vivisection Society (NEAVS) and the American Fund for Alternatives to Animal Research (AFAAR) for Postdoctoral Fellowship. X.L. acknowledges funding by the National Natural Science Foundation of China (No.31570947). X.Y. acknowledges funding by the National Natural Science Foundation of China(No. 81772712). X.H. acknowledges funding by the National Key Research and Development Program of China (Project No. 2018YFA0209500), the National Natural Science Foundation of China (21673197, 21621091), Young Overseas High-level Talents Introduction Plan, the 111 Project (B16029), the Natural Science Foundation of Fujian Province of China (No. 2018J06003) and Special Project of Strategic Emerging Industries from Fujian Development and Reform Commission. R.P.S acknowledges funding by Tecnologico de Monterrey and MIT Nanotechnology Program. F.R.P. was supported by the CONACYT postdoctoral fellowship (Grant No. 291018–173853). S.H. acknowledges SNSF for Early Mobility postdoctoral fellowship. Y.S.Z. acknowledges funding from the National Cancer Institute of the National Institutes of Health (K99CA201603) and the NEAVS. The authors would like to thank Prof. Samir Mitragotri for his generous help on microscopy.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Qingmeng Pi, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Plastic and Reconstructive Surgery Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200127, China.
Dr. Sushila Maharjan, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Research Institute for Bioscience and Biotechnology, Nakkhu-4, Lalitpur 44600, Nepal.
Prof. Xiang Yan, Department of Urology, Drum Tower Hospital, Medical School of Nanjing University, Institute of Urology, Nanjing University, Nanjing 210008, China Department of Urology, Anqing Petrochemical Hospital, Nanjing Gulou Hospital Group, Anqing 246002, China.
Prof. Xiao Liu, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Beijing Advanced Innovation Center for Biomedical Engineering, Key Laboratory for Biomechanics and Mechanobiology of Ministry of Education, School of Biological Science and Medical Engineering, Beihang University, Beijing 100083, China.
Dr. Bijay Singh, School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Anne Metje van Genderen, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Dr. Felipe Robledo-Padilla, ENCIT – Science Engineering and Technology School Tecnologico de Monterrey Monterrey 64849, Mexico
Prof. Roberto Parra-Saldivar, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; ENCIT – Science Engineering and Technology School Tecnologico de Monterrey Monterrey 64849, Mexico.
Dr. Ning Hu, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Biosensor National Special Laboratory, Key Laboratory of Biomedical Engineering of Education Ministry, Department of Biomedical Engineering, Zhejiang University, Hangzhou 310027, China.
Dr. Weitao Jia, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Orthopedic Surgery, Shanghai Jiaotong University Affiliated Sixth People’s Hospital, Shanghai Jiaotong University, Shanghai 200233, China.
Prof. Changliang Xu, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; The First Clinical Medical College, Nanjing University of Chinese Medicine, Jiangsu Collaborative Innovation, Center of Traditional Chinese Medicine Prevention, and Treatment of Tumor, Nanjing University of Chinese Medicine, Nanjing 210023, China.
Jian Kang, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Dr. Shabir Hassan, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Prof. Haibo Cheng, The First Clinical Medical College, Nanjing University of Chinese Medicine, Jiangsu Collaborative Innovation, Center of Traditional Chinese Medicine Prevention, and Treatment of Tumor, Nanjing University of Chinese Medicine, Nanjing 210023, China
Prof. Xu Hou, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; State Key Laboratory of Physical Chemistry of Solid Surface, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China; Research Institute for Soft Matter and Biomimetics, College of Physical Science and Engineering, Xiamen University, Xiamen 361005, China.
Prof. Ali Khademhosseini, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Bioengineering, Department of Chemical and Biomolecular Engineering, Henry Samueli School of Engineering and Applied Sciences, Department of Radiology, David Geffen School of Medicine, Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute (CNSI), University of California, Los Angeles, CA 90095, USA; Department of Bioindustrial Technologies, College of Animal Bioscience and Technology, Konkuk University, Seoul 05029, Republic of Korea.
Dr. Yu Shrike Zhang, Division of Engineering in Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
References
- [1].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [2].Khademhosseini A, Langer R, Nat. Protoc 2016, 11, 1775. [DOI] [PubMed] [Google Scholar]
- [3].Ozbolat IT, Yu Y, IEEE Trans Biomed Eng. 2013, 60, 691. [DOI] [PubMed] [Google Scholar]
- [4].Morgan JP, Delnero PF, Zheng Y, Verbridge SS, Chen J, Craven M, Choi NW, Diaz-Santana A, Kermani P, Hempstead B, Nat. Protoc 2013, 8, 1820. [DOI] [PubMed] [Google Scholar]
- [5].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [7].Hou X, Zhang YS, Trujillo-de Santiago G, Alvarez MM, Ribas J, Jonas SJ, Weiss PS, Andrews AM, Aizenberg J, Khademhosseini A, Nat. Rev. Mater 2017, 2, 17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cui H, Zhu W, Nowicki M, Zhou X, Khademhosseini A, Zhang LG, Adv. Healthcare Mater 2016, 5, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu Y, Zhang Y, Martin JA, Ozbolat IT, J. Biomech. Eng 2013, 135, 091011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Datta P, Ayan B, Ozbolat IT, Acta Biomater. 2017, 51, 1. [DOI] [PubMed] [Google Scholar]
- [11].Nishiyama Y, Nakamura M, Henmi C, Yamaguchi K, Mochizuki S, Nakagawa H, Takiura K, J. Biomech. Eng 2009, 131, 035001. [DOI] [PubMed] [Google Scholar]
- [12].Christensen K, Xu C, Chai W, Zhang Z, Fu J, Huang Y, Biotechnol. Bioeng 2015, 112, 1047. [DOI] [PubMed] [Google Scholar]
- [13].Blaeser A, Duarte Campos DF, Weber M, Neuss S, Theek B, Fischer H, Jahnen-Dechent W, Biores. Open Access 2013, 2, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xiong R, Zhang Z, Chai W, Huang Y, Chrisey DB, Biofabrication 2015, 7, 045011. [DOI] [PubMed] [Google Scholar]
- [15].Ouyang L, Highley CB, Sun W, Burdick JA, Adv. Mater 2017, 29, 1604983. [DOI] [PubMed] [Google Scholar]
- [16].Li Y, Liu F, Sun J, Chem. Commun 2009, 2730. [DOI] [PubMed] [Google Scholar]
- [17].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv. Mater 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [18].Zhang YS, Yue K, Aleman J, Mollazadeh-Moghaddam K, Bakht SM, Yang J, Jia W, Dell’Erba V, Assawes P, Shin SR, Ann. Biomed. Eng 2017, 45, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Klotz BJ, Gawlitta D, Rosenberg AJ, Malda J, Melchels FP, Trends Biotechnol. 2016, 34, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guan X, Avci-Adali M, Alarçin E, Cheng H, Kashaf SS, Li Y, Chawla A, Jang HL, Khademhosseini A, Biotechnol. J 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mehrali M, Thakur A, Pennisi CP, Talebian S, Arpanaei A, Nikkhah M, Dolatshahi-Pirouz A, Adv. Mater 2017, 29. [DOI] [PubMed] [Google Scholar]
- [22].Yang J, Zhang YS, Yue K, Khademhosseini A, Acta Biomater. 2017, 57, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A, Biomaterials 2015, 73, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Biomaterials 2016, 110, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell’Erba V, Miri AK, Albadawi H, Arneri A, Li X, Lab Chip 2016, 16, 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang YS, Duchamp M, Oklu R, Ellisen LW, Langer R, Khademhosseini A, ACS Biomater. Sci. Eng 2016, 2, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cristina C, Ryon SS, Vijayan M, Solange M, Marco C, Andrea B, Remzi DM, Mariella D, Ali K, Adv. Mater 2016, 28, 677.26606883 [Google Scholar]
- [28].Ali T, Hassani NA, Bahar A, Elmira AT, Mohsen A, Nasim A, David J, Ali K, Adv. Healthcare Mater 2015, 4, 2146. [Google Scholar]
- [29].Dolati F, Yu Y, Zhang Y, De Jesus AM, Sander EA, Ozbolat IT, Nanotechnology 2014, 25, 145101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Orabi H, Bouhout S, Morissette A, Rousseau A, Chabaud S, Bolduc S, Sci. World J 2013, 2013, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mundy AR, Postgrad. Med. J 2006, 82, 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Anthony A, Mikhail D, Alexey L, Petr G, Denis B, Andrey V, Yoo JJ, Tissue Eng J. Regener. Med 2017, 11, 3. [Google Scholar]
- [33].Raya-Rivera A, Esquiliano DR, Yoo JJ, Lopez-Bayghen E, Soker S, Atala A, Lancet 2011, 377, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rickard A, Dorokhov N, Ryerse J, Klumpp DJ, McHowat J, In Vitro Cell. Dev. Biol.: Anim 2008, 44, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van RF, Berx G, Cell. Mol. Life Sci 2008, 65, 3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zheng Y, Chang S, Boopathi E, Burkett S, John M, Malkowicz SB, Chacko S, In Vitro Cell. Dev. Biol.: Anim 2012, 48, 84. [DOI] [PubMed] [Google Scholar]
- [37].Nemeno-Guanzon JG, Lee S, Berg JR, Jo YH, Yeo JE, Nam BM, Koh Y-G, Lee JI, J. Biomed. Biotechnol 2012, 2012, 956345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang C, Cen L, Yin S, Liu Q, Liu W, Cao Y, Cui L, Biomaterials 2010, 31, 621. [DOI] [PubMed] [Google Scholar]
- [39].Nerem RM, Ensley AE, Am. J. Transplant 2004, 4, 36. [DOI] [PubMed] [Google Scholar]
- [40].Ilan N, Cheung L, Pinter E, Madri JA, J. Biol. Chem. 2000, 275, 21435. [DOI] [PubMed] [Google Scholar]
- [41].Lampugnani MG, Resnati M, Raiteri M, Pigott R, Pisacane A, Houen G, Ruco LF, Dejana E, J. Cell Biol 1992, 118, 1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lenting PJ, Christophe OD, Denis CV, Blood 2015, 125, 2019. [DOI] [PubMed] [Google Scholar]
- [43].Dubbin K, Hori Y, Lewis KK, Heilshorn SC, Adv. Healthcare Mater. 2016, 5, 2488. [DOI] [PubMed] [Google Scholar]
- [44].He Y, Yang F, Zhao H, Gao Q, Xia B, Fu J, Sci. Rep 2016, 6, 29977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kawasaki Y, Imaizumi T, Matsuura H, Ohara S, Takano K, Suyama K, Hashimoto K, Nozawa R, Suzuki H, Hosoya M, Pediatr. Nephrol 2008, 23, 913. [DOI] [PubMed] [Google Scholar]
- [46].Gerhardt H, Betsholtz C, Cell Tissue Res. 2003, 314, 15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.