ABSTRACT
Survivin (also known as BIRC5) is an evolutionarily conserved eukaryotic protein that is essential for cell division and can inhibit cell death. Normally it is only expressed in actively proliferating cells, but is upregulated in most, if not all cancers; consequently, it has received significant attention as a potential oncotherapeutic target. In this Cell Science at a Glance article and accompanying poster, we summarise our knowledge of survivin 21 years on from its initial discovery. We describe the structure, expression and function of survivin, highlight its interactome and conclude by describing anti-survivin strategies being trialled.
KEY WORDS: Survivin, BIRC5, Cancer, Mitosis, Apoptosis
Summary: A mini-review highlighting our current understanding of the structure, function and cancer implications of survivin biology 21 years on from its initial discovery.
Introduction
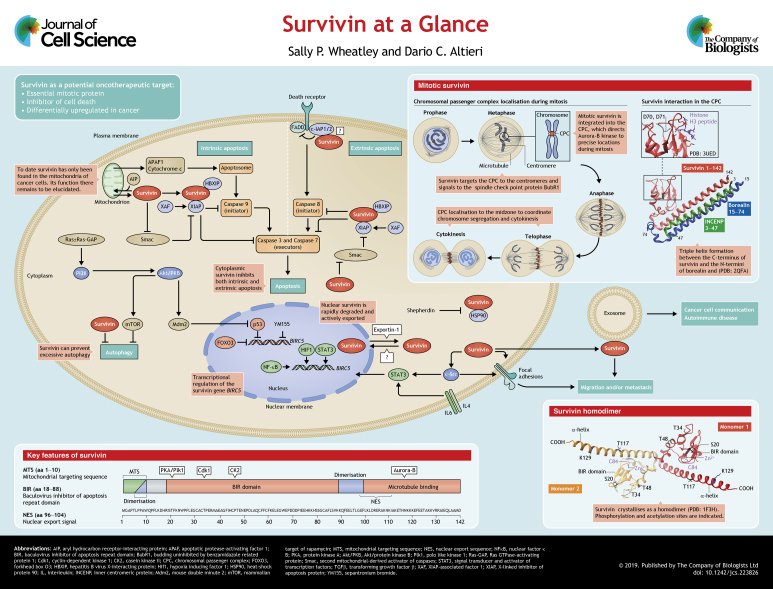
When survivin (BIRC5) was first described, its discovery sparked considerable interest from oncologists and cell biologists, an interest that persists today (Ambrosini et al., 1997). For oncologists seeking new anti-cancer targets, proteins that are required for cell proliferation feature high on their ‘most wanted’ list. Similarly appealing are proteins that interfere with programmed cell death (i.e. apoptosis), which is the intended response of tumour cells to traditional chemo- or radio-therapies. Therefore, as a protein that is both essential for mitosis and able to inhibit apoptosis, at first glance, survivin seemed a promising new target. Moreover, the most sought-after targets are those that are differentially expressed in cancer versus normal cells and, indeed, survivin is highly abundant in cancer (Velculescu et al., 1999), yet absent from most normal somatic cells. Thus in this respect too, it seemed an ideal candidate. Despite these desirable characteristics, rather disappointingly, a truly specific anti-survivin agent is yet to reach the clinic. Part of the challenge may be that survivin has no enzymatic activity of its own; instead, it achieves most of its tasks in association with other proteins and is probably best described as an adaptor protein that interacts with, or shuttles its partners to their destinations. This is certainly the case during mitosis, when it targets the chromosomal passenger complex (CPC) to the centromeres, thereby enabling aurora-B kinase to phosphorylate a number of proteins that ultimately ensure that the chromosomes are aligned properly before they are segregated at anaphase. Although it is less clear how survivin inhibits apoptosis, interactions with other members of the inhibitors of apoptosis (IAP) protein family (see Box 1), also appear to be key. In this Cell Science at a Glance article and accompanying poster, we aim to bring the reader up to date with the current understanding of this multi-tasking little protein.
Box 1. Apoptosis and IAPs.
Apoptosis is the primary form of programmed cell death and depends on cysteine proteinases, called caspases, to disassemble the cell in a controlled manner (see poster). There are two apoptotic pathways: the intrinsic and extrinsic pathway. Upon receiving endogenous stress signals or irradiation, mitochondria initiate the intrinsic cascade through loss of the mitochondrial outer membrane potential and release of cytochrome c release; this causes activation of the initiator caspase, caspase 9. In contrast, the extrinsic pathway is mitochondrion-independent and is triggered by the binding of ligands to receptors at the cell surface, for instance, the TRAIL-bound TNF receptor (TRAIL is also known as TNFSF10), which activates caspase-8 via FADD, which can be inhibited by cIAP1 and cIAP2. After stimulation of the initiator caspases (caspase 8 or 9), both pathways converge on the effector caspases 3 and 7, which cause cellular demise by cleaving downstream macromolecules.
As the name suggests, the IAP family of proteins prevent this form of programmed cell death. Inclusion in the IAP family is based on the presence of at least one baculovirus inhibitor of apoptosis repeat domain (BIR), a globular fold that has been originally found in insect viruses. Humans have eight IAPs, of which survivin is the smallest. Initially thought to bind to and inhibit caspase activity directly, current views suggest that only the canonical member of this family, XIAP, can efficiently and directly inhibit caspases in vivo (reviewed in Lalaoui and Vaux, 2018). However, XIAP can interact with other IAPs, including survivin; this can improve its stability to augment the inhibitory effect of XIAP. For XIAP, both the linker between two adjacent BIR domains and the BIR domains themselves contribute to the prevention of apoptosis: the former disables the catalytic cysteine residue of the effector caspases, while the latter can prevent dimerisation of the initiator caspases, which is critical for their activation. Upstream factors such as Smac, which is released from the mitochondria upon apoptotic stimulation, can inhibit IAPs by binding to the BIR domain; this prevents Smac binding to its caspase target, and there is evidence to suggest that survivin might inhibit this inhibitor (Song et al., 2004).
Structure, domains and key partners
Survivin is a small protein [142 amino acids (aa); 16.5 kDa] with multifunctional domains (see poster). Its N-terminal two-thirds comprise a globular baculovirus inhibitor of apoptosis repeat (BIR) domain (aa 20–90), the integrity of which depends on a Zn2+ finger that is created by C57, C60, C84 and H77 (Li et al., 1999); this defines survivin as an IAP (see Box 1). The C-terminal third is an extended α-helix (98–142). Survivin crystallises as a homodimer; the two monomers interact via its central linker region (aa 90–102), assisted by N-terminal residues L6 and W10 (Verdecia et al., 2000).
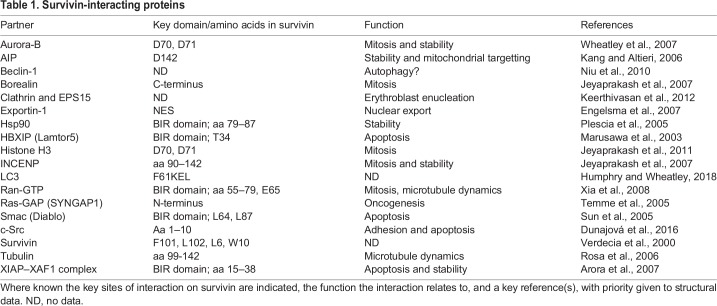
Survivin uses the same interface to interact with its mitotic partner borealin (Jeyaprakash et al., 2007), and its C-terminus forms a triple helical bundle with the N-termini of borealin and inner centromere protein (INCENP) (Jeyaprakash et al., 2007). Together with aurora-B kinase, these proteins constitute the CPC, an essential mitotic complex. Survivin also has multiple non-mitotic partners, which influence its stability, and its role in the inhibition of apoptosis, subcellular localisation and pro-oncogenic signalling, as well as its other roles (see Table 1). Survivin undergoes several post-translational modifications, including phosphorylation by protein kinase A (PKA) and polo-like kinase (Plk1), (Dohi et al., 2007; Colnaghi and Wheatley, 2010); cyclin-dependent kinase 1 (Cdk1), (O'Connor et al., 2000; Barrett, et al., 2009), casein kinase II (CKII), (Barrett et al., 2011) and aurora-B kinase (Wheatley et al., 2007). It is also acetylated (Wang, H. et al., 2010b) and ubiquitylated (Vong et al., 2005).
Table 1.
Survivin-interacting proteins
Expression
Survivin is expressed during development (Uren et al., 2000) and in proliferating adult cells (Li et al., 1998). Apart from activated T lymphocytes (Leung et al., 2007), erythroblasts (Keerthivasan et al., 2012) and self-renewing stem cells (Martini et al., 2016), it is absent from adult cells. Human survivin is encoded by the BIRC5 gene and located on the long arm of chromosome 17 (q25) (Ambrosini et al., 1997). It has a TATA-less promoter and four transcriptional elements: three cell cycle-dependent elements (CDEs), and a cell cycle homology region (CHR), which ensure its expression is minimal in G1 and maximal in mitosis (Li and Altieri, 1999). It is indirectly repressed by adenomatous polyposis coli protein (APC) (Zhang et al., 2001), retinoblastoma protein (pRB, also known as RB1) (Jiang et al., 2004) and phosphatase and tensin homologue (PTEN) and transforming growth factor β (TGFβ) (Martini et al., 2016) (Guha et al., 2009), and repressed directly by forkhead box O3 (FOXO3) (Hagenbuchner et al., 2012) and p53 (also known as TP53) (Hoffman et al., 2002) (see poster). Survivin expression is also under the influence of circadian rhythms (Siffroi-Fernandez et al., 2014), and is activated by many transcription factors, including hypoxia-inducing factor 1 (HIF1), nuclear factor κB cells (NFκB), signal transducer and activator of transcription factors (STATs), and β-catenin (for a full list, see Boidot et al., 2014).
BIRC5 has four exons and five introns, and encodes ten splice variants, seven with known function (Sah and Seniya, 2015). Although the isoform transcripts of survivin are frequently analysed in clinical studies, less information is available about the proteins; therefore, their significance as biomarkers in cancer remain unclear. The predominant wild-type form is referred to as survivin, after which 2β and ΔEx3 are the most common forms. Both deviate from survivin at the exon 2–exon 3 junction: 2β has a 26-aa insert that makes it pro-apoptotic; exon 3 is deleted in ΔEx3, causing a frameshift resulting in a different C-terminus. For further information regarding the different isoforms, the reader is referred to Sah and Seniya (2015).
Survivin is ubiquitylated and degraded by the 26S proteasome and has a half-life of 30 to 120 min. After mitosis, it is eliminated by midbody extrusion, and removal of any residual protein is mediated by the anaphase-promoting complex, an E3 ubiquitin ligase, that is activated by Cdc20 or Cdh1 (Connell, et al., 2008). Nuclear relocation of survivin reduces its half-life (Connell et al., 2008), and many of its interactions affect its stability. For instance, binding to X-linked inhibitor of apoptosis protein (XIAP) increases survivin turnover (Arora et al., 2007), whereas integration into the CPC stabilises it (Honda et al., 2003).
Localisation
Interphase
In interphase, survivin localises to the cytoplasm and/or the nucleus. The ratio between cytoplasmic and nuclear survivin has been assessed by many clinical researchers hoping to use it as a prognostic tool (Stauber et al., 2007). However, as with the survivin isoforms, data have been inconsistent, so the significance of this ratio remains uncertain. Survivin is trafficked out of the nucleus in an exportin-1-dependent manner (Stauber et al., 2006; Colnaghi et al., 2006). It has a centrally placed nuclear export sequence (NES) between the BIR domain and the C-terminal helix (Stauber et al., 2006; Colnaghi et al., 2006), which is masked by homodimerisation, and a bipartite NES in its C-terminus (Engelsma et al., 2007). While nuclear export is understood, how survivin enters the nucleus is not; it has no nuclear localisation signal and may depend on cofactor(s) to gain access (Villapol et al., 2008). Endogenous survivin is small enough to diffuse across the nuclear membrane, both as a monomer and a homodimer. However, the progressive nuclear accumulation of GFP–survivin, which is above the threshold size of diffusion, upon inhibition of exportin-1 (Connell, et al., 2008), suggests that it is actively imported (Colnaghi et al., 2006). Interestingly, Temme and co-workers noted that, in newly isolated fibroblasts, ectopically expressed survivin was nuclear but became progressively cytoplasmic with successive passages (Temme et al., 2005), which may depend on its association with heat-shock protein 90 (HSP90) and phosphorylation at T34 (Al-Khalaf and Aboussekhra, 2013). As age is one of the primary risk factors in cancer, this nuclear–cytoplasmic shift is intriguing.
Specifically in cancer cells, survivin is additionally detected in mitochondria (Dohi et al., 2004), and it is attractive to consider that mitochondrial survivin could be an ‘Achilles’ heel’ of cancer (Ausserlechner and Hagenbuchner, 2016). Note, however, that this pool of survivin, which is found in the inner mitochondrial membrane and matrix (Rivadeneira et al., 2015), is visually obscured by the cytoplasmic population, but can be readily detected by subcellular fractionation and immunoblotting, or by using nanobody trackers (Beghein et al., 2016). Mitochondrial import of survivin is directed through its proline-rich N-terminus (M1GAPTLPPAW10), which forms a canonical amphipathic α-helical mitochondrial targeting sequence (MTS) (Dunajová et al., 2016). Survivin can also be indirectly chaperoned into mitochondria by HSP90 and/or aryl hydrocarbon receptor-interacting protein (AIP) (Kang and Altieri, 2006). This pool increases with stress (Asumen et al., 2010), is released from the mitochondria into the cytosol in response to apoptotic stimuli and has enhanced anti-apoptotic activity compared to cytoplasmic survivin, the reason for which is unclear (Dohi et al., 2004).
Mitosis
In proliferating cells, survivin is first detected in G2 (Beardmore et al., 2004) when it targets the CPC to the centromeres through a direct interaction between its BIR-domain residues D70 and D71, and histone 3 that has been phosphorylated at T3 by haspin kinase (Jeyaprakash et al., 2011; Wang F. et al., 2010a; Kelly et al., 2010; Yamagishi et al., 2010). Several posttranslational modifications (PTMs) affect the centromeric association of survivin, most notably phosphorylation by aurora-B, and its ubiquitylation status, which is regulated by the de-ubiquitylating enzyme fat facet in mouse (FAM, also known as USP9X) (Vong et al., 2005). Survivin remains at the centromeres until the metaphase-anaphase transition, after which it travels to the midzone microtubules and the equatorial cortex, delineating the cleavage plane (see poster, mitotic survivin). How survivin is targeted to the midzone is unclear; however, this might occur through a direct association of its C-terminus with microtubules (Li et al., 1998). Its dynamic localisation during mitosis is regulated by all the key mitotic kinases (see poster).
Extracellular
Survivin has also been found on the surface of exosomes, which are constitutively secreted from cancer cells with secretion enhanced by oncotherapy; this suggests it may have prognostic potential in serum biopsies (Khan et al., 2011; Galbo et al., 2017). Interestingly, neighbouring cancer cells in culture can be coerced to proliferate and evade apoptosis by exosomally delivered survivin (Khan et al., 2011), demonstrating a role in cell–cell communication, which has already been reported in autoimmunity (see below).
Cellular functions of survivin
Cell death
Survivin protects cells against apoptotic and autophagic death. Localisation within the cytoplasm is crucial to the anti-apoptotic activity of survivin, as nuclear relocation abrogates it (Knauer et al., 2006; Connell, et al., 2008). However, prior mitochondrial residence augments this activity (Dohi et al., 2004). While there is no doubt that caspase activity is reduced by survivin expression, in both homodimeric and monomeric states (Pavlyukov et al., 2011), unlike other IAPs, survivin only has a single BIR domain and does not bind to caspases at physiological concentrations. The current consensus is that survivin cooperates with XIAP and hepatitis B virus X-interacting protein (HBXIP, also known as LAMTOR5) in a complex with XIAP-associated factor 1 (XAF1) (Marusawa et al., 2003) to affect the interaction of XIAP with caspases or to augment the effect of other IAP family members, such as c-IAP1 or c-IAP2 (also known as BIRC2 and BIRC3, respectively), which act further upstream in the extrinsic apoptotic pathway (Verhagen et al., 2001). Survivin may also prevent the release of apoptotic protease-activating factor 1 (APAF1) from the mitochondria, or sequester the IAP inhibitor second mitochondrial-derived activator of caspases (Smac; also known as Diablo), away from other IAPs (Song et al., 2004).
To maintain homeostasis, a basal level of autophagy operates to remove defective organelles and misfolded proteins. In response to stress, such as nutrient depletion, autophagy is induced to enable short-term survival. However, as a catabolic recycling system, excessive autophagy ultimately kills a cell, which in a cancer context would be tumour suppressive. Cytokine treatment, which hyperactivates the protein kinase B (also called AKT1, herein denoted Akt/PKB), phosphoinositide 3-kinase (PI3K) signalling pathway and increases survivin expression, can inhibit autophagic death (Roca et al., 2008); conversely, pharmacological inhibition of survivin with sepantronium bromide (YM155) increases it (Wang et al., 2011). We recently discovered that survivin binds to the autophagic regulator microtubule-associated protein light chain 3B (LC3B, also known as MAP1LC3B) through a canonical LC3-interacting region in its BIR domain; however, this interaction did not have any effect on its inhibition of autophagy (Humphry and Wheatley, 2018). Whether survivin inhibits excessive autophagy in association with Beclin-1 (Niu et al., 2010), or other autophagic factors, remains to be determined.
Mitosis
By targeting the CPC to the centromeres during prometaphase, survivin helps to ensure that chromosomes are properly aligned prior to anaphase. It achieves this by communicating with the spindle checkpoint tension sensor BubR1 (‘budding uninhibited by benzamidazole-related protein 1’, also known as BUB1B in mammals) via aurora-B kinase, which detect and detach misoriented chromosomes, respectively, allowing the cell further attempts to attach chromosomes correctly. As noted above, during prometaphase, phosphorylation of survivin by aurora-B ensures that its association with the centromeres remains dynamic until all chromosomes have oriented (Wheatley et al., 2007, 2001; Lens et al., 2003; Carvalho et al., 2003). Survivin can also affect mitotic spindle assembly by dampening microtubule dynamics (Rosa et al., 2006; Cheung et al., 2009), a phenomenon that is mediated by Ran-GTP and TPX2 (Xia et al., 2008). Finally, survivin directs cytokinesis by delineating the cleavage plane prior to actomyosin recruitment. The coordination of mitosis and cytokinesis is an essential and conserved role of survivin in all eukaryotes from yeast (where the survivin homologue is known as Bir1) (Rajagopalan and Balasubramanian, 2002) to human. Accordingly, loss or depletion of survivin leads to prometaphase defects, cytokinesis failure, mitotic catastrophe and increased apoptosis in all model systems (Lens et al., 2003; Carvalho et al., 2003; Rajagopalan and Balasubramanian, 2002; Yue et al., 2008; Speliotes et al., 2000; Jones et al., 2000). Furthermore, the mouse knockout is embryonic lethal at 2.5 days post-coitum (Uren et al., 2000).
Mitochondria
Mitochondrial residence of survivin appears to be an exclusively cancer-associated phenomenon and can affect cellular metabolism; however, the current data are contradictory. For instance, in neuroblastoma cells, which have an additional copy of chromosome 17q (Islam et al., 2000), survivin increases glycolysis (Hagenbuchner et al., 2016), whereas in prostate cancer and glioblastoma cells with high survivin expression, oxidative phosphorylation is increased (Rivadeneira et al., 2015). The metabolic adaptability of cancer cells makes understanding this aspect of survivin biology particularly challenging. In addition, mitochondria are highly dynamic organelles that continuously fuse and undergo fission in order to ensure that their health is maintained and any damaged parts, such as mitochondrial (mt)DNA-harbouring reactive oxygen species (ROS)-induced lesions, are eliminated. Aside from metabolism, evidence is amassing that survivin regulates mitochondrial dynamics, but exactly how this is achieved also remains to be elucidated (Hagenbuchner et al., 2012).
Migration and angiogenesis
Linking ATP demand to migration, we witnessed that mitochondria are recruited to actively migrating areas of adherent cells overexpressing survivin (Rivadeneira et al., 2015). Moreover, survivin can alter focal adhesion dynamics by regulating c-Src activity, which may promote migration (Dunajová et al., 2016) (see poster). Interestingly, communication with c-Src may be bidirectional: using a temperature-sensitive c-Src expression system in MDCK (epithelial) cells, in which cell-cell and cell-matrix interactions were disrupted, ultimately increased survivin expression through T-cell factor–β-catenin signalling (Capra and Eskelinen, 2017). Survivin also has a pro-angiogenic role, as it is downstream of vascular endothelial growth factor (VEGF) and might also contribute to vascular remodelling through inhibition of apoptosis (Daly et al., 2004; reviewed in Sanhueza et al., 2015).
Stemness
In embryonic stem cells, survivin knockdown decreases the expression of the key transcription factors associated with pluripotency, namely octamer-binding transcription factor 4 (Oct4, also known as POU5F1) and Nanog (Mull et al., 2014), suggesting that it is involved in stemness. This has pathogenic and potentially therapeutic implications, because survivin appears to prevent aneuploidy and formation of micronuclei in pluripotent stem cells that undergo neurogenesis (Sartore et al., 2011), whereas pharmacological targeting of survivin is sufficient to abolish pluripotent stem cell and teratoma formation (Lee et al., 2013). Furthermore, survivin is expressed in stem cells and rapid-amplifying progenitor cells in the intestinal crypts, where it helps to maintain gut homeostasis (Martini et al., 2016).
Survivin is constitutively expressed in cancer stem cells (CSCs), a discrete cell population within a particular cancer that retains and/or regains stem-like qualities. For example, in acute myeloid leukaemia, CSCs are thought to drive the malignant state and to be responsible for resistance to and relapse after treatment (Zhang et al., 2015). In this case, transcriptional de-repression of survivin, mediated by mitogen-activated protein kinase (MAPK) signalling, and the transcription factors Sp1 (specificity protein 1) and c-Myc, promote its constitutive expression (Zhang et al., 2015). Siddarth and colleagues (2016) reported that survivin expression is linked to the pre-metastatic state of breast cancer stem cells. By noting changes after shRNA-mediated knockdown of survivin, they found that its expression was not only linked to self-renewal, but also to epithelial-to-mesenchymal transition (EMT), invasion and metastasis through alterations in WNT/β-catenin signalling (Siddharth et al., 2016).
Survivin signalling
The signalling aspect of survivin biology is complex and incompletely understood, and may also differ depending on the cellular context. Evidence to date suggests that activation of Akt/PKB and PI3K occurs upstream of many events that involve survivin. These kinases regulate numerous cellular processes, including cell cycle, metabolism, apoptosis, angiogenesis and autophagy, by instructing downstream transcription and translation factors that alter protein expression. Upstream of Akt/PKB and PI3K, are β-catenin, mammalian target of rapamycin (mTOR), the MAPK cascade and Ras. Immunologically, the interleukins IL4 and IL6 instruct the STAT family of transcription factors to promote survivin expression. Survivin signalling pathways are outlined in the poster.
Survivin in cancer and other diseases
Undoubtedly the main clinical interest in survivin is in cancer, as it is the fourth most upregulated mRNA in the human cancer transcriptome (Velculescu et al., 1999), and its expression has been correlated with increased tumour resistance to a broad range of chemotherapy agents, radiation insensitivity and poor patient prognosis. Derangement of its natural cycle of expression is due principally to transcriptional de-repression, which causes continuous synthesis throughout the cell cycle (Siffroi-Fernandez et al., 2014) and/or altered splicing (Antonacopoulou et al., 2011). Although many single-nucleotide polymorphisms (SNPs) have been found in its promoter, mutations within the gene coding region are rare, but one that has been reported is a lysine to glutamic acid mutation, K129E (Jang et al., 2008). When assessed in cultured cells, the K129E variant caused mitotic defects by decreasing the affinity of survivin to borealin (Aljaberi et al., 2014). Clearly, with roles in mitosis, apoptosis suppression, autophagy, migration, metabolism and angiogenesis, there are many routes through which survivin can promote tumour cell survival and cancer metastasis. In addition to cancer, survivin has been implicated in rheumatoid arthritis (Bokarewa et al., 2005; Mera et al., 2008) and multiple sclerosis (Hebb et al., 2008). In these autoimmune disorders, survivin is secreted and its cytokine-dependent expression correlates with reduced apoptosis and inflammation.
Therapeutic targeting
Despite the critical role of survivin as a universal cancer gene that is pivotal for disease maintenance, targeting of this pathway for novel therapeutics so far has garnered only limited success. Lacking intrinsic catalytic activity, and having few deep ‘pockets’ that are suitable to accommodate small-molecule antagonists, survivin joins the vast majority of cancer genes that are considered ‘undruggable’ in the conventional sense (Dang et al., 2018). Against this backdrop, the best-studied survivin suppressor is YM155. YM155 is not technically a direct survivin inhibitor, but instead predicted to shut off transcription of the BIRC5 (survivin) gene, although this proposed mechanism is not universally accepted (Rauch et al., 2014). After encouraging phase I trials that showed manageable toxicity and hinted to clinical activity in heavily pre-treated patients (Tolcher et al., 2012), further clinical development of YM155 has been less successful and most combination regimens failed to meet their specified endpoints in different cancers, including advanced melanoma (Kudchadkar et al., 2015), HER2-negative metastatic breast cancer (Clemens et al., 2015), prostate cancer (Tolcher et al., 2012) and non-small cell lung cancer (Kelly et al., 2013). A potential exception may be advanced non-Hodgkin's lymphoma; here, the combination of YM155 with rituximab, a therapeutic monoclonal antibody against the CD20 surface molecule expressed on B cells, appeared well-tolerated and produced durable responses (Papadopoulos et al., 2016) in patients with aggressive and relapsed non-Hodgkin's lymphoma.
Alternative strategies to target the survivin pathway for novel cancer therapeutics continue to emerge. These include small molecule inhibitors of the survivin dimerisation interface (Qi et al., 2016), or adjacent cavities (Berezov et al., 2012), antibodies to a recently discovered cell surface pool of survivin (Fenstermaker et al., 2018), and survivin-directed short interfering RNA encapsulated in nanoparticles (Li et al., 2017). However, these approaches are still in their infancy and have yet to advance past preclinical evaluation.
Conversely, a third strategy aimed at generating survivin vaccines for cancer immunotherapy has successfully passed proof-of concept and has already produced encouraging results in the clinic (Kaneko et al., 2014). Specifically, several survivin-directed immunisation platforms have been developed that are well-tolerated in patients and give rise to robust immunological responses with initial evidence of clinical activity as both monotherapy or in combination in hard-to-treat malignancies; these are SurVaxM for malignant glioblastoma multiformis (Fenstermaker and Ciesielski, 2014) and the multi-epitope vaccine EMD640744 in solid tumours (Lennerz et al., 2014; Zhenjiang et al., 2018).
Conclusions and perspectives
In the wake of the 21st anniversary of its discovery, our knowledge of survivin has expanded exponentially, but we are yet to have a survivin-specific anti-cancer agent. However, now that we know survivin is a molecular collaborator extraordinaire, it is heartening that the development of drugs and peptides that target protein–protein interactions is gathering momentum (Dang et al., 2018). Incidentally, shepherdin, a competitive peptide that we developed to prevent survivin from binding to HSP90 has been used effectively in this capacity in the laboratory for several years (Plescia et al., 2005). Thus, now, with a relatively comprehensive understanding of survivin functions and a molecular inventory of its interactome, it seems that we are in a strong position to make headway in preventing this mischievous little protein from doing what it must.
Acknowledgements
We apologise to all the colleagues whose contributions to this exciting area of investigation could not be quoted for reason of space constraints.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work in this area is supported by National Institutes of Health (grant P01 CA140043 and R35 CA220446 to D.C.A.). Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.223826.supplemental
References
- Al-Khalaf H. H. and Aboussekhra A. (2013). Survivin expression increases during aging and enhances the resistance of aged human fibroblasts to genotoxic stress. Age 35, 549-562. 10.1007/s11357-011-9378-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljaberi A. M., Webster J. R. M. and Wheatley S. P. (2014). Analysis of the functional repertoire of a mutant form of survivin, K129E, which has been linked to lung cancer. Cancer Cell Int. 14, 78 10.1186/s12935-014-0078-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G., Adida C. and Altieri D. C. (1997). A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 3, 917-921. 10.1038/nm0897-917 [DOI] [PubMed] [Google Scholar]
- Antonacopoulou A. G., Floratou K., Bravou V., Kottorou A., Dimitrakopoulos F. I., Marousi S., Stavropoulos M., Koutras A. K., Scopa C. D. and Kalofonos H. P. (2011). The survivin −31 snp in human colorectal cancer correlates with survivin splice variant expression and improved overall survival. Cell Oncol. 34, 381-391. 10.1007/s13402-011-0038-4 [DOI] [PubMed] [Google Scholar]
- Arora V., Cheung H. H., Plenchette S., Micali O. C., Liston P. and Korneluk R. G. (2007). Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 282, 26202-26209. 10.1074/jbc.M700776200 [DOI] [PubMed] [Google Scholar]
- Asumen M. G., Ifeacho T. V., Cockerham L., Pfandl C. and Wall N. R. (2010). Dynamic changes to survivin subcellular localization are initiated by DNA damage. OncoTargets Ther. 3, 129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausserlechner M. J. and Hagenbuchner J. (2016). Mitochondrial survivin–an Achilles’ heel in cancer chemoresistance. Mol. Cell. Oncol. 3, e1076589 10.1080/23723556.2015.1076589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. M. A., Osborne T. P. and Wheatley S. P. (2009). Phosphorylation of survivin at threonine 34 inhibits its mitotic function and enhances its cytoprotective activity. Cell Cycle 8, 278-283. 10.4161/cc.8.2.7587 [DOI] [PubMed] [Google Scholar]
- Barrett R. M. A., Colanghi R. and Wheatley S. P. (2011). Threonine 48 in the BIR domain of survivin is critical to its mitotic and anti-apoptotic activities and can be phosphorylated by CK2 in vitro. Cell Cycle 10, 538-548. 10.4161/cc.10.3.14758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardmore V. A., Ahonen L. J., Gorbsky G. J. and Kallio M. J. (2004). Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J. Cell Sci. 117, 4033-4042. 10.1242/jcs.01242 [DOI] [PubMed] [Google Scholar]
- Beghein E., Van Audenhove I., Zwaenepoel O., Verhelle A., De Ganck A. and Gettemans J. (2016). A new survivin tracer tracks, delocalizes and captures endogenous survivin at different subcellular locations and in distinct organelles. Sci. Rep. 6, 31177 10.1038/srep31177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezov A., Cai Z., Freudenberg J. A., Zhang H., Cheng X., Thompson T., Murali R., Greene M. I. and Wang Q. (2012). Disabling the mitotic spindle and tumor growth by targeting a cavity-induced allosteric site of survivin. Oncogene 31, 1938-1948. 10.1038/onc.2011.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boidot R., Végran F. and Lizard-Nacol S. (2014). Transcriptional regulation of the survivin gene. Mol. Biol. Rep. 41, 233-240. 10.1007/s11033-013-2856-0 [DOI] [PubMed] [Google Scholar]
- Bokarewa M., Lindblad S., Bokarew D. and Tarkowski A. (2005). Balance between survivin, a key member of the apoptosis inhibitor family, and its specific antibodies determines erosivity in rheumatoid arthritis. Arthritis Res. Ther. 7, R349-R358. 10.1186/ar1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra J. and Eskelinen S. (2017). Correlation between E-cadherin interactions, survivin expression, and apoptosis in MDCK and ts-Src MDCK cell culture models. Lab. Investig. 97, 1453-1470. 10.1038/labinvest.2017.89 [DOI] [PubMed] [Google Scholar]
- Carvalho A., Carmena M., Sambade C., Earnshaw W. C. and Wheatley S. P. (2003). Survivin is required for stable checkpoint activation in response to loss of spindle tension in HeLa cells. J. Cell Sci. 116, 2987-2998. 10.1242/jcs.00612 [DOI] [PubMed] [Google Scholar]
- Cheung C. H. A., Chen H. H., Kuo C. C., Chang C. Y., Coumar M. S., Hsieh H. P. and Chang J. Y. (2009). Survivin counteracts the therapeutic effect of microtubule de-stabilizers by stabilizing tubulin polymers. Mol. Cancer 8, 43 10.1186/1476-4598-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M. R., Gladkov O. A., Gartner E., Vladimirov V., Crown J., Steinberg J., Jie F. and Keating A. (2015). Phase II, multicenter, open-label, randomized study of YM155 plus docetaxel as first-line treatment in patients with HER2-negative metastatic breast cancer. Breast Cancer Res. Treat. 149, 171-179. 10.1007/s10549-014-3238-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnaghi R. and Wheatley S. P. (2010). Liaisons between survivin and plk1 during cell division and cell death. J. Biol. Chem. 285, 22592-22604. 10.1074/jbc.M109.065003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnaghi R., Connell C. M., Barrett R. M. A. and Wheatley S. P. (2006). Separating the anti-apoptotic and mitotic roles of survivin. J. Biol. Chem. 281, 33450-33456. 10.1074/jbc.C600164200 [DOI] [PubMed] [Google Scholar]
- Connell C. M., Colnaghi R. and Wheatley S. P. (2008). Nuclear survivin has reduced stability and is not cytoprotective. J. Biol. Chem. 283, 3289-3296. 10.1074/jbc.M704461200 [DOI] [PubMed] [Google Scholar]
- Daly C., Wong V., Burova E., Wei Y., Zabski S., Griffiths J., Lai K. M., Lin H. C., Ioffe E., Yancopoulos G. D. et al. (2004). Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1). Genes Dev. 18, 1060-1071. 10.1101/gad.1189704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Reddy E. P., Shokat K. M. and Soucek L. (2018). Drugging the 'undruggable' cancer targets. Nature Reviews Cancer 17, 502-508. 10.1038/nrc.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi T., Beltrami E., Wall N. R., Plescia J. and Altieri D. C. (2004). Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J. Clin. Invest. 114, 1117-1127. 10.1172/JCI200422222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi T., Xia F. and Altieri D. C. (2007). Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol. Cell 27, 17-28. 10.1016/j.molcel.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunajová L., Cash E., Markus R., Rochette S., Townley A. R. and Wheatley S. P. (2016). The N-terminus of survivin is a mitochondrial-targeting sequence and Src regulator. J. Cell Sci. 129, 2707-2712. 10.1242/jcs.183277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsma D., Rodriguez J. A., Fish A., Giaccone G. and Fornerod M. (2007). Homodimerization antagonizes nuclear export of survivin. Traffic 8, 1495-1502. 10.1111/j.1600-0854.2007.00629.x [DOI] [PubMed] [Google Scholar]
- Fenstermaker R. A. and Ciesielski M. J. (2014). Challenges in the development of a survivin vaccine (SurVaxM) for malignant glioma. Expert Rev. Vaccines 13, 377-385. 10.1586/14760584.2014.881255 [DOI] [PubMed] [Google Scholar]
- Fenstermaker R. A., Figel S. A., Qiu J., Barone T. A., Dharma S. S., Winograd E. K., Galbo P. M., Wiltsie L. M. and Ciesielski M. J. (2018). Survivin monoclonal antibodies detect survivin cell surface expression and inhibit tumor growth in vivo. Clin. Cancer Res. 24, 2642-2652. 10.1158/1078-0432.CCR-17-2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbo P. M., Ciesielski M. J., Figel S., Maguire O., Qiu J., Wiltsie L., Minderman H. and Fenstermaker R. A. (2017). Circulating CD9/GFAP/survivin exosomes in malignant glioma patients following survivin vaccination. Oncotarget 8, 114722-114735. 10.18632/oncotarget.21773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M., Plescia J., Leav I., Li J., Languino L. R. and Altieri D. C. (2009). Endogenous tumor suppression mediated by PTEN involves survivin gene silencing. Cancer Res. 69, 4954-4958. 10.1158/0008-5472.CAN-09-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuchner J., Kuznetsov A. V., Obexer P. and Ausserlechner M. J. (2012). BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 32, 4748-4757. 10.1038/onc.2012.500 [DOI] [PubMed] [Google Scholar]
- Hagenbuchner J., Kiechl-Kohlendorfer U., Obexer P. and Ausserlechner M. J. (2016). BIRC5/Survivin as a target for glycolysis inhibition in high-stage neuroblastoma. Oncogene 35, 2052-2061. 10.1038/onc.2015.264 [DOI] [PubMed] [Google Scholar]
- Hebb A. L. O., Moore C. S., Bhan V., Campbell T., Fisk J. D., Robertson H. A., Thorne M., Lacasse E., Holcik M., Gillard J. et al. (2008). Expression of the inhibitor of apoptosis protein family in multiple sclerosis reveals a potential immunomodulatory role during autoimmune mediated demyelination. Mult. Scler. 14, 577-594. 10.1177/1352458507087468 [DOI] [PubMed] [Google Scholar]
- Hoffman W. H., Biade S., Zilfou J. T., Chen J. and Murphy M. (2002). Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 277, 3247-3257. 10.1074/jbc.M106643200 [DOI] [PubMed] [Google Scholar]
- Honda R., Körner R. and Nigg E. A. (2003). Exploring the functional interactions between aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325-3341. 10.1091/mbc.e02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry N. J. and Wheatley S. P. (2018). Survivin inhibits excessive autophagy in cancer cells but does so independently of its interaction with LC3. Open Biol. 7, bio037374 10.1242/bio.037374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Kageyama H., Takada N., Kawamoto T., Takayasu H., Isogai E., Ohira M., Hashizume K., Kobayashi H., Kaneko Y. and Nakagawara A (2000). High expression of Survivin, mapped to 17q25, is signicantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma Oncogene 19, 617-623. 10.1038/sj.onc.1203358 [DOI] [PubMed] [Google Scholar]
- Jang J. S., Kim K. M., Kang K. H., Choi J. E., Lee W. K., Kim C. H., Kang Y. M., Kam S., Kim I.-S., Jun J. E. et al. (2008). Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer 60, 31-39. 10.1016/j.lungcan.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A. and Conti E. (2007). Structure of a survivin-borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131, 271-285. 10.1016/j.cell.2007.07.045 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A. A., Basquin C., Jayachandran U. and Conti E. (2011). Structural basis for the recognition of phosphorylated histone H3 by the Survivin subunit of the chromosomal passenger complex. Structure 19, 1625-1634. 10.1016/j.str.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Saavedra H. I., Holloway M. P., Leone G. and Altura R. A. (2004). Aberrant regulation of survivin by the RB/E2F family of proteins. J. Biol. Chem. 279, 40511-40520. 10.1074/jbc.M404496200 [DOI] [PubMed] [Google Scholar]
- Jones G., Jones D., Zhou L., Steller H. and Chu Y. (2000). Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J. Biol. Chem. 275, 22157-22165. 10.1074/jbc.M000369200 [DOI] [PubMed] [Google Scholar]
- Kaneko N., Mitsuoka K., Amino N., Yamanaka K., Kita A., Mori M., Miyoshi S. and Kuromitsu S. (2014). Combination of YM155, a Survivin Suppressant, with Bendamustine and Rituximab: a new combination therapy to treat relapsed/refractory diffuse large B-cell lymphoma. Clin. Cancer Res. 20, 1814-1822. 10.1158/1078-0432.CCR-13-2707 [DOI] [PubMed] [Google Scholar]
- Kang B. H. and Altieri D. C. (2006). Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J. Biol. Chem. 281, 24721-24727. 10.1074/jbc.M603175200 [DOI] [PubMed] [Google Scholar]
- Keerthivasan G., Liu H., Gump J. M., Dowdy S. F., Wickrema A. and Crispino J. D. (2012). A novel role for survivin in erythroblast enucleation. Haematologica 97, 1471-1479. 10.3324/haematol.2011.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. E., Ghenoiu C., Xue J. Z., Zierhut C., Kimura H. and Funabiki H. (2010). Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235-239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. J., Thomas A., Rajan A., Chun G., Lopez-Chavez A., Szabo E., Spencer S., Carter C. A., Guha U., Khozin S. et al. (2013). A phase I/II study of sepantronium bromide (YM155, survivin suppressor) with paclitaxel and carboplatin in patients with advanced non-Small-Cell lung cancer. Ann. Oncol. 24, 2601-2606. 10.1093/annonc/mdt249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Jutzy J. M. S., Aspe J. R., Mcgregor D. W., Neidigh J. W. and Wall N. R. (2011). Survivin is released from cancer cells via exosomes. Apoptosis 16, 1-12. 10.1007/s10495-010-0534-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S. K., Krämer O. H., Knösel T., Engels K., Rödel F., Kovács A. F., Dietmaier W., Klein-Hitpass L., Habtemichael N., Schweitzer A. et al. (2006). Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 21, 207-216. 10.1096/fj.06-5741com [DOI] [PubMed] [Google Scholar]
- Kudchadkar R., Ernst S., Chmielowski B., Redman B. G., Steinberg J., Keating A., Jie F., Chen C., Gonzalez R. and Weber J. (2015). A phase 2, multicenter, open-label study of sepantronium bromide (YM155) plus docetaxel in patients with stage III (unresectable) or stage IV melanoma. Cancer Med. 4, 643-650. 10.1002/cam4.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaoui N, and Vaux D. L. (2018). Recent advances in understanding inhibitor of apoptosis proteins. F1000Res7, 1889 10.12688/f1000research.16439.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-O., Moon S. H., Jeong H.-C., Yi J.-Y., Lee T.-H., Shim S. H., Rhee Y.-H., Lee S.-H., Oh S.-J., Lee M.-Y. et al. (2013). Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc. Natl Acad. Sci. USA 110, E3281-E3290. 10.1073/pnas.1303669110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennerz V., Gross S., Gallerani E., Sessa C., Mach N., Boehm S., Hess D., Von Boehmer L., Knuth A., Ochsenbein A. F. et al. (2014). Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol. Immunother. 63, 381-394. 10.1007/s00262-013-1516-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S. M. A., Wolthuis R. M., Klompmaker R., Kauw J., Agami R., Brummelkamp T., Kops G. and Medema R. H. (2003). Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22, 2934-2947. 10.1093/emboj/cdg307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C. G., Xu Y., Mularski B., Liu H., Gurbuxani S. and Crispino J. D. (2007). Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J. Exp. Med. 204, 1603-1611. 10.1084/jem.20062395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. and Altieri D. C. (1999). Transcriptional analysis of human survivin gene expression. Biochem. J. 344, 305-311. 10.1042/0264-6021:3440305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ambrosini G., Chu E. Y., Plescia J., Tognin S., Marchisio P. C. and Altieri D. C. (1998). Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580-584. 10.1038/25141 [DOI] [PubMed] [Google Scholar]
- Li F., Ackermann E. J., Bennett C. F., Rothermel A. L., Plescia J., Tognin S., Villa A., Marchisio P. C. and Altieri D. C. (1999). Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1, 461-466. 10.1038/70242 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang L., Tang C. and Yin C. (2017). Co-delivery of doxorubicin and survivin shRNA-expressing plasmid via microenvironment-responsive dendritic mesoporous silica nanoparticles for synergistic cancer therapy. Pharm. Res. 34, 2829-2841. 10.1007/s11095-017-2264-6 [DOI] [PubMed] [Google Scholar]
- Martini E., Schneider E., Neufert C., Neurath M. F. and Becker C. (2016). Survivin is a guardian of the intestinal stem cell niche and its expression is regulated by TGF-β. Cell Cycle 15, 2875-2881. 10.1080/15384101.2016.1231286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusawa H., Matsuzawa S., Welsh K., Zou H., Armstrong R., Tamm I. and Reed J. C. (2003). HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22, 2729-2740. 10.1093/emboj/cdg263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mera S., Magnusson M., Tarkowski A. and Bokarewa M. (2008). Extracellular survivin up-regulates adhesion molecules on the surface of leukocytes changing their reactivity pattern. J. Leukoc. Biol. 83, 149-155. 10.1189/jlb.0507287 [DOI] [PubMed] [Google Scholar]
- Mull A. N., Klar A. and Navara C. S. (2014). Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Res. 12, 539-549. 10.1016/j.scr.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Niu T.-K., Cheng Y., Ren X. and Yang J.-M. (2010). Interaction of Beclin 1 with survivin regulates sensitivity of human glioma cells to TRAIL-induced apoptosis. FEBS Lett. 584, 3519-3524. 10.1016/j.febslet.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor D. S., Grossman D., Plescia J., Li F., Zhang H., Villa A., Tognin S., Marchisio P. C. and Altieri D. C. (2000). Regulation of apoptosis at cell division by p34(cdc2) phosphorylation of survivin. Proc. Natl Acad. Sci. USA 97, 13103-13107. 10.1073/pnas.240390697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos K. P., Lopez-Jimenez J., Smith S. E., Steinberg J., Keating A., Sasse C., Jie F. and Thyss A. (2016). A multicenter phase II study of sepantronium bromide (YM155) plus rituximab in patients with relapsed aggressive B-cell Non-Hodgkin lymphoma. Leuk. Lymphoma 57, 1848-1855. 10.3109/10428194.2015.1113275 [DOI] [PubMed] [Google Scholar]
- Pavlyukov M. S., Antipova N. V., Balashova M. V., Vinogradova T. V., Kopantzev E. P. and Shakhparonov M. I. (2011). Survivin monomer plays an essential role in apoptosis regulation. J. Biol. Chem. 286, 23296-23307. 10.1074/jbc.M111.237586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plescia J., Salz W., Xia F., Pennati M., Zaffaroni N., Daidone M. G., Meli M., Dohi T., Fortugno P., Nefedova Y. et al. (2005). Rational design of shepherdin, a novel anticancer agent. Cancer Cell 7, 457-468. 10.1016/j.ccr.2005.03.035 [DOI] [PubMed] [Google Scholar]
- Qi J., Dong Z., Liu J., Peery R. C., Zhang S., Liu J.-Y. and Zhang J.-T. (2016). Effective targeting of the survivin dimerization interface with small-molecule inhibitors. Cancer Res. 76, 453-462. 10.1158/0008-5472.CAN-15-1874 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S. and Balasubramanian M. K. (2002). Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics 160, 445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Hennig D., Schäfer C., Wirth M., Marx C., Heinzel T., Schneider G. and Krämer O. H. (2014). Survivin and YM155: how faithful is the liaison? Biochem. Biophys. Acta 45, 202-220. 10.1016/j.bbcan.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Rivadeneira D. B., Caino M. C., Seo J. H., Angelin A., Wallace D. C., Languino L. R. and Altieri D. C. (2015). Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci. Signal. 8, ra80 10.1126/scisignal.aab1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H., Varsos Z. and Pienta K. J. (2008). CCL2 protects prostate cancer PC3 cells from autophagic death via PI3K/AKT-dependent survivin up-regulation. J. Biol. Chem. 283, 25057-25073. 10.1074/jbc.M801073200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J., Canovas P., Islam A., Altieri D. C. and Doxsey S. J. (2006). Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 17, 1483-1493. 10.1091/mbc.e05-08-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah N. K. and Seniya C. (2015). Survivin splice variants and their diagnostic significance. Tumor Biol. 36, 6623-6631. 10.1007/s13277-015-3865-5 [DOI] [PubMed] [Google Scholar]
- Sanhueza C., Wehinger S., Castillo Bennett J., Valenzuela M., Owen G. I. and Quest A. F. (2015). The twisted survivin connection to angiogenesis. Mol. Cancer 14, 198 10.1186/s12943-015-0467-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartore R. C., Campos P. B., Trujillo C. A., Ramalho B. L., Negraes P. D., Paulsen B. S., Meletti T., Costa E. S., Chicaybam L., Bonamino M. H. et al. (2011). Retinoic acid-treated pluripotent stem cells undergoing neurogenesis present increased aneuploidy and micronuclei formation. PLoS ONE 6, e20667 10.1371/journal.pone.0020667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharth S., Das S., Nayak A. and Kundu C. N. (2016). SURVIVIN as a marker for quiescent-breast cancer stem cells—an intermediate, adherent, pre-requisite phase of breast cancer metastasis. Clin. Exp. Metastasis 33, 661-675. 10.1007/s10585-016-9809-7 [DOI] [PubMed] [Google Scholar]
- Siffroi-Fernandez S., Dulong S., Li X.-M., Filipski E., Gréchez-Cassiau A., Peteri-Brünback B., Meijer L., Lévi F., Teboul M. and Delaunay F. (2014). Functional genomics identify Birc5/Survivin as a candidate gene involved in the chronotoxicity of cyclin-dependent kinase inhibitors. Cell Cycle 13, 984-991. 10.4161/cc.27868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Liu S., He H., Hoti N., Wang Y., Feng S. and Wu M. (2004). A single amino acid change (Asp53-Ala53) converts survivin from anti-apoptotic to pro-apoptotic. Mol. Biol. Cell 15, 1287-1296. 10.1091/mbc.e03-07-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes E. K., Uren A., Vaux D. and Horvitz H. R. (2000). The survivin-like C. elegans BIR-1 protein acts with the aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell 6, 211-223. 10.1016/S1097-2765(00)00023-X [DOI] [PubMed] [Google Scholar]
- Stauber R. H., Rabenhorst U., Rekik A., Engels K., Bier C. and Knauer S. K. (2006). Nucleocytoplasmic shuttling and the biological activity of mouse survivin are regulated by an active nuclear export signal. Traffic 7, 1461-1472. 10.1111/j.1600-0854.2006.00486.x [DOI] [PubMed] [Google Scholar]
- Stauber R. H., Mann W. and Knauer S. K. (2007). Nuclear and cytoplasmic survivin: molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 67, 5999-6002. 10.1158/0008-5472.CAN-07-0494 [DOI] [PubMed] [Google Scholar]
- Sun C., Nettesheim D., Liu Z. and Olejniczak E. T. (2005). Solution structure of human survivin and its binding interface with Smac/DIABLO. Biochemistry 44, 11-17. 10.1021/bi0485171 [DOI] [PubMed] [Google Scholar]
- Temme A., Diestelkoetter-Bachert P., Schmitz M., Morgenroth A., Weigle B., Rieger M. A., Kiessling A. and Rieber E. P. (2005). Increased p21ras activity in human fibroblasts transduced with survivin enhances cell proliferation. Biochem. Biophys. Res. Commun. 327, 765-773. 10.1016/j.bbrc.2004.12.075 [DOI] [PubMed] [Google Scholar]
- Tolcher A. W., Quinn D. I., Ferrari A., Ahmann F., Giaccone G., Drake T., Keating A. and De Bono J. S. (2012). A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann. Oncol. 23, 968-973. 10.1093/annonc/mdr353 [DOI] [PubMed] [Google Scholar]
- Uren A. G., Wong L., Pakusch M., Fowler K. J., Burrows F. J., Vaux D. L. and Choo K. H. A. (2000). Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 10, 1319-1328. 10.1016/S0960-9822(00)00769-7 [DOI] [PubMed] [Google Scholar]
- Velculescu V. E., Madden S. L., Zhang L., Lash A. E., Yu J., Rago C., Lal A., Wang C. J., Beaudry G. A., Ciriello K. M. et al. (1999). Analysis of human transcriptomes. Nat. Genet. 23, 387-388. 10.1038/70487 [DOI] [PubMed] [Google Scholar]
- Verdecia M. A., Bowman M. E., Lu K. P., Hunter T. and Noel J. P. (2000). Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 7, 602-608. 10.1038/77929 [DOI] [PubMed] [Google Scholar]
- Verhagen A. M., Coulson E. J. and Vaux D. L. (2001). Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2, REVIEWS3009 10.1186/gb-2001-2-7-reviews3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S., Acarin L., Faiz M., Castellano B. and Gonzalez B. (2008). Survivin and heat shock protein 25/27 colocalize with cleaved caspase-3 in surviving reactive astrocytes following excitotoxicity to the immature brain. Neuroscience 153, 108-119. 10.1016/j.neuroscience.2008.01.054 [DOI] [PubMed] [Google Scholar]
- Vong Q. P., Cao K., Li H. Y., Iglesias P. A. and Zheng Y. (2005). Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310, 1499-1504. 10.1126/science.1120160 [DOI] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J. R., Niedzialkowska E., Banerjee B., Stukenberg P. T., Gorbsky G. J. and Higgins J. M. G. (2010a). Histone H3 Thr-3 phosphorylation by haspin positions aurora B at centromeres in mitosis. Science 330, 231-235. 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Holloway M. P., Ma L., Cooper Z. A., Riolo M., Samkari A., Elenitoba-Johnson K. S. J., Chin Y. E. and Altura R. A. (2010b). Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J. Biol. Chem. 285, 36129-36137. 10.1074/jbc.M110.152777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Chen Z., Diao X. and Huang S. (2011). Induction of autophagy-dependent apoptosis by the survivin suppressant YM155 in prostate cancer cells. Cancer Lett. 302, 29-36. 10.1016/j.canlet.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Wheatley S. P., Carvalho A., Vagnarelli P. and Earnshaw W. C. (2001). INCENP is required for proper targeting of survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11, 886-890. 10.1016/S0960-9822(01)00238-X [DOI] [PubMed] [Google Scholar]
- Wheatley S. P., Barrett R. M., Andrews P. D., Medema R. H., Morley S. J., Swedlow J. R. and Lens S. M. A. (2007). Phosphorylation by aurora-B negatively regulates survivin function during mitosis. Cell Cycle 6, 1220-1230. 10.4161/cc.6.10.4179 [DOI] [PubMed] [Google Scholar]
- Xia F., Canovas P. M., Guadagno T. M. and Altieri D. C. (2008). A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell. Biol. 28, 5299-5311. 10.1128/MCB.02039-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y. and Watanabe Y. (2010). Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239-243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yue Z., Carvalho A., Xu Z., Yuan X., Cardinale S., Ribeiro S., Lai F., Ogawa H., Gudmundsdottir E., Gassmann R. et al. (2008). Deconstructing survivin: comprehensive genetic analysis of survivin function by conditional knockout in a vertebrate cell line. J. Cell Biol. 183, 279-296. 10.1083/jcb.200806118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Otevrel T., Gao Z., Gao Z., Ehrlich S. M., Fields J. Z. and Boman B. M. T. (2001). Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 62, 8864-8867. [PubMed] [Google Scholar]
- Zhang Y., Chen H. X., Zhou S. Y., Wang S. X., Zheng K., Xu D. D., Liu Y. T., Wang X. Y., Wang X., Yan H. Z. et al. (2015). Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Mol. Cancer 14, 1-18. 10.1186/s12943-015-0326-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhenjiang L., Rao M., Luo X., Valentini D., Von Landenberg A., Meng Q., Sinclair G., Hoffmann N., Karbach J., Altmannsberger H.-M. et al. (2018). Cytokine networks and survivin peptide-specific cellular immune responses predict improved survival in patients with Glioblastoma Multiforme. EBioMedicine 33, 49-56. 10.1016/j.ebiom.2018.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]