Abstract
Scope:
The incidence of colorectal cancer (CRC), one of the most common cancers, is inversely proportional to dietary fiber intake. Butyrate, the fermentation end product of dietary fibers in the colon, is known for its antitumor effects. The objectives of this study were to evaluate the effects of butyrate on colorectal adenocarcinoma cell proliferation, differentiation, apoptosis, metabolism, and their mechanistic links using HT-29 and Caco-2 cells.
Methods and results:
Butyrate suppressed proliferation, potentiated differentiation and induced apoptosis in both HT-29 and Caco-2 cells, associated with enhanced expression of isocitrate dehydrogenase 1 (IDH1) and pyruvate dehydrogenase (PDH). Furthermore, butyrate upregulated acetyl-CoA and α-ketoglutarate (α-KG), concomitant with enhanced histone acetylation and DNA demethylation in the promoter of DNA mismatch repair (MMR) gene. Knocking down IDH1 abolished the positive effects of butyrate on CRC apoptosis and MMR protein expression, in junction with reduced α-KG content. Importantly, α-KG supplementation recovered the beneficial effects of butyrate in IDH1 deficient cells.
Conclusion:
In summary, butyrate inhibits indices of colorectal carcinogenesis in an α-ketoglutarate-dependent manner.
Keywords: α-ketoglutarate, butyrate, colorectal cancer, differentiation, DNA mismatch repair protein, IDH1
1. Introduction
Colorectal cancer (CRC) is the third leading cause of cancer death [1]. Genetic instability is considered as a hallmark of cancer cells to foster carcinogenesis [2]. The repair of genetic damage could preserve its integrity to prevent carcinogenesis [2]. DNA mismatch repair (MMR) proteins are critical for repairing DNA defects and eliciting cell apoptosis, an effective way to eliminate cells with genetic or epigenetic errors [3]. The inactivation of MMR proteins is linked to dysfunctional DNA repair, further leading to an increase in microsatellite instability, while the activation of MMR proteins is a therapeutic strategy for preventing carcinogenesis [4]. Mutations in MMR proteins occur in 15% of sporadic colorectal cancers and account for more than 90% of detected mutations in hereditary non-polyposis colorectal cancers [5].
Metabolism is highly associated with carcinogenesis [6]. The mutations in metabolic enzymes, such as those involved in the tricarboxylic acid cycle, may elicit carcinogenesis [7, 8]. Even though mutant isocitrate dehydrogenase (IDH1) did not occur frequently in colorectal cancer, the expression level of IDH1 was decreased in CRC tissues comparing to the paired normal tissue in patients [9], indicating that the inhibition of IDH1 is associated with the development of CRC. Emerging evidence suggests that metabolic enzymes and their metabolites might mediate carcinogenesis epigenetically [10–12]. Acetyl-CoA is considered to be the acetyl donor for acetylation [11], and α-ketoglutarate (α-KG) serves as a substrate for the catalytic activity of ten-eleven translocation (TET) enzymes, catalyzing the demethylation of 5-methylcytosine (5mC) to generate 5-hydroxymethylcytosine (5hmC) [13]. Addition of α-KG, the metabolite of IDH in the tricarboxylic acid cycle, increases the ratio of 5-hmC to 5-mC in chromaffin cells [14], and the intraperitoneal administration of α-KG impedes the growth and angiogenesis of transplanted tumor in mice [15].
Epidemiological studies link CRC incidence to dietary factors that are present in the lumen and directly contact with colonic epithelium, thereby affecting the growth, differentiation or death of epithelial cells [16]. The alcohol consumption and high meat intake are highly associated with the increased risk of CRC [17], while a low-fat and high-fiber diet reduces this risk [18]. Thus, dietary intervention is a promising method for CRC prevention or alleviation.
Butyrate, one of the short-chain fatty acids produced by gut microbiota through fermentation of dietary fibers, is known for its antitumor effects [19, 20]. Even though the exact underlying anti-tumor mechanisms have not been fully elucidated, increasing evidence points to its function as a histone deacetylase inhibitor (HDACi) [21, 22]. Through inhibiting HDAC activity, butyrate enhances nucleosomal DNA accessibility and upregulates gene expression due to enhanced histone acetylation [23], promoting the expression of key apoptotic mediators [24] and cell cycle inhibitors [25]. Furthermore, butyrate could alter the metabolic profile of CRC stem cells by promoting oxidative metabolism instead of glycolysis [26]. The metabolic changes might contribute to the alleviation of carcinogenesis. However, the underlying metabolic mechanisms responsible for the anti-tumor effects of butyrate remain undefined. The objectives of this study were to evaluate the effects of butyrate on proliferation, differentiation, apoptosis, and metabolism of colorectal adenocarcinoma cells, and to further unveil the role of metabolic enzymes in linking butyrate to its anti-tumor effects.
2. Materials and Methods
2.1. Cell culture
The Caco-2 cell line and HT-29 cell line were purchased from American Type Culture Collection (Manassas, VA) and grown at 37 °C with 5% CO2 in a humidified incubator in DMEM (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (GE, Fairfield, CT) and 1% penicillin-streptomycin (Life Technologies, Grand Island, NY). Caco-2 cells and HT-29 cells were seeded at a density of 2×105 per well on 12-well plates, and treated with 4 mM sodium butyrate (NaB, Sigma) for 2 d (for MTT assay) and 4 d (for other assays). Medium with/without NaB was refreshed every day.
For plasmid transfection, only Caco-2 cells were used. Briefly, Caco-2 cells were transfected with IDH1 knock-down (KD) plasmid (#62907, Addgene, Cambridge, MA), IDH1 wild-type (WT) plasmid (#62906, Addgene), or green fluorescent protein (GFP) plasmid (#13031, Addgene) using Lipofectamine 3000 (Life Technologies, Grand Island, NY) per manufacturer’s instructions. Caco-2 cells at 70% confluence were transfected with plasmids using a 2:4:4 ratio of DNA (μg): lipofectamine p3000 (μl): lipofectamine 3000 (μl). The medium was changed 12 h post transfection, then 400 μg/ml G418 (Amresco, Solon, OH) was added to IDH1 KD or IDH1 WT transfected cells in the following 7 d to select cells with transfection. After selection, the transfected cells were seeded onto 12-well plates at 2×105 cells per well to be treated with 4 mM NaB, or 4 mM NaB supplemented with 4 mM dimethyl-α-ketoglutarate, a membrane permeable α-ketoglutarate (α-KG), for 4 d.
2.2. MTT assay
About 1×104 HT-29 cells or Caco-2 cells were seeded in each well of a 96 well plate. Treated with 4 mM NaB for 48 h, cells were incubated with 5 mg/ml 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma) for 4 h. Formazan formed was dissolved in 100 μl of DMSO (VWR, Radnor, PA), and the absorbance at 540 nm was measured using a Synergy H1 microplate reader (BioTek, Winooski, VT).
2.3. Alkaline phosphatase assay
Alkaline phosphatase (AP) assay was performed as previously described [27]. Briefly, HT-29 cells or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well, and treated with 4 mM NaB for 4 d, then cells were rinsed with PBS, lysed and collected for AP activity measurement. AP activity was assayed by incubating with the substrate, 1 mg/ml pNPP (Sigma), at 37 °C for 30 min. The reaction was stopped by adding a final concentration of 3 mM NaOH. The absorbance at 405 nm was measured using a Synergy H1 microplate reader.
2.4. Immunoblotting analysis
Immunoblotting analysis was performed according to the procedures as previously described [27]. Briefly, HT-29 cells or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well, treated with 4 mM NaB for 4 d, then cells were lysed and collected for protein extraction. The extracts were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes, then incubated with antibodies and visualized using an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Band density was normalized to β-actin. Antibodies against villin, IDH1, PDH, PARP, caspase-3, MSH2, MLH1 were purchased from Cell Signaling Technology (Danvers, MA). Anti-E-cadherin antibody was purchased from Life Technologies. Anti-TET3 antibody was purchased from Thermo Scientific (Waltham, MA). Anti-Ac-H3 and anti-Ac-H4 antibodies were purchased from Santa Cruz (Dallas, Texas). Anti-β-actin antibody was purchased from DSHB (Iowa City, IA). IRDye 680 goat anti-mouse secondary antibody and IRDye 800 CW goat anti-rabbit secondary antibody were purchased from Li-Cor Biosciences (Lincoln, NE).
2.5. Immunofluorescence staining
HT-29 cells or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well, treated with 4 mM NaB for 4 d, fixed in ice-cold methanol for 10 min, blocked in 5% goat serum at room temperature for 1 h, then incubated with anti-PCNA (1:200, Santa Cruz, Dallas, Texas) overnight at 4 °C. Stained cells were then incubated with goat anti-rabbit Alexa Fluor 488 secondary antibody (1:1000, Cell Signaling Technology) at room temperature for 1 h. Fluorescence was examined using EVOS FL fluorescence microscope (Life Technologies) as previously described [27].
2.6. Acetyl-coenzyme A and α-ketoglutarate assays
HT-29 cells or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well, and treated with 4 mM NaB for 4d, then collected for α-ketoglutarate measurements using the α-ketoglutarate assay kit (Sigma). The relative content of α-ketoglutarate was assayed by incubating with the α-ketoglutarate converting enzyme at 37 °C for 30 min. The absorbance at 570 nm was measured using a Synergy H1 microplate reader.
Cytosolic acetyl-CoA was separated by subcellular fractionation, then measured using acetyl-coenzyme A assay kit (Sigma) following the manufacturer’s instruction. Briefly, HT-29 cells or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well, treated with 4 mM NaB for 4 d, then cells were incubated with fractionation buffer containing 250 mM sucrose, 20 mM HEPES (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA and 1 mM EGTA. The cell lysate was incubated on ice for 15 min, collected using a cell scraper and centrifuged at 6,000 g at 4 ºC for 10 min to isolate the supernatant as the cytosolic fraction. The relative content of cytosolic acetyl-CoA was assayed by incubating with the acetyl-CoA converting enzyme at 37 ºC for 30 min. The fluorescent excitation at 535 nm and emission at 587 nm were measured using a Synergy H1 microplate reader.
2.7. Hydroxymethyl-DNA and methylcytosine immunoprecipitation
Hydroxymethyl-DNA and methylcytosine immunoprecipitation was performed as previously described [28]. Briefly, HT-29 or Caco-2 cells were seeded onto 12-well plates at 2×105 cells per well and treated with 4 mM NaB for 4 d. Genomic DNA was isolated from cultured cells using phenol-chloroform method [29]. Isolated DNA (10 μg) was diluted in 300 μl TE buffer and sonicated (30% amplitude, 6×10s impulses with 1min pauses, Thermo Scientific FB120 Sonic Dismembrator) into fragments between 300-1000 bp. DNA was denatured by incubation in boiling water for 10 min then immediately cooled on an ice bath, followed by adding 1/10 volume of 10×IP buffer (100 mM Na-phosphate, pH 7, 1.4 M NaCl, 0.5% Triton X). Then, 1/50 5-hydroxymethylcytosine antibody (5hmC, Cell Signaling Technology), 5-methylcytosine antibody (5mC, Cell Signaling Technology), or normal rabbit IgG (Cell Signaling Technology) was added to denatured DNA. The DNA-antibody complex was incubated overnight at 4 °C, and pulled down with commercial pre-blocked Pierce™ magnetic protein A/G beads (Thermo Scientific, Waltham, MA). The captured beads were washed three times with 1×IP buffer and re-suspended in 250 μl digestion buffer (50 mM Tris HCl, pH 8, 10 mM EDTA, 0.5% SDS). Following treatment with proteinase K, DNA was purified using a ChIP DNA clean and concentrator kit (Zymo Research, Irvine, CA), which was then used as templates for quantitative PCR (qPCR). qPCRs were performed using SYBR Green supermixture (Bio-Rad) on a CFX96 RT-PCR detection system (Bio-Rad). Relative enrichment of detected regions was normalized to its respective input DNA. Primer information is shown in Supplementary Table S1.
2.8. Statistical analyses
Statistical data were analyzed as a complete randomized design using General Linear Model of Statistical Analysis System. Data are presented as mean ± standard error of the mean (SEM). Mean difference was separated by paired t-test. Statistical significance is considered as P ≤ 0.05.
3. Results
3.1. Butyrate suppresses proliferation, potentiates differentiation and induces apoptosis in colorectal adenocarcinoma cells
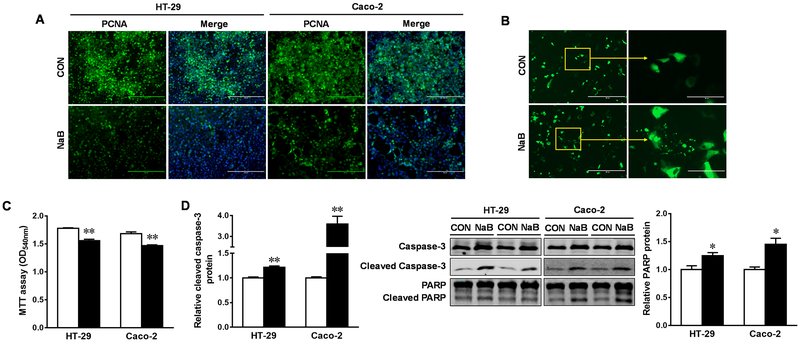
Given that 4 mM butyrate is a physiologically relevant concentration in the human colonic lumen [30, 31], 4 mM NaB was used in this study. Using PCNA staining and MTT analysis, we found that butyrate profoundly impeded DNA synthesis and the viability of HT-29 and Caco-2 cells, respectively (Figure 1A and C). Furthermore, butyrate-induced cell shrinkage and nuclear fragmentation in Caco-2 cells transfected with a GFP plasmid (Figure 1B), associated with enhanced cleavage of procaspase-3 and PARP, showing that butyrate remarkably stimulated apoptosis in both cell lines (Figure 1D). Butyrate potently increased the activity of AP and the expression of intestinal differentiation markers, E-cadherin and villin (Figure 2A and B), consistent with its anticarcinogenic effects (Figure 1).
Figure. 1. Butyrate suppresses proliferation and induces apoptosis in HT-29 and Caco-2 cells.
Cells were treated with 0 (□) or 4 mM sodium butyrate (NaB, ■) for 4 d. (A) Immunofluorescence staining of PCNA for cell proliferation. Scale bars are 200 μm. (B) Caco-2 cells were transfected with GFP plasmid, and observed under a fluorescent inverted microscope. Scale bars are 200 μm on left and 100 μm on right. (C) MTT analyses for cell survival. (D) Protein contents of cleaved caspase-3 and cleaved PARP. Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
Figure. 2. Butyrate promotes differentiation of HT-29 and Caco-2 cells.
Cells were treated with 0 (□) or 4 mM sodium butyrate (NaB, ■) for 4 d. (A) Alkaline phosphatase assay. (B) Protein contents of villin and E-cadherin. Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
3.2. Butyrate increases protein contents and activities of IDH1 and PDH in colorectal adenocarcinoma cells
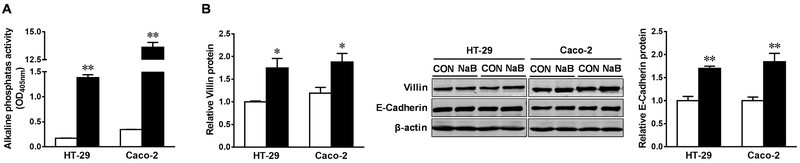
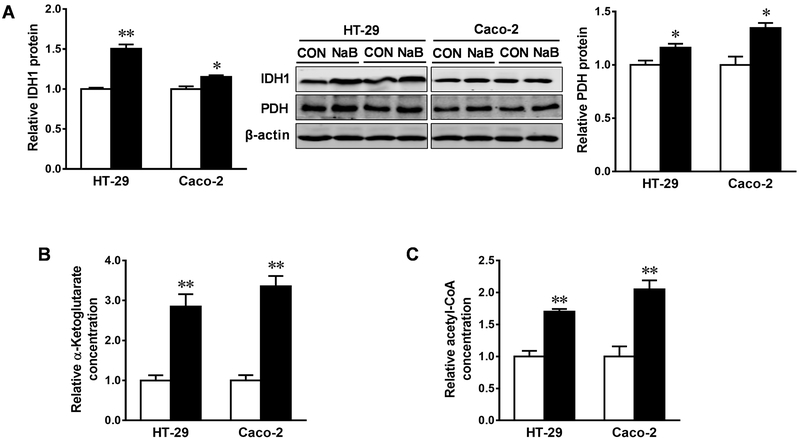
We further examined the possible alteration of metabolic enzymes elicited by butyrate. The inclusion of butyrate augmented the protein content of isocitrate dehydrogenase 1 (IDH1) and pyruvate dehydrogenase (PDH), key metabolic enzymes in the tricarboxylic acid cycle, in both HT-29 and Caco-2 cells (Figure 3A). Consistently, the activity of IDH1 and PDH were enhanced, as indicated by the elevated levels of their metabolites, α-ketoglutarate (α-KG) (Figure 3B) and cytosolic acetyl-CoA (Figure 3C), respectively.
Figure. 3. Butyrate increases protein contents and activities of IDH1 and PDH in HT-29 and Caco-2 cells.
Cells were treated with 0 (□) or 4 mM sodium butyrate (NaB, ■) for 4 d. (A) Protein contents of isocitrate dehydrogenase 1 (IDH1) and pyruvate dehydrogenase (PDH). (B) Relative contents of α-ketoglutarate. (C) Relative contents of cytosolic acetyl-CoA. Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
3.3. Butyrate triggers histone acetylation and DNA demethylation in colorectal adenocarcinoma cells
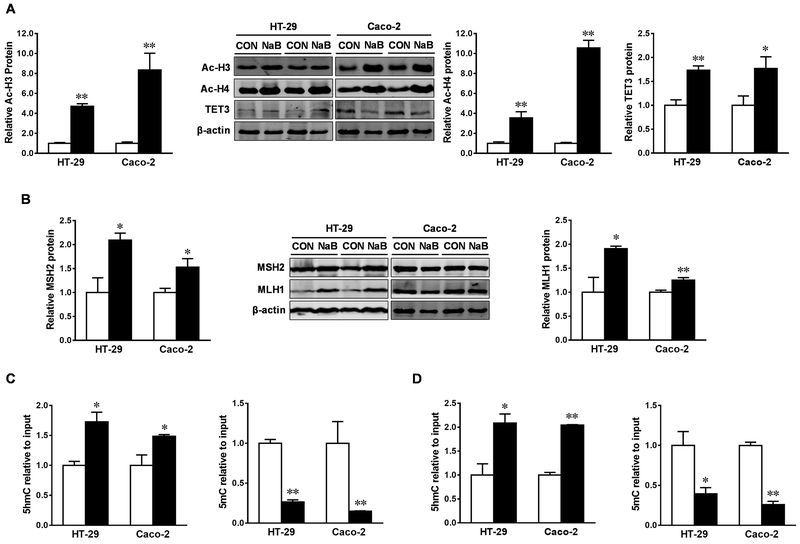
Consistent with increased cytosolic acetyl-CoA concentration, butyrate significantly increased the acetylation of histone H3 and H4 in both HT-29 and Caco-2 cells (Figure 4A), which is consistent with the function of butyrate as a histone deacetylase inhibitor (HDACi) [21]. DNA mismatch repair (MMR) proteins, MutS protein homolog 2 (MSH2) and MutL homolog 1 (MLH1), are inversely correlated to the incidence of tumorigenesis [32–34]. In this study, MSH2 and MLH1 were expressed in both colorectal adenocarcinoma cell lines, and their contents were upregulated in response to butyrate treatment, which were associated with the increased content of ten eleven translocation 3 (TET3) (Figure 4A and B). Furthermore, butyrate reduced 5mC and increased 5-hydroxymethylcytosine (5hmC), an intermediate of DNA demethylation, in the promoters of MSH2 (Figure 4C) and MLH1 (Figure 4D), in agreement with the anti-carcinogenic effects of butyrate.
Figure. 4. Butyrate triggers histone acetylation and DNA demethylation in HT-29 and Caco-2 cells.
Cells were treated with 0 (□) or 4 mM sodium butyrate (NaB, ■) for 4 d. (A) Protein contents of acetylated histone H3 (Ac-H3), acetylated histone H4 (Ac-H4) and Ten-eleven translocation 3 (TET3). (B) Protein contents of MutS homolog 2 (MSH2) and MutL homolog 1 (MLH1). 5hmC and 5mC modifications in the promoter of MSH2 (C) and MLH1 (D). Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
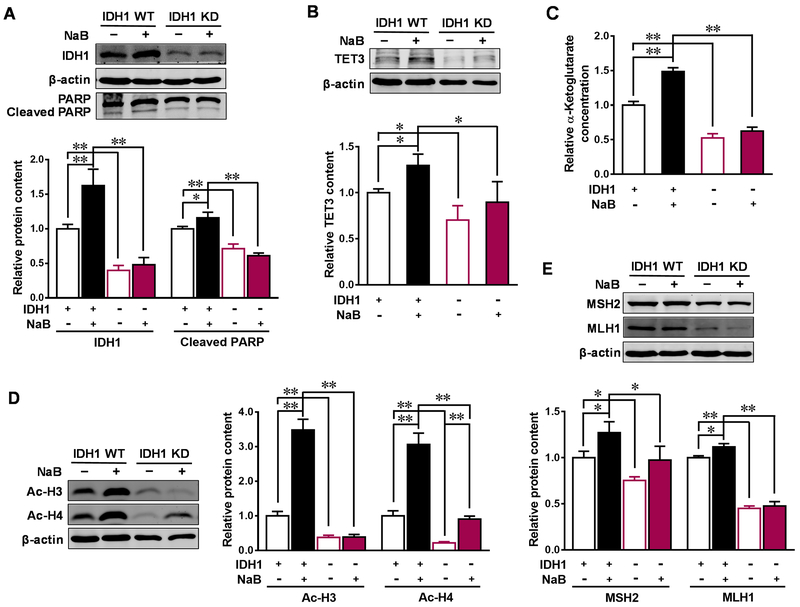
3.4. IDH1 mediates butyrate-induced antitumor effects in colorectal adenocarcinoma cells
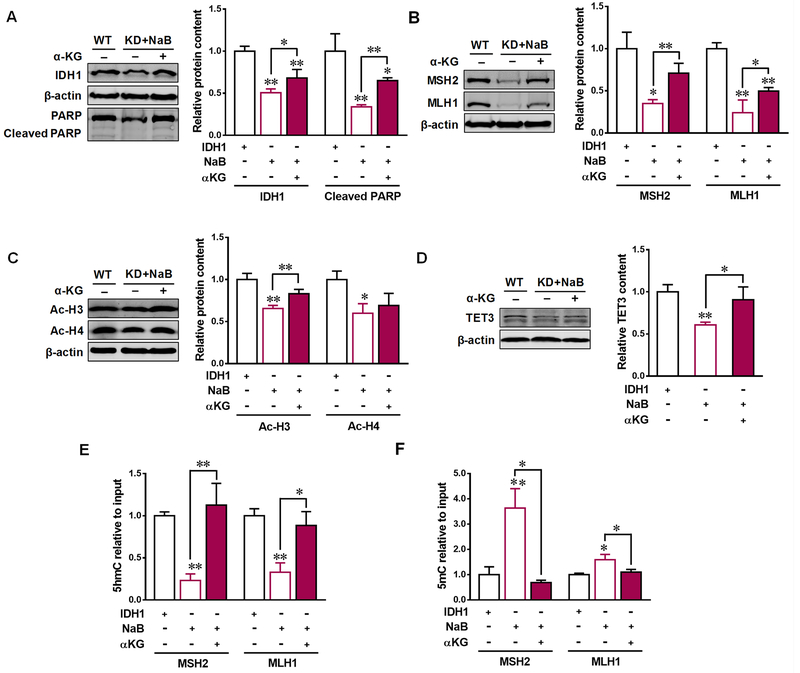
Aberrant mRNA expression of IDH1 is associated with the development of CRC [9]. To evaluate the role of IDH1 in mediating the anticarcinogenic effects of butyrate, we downregulated IDH1 expression by transfecting IDH1 KD plasmid into Caco-2 cells. In the meantime, IDH1 WT plasmid was used as the corresponding control. After transfection, approximately 50% of IDH1 expression was decreased in IDH1 KD cells (Figure 5A). The apoptotic marker, cleaved PARP, was decreased in IDH1 KD cells, and was unable to be elevated by butyrate (Figure 5A). As expected, the contents of its metabolite α-KG and subsequently downstream enzyme TET3 were downregulated in IDH1 KD cells. In addition, butyrate failed to increase the contents of α-KG and TET3 in Caco-2 cells with IDH1 KD plasmids (Figure 5B and C). Similarly, IDH1 KD diminished the upregulated effect of butyrate on acetylated histone H3 (Ac-H3) and acetylated histone H4 (Ac-H4) as well as the contents of MSH2 and MLH1 (Figure 5D and E). α-KG supplementation recovered the beneficial effects of butyrate in IDH1 KD cells. Addition of α-KG enhanced cleaved PARP and MMR contents (Figure 6A and B), which was correlated with the elevated contents of TET3 and Ac-H3 (Figure 6C and D). Furthermore, 5mC was downregulated and 5hmC was upregulated by α-KG in the promoters of MSH2 and MLH1 in NaB treated IDH1 KD cells supplemented with α-KG (Figure 6E and F). These data showed that butyrate-induced antitumor effects in colorectal adenocarcinoma cells are partially through upregulating IDH1.
Figure. 5. IDH1 mediates butyrate-induced anticarcinogenesis effects in Caco-2 cells.
Caco-2 cells were transfected with isocitrate dehydrogenase 1 (IDH1) knock-down (KD) plasmid or IDH1 wild-type (WT) plasmid to downregulate (−) or over-express (+) IDH1, then treated with 0 (−) or 4 mM sodium butyrate (NaB, +) for 4 d. (A) Protein contents of IDH1 and cleaved PARP. (B) Protein contents of ten-eleven translocation 3 (TET3). (C) Relative contents of α-ketoglutarate. (D) Protein contents of acetylated histone H3 (Ac-H3) and acetylated histone H4 (Ac-H4). (E) Protein contents of MutS homolog 2 (MSH2) and MutL homolog 1 (MLH1). Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
Figure. 6. α-ketoglutarate recovers the positive effects of butyrate in IDH KD Caco-2 cells.
Caco-2 cells were transfected with isocitrate dehydrogenase 1 (IDH1) knock-down (KD) plasmid or IDH1 wild-type (WT) plasmid to downregulate (−) or over-express (+) IDH1. IDH1 KD cells then were treated with 0 (−) or 4 mM sodium butyrate (NaB, +), or 4 mM NaB supplemented with 0 (−) or 4 mM dimethyl-α-ketoglutarate (a membrane permeable α-ketoglutarate, α-KG, +). (A) Protein contents of IDH1 and cleaved PARP. (B) Protein contents of ten-eleven translocation 3 (TET3). (C) Protein contents of acetylated histone H3 (Ac-H3) and acetylated histone H4 (Ac-H4). (D) Protein contents of MutS homolog 2 (MSH2) and MutL homolog 1 (MLH1). (E) 5hmC and (F) 5mC modifications in the promoter of MSH2 and MLH1. Mean ± SEM, n = 3, *: P < 0.05; **: P < 0.01.
4. Discussion
Apoptosis and differentiation are closely related processes, both of which reduce the chance of excessive proliferation to reduce the carcinogenic risk [35]. Radiotherapy and chemotherapy induce genetic defects causing apoptosis, while apoptotic dysfunction renders colorectal cancer cells resistant to those therapies [36]. In this study, butyrate suppressed proliferation and induced apoptosis and differentiation in HT-29 and Caco-2 cells. The increased differentiation might be due to upregulated activity of protein kinase C and c-Jun N-terminal kinase [37]. The induced apoptosis by butyrate is also observed in other colonic adenoma and carcinoma cell lines [22] partially facilitated by MMR, because MSH2 complex recognizes mismatched bases and recruits MLH1 complexes to either initiate DNA repair or induce apoptosis in response to DNA damage signaling during carcinogenesis [38]. Furthermore, HCT-116 cells deficient in MLH1 failed to induce apoptosis [39], indicating the critical role of MMR in colorectal apoptosis.
Due to the accumulation of epigenetic and genetic abnormalities, CRC develops in a sequential normal-adenoma-carcinoma process [40, 41]. Abnormalities in epigenetic changes, such as DNA methylation and histone deacetylation, occur more frequently compared to genetic mutations [42]. Thus, altering epigenetic modifications has a potential to ameliorate colorectal carcinogenesis [43]. Previous studies reported that butyrate and its derivatives inhibit oncogenesis, attributed to its HDACi role in regulating gene transcription [44, 45]. However, trichostatin A, a typical HDAC inhibitor, alone could not activate MLH1 in colon cancer cells [46]; trichostatin A combined with 5Aza-dC, a DNA demethylator, was sufficient for MLH1 activation [46]. Consistently, we found that butyrate leads to histone acetylation and reduces methylation in both MLH1 and MSH2 promoters in colonic epithelial cells. Similarly, the reduced methylation in MLH1 promoter was found in SW48 cells treated with butyrate [47]. The post-translational modifications in the N-terminal domain of histone influences chromatin structure and the accessibility to DNA [48], while DNA demethylation is a prerequisite for effective MMR transcription induced by histone acetylation [49], indicating that histone modifications combined with DNA demethylation allow long-term activation of MMR proteins. However, the antitumor effects of butyrate might depend on the methylation status of MMR genes, because butyrate-induced antineoplastic effects only function in CRC cells with hypermethylated MLH1, not in demethylated MLH1 [47]. MLH1 and MSH2 are commonly hyper-methylated in hereditary non-polyposis colorectal cancers [5] and normally expressed in patients with sporadic and microsatellite stable colorectal carcinomas [34, 50, 51]. MLH1 is reduced under hypoxic tumor microenvironment [32], and upregulation of MSH2 induced by vitamin D and calcium treatment is associated with reduced incidence of colorectal adenomas [33, 34], indicating that downregulation in MLH1 and MSH2 might predispose to tumorigenic conditions. In this study, both MLH1 and MSH2 were expressed in HT-29 and Caco-2 cell lines. Consistently, MLH1 is expressed in other colorectal adenocarcinoma cell lines, such as SW480 and LoVo [52].
Metabolic reprogramming is closely associated with carcinogenesis [53]. In a healthy colon, butyrate is metabolized into acetyl-CoA in mitochondria as the main fuel for colorectal cells [54, 55]. On the contrary, intestinal stem cells prefer aerobic glycolysis and metabolize butyrate to lactate in cancerous colon [56]. IDH is a metabolic enzyme in the tricarboxylic acid cycle, and its mutation was first discovered in patients with primary colorectal cancers [57] and commonly discovered in glioblastoma tumors [58], concomitantly with the reduced generation of IDH metabolite, α-KG [59]. Even though mutant IDH1 did not occur frequently in colorectal cancer, the expression level of IDH1 was decreased in CRC tissues compared to the paired normal tissue in patients [9]. Similarly, IDH1 downregulation leads to the reduction of apoptosis and MMR protein contents in this study, suggesting that the inhibition of IDH1 is associated with colorectal carcinogenesis.
IDH is epigenetically related to carcinogenesis [60]. The mutated IDH1 produces hydroxyl-glutarate, an oncometabolite, that competes with α-KG to bind to histone demethylases, TET proteins, thus prohibiting DNA and histone demethylations [59]. Conversely, the upregulated IDH1 and PDH promote the generation of α-KG and acetyl-CoA, which enter the nucleus and act as epigenetic modifiers [61]. In this study, IDH1 downregulation decreased α-KG and subsequently its downstream DNA demethylator, TET3. Besides DNA demethylation, dysfunctional IDH1 also blocks histone demethylation by downregulating Jumonji domain-containing histone demethylase [11].
In this study, we found that knocking down IDH1 diminished the anti-carcinogenetic effects of butyrate in junction with reduced α-KG contents and the TET enzyme, while α-KG supplementation rescued these beneficial effects of butyrate in IDH1 KD cells. Consistently, accumulation of succinate in chromaffin cells deficient of succinate dehydrogenase inhibits α-KG mediated demethylation, leading to hypermethylator phenotype [14]; intraperitoneal administration of α-KG impeded tumor growth in a murine tumor xenograft model [15]. α-KG increased the tolerance to genotoxic stress from ionizing radiation in MCF7 breast cancer cells [62], and inhibited angiogenesis in both hepatic [63] and [15] pulmonary cell lines, which might be due to the downregulation of hypoxia-inducible factors. Additionally, α-KG enhanced p21 and p27, and downregulated cyclin D1, thus inhibiting the proliferation of Caco-2, HT-29, and LS-180 cells [64], suggesting the protective role of α-KG in colon adenocarcinoma cells.
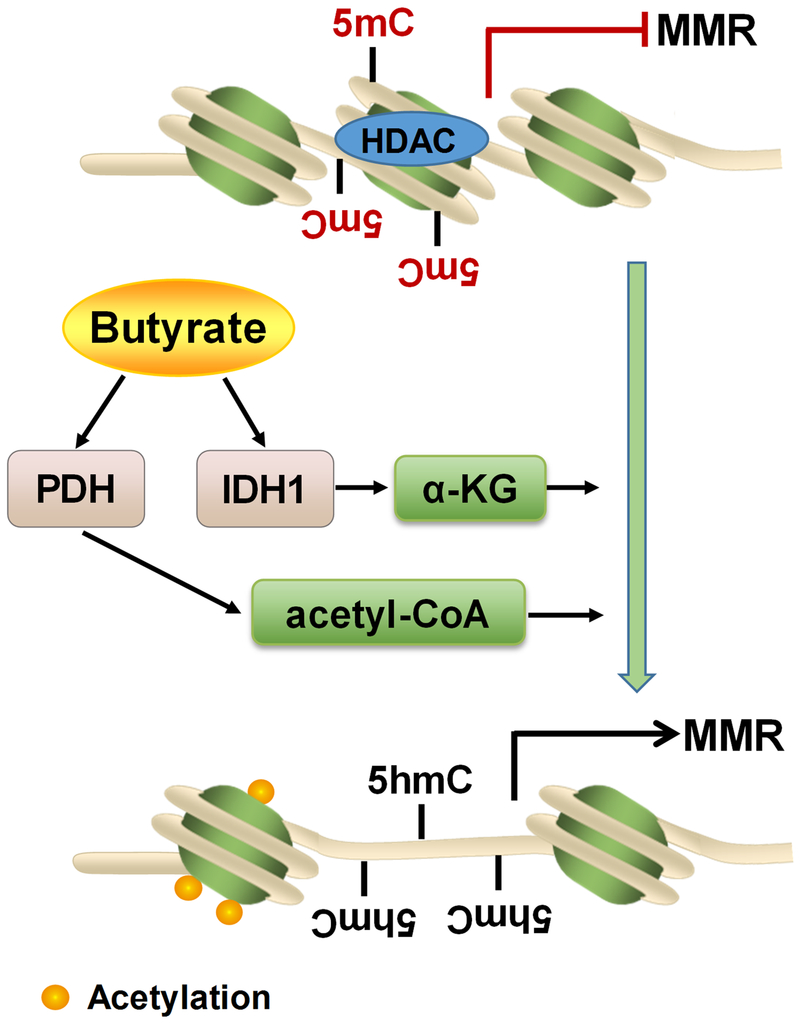
In summary, butyrate reduces proliferation, induces apoptosis and differentiation to inhibit colorectal carcinogenesis, which is associated with the upregulated expression and activity of metabolic enzymes, IDH1 and PDH. The metabolic alterations induced by butyrate lead to DNA demethylation and histone acetylation in the promoters of MMR genes (Fig. 6). These data deepen the current understanding about the link between butyrate and colorectal carcinogenesis, and provide new insights into the molecular mechanisms underlying the beneficial role of butyrate in intestinal health. Our finding is important because butyrate is a major fermentation end product of dietary fibers in the colon. Thus, butyrate as well as associated dietary fiber consumption can be used as a supportive preventive strategy for reducing the incidence of CRC.
Supplementary Material
Figure. 7. Schematic diagram of mechanisms linking butyrate to epigenetic modifications of colorectal adenocarcinoma cells.
Butyrate upregulates pyruvate dehydrogenase (PDH) and isocitrate dehydrogenase 1 (IDH1), which thereafter increases the contents of their metabolites, acetyl-CoA, and α-ketoglutarate (α-KG), respectively. α-KG promotes DNA demethylation in the promoter of DNA mismatch repair (MMR) genes, as indicated by the conversion of 5mC to 5hmC. Acetyl-CoA enhances the histone acetylation in colorectal cancer cells. Histone acetylation and DNA demethylation jointly promote MMR protein expression, thereby suppressing colorectal carcinogenesis.
Acknowledgements
This work was financially supported by National Institutes of Health (NIH) (R15HD073864); and Washington State University Agricultural Research Center Emerging Research Issues Competitive Grant (10A-3057-8640).
Abbreviations:
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- Ac-H3
acetylated histone H3
- Ac-H4
acetylated histone H4
- α-KG
α-ketoglutarate
- AP
Alkaline phosphatase
- CRC
colorectal cancer
- GFP
green fluorescent protein
- HDACi
histone deacetylase inhibitor
- IDH1
isocitrate dehydrogenase 1
- KD
knock-down
- MLH1
mutL homolog 1
- MSH2
mutS protein homolog 2
- MMR
mismatch repair
- NaB
sodium butyrate
- PDH
pyruvate dehydrogenase
- TET
ten-eleven translocation
- WT
wild-type
Footnotes
Competing interests
The authors declare no competing or financial interests.
5. References
- [1].Siegel R, DeSantis C, Jemal A, Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians 2014, 64, 104–117. [DOI] [PubMed] [Google Scholar]
- [2].Ferguson LR, Chen H, Collins AR, Connell M, Damia G, Dasgupta S, Malhotra M, Meeker AK, Amedei A, Amin A, Seminars in cancer biology, Elsevier; 2015, pp. S5–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fadeel B, Orrenius S, Apoptosis: a basic biological phenomenon with wide-ranging implications in human disease. Journal of internal medicine 2005, 258, 479–517. [DOI] [PubMed] [Google Scholar]
- [4].Vilar E, Gruber SB, Microsatellite instability in colorectal cancer—the stable evidence. Nature reviews Clinical oncology 2010, 7, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hegde MR, Roa BB, Genetic testing for hereditary nonpolyposis colorectal cancer (HNPCC). Current Protocols in Human Genetics 2009, 10.12. 11–10.12. 28. [DOI] [PubMed] [Google Scholar]
- [6].Dang CV, Links between metabolism and cancer. Genes & development 2012, 26, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].King A, Selak M, and, Gottlieb E, Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 2006, 25, 4675–4682. [DOI] [PubMed] [Google Scholar]
- [8].Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, IDH1 and IDH2 mutations in gliomas. New England Journal of Medicine 2009, 360, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li W-L, Xiao M-S, Zhang D-F, Yu D, Yang R-X, Li X-Y, Yao Y-G, Mutation and expression analysis of the IDH1, IDH2, DNMT3A, and MYD88 genes in colorectal cancer. Gene 2014, 546, 263–270. [DOI] [PubMed] [Google Scholar]
- [10].Katada S, Imhof A, Sassone-Corsi P, Connecting threads: epigenetics and metabolism. Cell 2012, 148, 24–28. [DOI] [PubMed] [Google Scholar]
- [11].Lu C, Thompson CB, Metabolic regulation of epigenetics. Cell metabolism 2012, 16, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Teperino R, Schoonjans K, Auwerx J, Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell metabolism 2010, 12, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y, Role of Tet proteins in 5mC to 5hmC conversion, ES cell self-renewal, and ICM specification. nature 2010, 466, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer cell 2013, 23, 739–752. [DOI] [PubMed] [Google Scholar]
- [15].Matsumoto K, Obara N, Ema M, Horie M, Naka A, Takahashi S, Imagawa S, Antitumor effects of 2-oxoglutarate through inhibition of angiogenesis in a murine tumor model. Cancer science 2009, 100, 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bultman SJ, Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Molecular nutrition & food research 2017, 61, 1500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M, The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. International journal of cancer 2009, 125, 171–180. [DOI] [PubMed] [Google Scholar]
- [18].Terry P, Giovannucci E, Michels KB, Bergkvist L, Hansen H, Holmberg L, Wolk A, Fruit, vegetables, dietary fiber, and risk of colorectal cancer. Journal of the National Cancer Institute 2001, 93, 525–533. [DOI] [PubMed] [Google Scholar]
- [19].Williams EA, Coxhead JM, Mathers JC, Anti-cancer effects of butyrate: use of micro-array technology to investigate mechanisms. Proceedings of the Nutrition Society 2003, 62, 107–115. [DOI] [PubMed] [Google Scholar]
- [20].Ruemmele FM, Dionne S, Qureshi I, Sarma D, Levy E, Seidman EG, Butyrate mediates Caco-2 cell apoptosis via up-regulation of pro-apoptotic BAK and inducing caspase-3 mediated cleavage of poly-(ADP-ribose) polymerase (PARP). Cell death and differentiation 1999, 6, 729–735. [DOI] [PubMed] [Google Scholar]
- [21].Davie JR, Inhibition of histone deacetylase activity by butyrate. The Journal of nutrition 2003, 133, 2485S–2493S. [DOI] [PubMed] [Google Scholar]
- [22].Hague A, Manning A, Hanlon K, Hart D, Paraskeva C, Huschtscha L, Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. International journal of cancer 1993, 55, 498–505. [DOI] [PubMed] [Google Scholar]
- [23].Gibson P, The intracellular target of butyrate’s actions: HDAC or HDON’T? Gut 2000, 46, 447–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Medina V, Edmonds B, Young GP, James R, Appleton S, Zalewski PD, Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Research 1997, 57, 3697–3707. [PubMed] [Google Scholar]
- [25].Siavoshian S, Segain J, Kornprobst M, Bonnet C, Cherbut C, Galmiche J, Blottiere H, Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut 2000, 46, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Blouin JM, Penot G, Collinet M, Nacfer M, Forest C, Laurent-Puig P, Coumoul X, Barouki R, Benelli C, Bortoli S, Butyrate elicits a metabolic switch in human colon cancer cells by targeting the pyruvate dehydrogenase complex. International Journal of Cancer 2011, 128, 2591–2601. [DOI] [PubMed] [Google Scholar]
- [27].Sun X, Yang Q, Rogers CJ, Du M, Zhu M-J, AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell Death & Differentiation 2017, 24, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Q, Liang X, Sun X, Zhang L, Fu X, Rogers CJ, Berim A, Zhang S, Wang S, Wang B, AMPK/α-ketoglutarate axis dynamically mediates DNA demethylation in the Prdm16 promoter and brown adipogenesis. Cell metabolism 2016, 24, 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mohn F, Weber M, Schübeler D, Roloff T-C, Methylated DNA immunoprecipitation (medip). DNA methylation: methods and protocols 2009, 55–64. [DOI] [PubMed] [Google Scholar]
- [30].Scheppach W, Weiler F, The butyrate story: old wine in new bottles? Current Opinion in Clinical Nutrition & Metabolic Care 2004, 7, 563–567. [DOI] [PubMed] [Google Scholar]
- [31].Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost F, BRUMMER RJ, Review article: the role of butyrate on colonic function. Alimentary pharmacology & therapeutics 2008, 27, 104–119. [DOI] [PubMed] [Google Scholar]
- [32].Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM, Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Molecular and cellular biology 2003, 23, 3265–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kesse E, Boutron-Ruault MC, Norat T, Riboli E, Clavel-Chapelon F, Dietary calcium, phosphorus, vitamin D, dairy products and the risk of colorectal adenoma and cancer among French women of the E3N-EPIC prospective study. International journal of cancer 2005, 117, 137–144. [DOI] [PubMed] [Google Scholar]
- [34].Sidelnikov E, Bostick RM, Flanders WD, Long Q, Fedirko V, Shaukat A, Daniel CR, Rutherford RE, Effects of calcium and vitamin D on MLH1 and MSH2 expression in rectal mucosa of sporadic colorectal adenoma patients. Cancer Epidemiology and Prevention Biomarkers 2010, 19, 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johnson LR, Regulation of gastrointestinal mucosal growth. Physiological Reviews 1988, 68, 456–502. [DOI] [PubMed] [Google Scholar]
- [36].Watson A, Apoptosis and colorectal cancer. Gut 2004, 53, 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Orchel A, Dzierżewicz Z, Parfiniewicz B, Wilczok T, Butyrate-induced differentiation of colon cancer cells is PKC and JNK dependent. Digestive diseases and sciences 2005, 50, 490–498. [DOI] [PubMed] [Google Scholar]
- [38].Berry SE, Davis TW, Schupp JE, Hwang H-S, de Wind N, Kinsella TJ, Selective radiosensitization of drug-resistant MutS homologue-2 (MSH2) mismatch repair-deficient cells by halogenated thymidine (dThd) analogues: Msh2 mediates dThd analogue DNA levels and the differential cytotoxicity and cell cycle effects of the dThd analogues and 6-thioguanine. Cancer research 2000, 60, 5773–5780. [PubMed] [Google Scholar]
- [39].Yanamadala S, Ljungman M, Potential Role of MLH1 in the Induction of p53 and Apoptosis by Blocking Transcription on Damaged DNA Templates1 1 NIH grant CA82376, a grant from the Gastrointestinal Oncology Program Fund of the University of Michigan Comprehensive Cancer Center, and a grant from the Biomedical Research Council at the University of Michigan Medical School. Molecular cancer research 2003, 1, 747–754. [PubMed] [Google Scholar]
- [40].Fearon ER, Vogelstein B, A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [DOI] [PubMed] [Google Scholar]
- [41].Lynch HT, de la Chapelle A, Hereditary colorectal cancer. N Engl J Med 2003, 348, 919–932. [DOI] [PubMed] [Google Scholar]
- [42].Fearon ER, Molecular genetics of colorectal cancer. Annual Review of Pathology: Mechanisms of Disease 2011, 6, 479–507. [DOI] [PubMed] [Google Scholar]
- [43].van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG, Colorectal cancer epigenetics: complex simplicity. Journal of Clinical Oncology 2011, 29, 1382–1391. [DOI] [PubMed] [Google Scholar]
- [44].Tong X, Yin L, Giardina C, Butyrate suppresses Cox-2 activation in colon cancer cells through HDAC inhibition. Biochemical and biophysical research communications 2004, 317, 463–471. [DOI] [PubMed] [Google Scholar]
- [45].Heidor R, Conti A, Ortega JF, Furtado KS, Silva RC, Tavares PE, Purgatto E, Ract JN, Paiva SA, Gioielli LA, The chemopreventive activity of butyrate-containing structured lipids in experimental rat hepatocarcinogenesis. Molecular Nutrition & Food Research 2016, 60, 420–429. [DOI] [PubMed] [Google Scholar]
- [46].Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB, Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature genetics 1999, 21, 103–107. [DOI] [PubMed] [Google Scholar]
- [47].Dronamraju SS, Coxhead JM, Kelly SB, Mathers JC, Differential antineoplastic effects of butyrate in cells with and without a functioning DNA mismatch repair. Nutrition and cancer 2009, 62, 105–115. [DOI] [PubMed] [Google Scholar]
- [48].Goel A, Boland CR, Epigenetics of colorectal cancer. Gastroenterology 2012, 143, 1442–1460. e1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Migheli F, Migliore L, Epigenetics of colorectal cancer. Clinical genetics 2012, 81, 312–318. [DOI] [PubMed] [Google Scholar]
- [50].Lanza G, Gafà R, Maestri I, Santini A, Matteuzzi M, Cavazzini L, Immunohistochemical pattern of MLH1/MSH2 expression is related to clinical and pathological features in colorectal adenocarcinomas with microsatellite instability. Modern Pathology 2002, 15, 741–749. [DOI] [PubMed] [Google Scholar]
- [51].Kruschewski M, Noske A, Haier J, Runkel N, Anagnostopoulos Y, Buhr HJ, Is reduced expression of mismatch repair genes MLH1 and MSH2 in patients with sporadic colorectal cancer related to their prognosis? Clinical and Experimental Metastasis 2002, 19, 71–77. [DOI] [PubMed] [Google Scholar]
- [52].Ma Y, Chen Y, Petersen I, Expression and promoter DNA methylation of MLH1 in colorectal cancer and lung cancer. Pathology-Research and Practice 2017, 213, 333–338. [DOI] [PubMed] [Google Scholar]
- [53].Ward PS, Thompson CB, Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell 2012, 21, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Roediger W, Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 1982, 83, 424–429. [PubMed] [Google Scholar]
- [55].Ardawi M, Newsholme E, Fuel utilization in colonocytes of the rat. Biochem. J 1985, 231, 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ, The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular cell 2012, 48, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, The consensus coding sequences of human breast and colorectal cancers. science 2006, 314, 268–274. [DOI] [PubMed] [Google Scholar]
- [58].Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-M, Gallia GL, An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Prensner JR, Chinnaiyan AM, Metabolism unhinged: IDH mutations in cancer. Nature medicine 2011, 17, 291. [DOI] [PubMed] [Google Scholar]
- [60].Prensner JR, Chinnaiyan AM, Metabolism unhinged: IDH mutations in cancer. Nature medicine 2011, 17, 291–293. [DOI] [PubMed] [Google Scholar]
- [61].Yun J, Johnson JL, Hanigan CL, Locasale JW, Interactions between epigenetics and metabolism in cancers. Frontiers in oncology 2012, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Efimova EV, Takahashi S, Shamsi NA, Wu D, Labay E, Ulanovskaya OA, Weichselbaum RR, Kozmin SA, Kron SJ, Linking Cancer Metabolism to DNA Repair and Accelerated Senescence. Molecular cancer research 2015, molcanres. 0263.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Matsumoto K, Imagawa S, Obara N, Suzuki N, Takahashi S, Nagasawa T, Yamamoto M, 2-oxoglutarate downregulates expression of vascular endothelial growth factor and erythropoietin through decreasing hypoxia-inducible factor-1α and inhibits angiogenesis. Journal of cellular physiology 2006, 209, 333–340. [DOI] [PubMed] [Google Scholar]
- [64].Rzeski W, Walczak K, Juszczak M, Langner E, PoŻarowski P, Kandefer-Szerszeń M, Pierzynowski SG, Alpha-ketoglutarate (AKG) inhibits proliferation of colon adenocarcinoma cells in normoxic conditions. Scandinavian journal of gastroenterology 2012, 47, 565–571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.