Abstract
A pheromone-binding protein from navel orange worm, Amyelois transiteella (Atra-PBP1) binds to non-polar pheromone molecules and facilitates the transport and delivery of pheromone to the membrane-bound pheromone receptors. We report complete NMR chemical shift assignments of Atra-PBP1 obtained at pH 4.5 and 25 °C (BMRB no. 15601).
Keywords: odorant-binding protein, pheromone signaling, olefaction, NMR
Biological Context
To find mates and successfully reproduce, male moths rely heavily on pheromone detection. Pheromone-binding proteins (PBPs) (Vogt and Riddiford, 1981) and odorant receptors (ORs) (Clyne et al., 1999, Vosshall et al., 1999) are essential for the uptake, delivery and detection of sex pheromones (Leal et al., 2005). Molecular interactions amongst pheromone molecules, PBPs and ORs lead to a remarkable selectivity and sensitivity of the olfactory system in insects. The pheromone-binding protein of the navel orange worm, Amyelois transitella (Atra-PBP1) binds tightly to pheromone molecules at neutral pH (but not at pH < 5). Atra-PBP1 undergoes a conformational transition at low pH (Damberger et al., 2007) to facilitate the transport and delivery of bound pheromone to the OR on the membrane surface, where the local pH is estimated to be low. Three-dimensional structures of a few PBPs are known (Damberger et al., 2007, Lautenschlager et al., 2007), but the atomic-level recognition of PBPs and ORs are not well understood. Three-dimensional structures of PBPs as a function of pH and bound ligand may unveil how ORs are activated by pheromones. We report here NMR assignments of Atra-PBP1 at pH 4.5, as a first step toward elucidating the atomic-level structural changes and regulatory mechanisms necessary for the activation of ORs by PBPs.
Methods and Experiments
Expression and Purification of Atra-PBP1.
Uniformly 15N-labeled and 13C,15N-labeled AtraPBP1 was expressed in E. coli and purified by ion-exchange and gel-filtration chromatography as described previously (Damberger et al., 2007). Typically, 5 mg of purified protein was obtained from a 1-liter culture. The identity and integrity of the final protein sample was confirmed by SDS-PAGE and LC-ESI/MS.
NMR Spectroscopy.
Samples for NMR analysis were prepared by dissolving 13C/15N-labeled Atra-PBP1 (0.5 mM) in 0.3 ml of a 95% H2O/5% [2H] H2O solution containing 10 mM [2H3] acetate at pH 4.5. All NMR experiments were performed at 25 °C on a Bruker Avance 600 MHz spectrometer equipped with a four-channel interface and triple-resonance cryogenic probe. The 15N-1H HSQC spectra (Fig. 1) were recorded on samples of 15N-labeled Atra-PBP1 (in 95% H2O, 5% 2H2O). The number of complex points and acquisition times were: 256, 180 ms (15N (F1)), and 512, 64 ms (1H (F2)). Assignment of backbone and side-chain resonances were obtained from the following spectra: HNCACB, HN(CO)CACB, HNCO, CBCACONH, HBHACONH, C(CO)NH-TOCSY, H(CCO)NH-TOCSY, HCCH-TOCSY, and H(CCH)-COSY. The NMR data were processed using NMRPipe and analyzed using NMRview.
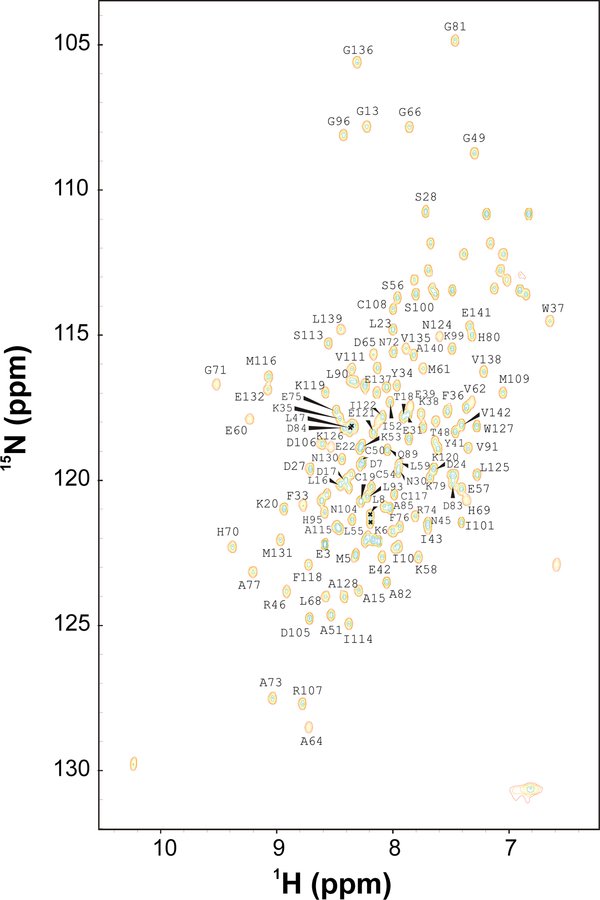
FIG. 1.
Two-dimensional 15N-1H HSQC spectrum of Atra-PBP1 recorded at 600-MHz 1H frequency. The protein sample was uniformly labeled with nitrogen-15. Side-chain amide proton resonances (NH2) are connected by dotted lines.
Assignments and Data Deposition
Figure 1 presents 1H/15N HSQC spectrum of Atra-PBP1 to illustrate representative backbone resonance assignments. NMR assignments were based on 3D heteronuclear NMR experiments performed on 13C/15N-labeled Atra-PBP1 (residues 1–142). The protein sample in this study consists of 142 native residues and does not contain any affinity tags or extra residues. All 142 residues exhibited strong NMR signals with uniform intensities, indicative of a well-defined three-dimensional protein structure. More than 95% the backbone resonances (1HN, 15N, 13Cα, 13Cβ, and 13CO) and ~85% of aliphatic side chain resonances were assigned, including stereospecific assignment of valine and leucine methyl groups. The chemical shift assignments (1H, 15N, 13C) of Atra-PBP1 have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 15601.
Acknowledgements
We thank Jeff de Ropp for technical support and help with NMR experiments. Work supported by NIH grant (EY012347) to J.B.A, NSF (0234769), USDA-NRI (2003-35302-13648), the Almond Board of California, and UC Davis NMR facility.
References
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J and Carlson JR (1999) Neuron, 22, 327–338. [DOI] [PubMed] [Google Scholar]
- Damberger FF, Ishida Y, Leal WS and Wuthrich K (2007) J. Mol. Biol, 373, 811–819. [DOI] [PubMed] [Google Scholar]
- Lautenschlager C, Leal WS and Clardy J (2007) Structure, 15, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Chen AM, Ishida Y, Chiang VP, Erickson ML, Morgan TI and Tsuruda JM (2005) Proc. Natl. Acad. Sci. U S A, 102, 5386–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG and Riddiford LM (1981) Nature, 293, 161–163. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A and Axel R (1999) Cell, 96, 725–736. [DOI] [PubMed] [Google Scholar]