Abstract
The physiological activity of xenon has long been recognized, though the exact nature of its interactions with biomolecules remains poorly understood. Xe is an inert noble gas, but can act as a general anesthetic, most likely by binding internal hydrophobic cavities within proteins. Understanding Xe-protein interactions, therefore, can provide crucial insight regarding the mechanism of Xe anesthesia and potentially other general anesthetic agents. Historically, Xe-protein interactions have been studied primarily through X-ray crystallography and NMR. In this chapter, we first describe our methods for preparing Xe derivatives of protein crystals and identifying Xe-binding sites. Second, we detail our procedure for 129Xe hyper-CEST NMR spectroscopy, a versatile NMR technique well-suited for characterizing the weak, transient nature of Xe-protein interactions.
Keywords: Xenon, protein, crystallography, NMR, hyper-CEST, spectroscopy, hyperpolarization
1. Introduction
Xenon, first discovered by British chemists Sir William Ramsay and Morris Travers in 1898, possesses a broad range of beneficial biological properties (Winkler et. al., 2016). Xe is clinically valuable, having been shown to act as a general anesthetic (Aziz, 2001), analgesic (Giacalone et al., 2013), and neuroprotective agent (David et al., 2010). In structural biology, Xe-lipid (Weinrich & Worcester, 2013; Yamamoto et al., 2012) and Xe-protein (Colloc’h et al., 2007; Abraini et al., 2014) interactions have been studied as a means of elucidating the mechanisms of general anesthesia, an enduring area of inquiry. Xe, unlike other general anesthetics such as nitrous oxide (N2O), propofol, or the halocarbons (e.g., sevoflurane), is a monoatomic species. Being a noble gas, Xe has a filled outer electron shell and is generally unreactive, although its inertness can be overcome under extreme conditions to form inorganic (Bartlett, 1962) and organic (Frohn & Bardin, 2001) compounds. Xenon has more than fifty isotopes, of which nine are stable. 133Xe, a byproduct of nuclear fission, serves as a marker for monitoring compliance with nuclear weapons testing bans (Bowyer, 2002). 135Xe is a robust neutron absorber used to control fission in nuclear reactors. The nuclei of two stable isotopes, 129Xe and 131Xe, have nuclear spins of ½ and 3/2, respectively, and are suitable as NMR/MRI probes.
Xe is the largest of the stable noble gases, with a van der Waals diameter of 4.3 Å. Its large atomic volume confers a large degree of polarizability – 4.0 Å3 for Xe, compared to just 0.2 Å3 for He and 1.5 Å3 for water (Rumble, 2017). This polarizability allows Xe to have interactions with solutes, and thus Xe can act as an inert solvent capable of dissolving biological and organic molecules (Rentzepis & Douglass, 1981). Conversely, xenon’s polarizability makes it more soluble in biologically-relevant fluids than the other noble gases (Table 1) (Weathersby & Homer, 1980).
Table 1:
Solubility of noble gases (mL/mL) at 37 °C
| Gas | Water | Plasma | Blood |
| He | 0.010 | 0.009 | 0.008 |
| Ne | 0.011 | N/A | 0.009 |
| Ar | 0.030 | 0.028 | 0.030 |
| Kr | 0.050 | 0.051 | 0.060 |
| Xe | 0.083 | 0.094 | 0.146 |
The anesthetic properties of the inert gases were first noted in 1939 by U.S. Navy physician Albert Behnke while investigating the cause of “drunkenness” presented by deep-sea divers. In 1946 Lawrence et al. predicted that Xe should also possess anesthetic properties based on its lipophilicity relative to the inert gases (Ar, N2), whose narcotic effects had already been recognized (Lawrence et al., 1946). Lawrence based his reasoning on the Meyer-Overton hypothesis – the near-perfect linear correlation (r = 0.99) between the partial pressure of inhaled anesthetic required to achieve general anesthesia (a.k.a. the median alveolar concentration, or MAC) and its olive-oil gas partition coefficient (Meyer, 1899; Overton, 1901; Campagna, Miller, & Forman, 2003). Indeed, Lawrence and coworkers were successful in demonstrating the anesthetic properties of Xe in mice, and five years later Xe was first used as an anesthetic in a clinical setting (Cullen & Gross, 1951).
Unlike most general anesthetics, which heighten the activity of inhibitory GABAA (γ-aminobutyric acid type-A) receptors, xenon’s anesthetic and analgesic properties arise from its non-competitive inhibition of excitatory N-methyl-D-aspartate (NMDA) receptors (Franks et al., 1998). There is renewed interest in using Xe as an anesthetic, largely due to its non-toxicity, circulatory stability, favorable perfusion rates, and potency (Xe has a MAC of around 71%) (Marx et al., 2000; Arch & Harper, 2011). Xe was approved for use as an anesthetic in Russia in 2000, Germany in 2005, and ultimately all of Europe in 2007 (Esencan et al., 2013). In 2007 Air Liquide introduced LENOXe™, Europe’s first Xe-based anesthetic. In addition to its efficacy and safety, Xe is also an environmentally-friendly alternative to halocarbon anesthetics such as sevoflurane, desflurane, and isoflurane. The primary disadvantage of xenon is its high price, roughly $10 per L, which stems from the relative scarcity of atmospheric Xe (only 87 ppb in dry air) (Neice & Zornow, 2016). Currently, Xe is prepared on an industrial scale as a byproduct of liquid oxygen and nitrogen production by fractional distillation of liquefied air. Increased Xe gas production, coupled with improved Xe-recovery methods such as the Felix Dual™ closed-circuit respirator (Air Liquide Medical Systems, France), have the potential to lower the price of Xe enough to make it marketable as a clinical anesthetic (Dingley et al., 2001; Sanders, 2003; Stoppe et al., 2013).
Xe is amenable to mechanistic study because of its chemical inertness, structural simplicity (just a lone atom!), and because its interactions with proteins can be characterized by a variety of techniques such as X-ray crystallography, hyperpolarized 129Xe NMR and other NMR methods, manometry, computational chemistry, and activity assays. Xenon is spherically symmetric with zero net charge, limiting its interactions to those arising from charge-induced, dipole-induced, and London (dispersion) forces. Xe is observed to occupy a wide range of sites in proteins, though typically it is found in small hydrophobic cavities, predominantly lined with short hydrophobic residues such as leucine, isoleucine, valine, and alanine (Prangé et al., 1998). These buried hydrophobic cavities are usually devoid of water (Wolfenden & Radzicka, 1994). Although the atomic packing density of proteins is comparable to the packing efficiency observed in small organic crystals (Richards, 1974), internal void spaces (a.k.a. cavities) are nearly always present in proteins larger than one hundred residues (Hubbard, Gross & Argos, 1994). Xenon interacts with proteins via non-covalent, low-energy van der Waals forces and is driven by the weak but favorable enthalpies of absorption (Ewing & Maestas, 1970; Tilton et al., 1986). Xe binding typically does not perturb protein structure, making it useful for overcoming the phase problem in X-ray crystallography (Mueller-Dieckmann et al., 2004; Quillin & Matthews, 2002; Quillin & Matthews, 2003; Schiltz et al., 1997; Schiltz, Fourme & Prangé, 2003; Vitali et al., 1991). Xe binding, however, can affect protein function by restricting the number of available conformational states. This Xe-mediated restriction is observed in the loss of rotational degrees of freedom of protein-bound waters (Schoenborn, et al., 1964). More recently, it has been proposed that xenon can reduce the biological function of proteins through the formation of nanobubbles at hydrophobic sites on or near the protein surface (Zhang et al., 2017).
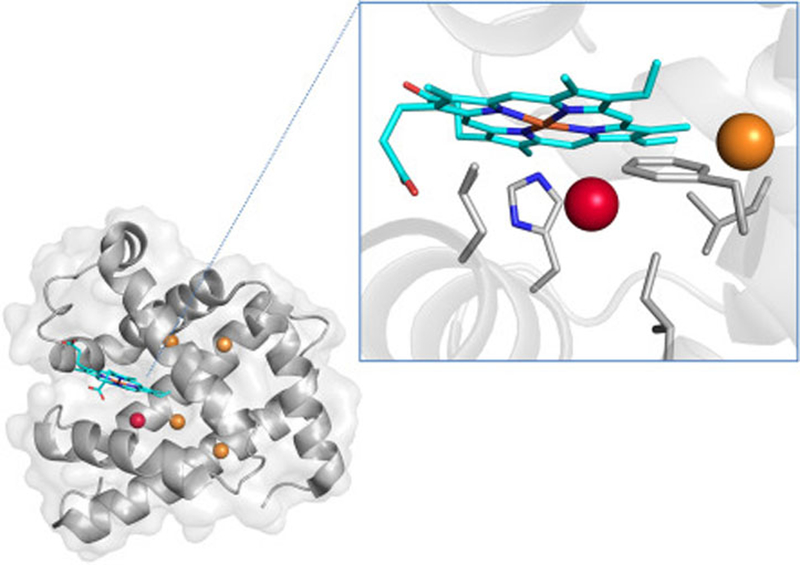
Myoglobin is the protein whose interactions with Xe have been most extensively characterized, and it serves as a useful case study for understanding protein-Xe interactions in general. Sperm whale myoglobin (Schoenborn, 1965) and horse hemoglobin (Schoenborn, 1965) were the first proteins whose structures were solved in the presence of Xe. At a Xe pressure of 2.5 bar, a single Xe atom was observed to bind inside a pre-existing void space in the interior of myoglobin. It was later shown that at 30 bar Xe pressure, four additional Xe atoms bind to myoglobin, though at low (<0.3) occupancies (Fig. 1) (Abraini et al., 2014). Computations based on crystallography coordinates determined favorable enthalpic contributions of 0.6 – 4.2 kcal/mol for Xe binding to myoglobin (Tilton et al., 1986). The 129Xe NMR spectrum of myoglobin in solution shows a single broadened peak near the Xe@solvent frequency, indicating that the population of myoglobin-bound xenon is small relative to the pool of dissolved xenon, and that exchange between the two Xe pools is fast on the NMR timescale (Miller et al., 1981). Increasing the concentration of myoglobin in solution moves the chemical shift of the lone NMR peak either upfield or downfield depending on the oxidation state and spin state of the heme Fe (Tilton & Kuntz, 1982; Rubin et al., 2002). In silico analysis of Xe-myoglobin interactions suggest that Xe exchange is controlled by complex gating processes in which structural fluctuations within the protein control the rate of Xe access to the buried interior binding cavity (Tilton et al., 1988; Teeter, 2004). Finally, although Xe binding minimally perturbs myoglobin structure, Xe (as well as nitrogen and cyclopropane) binding to myoglobin alters its function, specifically its binding affinity for carbon monoxide (Settle, 1973). It has been proposed that such functional inhibition may result from xenon reorienting the proximal histidine (H93), xenon disrupting the side chains that control ligand entry, and/or xenon rigidifying the protein (Tilton, Kuntz, & Petskom 1984).
Fig. 1.
Crystal structure of sperm whale myoglobin under 30 bar Xe pressure (PDB ID 4NXA). The Xe proximal to the heme has an occupancy of 0.8, whereas the other four Xe have occupancies less than 0.3. (Inset) Close-up of the major Xe binding site of myoglobin.
2. X-ray crystallography
2.1. Background
One prevailing hypothesis is that general anesthetics act on proteins by heightening the activity of inhibitory receptors and/or tempering the activity of excitatory receptors (Campagna, Miller & Forman, 2003; Colloc’h et al., 2007). The inhaled general anesthetics bind weakly and promiscuously to occupy internal cavities contained within numerous protein targets. These cavities, rather than merely being packing defects, confer a degree of flexibility necessary for protein function, thus molecules bound within these sites rigidify the protein and subsequently limit their activity (Hubbard, Gross & Argos, 1994; Hubbard & Argos, 1996; Vallone & Brunori, 2004. This theory can be neatly described as “calm molecules, calm minds” (Eckenhoff, 2001). Though structural studies with receptor protein complexes have proven challenging, experiments with simpler model proteins using Xe have been recognized as useful for understanding the binding modes of general anesthetics and the resulting molecular changes of the target proteins (Colloc’h et al., 2007).
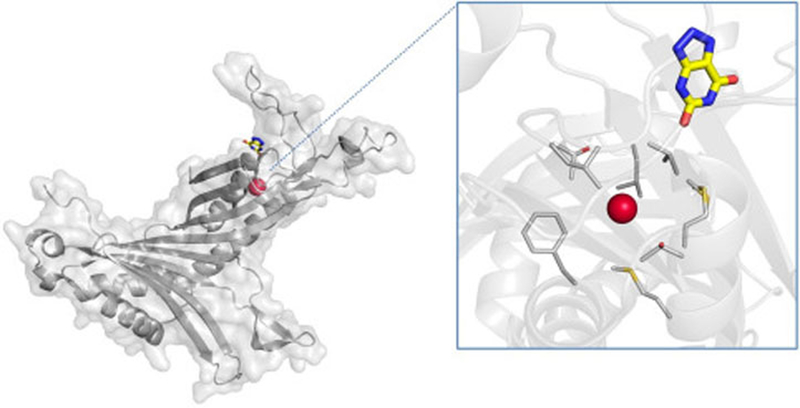
Urate oxidase, a globular protein possessing large hydrophobic cavities, has frequently been used as a model system for understanding anesthetic mechanisms (Colloc’h et al., 2007; Marassio et al., 2011; Abraini et al., 2014). Xe binding to urate oxidase was discovered in 1997 when 8 bar (0.8 MPa) Xe was employed as a heavy-atom for phasing (Colloc’h et al., 1997). Structural studies with Xe were repeated to improve the resolution to 1.75 Å, where it was observed that at 2 MPa Xe binds with 0.9 occupancy in a cavity near the active site (Fig. 2) (Colloc’h et al., 2007). As is typical, Xe binds to a preexisting cavity lined with hydrophobic residues and devoid of solvent in the native structure. The key conclusion drawn was that Xe binding expands the volume of the cavity by 23.5%, an unexpected result given the compression from the high Xe pressure. This volume expansion, in conjunction with the Xe binding site being near the active site of urate oxidase, supports the hypothesis that gaseous anesthetics function by expanding the volume of internal protein cavities, thereby leading to conformational rigidity and (reversible) disruption of protein (and subsequently cellular) function.
Fig. 2.
Crystal structure of urate oxidase complexed with the inhibitor 8-azaxanthine under 2.0 MPa Xe pressure (PDB ID 2IC0). Xe and 8-azaxanthine are shown bound to a single subunit of the urate oxidase homotetramer. (Inset) close-up of the Xe-binding pocket, with surrounding residues shown.
Comparisons of crystallographic studies performed with N2O and Xe show that both gaseous anesthetics typically bind at the same cavities and lead to volume expansion, suggesting that xenon’s interactions with proteins are generally indicative of gaseous anesthetics. Structural studies with Xe and N2O, however, have also revealed subtle differences about their respective binding to protein. Abraini et al. analyzed crystals of urate oxidase, lysozyme, myoglobin, and neuroglobin under varying pressures of Xe and N2O and observed that Xe, the more polarizable of the two species, prefers very hydrophobic environments, whereas N2O prefers mildly hydrophobic environments (Abraini et al., 2014). Additionally, it was found that Xe or N2O occupancies were primarily correlated to cavity volume, and not hydrophobicity. In short, while structural studies with Xe provide useful leads for understanding anesthetic mechanisms in general, caution should be taken to recognize the differences in binding site preference and binding affinities of different inhaled anesthetics.
The process of preparing Xe derivatives of crystallized protein is fairly straightforward and well-established, with numerous methodology papers having been published detailing the methodology of Xe derivatization and follow-up data analysis (Sauer, Schmidt & Kratky, 1997; Soltis et al., 1997; Schiltz, Prangé & Fourme, 1994; Stowell et al., 1996). X-ray diffraction data from Xe derivatives can be collected at room temperature or at 100 K under a cryostream of liquid nitrogen. In the first method, the native protein crystal is transferred to a sealed capillary tube connected to a reservoir of pressurized Xe, and X-ray diffraction data is collected from the protein crystal while under a set Xe pressure (Schiltz, Prangé & Fourme, 1994; Schiltz, Prangé & Fourme, 2003; Mizuno, Makino & Kumasaka, 2013). Although more specialized equipment is needed, the main advantage of this method is the ability to finely adjust and maintain the Xe pressure experienced by the protein crystal. This approach is preferred for quantitative structural studies in which Xe occupancies, B-factors, etc. are calculated as a function of Xe pressure (Abraini et al., 2014; Colloc’h, Marassio & Prangé, 2008). There are several papers detailing the methodology of this Xe derivatization technique (Stowell et al., 1996; Schiltz, Prangé & Fourme, 1994; Schiltz, Prangé & Fourme, 2003).
In the second method, the native protein crystal is pressurized with Xe and then immediately flash-frozen in liquid nitrogen, effectively immobilizing Xe bound to the protein. Though this method is somewhat imprecise in regards to determining the actual Xe gas pressure within the crystal (due to some Xe escaping between depressurization and freezing), the advantage of this method is the ease of the Xe derivatization process as well as the option to use standard cryoloops, goniometers, and synchrotron equipment. The cryocrystallography method is well-suited for preparing Xe derivatives for phasing purposes as well as identifying Xe binding sites within proteins. Our laboratory follows this method using a Xenon Chamber (Hampton Research) to study Xe binding to proteins and its implications for Xe NMR/MRI biosensing applications.
2.2. Freeze-trapping Xe derivatives
2.2.1. Equipment
Optical microscope for looping protein crystals
Cryocrystallography tools (e.g. tongs, magnetic transfer wand, etc.)
CrystalCap with a mounted CryoLoop (Hampton Research)
CrystalCap Vial with stand (Hampton Research)
1000 mL liquid nitrogen dewar
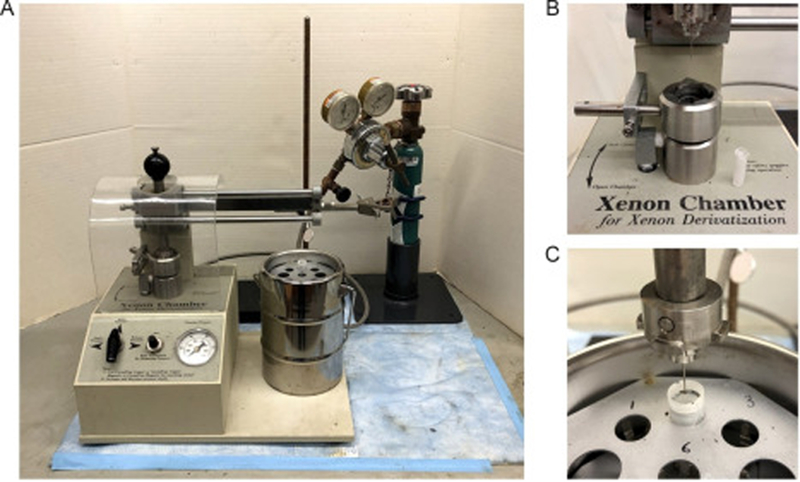
Xe derivatization apparatus such as the Xenon Chamber (Hampton Research, Alisa Viejo, CA, USA) (Fig. 3a), XCell (Oxford Cryosystems, Oxford, UK), and Cryo-Xe-Siter (Rigaku, The Woodlands, TX, USA)
Mini-Vial with Wick (Hampton Research)
Fig. 3.
(a) Xenon derivatization chamber manufactured by Hampton Research; (b) Xe pressurization chamber with a Mini-Vial with Wick on the side; (c) CrystalCap aligned with a CrystalCap Vial inside a 1000 mL dewar.
2.2.2. Reagents
Compressed Xe gas
Liquid nitrogen
2.2.3. Procedure
Crystallize the protein to be derivatized following standard techniques (e.g., vapor diffusion).
Transfer the protein crystal to a cryoprotectant solution or slowly introduce cyroprotectant to the mother liquor containing the protein crystal.
Collect the cryoprotected crystal in a CryoLoop attached to a CrystalCap.
Place the CrystalCap onto the magnetic base of the derivatization chamber plunger.
Insert the CrystalCap into the Xe pressurization chamber containing a Mini-Vial with Wick saturated with cryoprotectant solution (Fig. 3b). The Mini-Vial with Wick prevents the CryoLoop from drying out during the Xe derivatization process.
Seal the chamber and pressurize with up to 4.1 MPa Xe. If Xe pressure within the chamber exceeds 4.1 MPa, the pressure will vent through the safety valve.
Incubate the protein crystal with Xe for 15 to 60 min.
Slowly release Xe from the chamber so as not to disturb the protein crystal.
Immediately freeze the derivatized protein crystal by transferring the CrystalCap to a vial submerged in liquid nitrogen within the 1000 mL dewar (Fig. 3c). Shock freezing the crystal will immobilize Xe bound to the protein as well as Xe dissolved in the mother liquor.
2.2.4. Notes
The Xe pressure required to derivatize crystallized protein should be 5 to 10 times higher than the physiological concentration of Xe during anesthesia (~0.7 atm) (Miller, 2002; Colloc’h, Marassio & Prangé, 2008; Abraini et al., 2014).
Xe is soluble in water (~5 mM per atm) and quickly diffuses via solvent channels to binding sites within the protein crystal on a time scale of minutes (Schiltz, Prangé & Fourme, 1994). Later measurements show that Xe binding to crystallized protein is typically complete after ten minutes (Soltis et al., 1997).
After Xe depressurization, the diffusion of Xe from a tight binding site within a protein is roughly 5 min (Soltis et al., 1997). For a practiced Xe crystallographer, depressurizing and flash-freezing the derivatized protein crystal takes only a few seconds, so loss of Xe from the protein crystal is minimal.
2.3. Diffraction data collection and analysis
2.3.1. Software
iMosflm (Battye et al., 2011), AIMLESS (Evans, 2011) , and CCP4 suite of programs (Winn et al., 2011) are used for processing diffraction data.
Phaser (McCoy et al., 2007), PHENIX (Adams et al., 2010; Afonine et al., 2012) are used for phasing and model refinement.
COOT (Emsley et al., 2010) enables visualizing electron density maps and manual model manipulation.
2.3.2. Procedure
Collect diffraction data at the native wavelength of the synchrotron beamline, or at the Cu Kα wavelength (1.54 Å) if using a home-source X-ray instrument. However, if one is using Xe as a heavy-atom for de novo phasing, or if one aims to maximize anomalous scattering, diffraction data can be collected at longer wavelengths closer to the absorption edges of Xe: 2.29 Å (LI), 2.43 Å (LII), or 2.59 Å (LIII).
Generate isomorphous difference maps using |Fxenon| – |Fnative| as amplitudes and phase using the atomic coordinates of the native protein structure.
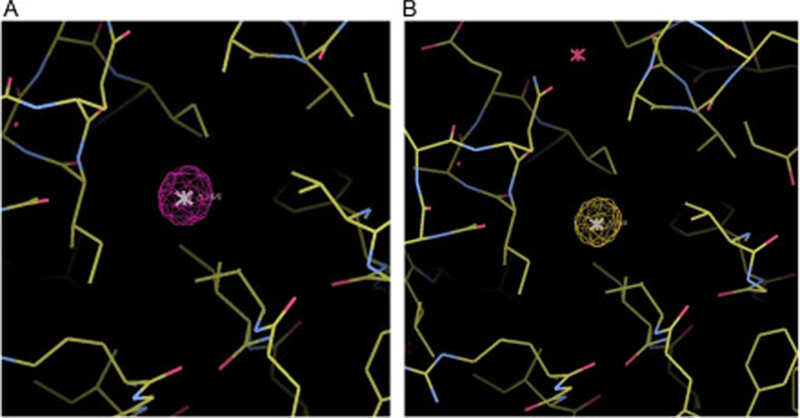
While viewing the protein model in COOT, add Xe atoms into the model where there are large spheres of positive electron density in the isomorphous difference map (Fig. 4a).
(Optional) Evaluate Xe binding by examining the anomalous maps (Fig. 4b).
Set the occupancies of the added Xe atoms to 0.5, and manually change the B-factors to roughly match those of the atoms from surrounding protein sidechains.
As additional rounds of refinement of the Xe derivative structure are performed, the occupancies and B-factors of Xe within the protein model should converge to their final values.
Fig. 4.
Major Xe binding site of TEM-1 β-lactamase derivatized with 1.2 MPa Xe (PDB ID 5HW1). (a) Isomorphous difference map contoured at 15σ; (b) anomalous map contoured at 5σ.
2.3.3. Notes
There are many other software programs that offer the same functionality as the ones listed above. The ones described here are simply the ones used by our laboratory.
Diffraction data of Xe-derivatized crystals can be collected at the Cu Kα wavelength (1.54 Å) or the native wavelength of the synchrotron because Xe has appreciable anomalous dispersion even at wavelengths far from its absorption edges. Sperm whale metmyoglobin was successfully phased from diffraction data collected at the Cu Kα wavelength, where f’ = −0.783, f’’ = 7.384 (Vitali et al., 1991).
Anomalous scattering is typically not used to identify protein-bound Xe, as bound Xe is readily identified from Fourier isomorphous difference maps (vide infra). Because Xe binding is weak and generally not structurally-perturbing, Xe derivatives are isomorphous to the native structure and thus the coordinates of bound Xe are readily identified from Fourier isomorphous difference maps (Prangé et al., 1998).
Crystallographic symmetry mates should be displayed when inspecting potential Xe binding sites on or near the protein surface. Sometimes Xe binding arises due to crystal packing (e.g., lysozyme, CytB), and as such the physiological relevance of these sites should be treated with caution.
3. Xe NMR
3.1. Background
The Xe-129 nuclide has a spin of ½ and is readily detectable by NMR. The large polarizable electron cloud of 129Xe gives it a ~200 ppm chemical shift range in common solvents and tissue (Miller et al., 1981). The magnetic signal of 129Xe can be increased by several orders of magnitude by spin exchange optical pumping (SEOP), a method that uses circularly-polarized light to excite the valence electrons of rubidium which then, in turn, collide with Xe in the gas state and polarizes the Xe nuclei (Walker & Happer, 1997; Barskiy et al., 2017). NMR studies of proteins with Xe have proven complementary to structural studies by X-ray crystallography. Whereas crystal structures provide a detailed but static view of Xe binding to specific sites in proteins, NMR can characterize specific and non-specific Xe-protein interactions as well as determine Xe exchange rates and binding affinities. Xenon in solution exchanges rapidly between dissolved protein and solvent. Thus, the 129Xe NMR spectrum of a protein solution reports a single peak whose chemical shift change relative to Xe in solvent alone (δobs) is an average of the chemical shifts of Xe at specific sites on or within the protein (δi) weighted by their respective binding constants (Ki), multiplied by protein concentration (cprotein) [Eq. (1)] (Locci et al., 2001; Lowery et al., 2003).
| (1) |
Dividing δobs by protein concentration gives a concentration-normalized chemical shift α (expressed in units of ppm/mM) that quantifies the sum of Xe-protein interactions. Xe NMR experiments with denatured proteins observed a linear dependence of the α value on protein size, with an average α of 0.005 ppm/mM per amino acid (Rubin et al., 2001). The α values of native proteins with known Xe-binding sites are typically between 1–3 ppm/mM (Rubin et al., 2002; Corda et al., 2004; Lowery et al., 2005). Conformational changes in proteins have been characterized by measuring changes of α in response to ligand-induced structural change (Rubin et al., 2001; Lowery et al., 2005). Beyond being used to measure global Xe-protein interactions, Xe NMR has also been used to measure the kinetics and thermodynamics of Xe-protein interactions. Xenon resonance peak line width analysis at varying Xe and protein concentrations determined Xe exchange rates for proteins such as myoglobin (Tilton & Kuntz, 1982) and T4 lysozyme (Desvaux et al., 2005). Additionally, observing chemical shift change as a function of Xe pressure through NOESY and HSQC experiments measured Xe binding affinities to T4 lysozyme (Desvaux et al., 2005), maltose binding protein (MBP) (Rubin et al., 2002), and lipid transfer protein 1 (LTP1) (Dubois et al., 2004).
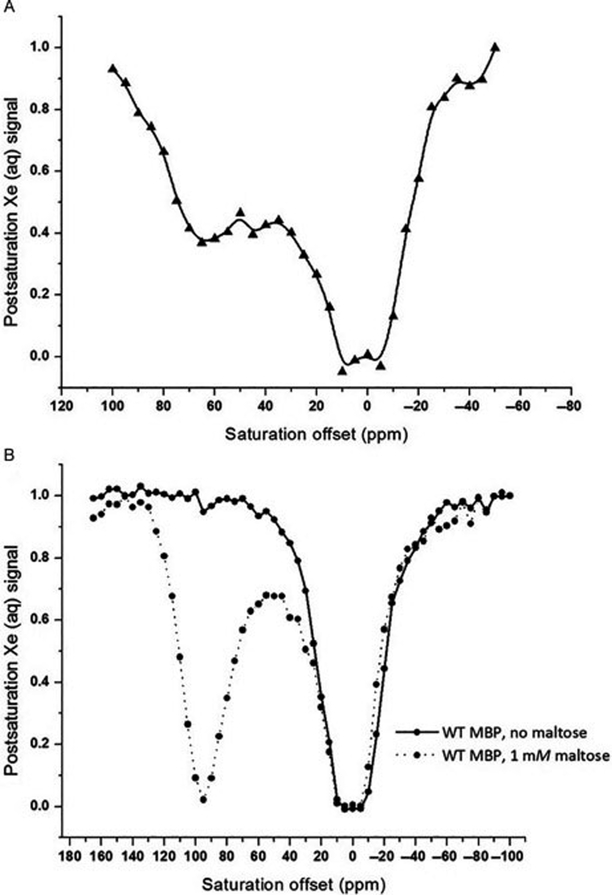
Though much information about Xe-protein interactions can be gleaned from direct detection NMR, a major shortcoming is the inability to resolve resonance peaks corresponding to Xe bound to protein. Moreover, large amounts of protein (typically high-μM to low-mM) are needed to observe any chemical shift changes and/or peak broadening. These limitations can be overcome through chemical exchange saturation transfer (CEST), an alternative contrast approach that takes advantage of the transient nature of Xe-protein interactions to heighten sensitivity (van Zijl & Yadav, 2011; Schröder, 2013). CEST used in combination with hyperpolarized 129Xe, a technique known as hyper-CEST, works by applying a saturation pulse that selectively depolarizes Xe bound to solute (e.g. protein, macrocycle). Depolarized Xe rapidly exchanges into solvent, depolarizing the reservoir of bulk Xe which, in turn, generates MR contrast. Our lab has employed hyper-CEST to study Xe interactions with spores (Bai et al., 2014) as well as the proteins TEM-1 β-lactamase (Bla) (Wang et al., 2016) and maltose binding protein (MBP) (Roose, Zemerov, & Dmochowski, 2017). Representative hyper-CEST z-spectra for 80 μM Bla and MBP in PBS are shown in Fig. 5. In both spectra, two peaks are resolved – one centered at 0 ppm, corresponding to Xe@H2O, and the other downfield corresponding to Xe@protein. The degree of downfield shift of the Xe@protein peak provides information as to the binding site of Xe, and the broadness of the peak gives an indication of the exchange kinetics. Additionally, in the case of MBP, the presence or absence of the downfield peak is a function of the conformational state of the protein. In the absence of maltose, MBP adopts an open conformation that does not generate CEST contrast, whereas adding maltose transitions the protein into a closed state that reports a distinct downfield saturation response. Our lab has used hyper-CEST NMR to monitor Xe-protein interactions primarily for biosensing and molecular imaging applications, but this technique has the potential to shed light on the anesthetic mechanisms of Xe as well.
Fig. 5.
Hyper-CEST z-spectra for Bla (a) and MBP with and without maltose (b). Spectra were obtained from 80 μM protein in PBS at 300 K.
The following sections will describe the procedure for generating hyperpolarized (hp) 129Xe using a gas mixture of 10% nitrogen, 88% helium, and 2% natural abundance xenon (Linde Group); as well as for running hyper-CEST NMR experiments using a 500 MHz Bruker BioDRX NMR spectrometer.
3.2. Hyperpolarization
Briefly, hyperpolarized (hp) Xe is generated via the spin exchange optical pumping (SEOP) method (Barskiy et al., 2011) using a home-built 129Xe hyperpolarizer, based on the commercial model IGI.Xe.2000 by GE (Wang et al., 2016). A Shark 65 W tunable ultra-narrow band laser (OptiGrate) set to 795 nm is used for optical pumping of Rb vapor contained in a glass cell. A water chiller (K-O Concepts) set to 16 °C supplies chilled water to the laser diodes. This setup is shown in Fig 6.
Fig. 6.
Overview of a home-built 129Xe hyperpolarizer. (1) Pressure gauge A; (2) Xe gas mixture tank; (3) N2 tank; (4) laser; (5) heat gun; (6) Rb cell; (7) laser control panel; (8) magnet (Helmholtz coil power supply); (9) chiller.
A standard procedure for preparing the SEOP setup and generating hp 129Xe is available in the literature (Witte et al., 2012).
3.3. Hyper-CEST
Turn on the module supplying chilled water to the laser diodes. Allow chilled water to circulate for at least 10 min before turning on the laser electronics.
If the pressure in the tubing leading from the Xe gas tank to the Rb cell is below 40 psi, the tubing should be degassed using the vacuum. It is important to keep this line pressurized to 40–50 psi to avoid leaking of O2 into the line. If the pressure is sufficient, allow Xe to enter the Rb cell.
Start heating the Rb cell to 180 °C. For best signal to noise, it is advised to heat the cell for 2–3 h before collecting spectra, especially if the cell has not been operated for several days.
Turn on the laser electronics, make sure the wavelength is set to 794.7 nm, and slowly increase the current to the laser diode while monitoring the laser emission profile.
While the current is being increased, degas the tubing leading from the polarizer outlet to the valve regulating Xe flow into the NMR tube.
After the laser is set to full power, turn on the power supply to the Helmholtz coils.
If necessary, change the current NMR probe to a 10-mm BBO NMR probe.
Insert a 90% H2O/10% D2O sample into the magnet.
Lock on the D2O signal and shim Z1, Z2, XZ0, XZ1, XZ1 and YZ1.
Run an acquisition and make sure the H2O peak is symmetric and that the FWHM is ~7–13 Hz.
Remove the 90% H2O/10% D2O sample and insert the Xe in ethanol sample.
Manually tune the probe to the Xe frequency (138.12 MHz).
Run an acquisition and make sure that the Xe signal is visible.
Turn off the vacuum and make sure that the only open valves are those leading from the Rb cell outlet to the tubing leading from the polarizer outlet.
Open the outlet valve of the Rb cell.
For convenience, place a webcam in front of the flow rate monitor so that the flow rate can be clearly monitored in the NMR room while performing experiments.
Add 2.5 mL of ddH2O to a 10 mm NMR tube and screw on the capillary cap onto the NMR tube. This cap should have the tubing leading from the polarizer as its inlet tubing and several long, thin capillaries for efficient bubbling of Xe as its outlet.
Pressurize the NMR tube with Xe. This can be done by rapidly opening and closing the valve leading from the polarizer outlet to the NMR tube several times.
Slowly, manually lower the NMR tube into the magnet until it touches a solid surface.
Open the NMR software and run a Xe stability test. To do this, apply a dSNOB-shaped saturation pulse at a frequency of 300 ppm and monitor the Xe(aq) signal. Run 20 acquisitions using a flow rate of approximately 0.7 SLM, making sure that this flow rate is stable throughout the acquisitions. Integrate the Xe(aq) peaks of all 20 experiments to determine whether or not the polarization is stable over time. If the intensities do not fluctuate by more than ± 0.1, move on to the next step. If they do, it may be necessary to heat the Rb cell for a longer period of time, or check to make sure that no leaks are present in the system. Note: the user-generated code to automatically run this stability test and process the data is available upon request.
Remove the NMR tube from the magnet and depressurize the tube.
After bubbling stops in the NMR tube, unscrew and remove the cap.
Prepare sample for hyper-CEST experiment. The sample volume should be 2.5 mL and 0.1% v/v Pluronic may be added to mitigate foaming. For protein z-spectra, start with a protein sample concentration of 80 μM.
Screw in the cap and pressurize the NMR tube as before.
To obtain a z-spectrum, apply a saturation train of 600 dSNOB-shaped pulses with bandwidth of 690 Hz at each frequency between the starting and ending chemical shift of interest. It is recommended to use a chemical shift step width of 5 ppm for preliminary data collection. Pulse length tpulse = 3.80 ms, field strength B1,max = 77 μT, saturation time Tsat = 1.52 s. The Xe(aq) signal will be displayed as the output. After data acquisition is complete, integrate all of the Xe(aq) peaks and plot the post-saturation Xe(aq) signal as a function of saturation frequency. Repeat 2x, or as necessary until 3 sets of similar data are obtained. Note: the user-generated code to automatically run a z-spectrum and process the data is available upon request.
Depressurize the tube, and recover the sample for further analysis, if necessary.
In the polarizer room, turn off the heat, the laser electronics, the power supply to the Helmholtz coils, and the chiller module.
After the temperature of the cell reaches <30 °C, close the inlet valve to the Rb cell, all valves between the Rb cell and the Xe tank, and finally the Xe tank.
4. Summary and Conclusions
The underlying physiological mechanisms of general anesthetics remain poorly understood despite their critical importance in modern medicine. Xenon, seen by some as the “anesthetic gas of the future” (Delhaye et al., 2010), is well positioned to serve as a proxy for unraveling some of the long-standing mysteries of general anesthesia due its utility as a probe in X-ray crystallography and NMR experiments. More recently, the hyper-CEST approach to NMR contrast has allowed even weak, transient Xe-protein interactions to be characterized with high spectral resolution, far beyond what has been observed in direct detection NMR experiments. Xe hyper-CEST, complemented by structural information from X-ray crystallography, has the potential to characterize Xe interactions with receptor proteins and ion channels implicated in general anesthesia but heretofore undetectable by conventional methods.
Abbreviations
- NMR
nuclear magnetic resonance
- MRI
magnetic resonance imaging
- MAC
median alveolar concentration
- GABAA
γ-aminobutyric acid type-A
- NMDA
N-methyl-D-aspartate
- PDB
protein data bank
- SEOP
spin exchange optical pumping
- NOESY
nuclear Overhauser effect spectroscopy
- HSQC
heteronuclear single-quantum correlation
- MBP
maltose binding protein
- LTP1
lipid transfer protein 1
- CEST
chemical exchange saturation transfer
- Bla
TEM-1 β-lactamase
- PBS
phosphate-buffered saline
5. References
- Abraini JH, Marassio G, David HN, Vallone B, Prangé T, & Colloc’h N (2014). Crystallographic studies with xenon and nitrous oxide provide evidence for protein-dependent processes in the mechanisms of general anesthesia. Anesthesiology, 121(5), 1018–1027. 10.1097/ALN.0000000000000435. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, … Zwart PH (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography, 66(2), 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, … Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography, 68(4), 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arch AM, & Harper NJN (2011). Xenon: An element of protection. Trends in Anaesthesia and Critical Care, 1(5–6), 238–242. 10.1016/j.tacc.2011.08.001. [DOI] [Google Scholar]
- Aziz TS (2001). Xenon in anesthesia. International Anesthesiology Clinics, 39(2), 1–14. 10.1097/00004311-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Bai Y, Wang Y, Goulian M, Driks A, & Dmochowski IJ (2014). Bacterial spore detection and analysis using hyperpolarized 129 Xe chemical exchange saturation transfer (Hyper-CEST) NMR. Chem. Sci, 5(8), 3197–3203. 10.1039/C4SC01190B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barskiy DA, Coffey AM, Nikolaou P, Mikhaylov DM, Goodson BM, Branca RT, … Chekmenev EY (2017). NMR hyperpolarization techniques of gases. Chemistry (Weinheim an Der Bergstrasse, Germany), 23(4), 725–751. 10.1002/chem.201603884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett N (1962). Xenon hexafluoroplatinate (V) Xe+[PtF6]-.Proceedings of the chemical society of London, (JUN), 218. [Google Scholar]
- Battye TGG, Kontogiannis L, Johnson O, Powell HR, & Leslie AGW (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica Section D Biological Crystallography, 67(4), 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer TW, Schlosser C, Abel KH, Auer M, Hayes JC, Heimbigner TR, … Weiss W (2002). Detection and analysis of xenon isotopes for the comprehensive nuclear-test-ban treaty international monitoring system. Journal of Environmental Radioactivity, 59(2), 139–151. 10.1016/S0265-931X(01)00042-X. [DOI] [PubMed] [Google Scholar]
- Campagna JA, Miller KW, & Forman SA (2003). Mechanisms of actions of inhaled anesthetics. The New England Journal of Medicine, 348(21), 2110–2124. 10.1056/NEJMra021261. [DOI] [PubMed] [Google Scholar]
- Colloc’h N, el Hajji M, Bachet B, L’Hermite G, Schiltz M, Prangé T, … Mornon JP (1997). Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nature Structural Biology, 4(11), 947–952. 10.1038/nsb1197-947. [DOI] [PubMed] [Google Scholar]
- Colloc’h N, Sopkova-de Oliveira Santos J, Retailleau P, Vivarès D, Bonneté F, Langlois d’Estainto B, … Abraini JH (2007). Protein crystallography under xenon and nitrous oxide pressure: comparison with in vivo pharmacology studies and implications for the mechanism of inhaled anesthetic action. Biophysical Journal, 92(1), 217–224. 10.1529/biophysj.106.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloc’h N, Marassio G, & Prangé T (2008). Protein-noble gas interactions investigated by crystallography on three enzymes - implication on anesthesia and neuroprotection mechanisms In Chandrasekeran A (Ed.), Current Trends in X-Ray Crystallography (pp. 285–308). Rijeka: InTech. [Google Scholar]
- Corda M, Era B, Fais A, & Casu M (2004). Structural investigation of pig metmyoglobin by 129Xe NMR spectroscopy. Biochimica et Biophysica Acta, 1674(2), 182–192. 10.1016/j.bbagen.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Cullen SC, & Gross EG (1951). The anesthetic properties of xenon in animals and human beings, with additional observations on krypton.Science,113(2942), 580–582. [DOI] [PubMed] [Google Scholar]
- David HN, Haelewyn B, Risso J-J, Colloc’h N, & Abraini JH (2010). Xenon is an inhibitor of tissue-plasminogen activator: adverse and beneficial effects in a rat model of thromboembolic stroke. Journal of Cerebral Blood Flow and Metabolis: Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 30(4), 718–728. 10.1038/jcbfm.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaye O, Robin E, Bazin J-E, Ripart J, Lebuffe G, & Vallet B (2010). Avantages décisifs, indications et limites de l’anesthésie au xénon. Annales Françaises d’Anesthésie et de Réanimation, 29(9), 635–641. 10.1016/j.annfar.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Desvaux H, Dubois L, Huber G, Quillin ML, Berthault P, & Matthews BW (2005). Dynamics of xenon binding inside the hydrophobic cavity of pseudo-wild-type bacteriophage T4 lysozyme explored through xenon-based NMR spectroscopy. Journal of the American Chemical Society, 127(33), 11676–11683. 10.1021/ja053074p. [DOI] [PubMed] [Google Scholar]
- Dingley J, Findlay GP, Foëx BA, Mecklenburgh J, Esmail M, & Little RA (2001). A closed xenon anesthesia delivery system. Anesthesiology, 94(1), 173–176. 10.1097/00000542-200101000-00034. [DOI] [PubMed] [Google Scholar]
- Dubois L, Da Silva P, Landon C, Huber JG, Ponchet M, Vovelle F, … Desvaux H (2004). Probing the hydrophobic cavity of lipid transfer protein from Nicotiana tabacum through xenon-based NMR spectroscopy. Journal of the American Chemical Society, 126(48), 15738–15746. 10.1021/ja046195i. [DOI] [PubMed] [Google Scholar]
- Eckenhoff RG (2001). Promiscuous ligands and attractive cavities: how do the inhaled anesthetics work? Molecular Interventions, 1(5), 258–268. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14993365. [PubMed] [Google Scholar]
- Emsley P, & Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography, 60(12), 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esencan E, Yuksel S, Tosun YB, Robinot A, Solaroglu I, & Zhang JH (2013). XENON in medical area: emphasis on neuroprotection in hypoxia and anesthesia. Medical Gas Research, 3(1), 4 10.1186/2045-9912-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR (2011). An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallographica. Section D, Biological Crystallography, 67(4), 282–292. 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing GJ, & Maestas S (1970). The thermodynamics of absorption of xenon by myoglobin. The Journal of Physical Chemistry, 74(11), 2341–2344. 10.1021/j100705a018. [DOI] [PubMed] [Google Scholar]
- Franks NP, Dickinson R, de Sousa SL, Hall AC, & Lieb WR (1998). How does xenon produce anaesthesia? Nature, 396(6709), 324 10.1038/24525. [DOI] [PubMed] [Google Scholar]
- Frohn H-J, & Bardin VV (2001). Preparation and reactivity of compounds containing a carbon−xenon bond. Organometallics, 20(23), 4750–4762. 10.1021/om010490j. [DOI] [Google Scholar]
- Giacalone M, Abramo A, Giunta F, & Forfori F (2013). Xenon-related analgesia: a new target for pain treatment. The Clinical Journal of Pain, 29(7), 639–643. 10.1097/AJP.0b013e31826b12f5. [DOI] [PubMed] [Google Scholar]
- Hermans J, & Shankar S (1986). The free energy of xenon binding to myoglobin from molecular dynamics simulation. Israel Journal of Chemistry, 27(2), 225–227. 10.1002/ijch.198600032. [DOI] [Google Scholar]
- Hubbard SJ, & Argos P (1996). A functional role for protein cavities in domain: domain motions. Journal of Molecular Biology, 261(2), 289–300. 10.1006/jmbi.1996.0460. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Gross KH, & Argos P (1994). Intramolecular cavities in globular proteins. Protein Engineering, 7(5), 613–626. http://www.ncbi.nlm.nih.gov/pubmed/8073031. [DOI] [PubMed] [Google Scholar]
- In Rumble JR (2017).CRC handbook of chemistry and physics: A ready-reference book of chemical and physical data. [Google Scholar]
- Lawrence JH, Loomis WF, Tobias CA, & Turpin FH (1946). Preliminary observations on the narcotic effect of xenon with a review of values for solubilities of gases in water and oils.The Journal of physiology,105(3), 197–204. [PMC free article] [PubMed] [Google Scholar]
- Locci E, Dehouck Y, Casu M, Saba G, Lai A, Luhmer M, … Bartik K (2001). Probing proteins in solution by 129Xe NMR spectroscopy. Journal of Magnetic Resonance, 150(2), 167–174. 10.1006/jmre.2001.2325. [DOI] [PubMed] [Google Scholar]
- Lowery TJ, Doucleff M, Ruiz EJ, Rubin SM, Pines A, & Wemmer DE (2005). Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR. Protein Science : A Publication of the Protein Society, 14(4), 848–855. 10.1110/ps.041231005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery TJ, Rubin SM, Ruiz EJ, Spence MM, Winssinger N, Schultz PG, … Wemmer DE (2003). Applications of laser-polarized 129Xe to biomolecular assays. Magnetic Resonance Imaging, 21(10), 1235–1239. 10.1016/j.mri.2003.08.025. [DOI] [PubMed] [Google Scholar]
- Marassio G, Prangé T, David HN, Sopkova-de Oliveira Santos J, Gabison L, Delcroix N, … Colloc’h N (2011). Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study. The FASEB Journal, 25(7), 2266–2275. 10.1096/fj.11-183046. [DOI] [PubMed] [Google Scholar]
- Marx T, Schmidt M, Schirmer U, & Reinelt H (2000). Xenon anaesthesia. Journal of the Royal Society of Medicine, 93(10), 513–517. 10.1177/014107680009301005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, & Read RJ (2007). Phaser crystallographic software. Journal of Applied Crystallography, 40(4), 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H (1899). Zur theorie der alkoholnarkose.Naunyn-Schmiedeberg’s Archives of Pharmacology,42(2), 109–118. [Google Scholar]
- Miller KW (2002). The nature of sites of general anaesthetic action. British Journal of Anaesthesia, 89(1), 17–31. 10.1093/bja/aef167. [DOI] [PubMed] [Google Scholar]
- Miller KW, Reo NV, Schoot Uiterkamp AJ , Stengle DP, Stengle TR, & Williamson KL (1981). Xenon NMR: chemical shifts of a general anesthetic in common solvents, proteins, and membranes. Proceedings of the National Academy of Sciences of the United States of America, 78(8), 4946–4949. 10.1073/pnas.78.8.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Makino M, & Kumasaka T (2013). A convenient tool for gas derivatization using fine-needle capillary mounting for protein crystals. Journal of Synchrotron Radiation, 20(6), 999–1002. 10.1107/S0909049513021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C, Polentarutti M, Djinovic Carugo K, Panjikar S, Tucker PA, & Weiss MS (2004). On the routine use of soft X-rays in macromolecular crystallography. Part II. Data-collection wavelength and scaling models. Acta Crystallographica Section D Biological Crystallography, 60(1), 28–38. 10.1107/S0907444903020833. [DOI] [PubMed] [Google Scholar]
- Neice AE, & Zornow MH (2016). Xenon anaesthesia for all, or only a select few? Anaesthesia, 71(11), 1267–1272. 10.1111/anae.13569. [DOI] [PubMed] [Google Scholar]
- Overton E (1901). Studien uber die Narkose.Fischer, Jena,45. [Google Scholar]
- Prangé T, Schiltz M, Pernot L, Colloc’h N, Longhi S, Bourguet W, & Fourme R (1998). Exploring hydrophobic sites in proteins with xenon or krypton. Proteins, 30(1), 61–73. . [DOI] [PubMed] [Google Scholar]
- Quillin ML, & Matthews BW (2002). Generation of noble-gas binding sites for crystallographic phasing using site-directed mutagenesis. Acta Crystallographica Section D Biological Crystallography, 58(1), 97–103. 10.1107/S0907444901018145. [DOI] [PubMed] [Google Scholar]
- Quillin ML, & Matthews BW (2003). Selling candles in a post-Edison world: phasing with noble gases bound within engineered sites. Acta Crystallographica Section D Biological Crystallography, 59(11), 1930–1934. 10.1107/S0907444903018018. [DOI] [PubMed] [Google Scholar]
- Rentzepis PM, & Douglass DC (1981). Xenon as a solvent. Nature, 293(5828), 165–166. 10.1038/293165a0. [DOI] [Google Scholar]
- Richards FM (1974). The interpretation of protein structures: total volume, group volume distributions and packing density. Journal of Molecular Biology, 82(1), 1–14. 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Roose BW, Zemerov SD, & Dmochowski IJ (2017). Nanomolar small-molecule detection using a genetically encoded 129 Xe NMR contrast agent. Chem. Sci, 8(11), 7631–7636. 10.1039/C7SC03601A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SM, Spence MM, Goodson BM, Wemmer DE, & Pines A (2000). Evidence of nonspecific surface interactions between laser-polarized xenon and myoglobin in solution. Proceedings of the National Academy of Sciences, 97(17), 9472–9475. 10.1073/pnas.170278897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SM, Lee SY, Ruiz EJ, Pines A, & Wemmer DE (2002). Detection and characterization of xenon-binding sites in proteins by 129Xe NMR spectroscopy. Journal of Molecular Biology, 322(2), 425–440. 10.1016/S0022-2836(02)00739-8. [DOI] [PubMed] [Google Scholar]
- Rubin SM, Spence MM, Dimitrov IE, Ruiz EJ, Pines A, & Wemmer DE (2001). Detection of a conformational change in maltose binding protein by 129Xe NMR spectroscopy. Journal of the American Chemical Society, 123(35), 8616–8617. 10.1021/ja0110325. [DOI] [PubMed] [Google Scholar]
- Rubin SM, Spence MM, Pines A, & Wemmer DE (2001). Characterization of the effects of nonspecific xenon-protein interactions on 129Xe chemical shifts in aqueous solution: further development of xenon as a biomolecular probe. Journal of Magnetic Resonance, 152(1), 79–86. 10.1006/jmre.2001.2389. [DOI] [PubMed] [Google Scholar]
- Sanders RD (2003). Xenon: no stranger to anaesthesia. British Journal of Anaesthesia, 91(5), 709–717. 10.1093/bja/aeg232. [DOI] [PubMed] [Google Scholar]
- Sauer O, Schmidt A, & Kratky C (1997). Freeze-trapping isomorphous xenon derivatives of protein crystals. Journal of Applied Crystallography, 30(4), 476–486. 10.1107/S0021889897000745. [DOI] [Google Scholar]
- Schiltz M, Fourme R, & Prangé T (2003). Use of noble gases xenon and krypton as heavy atoms in protein structure determination. Methods in Enzymology, 374, 83–119. 10.1016/S0076-6879(03)74004-X. [DOI] [PubMed] [Google Scholar]
- Schiltz M, Kvick Å ;., Svensson OS, Shepard W, La Fortelle E, Prangé T, … Fourme R (1997). Protein crystallography at ultra-short wavelengths: feasibility study of anomalous-dispersion experiments at the xenon K-edge. Journal of Synchrotron Radiation, 4(5), 287–297. 10.1107/S0909049597008571. [DOI] [PubMed] [Google Scholar]
- Schiltz M, Prangé T, & Fourme R (1994). On the preparation and X-ray data collection of isomorphous xenon derivatives. Journal of Applied Crystallography, 27(6), 950–960. 10.1107/S0021889894005923. [DOI] [Google Scholar]
- Schoenborn BP (1965). Binding of xenon to horse haemoglobin. Nature, 208(5012), 760–762. 10.1038/208760a0. [DOI] [PubMed] [Google Scholar]
- Schoenborn BP, Featherstone RM, Vogelhut PO, & Süsskind C (1964). Influence of xenon on protein hydration as measured by a microwave absorption technique. Nature, 202(4933), 695–696. 10.1038/202695a0. [DOI] [PubMed] [Google Scholar]
- Schoenborn BP, Watson HC, & Kendrew JC (1965). Binding of xenon to sperm whale myoglobin. Nature, 207(992), 28–30. 10.1038/207028a0. [DOI] [PubMed] [Google Scholar]
- Schröder L, Lowery TJ, Hilty C, Wemmer DE, & Pines A (2006). Molecular imaging using a targeted magnetic resonance hyperpolarized biosensor. Science, 314(5798), 446–449. 10.1126/science.1131847 [DOI] [PubMed] [Google Scholar]
- Schröder L (2013). Xenon for NMR biosensing – inert but alert. Physica Medica, 29(1), 3–16. 10.1016/j.ejmp.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Settle W (1973). Function of the myoglobin molecule as influenced by anesthetic molecules In Featherstone RM (Ed.), A Guide to Molecular Pharmacology and Toxicology (pp. 477–493). New York, NY: Marcel Dekker. [Google Scholar]
- Soltis SM, Stowell MHB, Wiener MC, Phillips GN, & Rees DC (1997). Successful flash-cooling of xenon-derivatized myoglobin crystals. Journal of Applied Crystallography, 30(2), 190–194. 10.1107/S0021889896012939. [DOI] [Google Scholar]
- Stoppe C, Rimek A, Rossaint R, Rex S, Stevanovic A, Schälte G, … Coburn M (2013). Xenon consumption during general surgery: a retrospective observational study. Medical Gas Research, 3(1), 12 10.1186/2045-9912-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell MHB, Soltis SM, Kisker C, Peters JW, Schindelin H, Rees DC, … Whitby FG (1996). A simple device for studying macromolecular crystals under moderate gas pressures (0.1–10 MPa). Journal of Applied Crystallography, 29(5), 608–613. 10.1107/S0021889896004712. [DOI] [Google Scholar]
- Teeter MM (2004). Myoglobin cavities provide interior ligand pathway. Protein Science, 13(2), 313–318. 10.1110/ps.03334304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton RF, & Kuntz ID (1982). Nuclear magnetic resonance studies of xenon-129 with myoglobin and hemoglobin. Biochemistry, 21(26), 6850–6857. 10.1021/bi00269a035. [DOI] [PubMed] [Google Scholar]
- Tilton RF, Kuntz ID, & Petsko GA (1984). Cavities in proteins: structure of a metmyoglobin-xenon complex solved to 1.9 A. Biochemistry, 23(13), 2849–2857. 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- Tilton RF, Singh UC, Kuntz ID, & Kollman PA (1988). Protein-ligand dynamics. Journal of Molecular Biology, 199(1), 195–211. 10.1016/0022-2836(88)90389-0. [DOI] [PubMed] [Google Scholar]
- Tilton RF, Singh UC, Weiner SJ, Connolly ML, Kuntz ID, Kollman PA, … Case DA (1986). Computational studies of the interaction of myoglobin and xenon. Journal of Molecular Biology, 192(2), 443–456. 10.1016/0022-2836(86)90374-8. [DOI] [PubMed] [Google Scholar]
- Vallone B, & Brunori M (2004). Roles for holes: are cavities in proteins mere packing defects? The Italian Journal of Biochemistry, 53(1), 46–52. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/15356962 [PubMed] [Google Scholar]
- van Zijl PCM, & Yadav NN (2011). Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magnetic Resonance in Medicine, 65(4), 927–948. 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali J, Robbins AH, Almo SC, & Tilton RF (1991). Using xenon as a heavy atom for determining phases in sperm whale metmyoglobin. Journal of Applied Crystallography, 24(5), 931–935. 10.1107/S0021889891005897. [DOI] [Google Scholar]
- Walker TG, & Happer W (1997). Spin-exchange optical pumping of noble-gas nuclei. Reviews of Modern Physics, 69(2), 629–642. 10.1103/RevModPhys.69.629 [DOI] [Google Scholar]
- Wang Y, Roose BW, Philbin JP, Doman JL, & Dmochowski IJ (2016). Programming a molecular relay for ultrasensitive biodetection through 129Xe NMR. Angewandte Chemie (International Ed. in English), 55(5), 1733–1736. 10.1002/anie.201508990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Roose BW, Palovcak EJ, Carnevale V, & Dmochowski IJ (2016). A genetically encoded β-lactamase reporter for ultrasensitive 129Xe NMR in mammalian cells. Angewandte Chemie (International Ed. in English), 55(31), 8984–8987. 10.1002/anie.201604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weathersby PK, & Homer LD (1980). Solubility of inert gases in biological fluids and tissues: a review. Undersea Biomedical Research, 7(4), 277–296. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/6262972. [PubMed] [Google Scholar]
- Weinrich M, & Worcester DL (2013). Xenon and other volatile anesthetics change domain structure in model lipid raft membranes. The Journal of Physical Chemistry. B, 117(50), 16141–16147. 10.1021/jp411261g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DA, Thornton A, Farjot G, & Katz I (2016). The diverse biological properties of the chemically inert noble gases. Pharmacology and Therapeutics, 160, 44–64. 10.1016/j.pharmthera.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, … Wilson KS (2011). Overview of the CCP4 suite and current developments. Acta Crystallographica Section D Biological Crystallography, 67(4), 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C, Kunth M, Döpfert J, Rossella F, Schröder L (2012). Hyperpolarized xenon for NMR and MRI applications. Journal of Visualized Experiments. 67, 4268 https://doi.org/10.379¼268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, & Radzicka A (1994). On the probability of finding a water molecule in a nonpolar cavity. Science, 265(5174), 936–937. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8052849. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Akimoto T, Shimizu H, Hirano Y, Yasui M, & Yasuoka K (2012). Diffusive nature of xenon anesthetic changes properties of a lipid bilayer: molecular dynamics simulations. The Journal of Physical Chemistry. B, 116(30), 8989–8995. 10.1021/jp303330c. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Cheng J, Wang L, Wang X, Zhang M, … Fang H (2017). Inert gas deactivates protein activity by aggregation. Scientific Reports, 7(1), 10176 10.1038/s41598-017-10678-3. [DOI] [PMC free article] [PubMed] [Google Scholar]