Abstract
Swine influenza A virus (SwIV) infection has considerable economic and animal welfare consequences and, because of the zoonotic potential, can also have public health implications. The 2009 pandemic H1N1 ‘swine-origin’ infection is now endemic in both pigs and humans. In Europe, avian-like H1avN1, human-like H1huN2, human-like swine H3N2 and, since 2009, pandemic H1N1 (pH1N1) lineage viruses and reassortants, constitute the dominant subtypes. In this study, we used a swine pH1N1 challenge virus to investigate the efficacy of whole inactivated virus vaccines homologous or heterologous to the challenge virus as well as a commercial vaccine. We found that vaccine-mediated protection was most effective when vaccine antigen and challenge virus were homologous and correlated with the specific production of neutralising antibodies and a cellular response to the challenge virus. We conclude that a conventional whole inactivated SwIV vaccine must be antigenically matched to the challenge strain to be an effective control measure.
Keywords: Swine influenza A virus, Pandemic 2009 influenza A virus, H1N1, Whole inactivated virus vaccine, Immune response
1. Introduction
Swine Influenza A virus (SwIV) infection is of economic importance to the pig industry and may also affect public health, because of the zoonotic potential. In particular, infection of humans with the 2009 pandemic H1N1 (pH1N1) ‘swine-origin’ virus has been a serious ‘One Health’ concern in recent years. This virus is now endemic in pigs and humans [1].
Pigs can be infected by both avian- and mammalian-origin influenza A viruses and are a key intermediate host species in the adaptation of avian influenza viruses to mammals, including humans. In addition, SwIV strains display greater genetic diversity in comparison to influenza A viruses circulating in the human population at any one time. This increased genetic diversity is likely linked to the permissiveness of swine to a wider spectrum of viruses and the frequent re-introductions of strains from the human population [1], [2]. Vaccination of swine could be a key element for mitigating influenza A disease risks to both human and animal health [1], [2], [3], [4].
SwIV vaccine composition and application for controlling disease varies [2], [4]. Lack of vaccine use by swine producers is attributed to the short lifespan of meat production pigs, the requirement to vaccinate in the absence of maternally-derived antibodies and the need for conventional whole inactivated virus (WIV) vaccines to be antigenically matched to circulating virus lineages in order to be effective [2], [4]. Because of the genetic diversity of circulating SwIVs, vaccines containing a defined number of antigens can only provide limited cross-protective immunity [1], [2]. In addition WIV vaccine efficacy is constrained as it depends largely on the generation of neutralising IgG antibodies to the highly variable regions of the major viral envelope hemagglutinin (HA) antigen [4]. Such vaccines reduce clinical disease following infection by antigenically matched strains, but complete sterile immunity is rarely observed [2], [4].
The most recently available SwIV vaccines in the EU [4] are a trivalent vaccine, licensed in 2010, which incorporates H1avN1 and H3N2 antigens isolated in 2003 as well as a reassortant antigen (H1huN2) with human-origin H1 and N2 gene segments. This vaccine, Gripovac®3 is now licenced as Respiporc Flu3 (web references 1, 2). A veterinary monovalent vaccine against the swine pH1N1 influenza A virus was licenced in the UK in 2017 (Respiporc FLUpan H1N1, web reference 3). Manufacturers are required to follow the full licensing procedure in order to change vaccine strains, which creates a regulatory hurdle for updating of SwIV vaccine composition. Therefore, commercially available vaccines do not always antigenically match contemporary circulating strains [4]. Current research efforts are directed at developing broadly cross-protective vaccines capable of stimulating both humoral and cellular arms of the immune system.
To understand the strengths and limitations of current SwIV vaccines as a disease control tool, this study assessed the protective efficacy and reduction in virus shedding provided by three different vaccines upon challenge of pigs with a swine pandemic 2009 influenza A virus isolate and the humoral and cellular immune responses. The three vaccines were a commercially available trivalent vaccine (Gripovac®3) and two monovalent WIV vaccines antigenically homologous or heterologous to the challenge strain.
2. Materials and methods
Influenza A virus strains used to generate the whole inactivated virus (WIV) vaccines were a pandemic 2009 H1N1 isolate, A/swine/England/1353/2009 (pH1N1) [3], and a Eurasian avian-like H1N1 isolate, A/swine/England/453/2006 (H1avN1) [5]. Vaccine antigen was prepared from virus grown in SPF embryonated chicken eggs and inactivated with beta-propiolactone (BPL) [6].The antigenic content was quantified and expressed in HA units per ml (HAU/ml). WIV vaccines were formulated in an oil-in-water adjuvant, TS6 (web reference 4, CEVA), and were formulated in 1 ml with a 1:2 antigen to TS6 ratio. Gripovac®3, is a WIV vaccine incorporating three European-origin SwIV subtypes, H1avN1 and H1huN2 and H3N2, isolated between 2000 and 2003, formulated in carbomer anionic polymer adjuvant (Table 1). An H1huN2 subtype antigen A/Sw/Eng/438207/94, was used to monitor the immune response to Gripovac®3.
Table 1.
Summary of pig groups and vaccines administered.
| Group | Vaccine virus subtype | Strain(s) | Vaccination | Boost | Adjuvant |
|---|---|---|---|---|---|
| 1 | Homologous | ||||
| pH1N1 | A/swine/England/1353/2009 | 1024 HAUa | 3072 HAU | TS6 | |
| 2 | Heterologous | ||||
| H1avN1 | A/swine/England/453/2006 | 1024 HAU | 3072 HAU | TS6 | |
| 3 | Gripovac®3 | ||||
| H1avN1 | A/swine/Haselünne/IDT2617/2003 | 10.22 GMNUb | 10.22 GMNUb | Carbomer | |
| H1huN2 | A/swine/Bakum/1832/2000 | 12.34 GMNU | 12.34 GMNU | 971 P NF | |
| H3N2 | A/swine/Bakum/IDT1769/2003 | 10.53 GMNU | 10.53 GMNU | ||
| 4 | Adjuvant control | TS6 in PBS |
HAU, log2 hemagglutination units.
GMNU, log2 geometric mean of neutralising units induced in Guinea pigs after immunisation twice with 0.5 ml of this vaccine.
All in vivo studies were conducted at APHA following ethical approval according to the U.K. Animals (Scientific Procedures) Act, 1986 and the ARRIVE guidelines. Sixteen Landrace cross female pigs of high health status were verified to be negative for influenza A virus infection by matrix gene real-time RT-PCR [7] and for antibodies by an HA inhibition (HI) test [8]. Pigs were randomised into four groups (n = 4) and vaccinated at 6.5 weeks (0 days post vaccination (dpv)) and 21dpv. The vaccines (Table 1) were administered into the trapezius muscle, 25–30 mm posterior to the ear using a 1 in., 19G needle. Antigen content was increased for the boost when no adverse reaction to the primary vaccination occurred.
Pigs were challenged intranasally, as in previous studies [9], [10], [11]. On 69dpv, a dose of 1x107 TCID50 pH1N1 virus was administered to each pig using a MAD300 (Teleflex) which delivers an atomised spray of droplets 30–100 μm in diameter [12]. This non-invasive method of challenging alert pigs was used, as previously [5], [11], [12], [13], to mimic the natural route infection. Animals were monitored daily following challenge on 0 days post-infection (dpi) according to a clinical scoring system [13]. Animals were euthanized at 10dpi or 79dpv with an overdose of intravenous pentobarbital sodium.
Serum samples were obtained pre-vaccination, weekly between boost and challenge and at 1, 3, 7 and 9 dpi and stored at −80 °C. Heparin anticoagulated blood samples were taken for peripheral blood mononuclear cells (PBMC) isolation at 0, 14, 28, and 63dpv and 3 and 9dpi. Four nasal swabs (two per nostril) were taken at 14, 28 and 56dpv, before challenge and daily after challenge until euthanasia. Dry swabs were stored at −80 °C. Daily nasal swabs were processed together by immersion in 2 ml of Leibovitz L-15 medium, containing 1% FBS, 100U/ml penicillin and 100 μg/ml streptomycin (Gibco). Swab suspensions were aliquoted and stored at −80 °C for analysis.
RNA extracted from nasal swab suspensions using the RNeasy® mini kit (Qiagen) was quantified by RRT-qPCR [7] for the influenza A virus M gene using an MxPro 3000P instrument and MxPro analysis software (Agilent). RNA quantity was expressed as relative equivalent units (REU) of RNA using a standard 10-fold dilution series of RNA purified from the virus stock used for challenge, with known titre. Samples with ‘no Cycle threshold (Ct)’ or negative REU value were assigned a value of 0 once log10 transformed. Although REU values measure the amount of viral RNA present and not infectivity, it may be inferred from the linear relationship with the dilution series that they are proportional to the amount of virus present. Area under the curve (AUC) was calculated using GraphPad Prism software to evaluate the total shedding of viral RNA. Vaccine groups comparisons for height of shedding peak, AUC and cellular response, used a pairwise permutational t-test from the package ‘RVAideMemoire 0.9-55′ in R3.1.1. We used 1000 permutations and the p-value was adjusted for multiple testing [14].
Antibody titres were measured using HI and virus neutralisation (VN) [11]. HI antigens were the homologous and heterologous vaccines as well as an H1huN2 SwIV strain, A/Sw/Eng/438207/94. VN was measured against the pH1N1 challenge virus. Results are expressed as the Geometric Mean Ratio (GMR) for each group, calculated relative to the baseline titre. PBMC were isolated from heparinized venous blood and cryopreserved. Samples were retrieved at the same time to ensure consistency of handling and analysed using a Porcine IFN-γ ELISpot assay [15].
3. Results
We evaluated the protective efficacy of three vaccine candidates: homologous inactivated, heterologous inactivated (both in TS6 adjuvant) and the commercial trivalent Gripovac®3 vaccine delivered as intra-muscular vaccinations 21 days apart. Control animals received TS6 adjuvant only (Table 1). Animals were challenged 69 days after the first vaccination. Vaccination and challenge produced no adverse reactions in any group and clinical scores transiently reached no more than 3 out of a possible maximum of 20 for 2 animals (data not shown).
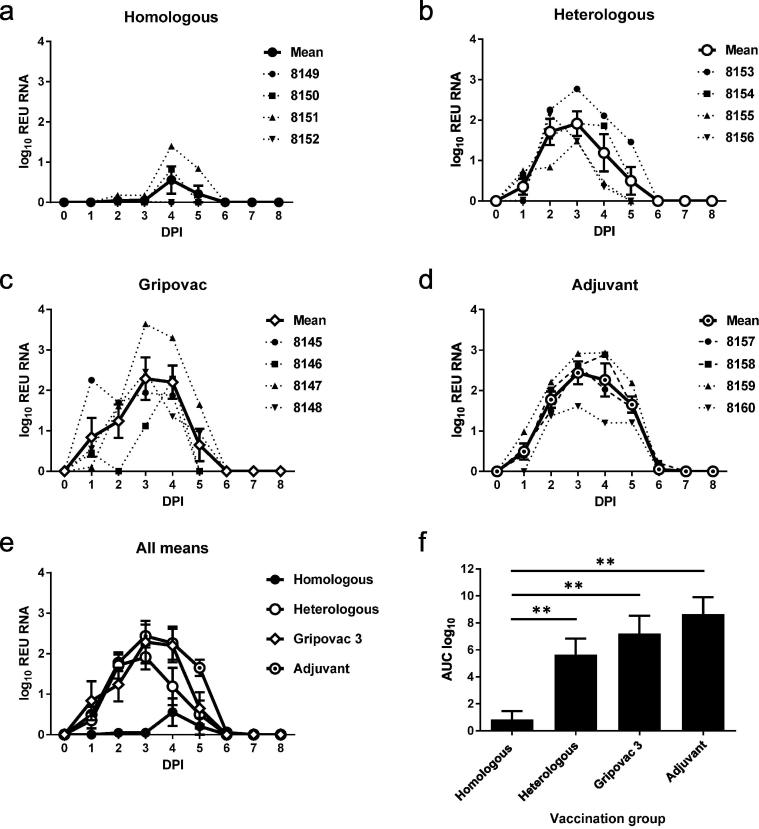
To assess viral RNA shedding, viral RNA present in daily nasal swab samples was quantified (Fig. 1a–d). Viral RNA, expressed as REU, correlates with the amount of infectious virus, if the infectious titre is above approximately 100pfu/ml (Fig. S1). Despite variation between individual animals, viral RNA shedding, measured by the AUC (Fig. 1e and f), was significantly reduced only in the pigs vaccinated with the homologous WIV vaccine. When compared to pigs which received the homologous WIV vaccine, shedding was significantly elevated by 88.6% (P = 0.045) in the heterologous WIV vaccine group, 91.0% (P = 0.048) in the Gripovac®3 vaccine group or 92.5% (P = 0.045) in the adjuvant control group. The peak of viral shedding was delayed and significantly lower in the homologous WIV group when compared to the heterologous WIV vaccine (P = 0.042), Gripovac®3 vaccine (P = 0.039) or adjuvant (P = 0.039) groups.
Fig. 1.
Viral shedding. Viral shedding levels in nasal swabs were assessed daily by RRT-qPCR and amounts are expressed as log10 Relative equivalent units (REU) of viral RNA. Individual pig and group mean REU graphs for groups receiving (a) homologous WIV vaccine, (b) heterologous WIV vaccine, (c) Gripovac®3 or (d) TS6 adjuvant control alone are shown. Mean virus shedding (e) and area under the curve (AUC) analysis (f) revealed that virus shedding was significantly reduced (**P < 0.05) in the group that had been vaccinated with the homologous WIV vaccine. Vertical bars represent ±SEM.
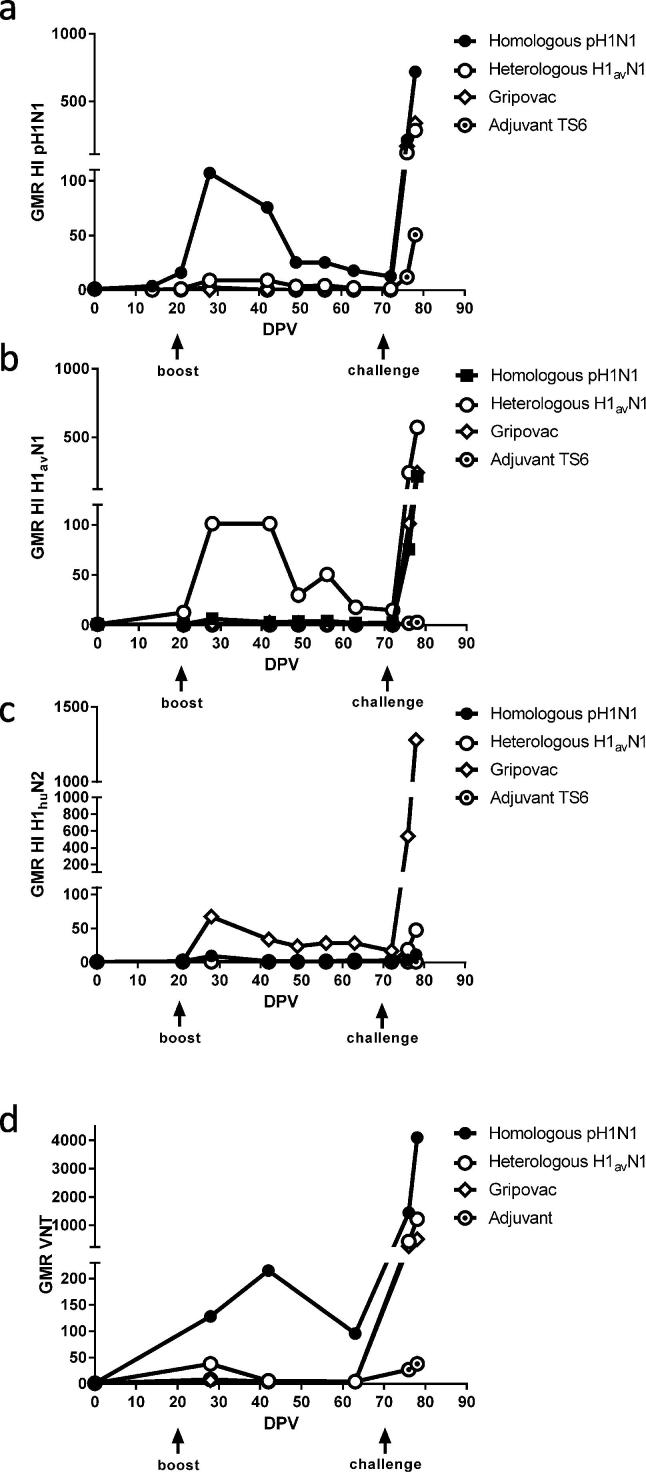
The serum antibody HI titres were obtained for all samples using the same antigen in each WIV vaccine and a SwIV H1huN2 representing one antigen in Gripovac®3 (Fig. 2a–c). A specific humoral response to each vaccine antigen was detected in the group vaccinated with the cognate antigen peaking 7d post-boost. The response was strongest against the TS6-adjuvanted formulations (Fig. 2a and b) and lower in the Gripovac®3 group Fig. 2c. Following challenge with the pH1N1 virus, responses to the cognate vaccine antigen were rapidly boosted by 9dpi in all vaccine groups compared to the adjuvant only group, confirming that there was efficient priming by all vaccines.
Fig. 2.
Serological analysis. Longitudinal serum samples were assessed by hemagglutination inhibition (HI) against (a) the homologous antigen, pH1N1 (A/swine/England/1353/2009), (b) the heterologous antigen, H1avN1 (A/swine/England/453/2006) and (c) a representative H1huN2 antigen, A/swine/England/438207/1994. The Geometric Mean Ratio (GMR) for samples relative to the 0dpv sample for each corresponding animal is shown. Virus neutralisation (VN) titres (d) were evaluated for serum samples using the homologous pH1N1 challenge strain A/swine/England/1353/2009.
Neutralising antibody responses to the pH1N1 challenge virus differed between the vaccinated groups (Fig. 2d). The homologous WIV vaccine elicited high titres of neutralising antibodies to the challenge strain, but with different kinetics to the HI antibody titre. In the other vaccine groups, although pH1N1 virus infection elicited a humoral response, the response was directed against the vaccine antigen with a lower specific neutralising ability against the challenge strain.
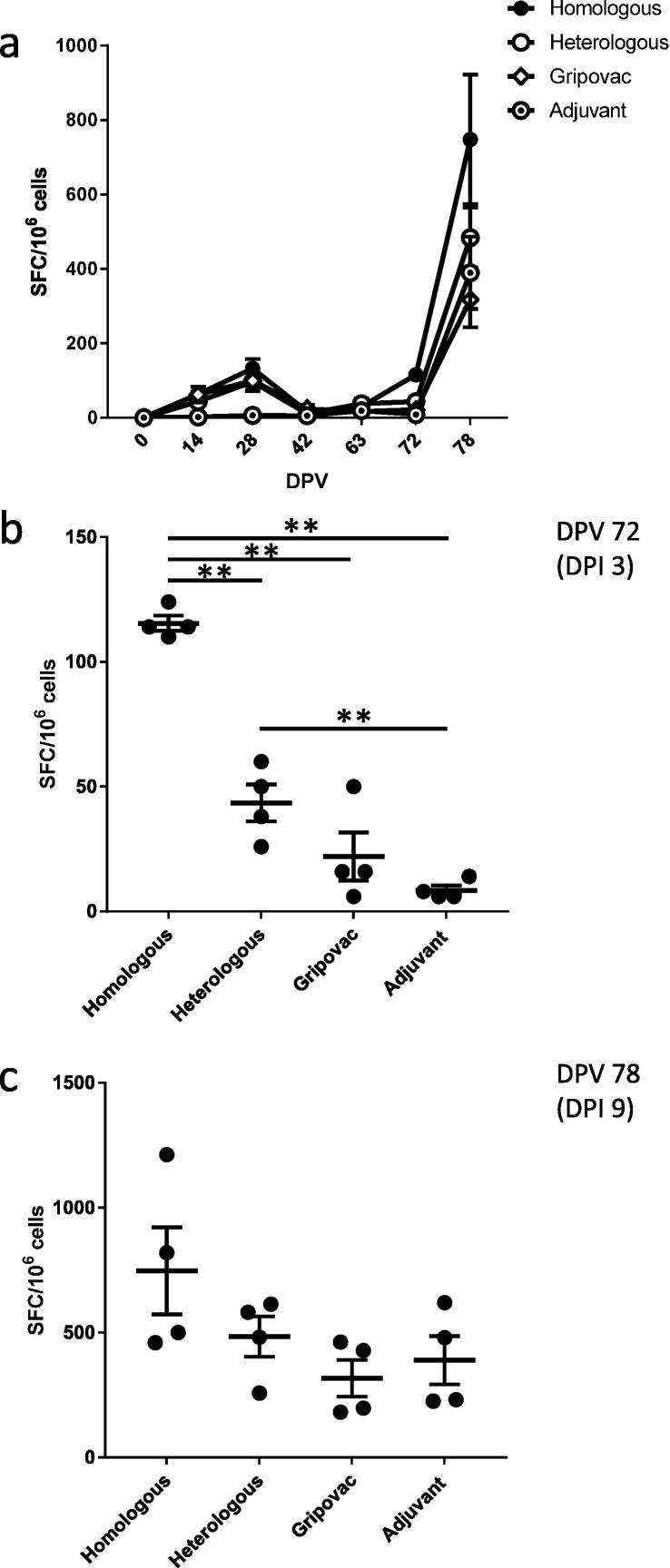
Analysis of the cellular response (Fig. 3a) showed, as expected, that the homologous WIV vaccine group produced the highest virus-specific responses whereas lower responses were seen in the remaining groups. After challenge, the highest response was observed in the homologous WIV vaccine group when stimulated by vaccination and homologous virus challenge within 3 days (Fig. 3b) and was significant (p = 0.049) relative to all other groups using a pairwise permutation test. The heterologous WIV vaccine group also displayed a significantly different response (p = 0.049) relative to the adjuvant control group. There was no significant difference in the response measured between groups at 9dpi (Fig. 3c). This result likely reflected the primary cellular response to infection rather than a vaccine-stimulated response.
Fig. 3.
Cellular response. IFN-γ ELISpot analysis of the PBMC response is expressed as SFC/106 cells to the pH1N1 A/swine/England/1353/2009 antigen (a) for the duration of the study or specifically for (b) 72dpv (3dpi) and (c) 78dpv (9dpi). Horizontal lines indicate significant differences (**p = 0.049) between groups and vertical bars represent ±SEM.
4. Discussion
This study confirms that a conventional WIV vaccine needs to be antigenically matched to the challenge virus lineage in order to be efficacious, as reported previously [11], [16]. A 10 week interval separated the first vaccination and virus challenge to be representative of the lifespan of a food-production pig. Intranasal challenge with a pH1N1 strain produced minimal clinical signs, as observed previously in mature animals [11], [12]. Viral shedding was observed in all groups but in the homologous vaccine group, was significantly reduced as indicated by the AUC analysis of the viral RNA shedding profiles, in keeping with other pH1N1 vaccination-challenge studies [11], [16]. In a field situation, it is unknown whether this level of shedding would support onward transmission of the virus, although successful control of a pH1N1 outbreak with a homologous vaccine has been reported [17]. The Gripovac®3 and heterologous vaccines, which contained an H1avN1 antigen, did not significantly reduce shedding following infection with the divergent lineage pH1N1 SwIV, as reported previously for Gripovac®3 [11].
Humoral and cellular immune responses to influenza virus infection were elicited following challenge, indicating that the WIV vaccines efficiently primed the immune system. We found that vaccinated pigs developed hemagglutinating antibodies to the corresponding vaccine antigen in all cases. Following pH1N1 challenge, HI titre was elevated against the original priming (vaccine) antigen, rather than the challenge strain. This result could be indicative of back-boosting (reviewed in [2]) and corresponds to previous observations with vaccination-challenge studies [10]. The neutralising antibody response distinguished the homologous vaccine group from the other groups, although greater levels of cross-reactive neutralising antibodies have been detected in other studies [9], [10], [11]. In our study, the neutralising antibody response correlated with the decrease in nasal shedding of viral RNA and is potentially a better predictor of vaccine efficacy.
The cellular immune response to vaccination and challenge is not frequently assessed [4]. In this study the homologous vaccinated-challenge group developed a robust cellular response to the cognate antigen, as expected, and the post-challenge kinetics mirrored that of the humoral response. This indicates that the cellular immune response may also have played a role in vaccine-mediated protection, but the experiments performed here cannot determine the relative contribution of either humoral or cellular immunity to protection.
During this study, clinical signs corresponding to vaccine-associated enhanced respiratory disease (VAERD) [18] were not observed. VAERD has been reported in pigs when WIV vaccines in an oil-in-water adjuvant has been used followed by challenge with a virus incorporating an antigenically related HA envelope protein. Recently, VAERD has been linked to factors including the production of non-neutralising HA-specific antibodies without inhibitory anti-NA antibodies [2] and the formulation of oil-in-water adjuvants [19]. It is unknown which aspects of the current study design may have avoided disease exacerbation.
Pigs are a key species and an important veterinary host for the study and control of mammalian influenza viruses. Inter-species transmission and reassortment events frequently occur between pigs and humans, increasing the risk of emergence of novel viruses. For SwIV vaccination to serve as an effective control measure in reducing disease burden and pandemic influenza risk, a structured approach is needed for regular updating of vaccine composition [1].
5. Conclusions
Currently available WIV vaccines need to be antigenically matched to circulating influenza virus lineages in order to be efficacious. Improvement in vaccination strategies that reduce swine influenza A virus shedding and therefore the likelihood of onward transmission of infection, would be beneficial to swine health and the agricultural sector. Our results reinforce the need for the development of broadly cross-reactive influenza virus vaccines.
Acknowledgments
Acknowledgements
This work was funded by the Biotechnology and Biological Sciences Research Council sLoLa Grant BB/L001330/1. We are grateful to the Animal Sciences Unit staff at the Animal and Plant Health Agency (APHA) for ensuring excellent animal care. We wish to thank Dr. Catherine Charreyre from Boehringer-Ingleheim for her consultancy. Gripovac®3 vaccine and TS6 adjuvant were kindly supplied by Merial Animal Health.
Conflict of interest statement
The authors declare they have no conflict of interest.
Sarah Gilbert is co-founder of Vaccitech, a company developing influenza vaccines for use in humans.
Footnotes
Correspondence of REU and infectious titre for the pH1N1 challenge strain is shown in Supplementary Fig. S1. The results indicate that there is a correlation between quantification measures at titres exceeding 100 pfu/ml but below this value, REU data can be obtained using the more sensitive RRT-qPCR assay, when culture of infectious virus can be inconsistent. This observation could indicate the presence of defective, non-infectious virus particles but could also be due to the confounding effect of neutralising antibodies, which can prevent the detection of infectious virus. Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2019.02.078.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Correlation between infections titre (pfu/ml) and RRT-qPCR measure of relative equivalent units (REU) log10 in nasal swabs from pigs infected with the pH1N1 strain A/sw/Eng/1353/2009.
References
- 1.Lewis N.S., Russell C.A., Langat P., Anderson T.K., Berger K., Bielejec F. The global antigenic diversity of swine influenza A viruses. eLife. 2016;5:e12217. doi: 10.7554/eLife.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent A.L., Perez D.R., Rajao D., Anderson T.K., Abente E.J., Walia R.R. Influenza A virus vaccines for swine. Vet Microbiol. 2017;206:35–44. doi: 10.1016/j.vetmic.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson S.J., Langat P., Reid S.M., Lam T.T., Cotten M., Kelly M. Molecular epidemiology and evolution of influenza viruses circulating within european swine between 2009 and 2013. J Virol. 2015;89:9920–9931. doi: 10.1128/JVI.00840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Reeth K., Ma W. Swine influenza virus vaccines: to change or not to change-that's the question. Curr Top Microbiol Immunol. 2013;370:173–200. doi: 10.1007/82_2012_266. [DOI] [PubMed] [Google Scholar]
- 5.Murcia P.R., Hughes J., Battista P., Lloyd L., Baillie G.J., Ramirez-Gonzalez R.H. Evolution of an Eurasian avian-like influenza virus in naive and vaccinated pigs. PLoS Pathogens. 2012;8:e1002730. doi: 10.1371/journal.ppat.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budowsky E.I., Friedman E.A., Zheleznova N.V. Noskov FS. Principles of selective inactivation of viral genome. VI. Inactivation of the infectivity of the influenza virus by the action of beta-propiolactone. Vaccine. 1991;9:398–402. doi: 10.1016/0264-410x(91)90125-p. [DOI] [PubMed] [Google Scholar]
- 7.Nagy A., Vostinakova V., Pirchanova Z., Cernikova L., Dirbakova Z., Mojzis M. Development and evaluation of a one-step real-time RT-PCR assay for universal detection of influenza A viruses from avian and mammal species. Arch Virol. 2010;155:665–673. doi: 10.1007/s00705-010-0636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitikoon P., Gauger P.C., Vincent A.L. Hemagglutinin inhibition assay with swine sera. Methods Mol Biol (Clifton, NJ) 2014;1161:295–301. doi: 10.1007/978-1-4939-0758-8_24. [DOI] [PubMed] [Google Scholar]
- 9.Dürrwald R., Krumbholz A., Baumgarte S., Schlegel M., Vahlenkamp T.W., Selbitz H.-J. Swine influenza A vaccines, pandemic (H1N1) 2009 virus, and cross-reactivity. Emerg Infect Dis. 2010;16:1029–1030. doi: 10.3201/eid1606.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kyriakis C.S., Olsen C.W., Carman S., Brown I.H., Brookes S.M., Doorsselaere J.V. Serologic cross-reactivity with pandemic (H1N1) 2009 virus in pigs, Europe. Emerg Infect Dis. 2010;16:96–99. doi: 10.3201/eid1601.091190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffen W.L., Stockhofe N., Weesendorp E., van Zoelen-Bos D., Heutink R., Quak S. Efficacy of a pandemic (H1N1) 2009 virus vaccine in pigs against the pandemic influenza virus is superior to commercially available swine influenza vaccines. Vet Microbiol. 2011;152:304–314. doi: 10.1016/j.vetmic.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Hemmink J.D., Morgan S.B., Aramouni M., Everett H., Salguero F.J., Canini L. Distinct immune responses and virus shedding in pigs following aerosol, intra-nasal and contact infection with pandemic swine influenza A virus, A(H1N1)09. Vet Res. 2016;47:103. doi: 10.1186/s13567-016-0390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brookes S.M., Nunez A., Choudhury B., Matrosovich M., Essen S.C., Clifford D. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non-immune pigs. PloS One. 2010;5:e9068. doi: 10.1371/journal.pone.0009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 15.Franzoni G., Kurkure N.V., Essler S.E., Pedrera M., Everett H.E., Bodman-Smith K.B. Proteome-wide screening reveals immunodominance in the CD8 T cell response against classical swine fever virus with antigen-specificity dependent on MHC class I haplotype expression. PloS One. 2013;8:e84246. doi: 10.1371/journal.pone.0084246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent A.L., Ciacci-Zanella J.R., Lorusso A., Gauger P.C., Zanella E.L., Kehrli M.E., Jr. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine. 2010;28:2782–2787. doi: 10.1016/j.vaccine.2010.01.049. [DOI] [PubMed] [Google Scholar]
- 17.Mughini-Gras L., Beato M.S., Angeloni G., Monne I., Buniolo F., Zuliani F. Control of a reassortant pandemic 2009 H1N1 influenza virus outbreak in an intensive swine breeding farm: effect of vaccination and enhanced farm management practices. PLoS Curr. 2015;7 doi: 10.1371/currents.outbreaks.4211b8d6cedd8c870db723455409c0f8. [ecurrents.outbreaks.4211b8d6cedd8c870db723455409c0f8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauger P.C., Vincent A.L., Loving C.L., Henningson J.N., Lager K.M., Janke B.H. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol. 2012;49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 19.Souza C.K., Rajao D.S., Sandbulte M.R., Lopes S., Lewis N.S., Loving C.L. The type of adjuvant in whole inactivated influenza a virus vaccines impacts vaccine-associated enhanced respiratory disease. Vaccine. 2018 doi: 10.1016/j.vaccine.2018.08.072. [DOI] [PubMed] [Google Scholar]
Web references
- 1.Gripovac ®3 vaccine [last accessed 23 June 2018] http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/veterinary/medicines/000157/vet_med_000206.jsp&mid=WC0b01ac058001fa1c.
- 2.Respiporc Flu3 vaccine [last accessed 23 June 2018] http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/veterinary/medicines/000153/vet_med_000205.jsp&mid=WC0b01ac058001fa1c.
- 3.Respiporc FLUpan H1N1 [last accessed 23 June 2018] http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/veterinary/medicines/003993/vet_med_000345.jsp&mid=WC0b01ac058001fa1c.
- 4.TS6 adjuvant is licensed to CEVA and covered by patent WO2005009462A3. https://patents.google.com/patent/WO2005009462A3/en.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between infections titre (pfu/ml) and RRT-qPCR measure of relative equivalent units (REU) log10 in nasal swabs from pigs infected with the pH1N1 strain A/sw/Eng/1353/2009.