Abstract
DNA polymerase (pol) δ is essential for both leading and lagging strand DNA synthesis during chromosomal replication in eukaryotes. Pol δ has been implicated in the Okazaki fragment maturation process for the extension of the newly synthesized fragment and for the displacement of the RNA/DNA segment of the preexisting downstream fragment generating an intermediate flap structure that is the target for the Dna2 and flap endonuclease-1 (Fen 1) endonucleases. Using a single-stranded minicircular template with an annealed RNA/DNA primer, we could measure strand displacement by pol δ coupled to DNA synthesis. Our results suggested that pol δ alone can displace up to 72 nucleotides while synthesizing through a double-stranded DNA region in a distributive manner. Proliferating cell nuclear antigen (PCNA) reduced the template dissociation rate of pol δ, thus increasing the processivity of both synthesis and strand displacement, whereas replication protein A (RP-A) limited the size of the displaced fragment down to 20–30 nucleotides, by generating a “locked” flap DNA structure, which was a substrate for processing of the displaced fragment by Fen 1 into a ligatable product. Our data support a model for Okazaki fragment processing where the strand displacement activity of DNA polymerase δ is modulated by the concerted action of PCNA, RP-A and Fen 1.
DNA polymerase (pol) δ is the major pol involved in chromosomal replication in eukaryotes (reviewed in ref. 1). It is essential for both leading and lagging strand synthesis in the in vitro reconstituted SV40 replication system (2) and in eukaryotic cells. The current view of DNA replication in eukaryotes predicts that pol α/primase synthesizes the first RNA/DNA primer on the leading strand and at each Okazaki fragment on the lagging strand. Then replication factor C (RF-C) binds to the 3′-OH end of the nascent DNA strand and loads proliferating cell nuclear antigen (PCNA), thereby displacing pol α (3). The displacement of pol α occurs after 30 nt (4), likely by RF-C action (5). Next, pol δ is attracted and this event is called DNA polymerase switch. Beside its role in DNA replication, pol δ has been shown to be involved in DNA repair processes such as long patch base excision repair, nucleotide excision repair, and mismatch repair (reviewed in refs. 6 and 7). Pol δ is a heteromultimeric enzyme composed of one major catalytic subunit of 125 kDa and three small accessory subunits (8); by itself pol δ is a poorly processive enzyme due to its unstable interaction with the DNA substrate, but a physical interaction with the processivity factor PCNA leads to a catalytically competent processive holoenzyme (9). The consequence of such an interaction is a marked increase in the processivity (10). Pol δ has intrinsic DNA polymerization and 3′ → 5′ exonucleolytic activities. In addition, a limited strand displacement activity has been described for the pol δ/PCNA/RF-C holoenzyme on a gapped M13 DNA template (11). Intrinsic strand-displacement activity of pols has been inversely correlated to their tendency to perform template slippage during replication (12). However, in the case of pol δ, it is not known whether the observed strand displacement activity depends on PCNA/RF-C or it is an intrinsic property of the enzyme. The endonuclease flap endonuclease-1 (Fen 1) has been shown to interact with PCNA (13, 14) and has been proposed to work in a coordinated fashion with pol δ, replication protein A (RP-A), and DNA ligase 1 in Okazaki fragment maturation (15). In the current model, RNase H is proposed to cut and degrade the initiator RNA of the primer DNA, leaving a single ribonucleotide at the RNA–DNA junction that is subsequently removed by the 5′ → 3′ exonuclease activity of Fen 1 (16–19). In mammalian cells, pol δ, RP-A, PCNA, RF-C, RNase H, Fen 1, and DNA ligase I are necessary and sufficient to reconstitute lagging strand synthesis in vitro (20). Fen 1 contains a 5′ → 3′ exonuclease activity and a structure-specific endonuclease activity that cleaves the 5′ unannealed single-stranded (ss)-DNA or RNA at the duplex junction (21, 22). In addition, mammalian RNase H can cleave 5′ of the last ribonucleotide of ssRNA–DNA hybrid molecules (23). These findings suggest that Okazaki fragment maturation is likely to require formation of a 5′ tail before the action of Fen 1 and/or RNase H (24). Recently, it has been shown that the helicase/endonuclease Dna2 in yeast cells interacts with Fen 1 and can substitute for RNase H, leading to a novel model for Okazaki fragment processing, involving the sequential action of Fen 1 and Dna2 enzymes for the removal of primer RNA and DNA (15, 22). Interestingly, in this model the action of pol δ is not only critical for the extension of the newly synthesized Okazaki fragment, but also for the displacement of the RNA segment of the preexisting downstream Okazaki fragment, thus creating an intermediate flap structure that is the target for the subsequent concerted action of Dna2 and Fen 1. This process could also have the advantage of removing entirely the RNA–DNA hybrid fragment synthesized by pol α/primase, potentially containing nucleotide misincorporations due to the lack of a proofreading exonuclease activity of pol α/primase, and substituting it with a more accurate copy synthesized by pol δ.
In this paper we have measured the strand displacement by pol δ coupled to DNA synthesis, using a single-stranded minicircular template with an annealed primer. We found that pol δ can synthesize through a double-stranded DNA. PCNA and RP-A can control the strand displacement activity of pol δ by either enhancing the processivity (PCNA) or by limiting the size of the displaced fragment down to 20–30 nt (RP-A). In addition, Fen 1 can exactly process the displaced fragment in the presence of RP-A into a ligatable product. Based on these results, we propose a model in which the concerted action of RP-A, PCNA, and Fen 1 can modulate the strand displacement activity of pol δ.
Materials and Methods
Chemicals.
[α-32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq) and [γ-32P]ATP (3,000 Ci/mmol) were from Amersham Pharmacia. Unlabeled dNTPs and poly(dA)/oligo(dT)10:1 were from Amersham Pharmacia. All other reagents were of analytical grade and purchased from Merck or Fluka. T4 DNA kinase and T4 DNA ligase were from New England Biolabs
Construction of the Minicircle Template-Primer.
The sequence of the 72-mer template was: 5′-CTTCTAGTTGTGAATTCGGCACTGGCCGTCGTATGCTCTTGGTTGTAGGATCCCAGCACATTGAAGGATGCA-3′. Bold letters indicated the sequence annealed to the 17-mer primer. The 72-mer primer was converted into a single-stranded minicircle as described by using a 39-mer scaffold oligonucleotide (5′-CAGTGCCGAATTCACAACTAGAAGTGCATCCTTCAATGT-3′) complementary to the first 5′-end 24 bases and the last 3′-end 15 bases of the 72-mer primer (25). The 72-mer primer was first phosphorylated by using T4 polynucleotide kinase followed by annealing with the 39-mer bridging oligonucleotide that brought together the ends of the 72-mer primer for subsequent ligation using T4 DNA ligase. The 72-mer primer minicircle was then purified from a 10% polyacrylamide/7 M urea gel. After elution from the gel and ethanol precipitation, its concentration was determined spectrophotometrically. The annealing mixture, in a final volume of 10 μl, contained 20 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl, 0.5 pmol of minicircle, and 0.8 pmol of linear 17-mer DNA primer [5′-d(CATACGACGGCCAGTGC)-3′] or the RNA/DNA 27-mer hybrid [5′-r(CAACCAAGAG)d(CATACGACGGCCAGTGC)-3′]. The mixture was heated to 98°C for 3 min and then left to slowly cool down to room temperature. When indicated, the 17-mer primer was 5′-end-labeled before annealing with [γ-32P]ATP and polynucleotide kinase, according to the manufacturer's protocol (New England Biolabs).
Enzymes and Proteins.
Calf thymus pol δ and pol α were purified as described (26). The pol δ used in this study was 2,200 units/ml (0.08 mg/ml/27,500 units/mg), and pol α was 580 units/ml. 1 unit of pol activity corresponds to the incorporation of 1 nmol of total dTMP into acid-precipitable material for 60 min at 37°C in a standard assay containing 0.5 μg (nucleotides) of poly(dA)/oligo(dT)10:1 and 20 μM dTTP. Recombinant human wt PCNA was prepared as described (27). Recombinant human RP-A and Fen 1 were isolated as described (28, 29). Mutants Fen 1 D86A and Fen 1 ΔC (lacking amino acid 360–380), were prepared as described (29). Human RF-C was purified from HeLa cells nuclei according to ref. 30.
Enzymatic Assays.
Pol δ activity was assayed on poly(dA)/oligo(dT) as described (30, 31). For studies with the singly primed minicircular d17:d72 oligodeoxynucleotide as template, a final volume of 10 μl contained 50 mM Tris⋅HCl (pH 7.6), 0.25 mg/ml BSA, 1 mM DTT, 6 mM MgCl2, 12 nM (3′-OH ends) of primed minicircular DNA template, 1 μM [α-32P]dCTP (3,000 Ci/mmol), and 50 μM unlabeled dATP, dGTP, and dTTP. When the minicircular DNA template was used with a 5′-32P-labeled d17 primer, the reaction conditions were the same as above, except that 50 μM of all four unlabeled dNTPs were added. For single-turnover experiments, 600 nM (3′-OH ends) of poly(dA)/oligo(dT)10:1 was added as the trapping agent. Enzymes and proteins were added as indicated in the figure legends. All reactions were incubated for 15 min at 37°C, unless otherwise stated in the figure legends, and samples were mixed with denaturing gel loading buffer (95% vol/vol Formamide/10 mM EDTA/0.25 mg/ml bromophenol blue/0.25 mg/ml xylene cyanol), heated at 95°C for 5 min, and then subjected to electrophoresis on a 7 M Urea/14% polyacrylamide gel. Quantification of the reaction products on the gel was performed by using a Molecular Dynamics PhosphoImager and IMAGE QUANT software.
Kinetic Parameters Calculations.
The kburst (kb) and ksteady-state (kss) values for time-dependent nucleotide incorporation by pol δ on the minicircular template were determined according to the exponential equation:
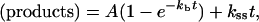 |
where A is the burst amplitude and t is time.
Results
Pol δ Can Displace a Newly Synthesized Strand on a Minicircle Template.
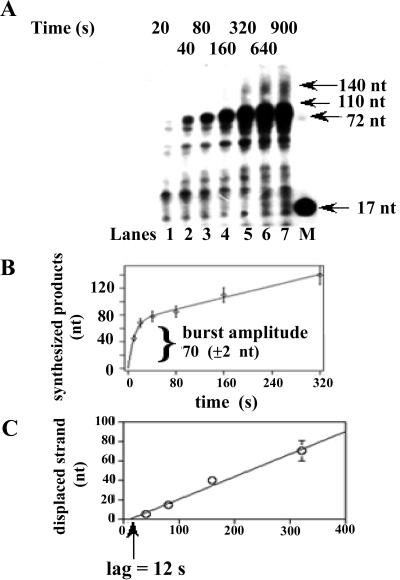
On the 72-mer circularized oligonucleotide template with a 17-mer complementary primer, full-length synthesis by pol δ generates a 72-nt product, whereas the appearance of longer synthesized DNA molecules is diagnostic of continuous elongation through the double-stranded region coupled to strand displacement of the newly synthesized strand (Fig. 1). In Fig. 2A the time-dependent elongation of the 17-mer primer is shown. Full-length products accumulated within the first 20-40 s of the reaction, followed by longer products up to 140 nt, corresponding to two complete consecutive rounds of replication. Incubation times up to 20 min did not reveal the appearance of longer products (data not shown). The time course of nucleotide incorporation was biphasic (Fig. 2B), with a faster rate of 3.8 nt × s−1, followed by a slower rate of 0.25 nt × s−1. The transition point between the two modes of synthesis (i.e., normal primer elongation and strand-displacement coupled synthesis) was estimated from the burst amplitude of the biphasic curve and was 70 (±2) nt. As shown in Fig. 2C, the length of the displaced strand increased with time at a rate of 0.22 nt × s−1. The plot was linear but the intersection point of the line representing the best interpolation of the data with the time axis was consistent with a lag of 12 s before strand displacement started. This likely represents the maximal rate of synthesis of pol δ during primer elongation across the ssDNA region. The number of incorporation events before the beginning of strand displacement was 70 (burst amplitude) − 17 (primer length), that is 53. The ratio between the latter value and the lag value (12 s) gave again an estimated rate of primer elongation of 4.1 nt × s−1, in good agreement with the value calculated from the linear part of the curve shown in Fig. 2B. Taken together, these results suggested that the faster rate corresponded to synthesis across ssDNA, whereas the rate of DNA synthesis through double-stranded (ds)-DNA was limited by the rate of strand displacement. No strand displacement activity was observed under these conditions with pol δ in the absence of either MgCl2 or dNTPs (data not shown). Finally, as expected, pol α produced full-length products but was inactive in strand displacement (data not shown).
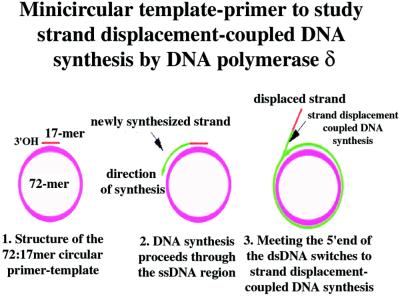
Figure 1.
Minicircular template-primer to study strand displacement-coupled DNA synthesis by pol δ. See text for details.
Figure 2.
Pol δ shows strand displacement activity on the 72:17-mer minicircular template-primer. (A) Pol δ (0.1 unit), was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer under the conditions described in Materials and Methods. Reactions were stopped at the indicated times and the products resolved on a 14% PAA/7 M Urea gel. Lane M: 5′-labeled linear 70-mer and 17-mer were loaded as markers. (B) Sizes of the synthesized products in nt were calculated from their relative electrophoretic mobility and plotted versus time. Data points were fitted to a mixed exponential equation by computer simulation. (C) Length (in nt) of the displaced strand was calculated from the size of the synthesized products and plotted versus time. The solid line represents the best linear interpolation of the data points.
RP-A Limits the Size of the DNA Strand Displaced by pol δ.
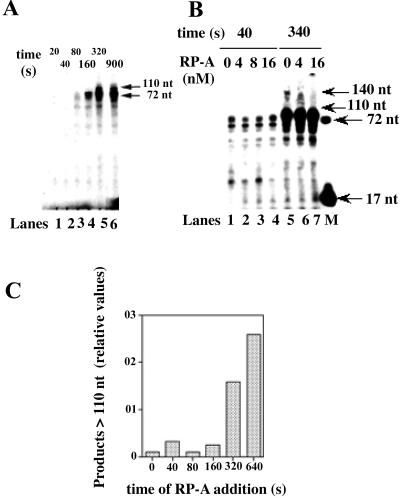
When tested on the strand displacement activity of pol δ, RP-A not only reduced the overall rate of DNA synthesis but also limited the size of the displaced fragments to ≈30 nt, as visualized by the accumulation of products of about 100 nt (Fig. 3A, lanes 5 and 6). Again, the time-dependent course of DNA synthesis was biphasic, with a faster phase of 1.1 nt × s−1, corresponding to the synthesis of the first 72 nt (burst amplitude of 69 ± 5 nt) and a slower phase of 0.068 nt × s−1. The rate of strand displacement was 0.07 nt × s−1, further suggesting that the rate of synthesis through the dsDNA was limited by the rate of strand displacement. Titration of increasing amounts of RP-A in the reaction showed that the accumulation of the 100-nt-long product and the concomitant reduction of a 140-nt-long product within the first 340 s of incubation was RP-A-dependent (Fig. 3B, lanes 5–7). The same concentrations of RP-A, however, did not affect the synthesis of the 72-nt product synthesized in the first 40 s of the reaction (Fig. 3B, lanes 1–4). Interestingly, the concentration of RP-A that gave the maximal effect (16 nM) was nearly equimolar to the concentration of the template molecules (12 nM). As shown in Fig. 2A, reaction products longer than 100 nucleotides in the absence of RP-A started to accumulate after 320 s of incubation. Fig. 3C shows the quantitative analysis by scanning densitometry of the reaction products of an experiment, in which a fixed amount of RP-A was added at different times during the reaction. The relative amounts of the products longer than 100 nt have been plotted in dependence of the time of addition of RP-A. The results shown in Fig. 3C clearly indicated that RP-A limited the size of the displaced fragment only if added within the first 320 s of incubation; that is, before the strand displaced by pol δ exceeded a critical length of about 30 nt. When tested under our assay conditions, RP-A alone showed no displacement of the primer (data not shown). Moreover, when tested in combination with Escherichia coli pol I (Klenow fragment), an enzyme possessing strong strand displacement activity, RP-A did not show any effect on the size of the displaced products, indicating that the observed effect with pol δ was specific (data not shown).
Figure 3.
RP-A limits the size of the DNA strand displaced by pol δ. (A) Pol δ (0.1 unit), was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer and 16 nM RP-A (as heterotrimer), under the conditions described in Materials and Methods. Reactions were stopped at the indicated time points and the products were resolved on a 14% PAA/7 M Urea gel. Lane M: 5′-labeled linear 70-mer and 17-mer were loaded as markers. (B) Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer and increasing amounts of RP-A under the conditions described in Materials and Methods. Parallel reactions were stopped after 40 s (lanes 1–4) or 340 s (lanes 5–7) and the products were resolved on a 14% PAA/7 M Urea gel. Lane M: 5′-labeled linear 70-mer and 17-mer were loaded as markers. (C) Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer under the conditions described in Materials and Methods. Sixteen nanomoles of nM RP-A were added either at the beginning of the reaction or at the indicated times. Reactions were stopped after 5 min and the products were resolved on a 14% PAA/7 M Urea gel. Densitometric analysis of the bands corresponding to the products longer than 90 nt allowed the calculation of their relative amounts. Band intensities (I) were corrected for background and normalized to the total through the equation: relative I = (In/(In-1 + . . . + In-i))/Σ(Ii)i = 1… n, where In is the intensity at the position of interest (n), (In-1 + . . . + In-i) is the sum of the intensities of all bands at positions below n, and Σ(Ii)i = 1… n is the sum of the intensities of all of the bands in the lane. These values were then plotted in dependence of the times of addition of RP-A.
Strand Displacement Activity by pol δ Is Highly Distributive.
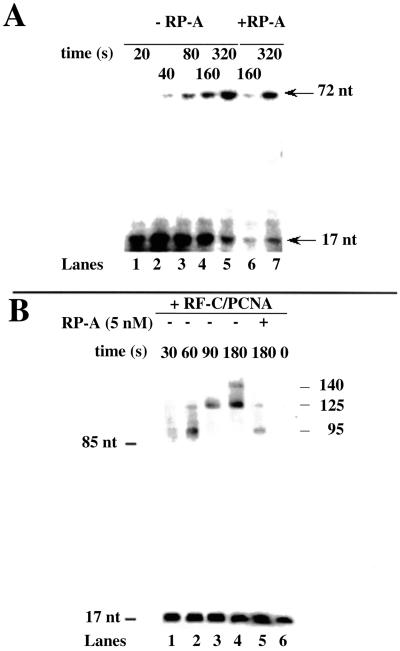
To obtain some insights on the mechanism by which pol δ displaces a newly synthesized strand, the kinetic of DNA synthesis by pol δ on the minicircle template was followed under single turnover conditions, in the presence of a 5′-32P-labeled 17-mer primer annealed to the 72-mer minicircle. Preformed pol δ/template–primer complexes were incubated in the presence of a large excess of trapping template poly(dA)/oligo(dT). Under these conditions, only the products synthesized during a single binding event by pol δ can be visualized. A shown in Fig. 4A, pol δ alone could processively synthesize a full length 72-nt product, but no strand displacement synthesis was detected (lanes 1–5). When a similar experiment was performed in the presence of RP-A, no strand displacement by pol δ could be seen either (Fig. 4A, lanes 6 and 7). These results suggested that the strand displacement activity of pol δ previously observed was due to its distributive mode of action and was strongly dependent on multiple dissociation-reassociation events.
Figure 4.
PCNA increases the processivity of strand displacement-coupled DNA synthesis by pol δ. (A) Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72–5′-32P-labeled 17-mer template-primer under the conditions described in Materials and Methods for single turnover synthesis. Reactions were stopped at the indicated time points and the products were resolved on a 14% PAA/7 M Urea gel. Linear 5′-labeled 70- and 17-mer markers migrated at the positions indicated by arrows. (B) Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer in the absence or presence of RP-A (16 nM), RF-C (0.02 unit), and PCNA (1 μM as trimer), under the conditions described in Materials and Methods. Reactions were stopped at the indicated times and the products were resolved on a 14% PAA/7 M Urea gel. Arrows indicate the position of 85- and 17-mer markers, as well as the length of the different products synthesized.
PCNA Stimulates the Processivity of the Strand Displacement Activity by pol δ.
Next, RF-C and PCNA were included in the reaction. The sp minicircle was indeed a poor substrate for the RF-C-dependent loading of PCNA, thus requiring the utilization of high concentrations of both RF-C and PCNA, which were, however, not saturating, as determined by titration experiments (data not shown). It is well known that, in the presence of saturating amount of PCNA, pol δ becomes highly processive, being able to synthesize the 55-nt strand complementary to ssDNA region of the template without dissociating (11). As shown above (Figs. 2A and 4A), the strand displacement activity of pol δ was found to be highly distributive; thus, it might be anticipated that reduction of pol δ dissociation from the template would result in limited strand displacement. Indeed, as shown in Fig. 4B, time-dependent course of pol δ catalyzed reaction on the sp 17:72-mer primer/template in the presence of PCNA/RF-C showed a more processive synthesis (as indicated by the absence of intermediate pausing sites) and faster kinetics of strand displacement (compare with Fig. 2A). For this experiment, a 5′-32P-labeled 17-mer primer was used in place of labeled nucleotides, to more clearly visualize the reaction products. The rate of strand displacement DNA synthesis was calculated to be 0.5 nt × s−1, 2.5-fold higher than in the absence of RF-C/PCNA, and the size of the longest products was about 140 nt, consistent with two consecutive rounds of replication (see also Fig. 2A). However, the pol δ holoenzyme was still responsive to RP-A, because its addition limited the size of the displaced products down to about 30 nt, even after prolonged incubation (Fig. 4B, lane 5). Incubation up to 1,800 sec did not produce longer products in the presence of RP-A (data not shown).
DNA Displaced by pol δ Can Be Cleaved by Fen 1 in Dependence of RP-A and Ligated by DNA Ligase.
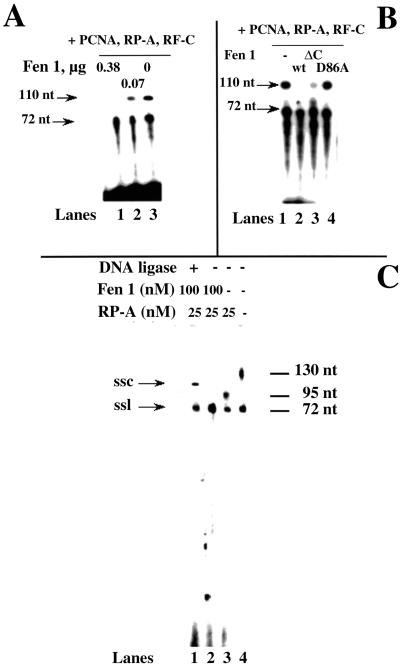
Finally, we addressed whether Fen 1 could process the displaced fragment by pol δ. As shown in Fig. 5A, incorporation of radioactively labeled nucleotides by pol δ on the sp 17:72-mer primer template, was followed in the presence of RP-A (12 nM), PCNA, RF-C, and increasing amounts of Fen 1. Under these conditions, pol δ was able to displace up to 30–35 nt, as indicated by the accumulation of products no longer than 110 nt (Fig. 5A, lane 3). Addition of Fen 1 caused the decrease of the 110-nt band, whereas the product corresponding to the full-length synthesis (72 nt) was unaffected (Fig. 5A, lanes 1 and 2). In another experiment (Fig. 5B), Fen 1 wild type was compared with two mutants, one carrying the D86A mutation, which targets one of the essential catalytic acidic residues, and the other carrying a deletion of the C-terminal DNA binding domain (29). As expected, the D86A mutant was unable to cleave the ssDNA flap, leaving the 110-nt product intact (Fig. 5B, lane 4). On the other hand, the DNA-binding mutant was over 70% active compared with the wild type (Fig. 5B, lane 3; as derived by quantification of the remaining 110-nt product). Our previous studies have shown that this mutant cannot cleave flap DNA structures by itself, because of its inability to bind DNA. However, in the presence of PCNA, which is able to bind Fen 1 and targets it to the DNA flap, its activity is partially restored (29). These results suggested that the ssDNA flap, resulting from the strand displacement activity of pol δ, can be precisely processed by Fen 1 in dependence of PCNA, leaving a newly synthesized full-length 72-nt strand. To reconstitute a more physiological system, we used a 27-mer RNA/DNA hybrid oligonucleotide as the primer, mimicking the initiator DNA of an Okazaki fragment (Fig. 5C). With this hybrid primer we also could detect strand displacement (Fig. 5C, lane 4), size-limiting by RP-A (lane 3), and Fen 1 processing (lane 2). Moreover, after Fen 1 processing, DNA ligase was added (lane 1), resulting in the conversion of the ss linear 72-mer primer into a ss circular DNA with reduced electrophoretic mobility, indicating that the product of Fen 1 processing was ligatable.
Figure 5.
Fen 1 can process the ssDNA flap generated by pol δ strand displacement activity into ligatable products. (A) Dose-dependent processing of the ssDNA displaced flap by Fen 1. Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer, 16 nM RP-A (as heterotrimer), RF-C (0.02 units), PCNA (1 μM as trimer), and in the absence (lane 1) or presence of 0.2 μM (lane 2) or 1 μM (lane 3) of Fen 1, under the conditions described in Materials and Methods. Reactions were stopped after a 10-min incubation and the products were resolved on a 14% PAA/7 M Urea gel. Sizes of the DNA products are indicated at the left. (B) Ability of Fen 1 wild type and mutants to process the ssDNA displaced flap. Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:17-mer template-primer, 16 nM RP-A (as heterotrimer), RF-C (0.02 units), PCNA (1 μM as trimer), and in the absence (lane 1) or presence of 1 μM of Fen 1 wild type (lane 2), Fen 1 ΔC (lane 3), or Fen 1 D86A (lane 4), under the conditions described in Materials and Methods. Reactions were stopped after a 10-min incubation and the products were resolved on a 14% PAA/7 M Urea gel. Size of the DNA products are indicated at the left. (C) Fen 1 generates ligatable products. Pol δ (0.1 unit) was incubated in the presence of 12 nM (3′-OH ends) 72:27-mer template-RNA/DNA hybrid primer, 25 nM RP-A (as heterotrimer), RF-C (0.1 unit), and PCNA (0.2 μM as trimer) and in the absence (lanes 3 and 4) or presence (lanes 1 and 2) of 100 nM sFen 1, under the conditions described in Materials and Methods. Reactions were stopped after a 10-min incubation and the products were resolved on a 14% PAA/7 M Urea gel. Size of the DNA products are indicated at the left. Lane 1: after a 10-min incubation, sample was heat-inactivated for 20 min at 75°C, cooled down at 37°C, and then T4DNA ligase was added along with ligase reaction buffer. After a 30-min incubation at 37°C, sample was stopped and processed as above. ssl, single-stranded linear DNA; ssc, single-stranded circular DNA.
Discussion
Eukaryotic pol α does not have 3′ → 5′ exonuclease proofreading activity, showing a higher misincorporation rate than pol δ and pol ɛ, the other two major replicative enzymes in eukaryotic cells (32). This inherent infidelity of pol α poses a problem to the cell, which apparently takes up the risk of accumulating dangerous mutations during DNA replication. Genetic and biochemical data indicate that a multiprotein machinery can remove the first 30 nt of each Okazaki fragment synthesized by pol α and potentially containing mistakes, by the action of Dna2 helicase and Fen 1 endonuclease. The resulting gap is filled in by the error-free pol δ or pol ɛ together with the accessory proteins RP-A, PCNA, and RF-C and sealed by DNA ligase I (15, 22, 33). The limited strand displacement activity of pol δ (11) might be important in this process. This strand displacement activity of pol δ was tested on the minicircular template that allowed the discrimination between the elongation and the strand displacement-coupled modes of DNA synthesis by pol δ, based on the size of the synthesized products (Fig. 1). We showed that the strand displacement activity was intrinsic to pol δ, because it occurred in the absence of any auxiliary protein and could displace up to 70 nt (Fig. 2A). The DNA synthesis across a dsDNA region imposed a severe block to pol δ, resulting in a reduced incorporation rate (Fig. 2 B and C) and was highly distributive, depending on multiple dissociation/reassociation events of pol δ (Figs. 2A and 4A). PCNA and RF-C increased the processivity of the strand displacement activity of pol δ (Fig. 4B), whereas RP-A was able to limit the size of the displaced fragment down to about 30 nt (Fig. 3A), without affecting the synthesis of the shorter products (Fig. 3B). This block occurred at a concentration of RP-A that was nearly equimolar to the one of the template, but only if RP-A was present before the displaced strand exceeded the critical size of 30 nt (Fig. 3C). Fen 1 was able to cleave the displaced 30-nt fragment in the presence of RP-A, PCNA, and RF-C, rendering a 72-nt full-length product (Fig. 5 A and B). This reaction absolutely required the catalytic activity of Fen 1, because the active-site D86A mutant was unable to process the flap (Fig. 5B). Moreover, the DNA-binding mutant Fen 1 ΔC retained significant flap processing activity in this system (Fig. 5B). This mutant was shown to be impaired in processing a DNA flap structure, but its activity was partially rescued by PCNA (29). In fact, PCNA and Fen 1 ΔC can still physically interact and it is through this interaction that PCNA tethers Fen 1 to the DNA, thus allowing it to cleave the flap. Thus, the results obtained in our system suggested that RP-A plays a critical role in regulating the extent of unwinding of the Okazaki fragment. The block imposed to pol δ by RP-A might be important to prevent uncontrolled displacement synthesis catalyzed by pol δ, which could lead to unnecessary extensive degradation of a preexisting Okazaki fragment and the formation of excessively long ssDNA segments, potentially forming secondary structures that would be resistant to cleavage by Fen 1 (34). This system was shown to work also with an RNA/DNA hybrid oligonucleotide, whose processing by Fen 1 resulted into a ligatable product (Fig. 5C), indicating a potential physiological significance.
A Model for Okazaki Fragment Processing.
Based on the results shown here, we propose a model for the concerted action of pol δ, PCNA, RP-A, and Fen 1 in Okazaki fragment processing. When the growing chain of an Okazaki fragment elongated by pol δ/PCNA meets the 5′ end of the previously synthesized fragment, pol δ can invade the double-stranded region displacing the RNA/DNA fragment synthesized by pol α/primase and replacing it with a faithful DNA copy. PCNA can limit the tendency of pol δ to dissociate on encountering a double-stranded region, thus increasing its processivity. RP-A, on the other hand, acts by ensuring that the displaced fragment does not exceed a critical size of about 30 nt, corresponding to the stretch synthesized by pol α/primase. The size of the displaced fragment corresponded to the length of DNA necessary to bind RP-A (35), suggesting that, as soon as the single-stranded tail generated during strand displacement by pol δ reaches the right size, it can be bound by RP-A. This leads to a “locked” complex and imposes a structural block, avoiding further displacement by pol δ. The model predicts that one flap is generated per molecule of template and that this flap, due to its limited size, is able to bind only one molecule of RP-A. Indeed, maximal block of pol δ strand displacement by RP-A was reached at a concentration nearly equimolar to the one of the template (Fig. 3B). The flap structure, which is now stabilized, is then an ideal substrate for Fen 1, which can cleave it, leaving a nick to be sealed by DNA ligase (Fig. 5C). The fact that Fen 1 and PCNA can physically interact and that this interaction can stimulate Fen 1 activity (13, 36), together with the results with the Fen 1 DNA binding mutant ΔC, suggested that Fen 1 is targeted to the ssDNA flap by its interaction with PCNA. This does not exclude an additional role of Dna2 in this process, which might be able to substitute or assist Fen 1 in the cleavage reaction.
It is intriguing to note that many of the proteins used for this study have evolved a similar DNA substrate specificity: (i) the size of the RNA/DNA primer synthesized by pol α/primase has been shown to be critical for the pol switching process, being the limiting factor for RF-C binding and subsequent displacement of pol α/primase, because RF-C has been shown to preferentially bind to a newly synthesized 30-nt-long primer (4); (ii) RP-A preferentially binds to ssDNA tracts of 10–30 nt (35); and (iii) recent data showed that maximal endonucleolytic efficiency of Fen 1 occurs in the presence of ssDNA flaps from 20–40 nt in length (37). The results presented here suggest that these common DNA binding preferences could be important for yet another critical event in the DNA replication process, namely Okazaki fragment processing.
In agreement with our findings, it has been recently shown that the endonucleases Dna2 and Fen1 act sequentially to facilitate the complete removal of the primer RNA and that the sequential action of these enzymes is governed by RP-A (38).
Acknowledgments
This work was supported by European Union–Training and Mobility of Researchers Programme Grant ERBMRXCT 970125 (to S.S. and U.H.), the Consiglio Nazionale delle Ricerche Target Project on Biotechnology (to S.S.), Grant 4373 from the Association pour la Recherche sur le Cancer (to G.V.), and the Kanton of Zürich (to U.H. and G.M.). I.F. was supported by a fellowship from the European Union–Training and Mobility of Researchers Programme Network (Grant ERBMRXCT 970125).
Abbreviations
- pol
polymerase
- PCNA
proliferating cell nuclear antigen
- RP-A
replication protein A
- Fen 1
flap endonuclease-1
- RF-C
replication factor C
- ss
single-stranded
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hindges R, Hübscher U. Biol Chem. 1997;378:345–362. doi: 10.1515/bchm.1997.378.5.345. [DOI] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 3.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 4.Mossi R, Keller R C, Ferrari E, Hübscher U. J Mol Biol. 2000;295:803–814. doi: 10.1006/jmbi.1999.3395. [DOI] [PubMed] [Google Scholar]
- 5.Maga G, Stucki M, Spadari S, Hübscher U. J Mol Biol. 2000;295:791–801. doi: 10.1006/jmbi.1999.3394. [DOI] [PubMed] [Google Scholar]
- 6.Burgers P M J. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 7.Stucki M, Stagliar I, Jonsson Z O, Hübscher U. Prog Nucleic Acid Res Mol Biol. 2000;65:261–298. doi: 10.1016/s0079-6603(00)65007-9. [DOI] [PubMed] [Google Scholar]
- 8.Hübscher U, Nasheuer H-P, Syväoja J. Trends Biochem Sci. 2000;25:143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 9.Mozzherin D J, Tan C-K, Downey K M, Fisher P A. J Biol Chem. 1999;274:19862–19867. doi: 10.1074/jbc.274.28.19862. [DOI] [PubMed] [Google Scholar]
- 10.Einolf H J, Guengerich F P. J Biol Chem. 2000;275:16316–16322. doi: 10.1074/jbc.M001291200. [DOI] [PubMed] [Google Scholar]
- 11.Podust V N, Podust L M, Muller F, Hübscher U. Biochemistry. 1995;34:5003–5010. doi: 10.1021/bi00015a011. [DOI] [PubMed] [Google Scholar]
- 12.Canceill D, Viguera E, Ehrlich S D. J Biol Chem. 1999;274:27481–27490. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Li J, Harrington J, Lieber M R, Burgers P M. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 14.Gomes X V, Burgers P M. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H Y, Choi E, Bae S H, Lee K H, Gim B S, Kim H D, Park C, MacNeill S A, Seo Y S. Genetics. 2000;155:1055–1067. doi: 10.1093/genetics/155.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Rumbaugh J A, Murante R S, Lin R J, Rust L, Bambara R A. Biochemistry. 1996;35:9266–9277. doi: 10.1021/bi9603074. [DOI] [PubMed] [Google Scholar]
- 17.Murante R S, Rumbaugh J A, Barnes C J, Norton J R, Bambara R A. J Biol Chem. 1996;271:25888–25897. doi: 10.1074/jbc.271.42.25888. [DOI] [PubMed] [Google Scholar]
- 18.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 19.Rumbaugh J A, Murante R S, Shi S, Bambara R A. J Biol Chem. 1997;272:22591–22599. doi: 10.1074/jbc.272.36.22591. [DOI] [PubMed] [Google Scholar]
- 20.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 21.Zhu F X, Biswas E E, Biswas S B. Biochemistry. 1997;36:5947–5954. doi: 10.1021/bi962889v. [DOI] [PubMed] [Google Scholar]
- 22.Bae S H, Seo Y S. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 23.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murante R S, Henricksen L A, Bambara R A. Proc Natl Acad Sci USA. 1998;95:2244–2249. doi: 10.1073/pnas.95.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Chastain P D, Jr, Kusakabe T, Griffith J D, Richardson C C. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 26.Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hübscher U. J Biol Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 27.Jonsson Z O, Hindges R, Hübscher U. EMBO J. 1998;17:2412–24125. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 29.Stucki M, Jonsson Z O, Hübscher U. J Biol Chem. 2001;276:7843–7849. doi: 10.1074/jbc.M008829200. [DOI] [PubMed] [Google Scholar]
- 30.Hübscher U, Mossi R, Ferrari E, Stucki M, Jonson Z O. In: Functional Analysis of DNA Replication Accessory Proteins. Cotterill S, editor. Oxford: Oxford Univ. Press; 1999. pp. 119–137. [Google Scholar]
- 31.Hübscher U, Kornberg A. Proc Natl Acad Sci USA. 1979;76:6284–6288. doi: 10.1073/pnas.76.12.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 33.Rumbaugh J A, Henricksen L A, DeMott M S, Bambara R A. J Biol Chem. 1999;274:14602–14608. doi: 10.1074/jbc.274.21.14602. [DOI] [PubMed] [Google Scholar]
- 34.Henricksen L A, Tom S, Liu Y, Bambara R A. J Biol Chem. 2000;275:16420–16427. doi: 10.1074/jbc.M909635199. [DOI] [PubMed] [Google Scholar]
- 35.Blackwell L J, Borowiec J A. Mol Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tom S, Henricksen L A, Bambara R A. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 37.Negritto M C, Qiu J, Ratay D O, Shen B, Bailis A M. Mol Cell Biol. 2001;21:2349–2358. doi: 10.1128/MCB.21.7.2349-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae S H, Bae K H, Kim J A, Seo Y S. Nature (London) 2001;26:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]