Abstract
Mayaro virus (MAYV; Togaviridae; Alphavirus) has drawn increasing attention as an arthropod-borne virus with potential to cause outbreaks among the human populations of the Western Hemisphere. In the tropical regions of Central and South America, the virus exists in sylvatic cycles between mosquitoes and primate reservoirs such as marmosets. Although forest-dwelling mosquitoes are regarded as important vectors for MAYV, it has been shown previously that the virus can infect and potentially be transmitted by the mosquitoes, Aedes aegypti and Aedes albopictus (Diptera: Culicidae). Here, we compare the infection and transmission efficiencies of two MAYV strains, IQT 4235 from Iquitos, Peru (‘IQT’) and the type strain of MAYV from Trinidad, TRVL 4675 (‘TRVL’) in two laboratory-adapted Ae. aegypti strains, Higgs White Eye and Orlando. The TRVL strain was less efficiently transmitted by both mosquito strains than MAYV IQT. Based on the full-length nucleotide sequences of the two viral genomes, we show that the TRVL prototype strain of MAYV is phylogenetically ancestral and more distantly related to the IQT strain. The TRVL strain efficiently infected wild-type Ae. albopictus from Missouri and readily disseminated in those. Considering scenarios in which natural MAYV transmission cycles may overlap with those of chikungunya virus (CHIKV; Togaviridae; Alphavirus), we assessed the effects of mixed infections of the two viruses in mosquitoes based on coinfection or superinfection. Although coinfection had no measurable effect on the transmission potential of either virus, we observed superinfection exclusion for CHIKV in MAYV-infected mosquitoes but not for MAYV in CHIKV-infected mosquitoes.
Keywords: Mayaro virus, chikungunya virus, mosquito, saliva, superinfection exclusion
Mayaro virus (MAYV) is an alphavirus (Togaviridae; Alphavirus) of the Semliki Forest virus antigenic complex, which was originally isolated in 1954 from the serum of a forest worker living in Mayaro County of Trinidad Island (Anderson et al. 1957, Casals and Whitman 1957, Causey and Maroja 1957, Aitken et al. 1960). The virus isolate was originally designated Tr4675 and is now referred to as the MAYV prototype strain, TRVL 4675 (Casals and Whitman 1957, Powers et al. 2006). Like other medically important alphaviruses, MAYV is mosquito-borne. Forest- and aquatic plant-dwelling mosquitoes such as Mansonia venezuelensis (Diptera: Culicidae), Haemagogus janthinomys, Sabethes spp., and Culex spp. have been implicated as potential sylvan vectors (Aitken et al. 1960, Izurieta et al. 2011). New World primates of the families Cebidae and Callithricidae, the latter of which include marmosets, are considered to be important reservoirs for the virus (Hoch et al. 1981, Izurieta et al. 2011). MAYV has also been isolated from a migrating bird (Calisher et al. 1974). Furthermore, several equids in the Pantanal region of Brazil as well as anteaters, armadillos, opossums, and rodents in French Guiana have been found seropositive for the virus (de Thoisy et al. 2003, Pauvolid-Corrêa et al. 2015). MAYV represents an interesting phenomenon as it is, based on its geographic origin, considered to be a New World alphavirus with biological characteristics typical for Old World alphaviruses. Such characteristics include primates as animal reservoirs and a nonneurotropic tropism of the virus in the human host. MAYV infections have been known for decades among people living in tropical and rural areas of South and Central America predominantly in the vicinity of rain forest areas. The virus causes disease symptoms in humans similar to chikungunya virus (CHIKV, Togaviridae; Alphavirus) including febrile illness, rash, and (occasionally long-lasting) arthralgia (Tesh et al. 1999).
Like other alphaviruses, MAYV has a ~12 kb single-stranded positive sense RNA genome, with nine genes encoding four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) and five structural proteins (CP, E3, E2, 6k/TF, and E1; Powers et al. 2006, Mavian et al. 2017). MAYV strains typically exhibit a relatively narrow genetic divergence of up to 17% at the nucleotide level clustering them into three distinct genotypes named L (limited), N (new), and D (widely dispersed). Genotype L is restricted to isolates from (Belterra) Brazil, N to an isolate from Peru, and D containing isolates of wide geographical distribution (Powers et al. 2006, Auguste et al. 2015). In 2015, a MAYV case was reported in Haiti, which is the first time in the Antilles (Lednicky et al. 2016). This virus isolate represented a recombinant between a D and an L genotype virus (Mavian et al. 2017). Authors of these studies also speculated whether MAYV could have the potential to become the next mosquito-borne virus, following CHIKV in 2013 and Zika virus (ZIKV; Flaviviridae; Flavivirus) in 2015, to cause large-scale disease outbreaks among human populations of the Americas and the Caribbean (Auguste et al. 2015, Lednicky et al. 2016, Mavian et al. 2017). A critical aspect facilitating a switch from a sylvatic to an urban arboviral disease cycle is the abundant availability of urban vectors, which would be highly susceptible to the sylvan arbovirus. Aedes aegypti and Aedes albopictus (Diptera: Culicidae) are two mosquito species that are typically involved in urban arboviral transmission cycles of dengue virus type 1–4 (DENV1-4; Flaviviridae; Flavivirus), ZIKV, and CHIKV. Previously, an Ae. albopictus population from Brazil has been shown to be susceptible to MAYV TRVL 4675, albeit at a relatively low infection rate (Smith and Francy 1991). It was suggested that these mosquitoes still had the potential to act as secondary or bridging vectors expanding typical transmission cycles of MAYV. Furthermore, an Ae. aegypti population from Peru has been shown to be susceptible to MAYV IQT 4235, a strain isolated in 1997 from a febrile patient in the Loreto region of Peru (Long et al. 2011). A recent publication describes the infection and transmission potentials of Ae. albopictus and Ae. aegypti strains from Miami and Jacksonville, FL for MAYV TRVL 4675 (Wiggins et al. 2018). In their study, Ae. albopictus had a significantly higher rate of susceptibility to orally acquired MAYV and produced a significantly higher number of saliva samples containing the virus than Ae. aegypti. More recently, we compared the midgut dissemination efficiencies between CHIKV 37997 and MAYV TRVL 4675 in Ae. aegypti based on the detection of plus-strand and minus-strand viral RNAs, showing that both viruses disseminated from the mosquito midgut at similar time points (Kantor et al. 2018).
In this study, we compare the vector competence of two laboratory-adapted Ae. aegypti strains, Higgs White Eye (‘HWE’) and Orlando (‘ORL’), for the two genotype D MAYV strains TRVL 4675 (‘TRVL’) and IQT 4235 (‘IQT’). We also tested the vector competence of a locally caught Ae. albopictus population for MAYV, confirming that geographically distinct mosquito populations can be highly susceptible for exotic arboviruses. So far, only partial genome sequences have been available for both MAYV strains, TRVL and IQT. We present the full-length genomes of both virus strains and compare their phylogenetic relationships with other full-length MAYV genomes. Current outbreaks of CHIKV in South and Central America increase the likelihood that MAYV and CHIKV may be able to cocirculate in common regions. In line with such a scenario, incidents of coinfections in patients involving CHIKV and flaviviruses such as DENV1-4 or ZIKV have been recently reported from the Americas and the Caribbean (Dupont-Rouzeyrol et al. 2015, Furuya-Kanamori et al. 2016, Villamil-Gomez et al. 2016, Waggoner et al. 2016, Zambrano et al. 2016). Thus, we decided to investigate how mixed infections between MAYV and CHIKV, when acquired by mosquitoes as coinfections or superinfections, would affect their ability to cotransmit both viruses either singly or together.
Materials and Methods
Mosquitoes
Aedes aegypti mosquitoes of the Orlando (‘ORL’; Travis et al. 1946) and Higgs White Eye (‘HWE’; Wendell et al. 2000) strains were reared and maintained in a BSL2 insectary at 28°C, 75–80% relative humidity, and a 12:12 (L:D) h cycle (Dong et al. 2016). For colony maintenance, mosquitoes received artificial bloodmeals consisting of defibrinated sheep blood (Colorado Serum Company, Denver, CO). Raisins were provided to the adult mosquitoes as a sugar source. For each MAYV challenge experiment, mated 1-wk old Ae. aegypti females were placed in 1-liter modified ice cream cartons (100 females/carton) each covered with a netting and moved to the BSL3 laboratory.
The Ae. albopictus colony (‘CoMO’) was newly established in 2016 from a locally (Columbia, MO) captured single female, which was infected with Wolbachia (data not shown). Aedes albopictus CoMO were hatched in distilled water following a deoxygenation step to generate higher hatch rates. Briefly, a 1-liter mason jar was filled with 750-ml distilled water and autoclaved. Immediately after removal from the autoclave, the lid was sealed tight and allowed to cool to establish a vacuum and deoxygenate the water. Egg papers were placed in the water immediately after the vacuum was broken and allowed to hatch for 3 h. Aedes albopictus mosquitoes were reared and maintained via sibling mating of offspring and F8 females were used for the MAYV challenge experiments.
Viruses
MAYV strains IQT 4235 from Loreto, Peru and the prototype strain TRVL 4675 from Trinidad (full-length sequences submitted to GenBank: MK070491 and MK070492, respectively) were used in this study. The IQT 4235 strain was isolated in 1997 from a human and has been passaged twice in C6/36 (Ae. albopictus) cells and once in Vero (monkey kidney) cells before being used in this study (Powers et al. 2006, Long et al. 2011). We prepared virus stocks by passaging the virus twice in Vero cells. At 30 and 39 h postinfection, titers in Vero cells reached 4.6 × 106 plaque forming unites (PFU)/ml and 3.3 × 106 PFU/ml, respectively. The TRVL 4675 prototype strain was isolated in 1954 and has been passaged once in suckling mice and once in Vero cells before use in this study (Powers et al. 2006). Virus stocks were prepared via passage in Vero cells in which titers reached 6.0 × 106 PFU/ml at 30 h postinfection. For our experiments, we used IQT and TRVL stocks that had been collected at 30 h postinfection.
The 37997 strain of CHIKV (GenBank: AY726732.1) was isolated in 1983 in Senegal from Aedes furcifer (Diptera: Culicidae) and belongs to the West African genotype. Prior to use in our studies, the virus has been passed once in Aedes pseudoscutellaris (Diptera: Culicidae) AP-61 cells and twice in Vero cells (Vanlandingham et al. 2005). CHIKV stocks were generated in Vero cells with titers ranging from 2.0 × 106 to 3.3 × 106 PFU/ml.
MAYV and CHIKV Infections of Mosquitoes Via Artificial Bloodmeals and Intrathoracic Injections
Prior to mosquito bloodfeeding, ~90% confluent Vero cells, which had been seeded in a T25 flask in presence of Dulbecco’s Modified Eagle Medium (DMEM) complemented with 7% FBS, were infected with MAYV or CHIKV stock at a multiplicity of infection (m.o.i.) of 0.01. Virus-infected cells were incubated for 30–36 h postinfection until 60–70% of the cells showed severe cytopathic effects (CPE) and started to float. Cell culture media was then collected and mixed with an equal amount of defibrinated sheep blood including 10 mM ATP. One-week-old Ae. aegypti or Ae. albopictus females that had been deprived of sugar (raisins) for 24 h were fed for 1 h on this mixture at 37°C using a single, water-jacketed glass feeder per mosquito carton. After feeding, fully engorged females were selected and maintained by providing raisins and water until further analysis.
In the MAYV-CHIKV coinfection experiment, HWE females were exposed to bloodmeals that contained either MAYV-IQT (titer: 3.3 × 106 PFU/ml), CHIKV 37997 (titer: 2.7 × 106 PFU/ml), or both viruses in combination. In the superinfection experiment, a group of HWE mosquitoes received an initial bloodmeal containing either MAYV-IQT or CHIKV 37997 followed by a subsequent bloodmeal at 6 d postinfectious bloodmeal (pibm) containing the virus (either MAYV or CHIKV), which had not been initially acquired. Controls consisted of mosquito groups that were initially fed with bloodmeals containing either MAYV or CHIKV followed by noninfectious bloodmeals (noninfected cell culture supernatant mixed with defibrinated sheep blood at a 1:1 ratio) at 6 d pibm.
To infect Ae. albopictus CoMO females with MAYV TRVL via injections, 210 nl (= 700 PFU/mosquito) of MAYV TRVL was intrathoracically injected into each 1-wk-old female using the Nanoject II injection system (Drummond Scientific, Broomall, PA). Saliva samples of these females were collected at 3 d postintrathoracic injection and processed as described below.
Saliva Collection From Individual Mosquitoes
Saliva was collected from individual MAYV-challenged Ae. aegypti mosquitoes at 2, 4, and 7 d pibm using a forced salivation method (Dong et al. 2016). The proboscis of each female whose wings and legs had been clipped off was inserted into a 1-mm glass capillary tube filled with 3–5 µl Cargille Type B immersion oil (Cargille Laboratories, Cedar Groove, NJ). After 40 min, saliva was collected from capillaries in which droplets of saliva exuding from the proboscis were visible. Saliva samples were recovered from each capillary tube by centrifugation at 3,000 × g for 5 min in a 1.5-ml microfuge tube containing 200 µl of sample processing buffer. After addition of another 200 µl of sample processing buffer, each sample was vortexed and filtered using a 0.2-µm Acrodisc HT Tuffryn syringe filter (Pall Life Sciences, East Hills, NY). Virus from saliva samples was amplified in Vero cells seeded onto 24-well plates at ~90% confluency. Cell growth medium was removed before cells were inoculated with 180 µl of saliva sample for 1 h at 37°C. Thereafter, 1 ml of DMEM supplemented with 7% FBS was added to each well. Cells were observed daily for the development of CPE until 7 d pibm.
In the MAYV-CHIKV co- and superinfection experiments, saliva samples of mosquitoes of both experimental groups were collected at 12 d pibm as described above except that 50 μl of saliva samples from five individuals were pooled together and then diluted in 750-μl TRIzol LS (Thermo Fisher, Waltham, MA) for total RNA extraction according to the manufacturer’s instructions.
Virus Detection in Individual Mosquito Tissues by Plaque Assays
At various time points postoral challenge, midguts and carcasses (or head tissues) of mosquitoes were dissected and stored individually at −80°C until further testing. After addition of processing buffer (0.5–1 ml DMEM, 7% FBS, 1% HEPES), each individual midgut and carcass was homogenized with a pestle prior to sterile filtering using 0.2-μm Acrodisc HT Tuffryn syringe filters (Pall Life Sciences). Virus titers from individual midguts and carcasses were determined by plaque assays in Vero cells as described previously (Dong et al. 2016). Following a 3-d incubation period of the plaque assays at 37°C (under 5% CO2 supplement), 150 μl/well of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Sigma–Aldrich, St. Louis, MO) was added to each 24-well plate as substrate. After 24 h MTT incubation, plaques were counted and recorded as PFU/ml. All MAYV infection and detection assays were carried out in a Biosafety Level 3 laboratory within the Laboratory for Infectious Disease Research (LIDR) of the University of Missouri.
Quantitative (q)RT–PCR for the Virus-Specific Detection in Samples Containing Mixed Virus Infections
cDNA synthesis was performed with 500 ng of extracted total RNA using the Superscript III First-Strand Synthesis System (Invitrogen, Waltham, MA) and CHIKV- or MAYV-specific primers (Table 1). The multiplex quantitative polymerase chain reaction (qPCR) reaction was performed using the iTaq Universal Probes Supermix (Bio-Rad, Hercules, CA) with a total reaction volume of 20 μl. Each reaction contained 2 μl of cDNA template, virus specific primer at a final concentration of 900 nM, and a virus-specific probe at a final concentration of 250 nM (Table 1). Quantitative reverse transcriptase (qRT)-PCR reactions were conducted using a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA). The 40-cycle qRT–PCR program was as follows: denaturing at 95°C for 15 s followed by a 1 min annealing and extension step at 60°C. DNA probes specific for either CHIKV or MAYV contained two distinct reporters 5′FAM or 5′JOE, respectively, and two quenchers, ZEN located in the center of the probe and IABkFQ at the probe’s 3′end. qRT–PCR results were analyzed based on standard curves that were generated by cloning a CHIKV- or MAYV-derived cDNA segment into the pCR-2.1-TOPO TA vector (Invitrogen). DNA plasmid copy numbers were calculated based on plasmid sizes and concentrations. Based on Ct values of the negative controls, Ct values that measured above 36.3 were deemed undetectable and considered negative for viral RNA. All samples consisted of three independent biological replicates.
Table 1.
Oligonucleotide primers and DNA probes for the specific detection of MAYV and CHIKV in mixed infections
| 5′→3′ | Nucleotide position | |
| Primers for cDNA synthesis | ||
| CHIKV | ACGAAACCACTGTATCACAGCG | 921-900 |
| MAYV | CGGTTTCATTCTCTTCTTCCTC | 781-760 |
| Primers for qPCR | ||
| CHIKV F | CGTACTGTTCTCAGTCGGGTC | 791-811 |
| CHIKV R | GATGGAACACTGAAGGTAAGTGC | 873-851 |
| MAYV F | CCTACCCAACATATGCAACCA | 648-668 |
| MAYV R | AAGGTGTCCCTCAGTCAGT | 742-724 |
| Probes | ||
| CHIKV | TACCCGGAGAGCCGTAAGCTTCTTA | 819-843 |
| MAYV | ACGAACAGGTGTTGAAAGCCAGGA | 678-701 |
Sequencing of Viral Genomes and Phylogenetic Analysis
MAYV IQT and MAYV TRVL were propagated in Vero cells until their titers reached >106 PFU/ml. For each virus, infectious cell culture supernatant was harvested and viral RNA was isolated from 280 μl of sample using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). cDNA synthesis was performed using 1 μg of total RNA and the Superscript III First-Strand Synthesis System (Invitrogen) in combination with an oligo (dT)20 primer. Genome segments were amplified using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen) and primer pairs designed to walk through the genome of each MAYV strain. PCR products were purified and Sanger sequenced at the DNA Core of the University of Missouri. 3′ RACE was performed with the 3′ RACE System for Rapid Amplification of cDNA Ends kit (Invitrogen). Sequences were then assembled and aligned using the DNASTAR LaserGene Molecular Biology suite software, version 15 (Madison, WI). Phylogenetic analysis was conducted by aligning the full-length nucleotide sequences of 26 MAYV strains and comparing them using the Maximum Likelihood method based on the General Time Reversible model. Bootstrap values of 1000 were applied to the analysis performed in MEGA X (Kumar et al. 1994).
Statistical Analyses
Statistical analyses were performed using the GraphPad Prism (version 5) software suite (San Diego, CA). Median titers were analyzed using the Mann–Whitney U test followed by the Kruskal–Wallis test. Infection and dissemination rates were compared using Fisher exact test. In the qRT–PCR experiments, data analysis was conducted using one-way analysis of variance (ANOVA) followed by Dunn’s multiple comparisons test. All tests were considered significant at P ≤ 0.05.
Results
Phylogenetic Characterization of MAYV Strains IQT and TRVL
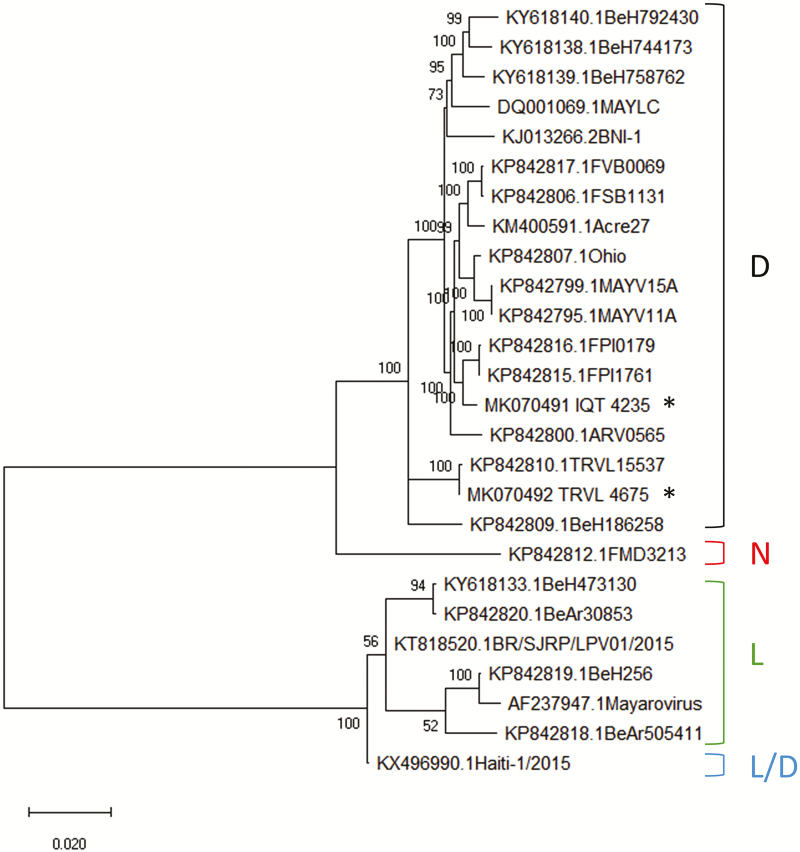
Sequencing of the complete genomes of both MAYV strains, IQT and TRVL, confirmed that both were of the D-genotype. Their nucleotide sequences differed by 2.8% (320 nucleotides) from each other. A phylogenetic analysis was conducted using the full-length nucleotide sequences of 26 geographically diverse MAYV strains (including TRVL [4675] and IQT [4235]). The analysis revealed that similar to TRVL 15537, another strain from Trinidad, MAYV TRVL [4675] is ancestral to other genotype D MAYV strains including IQT, which clustered with two other strains from Peru (Fig. 1). When comparing the 26 MAYV strains, 10 amino acid residues within nsP1, nsP2, and nsP3 were specific for the two strains from Trinidad, TRVL [4675] and TRVL 15537, the latter isolated in 1957 from a Ma. venezuelensis mosquito caught in the Rio Grande Forest (Powers et al. 2006; Table 2). Unique for the TRVL [4675] strain from 1954 were single amino acid substitutions in nsP1 and nsP2. The IQT strain had single unique amino acid substitutions in nsP1 and nsP3. Fewer amino acid substitutions were observed in the polypeptides encoding the structural proteins of the viruses as the E2 amino acid sequence of the IQT strain had no unique residue and that of the TRVL [4675] strain had only a single unique amino acid substitution. However, both strains isolated from Trinidad shared seven specific amino acid substitutions in E2 and E1.
Fig. 1.
Phylogenetic analysis of full-length RNA genomes of geographically diverse MAYV strains/isolates. The full-length nucleotide sequences of 26 MAYV strains including those of IQT (*) and TRVL (*) representing the genotypes Widely Dispersed (D), New (N), and Limited (L) were phylogenetically analyzed using the Maximum Likelihood algorithm based on the General Time Reversible model with bootstrap values of 1000. For each virus strain, the GenBank accession number is indicated followed by its strain designation. The analysis was conducted in MEGA X.
Table 2.
Amino acid substitutions specific for MAYV strains TRVL 4675, TRVL 15537, and IQT 4235
| nsP1 | nsP2 | nsP3 | nsP4 | C | E3 | E2 | 6K/TF | E1 | |
| TRVL 4675/ TRVL 15537 | T317IT425IS454N | V40AK497RT770S | F284LD372VR436SE463D | T59KH130RI175V | D50ET136AS343FT396I | ||||
| TRVL 4675 | V386M | K41R | T196I | ||||||
| IQT 4235 | S445A | T349I |
Midgut and Carcass Infections of HWE and ORL Mosquitoes With the IQT and TRVL Strains of MAYV
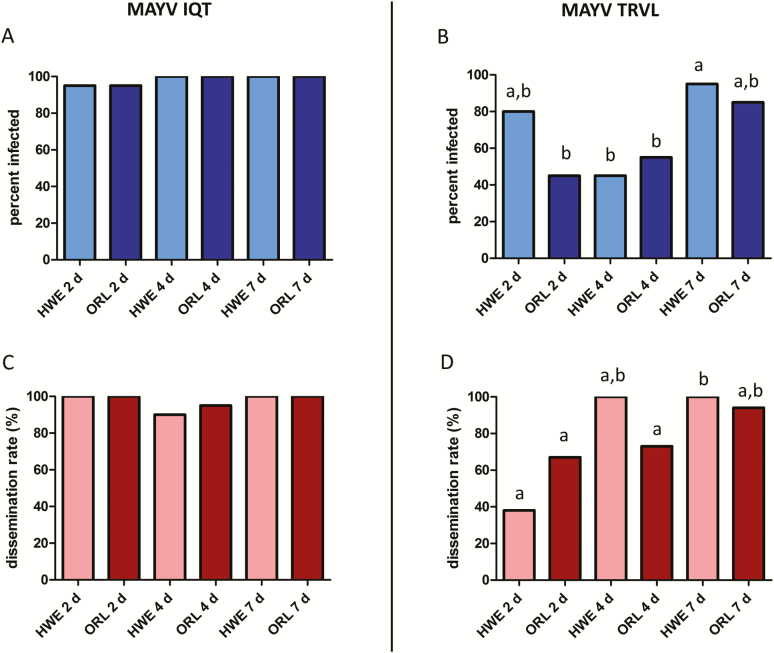
Comparison of the vector competence of Ae. aegypti strains HWE (n = 20 for each virus strain) and ORL (n = 20 for each virus strain) for MAYV IQT and TRVL clearly exposed different levels of susceptibility for the two viruses (Figs. 2 and 3). Importantly, input virus titers (~105 PFU/ml) in individual HWE and ORL whole-body mosquitoes (n = 10 for each virus and mosquito strain) were similar for both virus strains immediately after bloodmeal ingestion (0 h; Fig. 3A and B). At 2 d pibm, MAYV IQT produced significantly higher (P = 0.0012) infection rates in midguts of ORL mosquitoes than the TRVL strain and at 4 d pibm, infection rates in midguts of both mosquito strains were significantly higher (HWE: P = 0.0001; ORL: P = 0.0012) for MAYV IQT than for the TRVL strain (Fig. 2A and B). Midgut infection rates reached similar levels (85–100%) for both virus strains in HWE and ORL mosquitoes at 7 d pibm. At 2 d pibm, dissemination rates from the midguts of HWE and ORL mosquitoes, based on the number of infected carcasses of each treatment group divided by the number of infected midguts, were significantly lower (HWE: P < 0.0001; ORL: P = 0.0256) for MAYV TRVL than for the IQT strain (Fig. 2C and D). However, similar dissemination rates for both viruses and mosquito strains were observed at 4 and 7 d pibm.
Fig. 2.
Midgut infection and dissemination rates of MAYV IQT and TRVL in Ae. aegypti HWE and ORL. Midgut infection rates of (A) MAYV IQT and (B) MAYV TRVL and dissemination rates of (C) MAYV IQT and (D) MAYV TRVL in HWE (n = 20) and ORL (n = 20) mosquitoes at 2, 4, and 7 d pibm as analyzed by plaque assays in Vero cells. Virus dissemination rates from midguts were calculated based on the number of infected carcasses of each treatment group divided by the number of infected midguts. Different letters indicate infection rates that were significantly different from each other (Fisher exact test). Significantly different comparisons in (B and D): P values ranged from < 0.0001 to 0.0484.
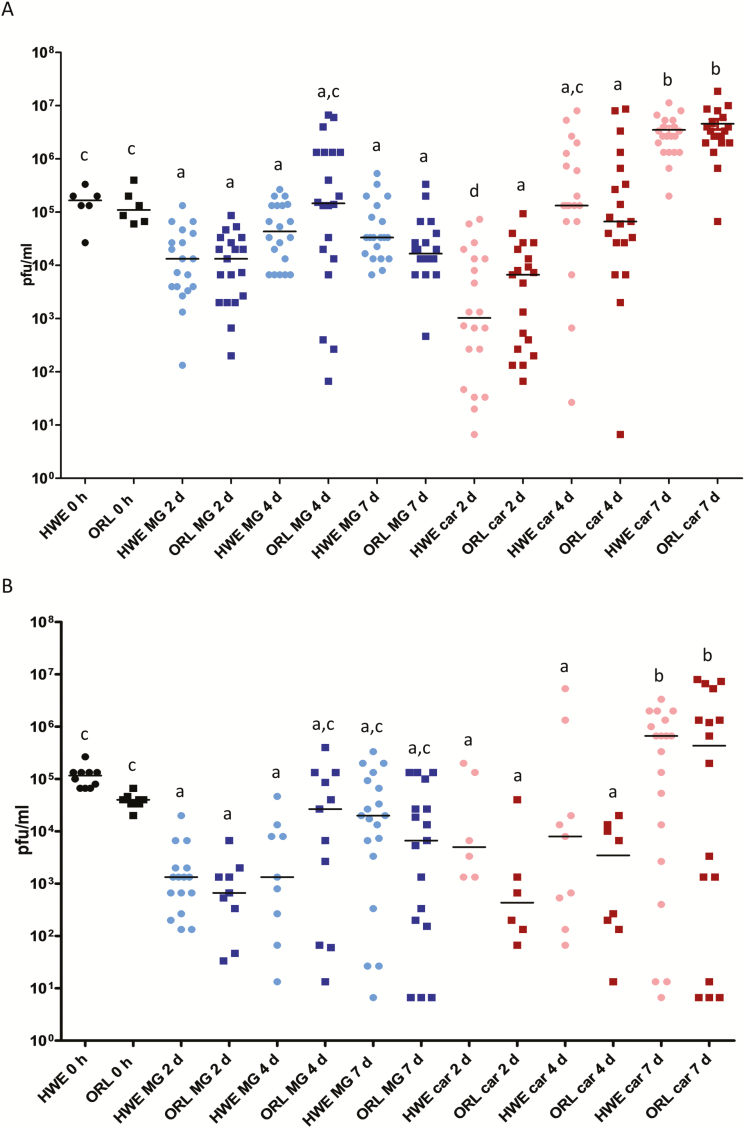
Fig. 3.
Intensities of MAYV IQT and TRVL infections in midguts and carcasses of Ae. aegypti HWE and ORL. (A) MAYV IQT (artificial bloodmeal titer: 2.0 × 106 plaque forming units (PFU)/ml) and (B) MAYV TRVL (artificial bloodmeal titer: 5.0 × 106 PFU/ml) titers in midguts (n = 20) and carcasses (n = 20) of individual HWE and ORL females analyzed at 2, 4, and 7 d pibm by plaque assays in Vero cells. Each data point represents the MAYV titer of an individual midgut or carcass. For 0 h, only whole-body females were assayed. Only infected mosquitoes were included in the analysis. Black bars indicate medians. Different letters indicate median titers that were significantly different from each other (Mann–Whitney U test). Significantly different comparisons: P values ranged from < 0.0001 to 0.0016 in (A) and from < 0.0001 to 0.0004 in (B).
In both mosquito strains, median virus titers in midguts and carcasses were about 1 log PFU/ml higher for MAYV IQT than for TRVL during the 7-d time course (Fig. 3A and B). In midgut tissues, median titers for both viruses did not change significantly over time although there was a tendency of increased median TRVL titers after 2 d pibm. However, both virus strains produced significantly increased (P < 0.0001) median virus titers in carcasses at 7 d pibm, indicating productive infection in secondary tissues. Interestingly, the median input titers of both viruses at time point zero (~105 PFU/ml) were not exceeded in midguts during the 7-d observation period and like-wise not in secondary tissues before 7 d pibm. Taken together, infection patterns varied more strongly between the two MAYV strains than between the two mosquito strains. Overall, MAYV TRVL was less infectious in HWE and ORL mosquitoes than MAYV IQT.
MAYV IQT and TRVL Head Tissue Infections and Prevalence of the Viruses in Saliva of HWE and ORL Mosquitoes
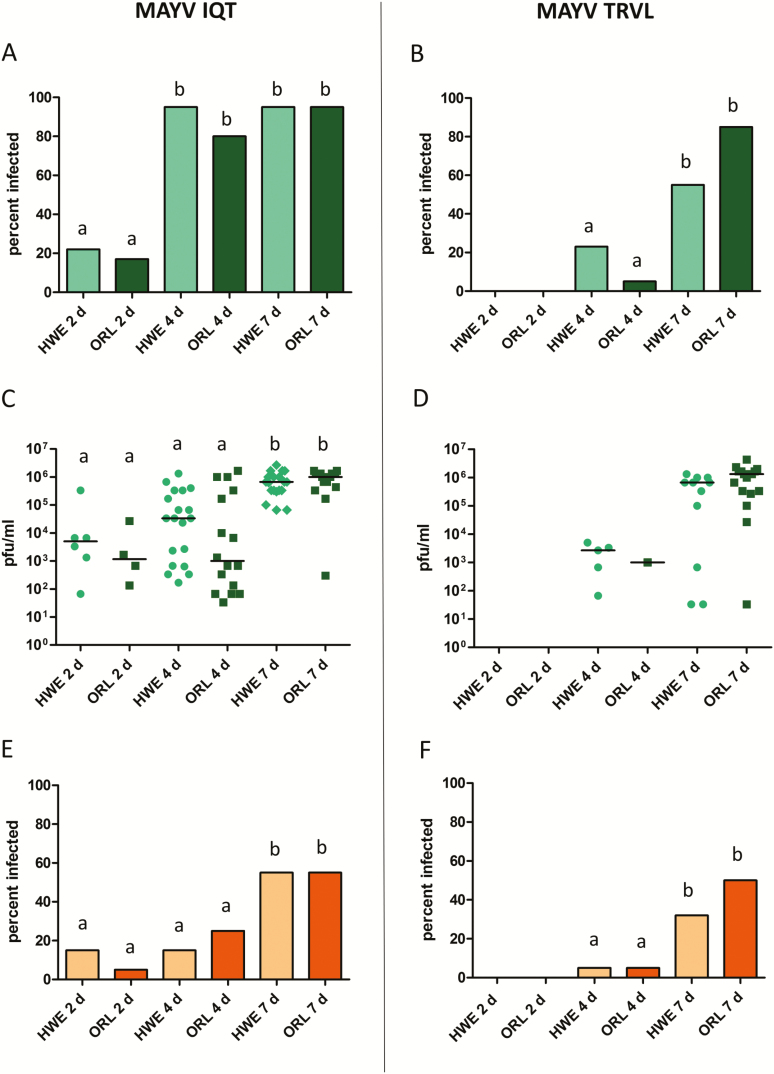
Arbovirus infection of head tissue following oral virus acquisition is an indication that the virus has systemically infected the mosquito body. From 4 d pibm onwards, at least 80% of the head tissues of HWE and ORL mosquitoes (n = 20 for each virus and mosquito strain) were infected with the IQT strain of MAYV, which developed its maximal median titer in head tissue at 7 d pibm (Fig. 4A and C). The proportion of MAYV IQT containing saliva samples (n = 20 for each virus and mosquito strain) increased significantly for HWE (P < 0.001) and ORL (P < 0.001) mosquitoes between 4 and 7 d pibm (Fig. 4E). In contrast, MAYV TRVL was less infectious in both mosquito strains, as it was undetectable in both head tissue and saliva of HWE and ORL mosquitoes until 4 d pibm (Fig. 4B, D, and F). Based on head tissue infection rates, virus titers, and proportion of virus containing saliva samples, MAYV TRVL did not develop its maximum level of infection in both mosquito strains before 7 d pibm. At the individual level, there was only a weak link between MAYV titers in head tissue and successful virus excretion into saliva. The minimal extrinsic incubation periods (EIP) for MAYV IQT and TRVL in both mosquito strains were 2 and 4 d pibm, respectively.
Fig. 4.
Prevalence and intensities of infection of MAYV IQT and TRVL in head tissues of Ae. aegypti HWE and ORL, and prevalence of the viruses in HWE and ORL saliva samples. (A) Prevalence of MAYV IQT and (B) MAYV TRVL infections in head tissues of HWE (n = 20) and ORL (n = 20) mosquitoes at 2, 4, and 7 d pibm. Different letters indicate infection rates that were significantly different from each other as analyzed by Fisher exact test. Significantly different comparisons: P values ranged from 0.0012 to 0.0484. (C) MAYV IQT and (D) MAYV TRVL titers in head tissues of individual HWE (n = 20) and ORL (n = 20) females analyzed at 2, 4, and 7 d pibm by plaque assays in Vero cells. Each data point represents the MAYV titer of an individual head tissue. Only infected mosquitoes were included in the analysis. Black bars indicate medians. Different letters indicate median titers that were significantly different from each other (Mann–Whitney U test). Significantly different comparisons: P values ranged from < 0.0001 to 0.0045. Prevalence of (E) MAYV IQT and (F) MAYV TRVL in saliva samples (n = 20) collected from each mosquito strain at 2, 4, and 7 d pibm. Significantly different comparisons: P values ranged from 0.0033 to 0.0436 (Fisher exact test).
MAYV TRVL Infection Patterns in Ae. albopictus From Missouri
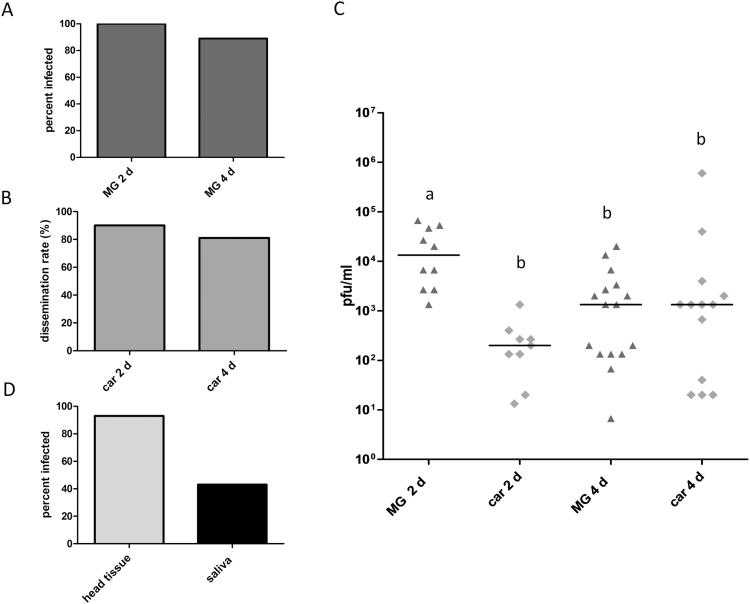
We orally challenged Ae. albopictus CoMO females with MAYV TRVL. Between 2 d (n = 10) and 4 d pibm (n = 18), at least 89% of the midguts were infected indicating that there was virtually no midgut infection barrier for the virus (Fig. 5A). Midgut dissemination rates in CoMO mosquitoes exceeded 80% at 2 and 4 d pibm (Fig. 5B). Median midgut titers ranged between 103 and 104 PFU/midgut, whereas median carcass titers were up to 103 PFU/carcass (Fig. 5C). Following intrathoracic injection of 700 PFU/mosquito MAYV TRVL, median head tissue titers reached ~104 PFU/ml at 72 h postinjection (data not shown). At that time point, 43% of the injected mosquitoes (14/15 with head tissue infections) released the virus along with their saliva (Fig. 5D).
Fig. 5.
Prevalence and intensities of infection of MAYV TRVL in midguts, carcasses, and head tissues of Ae. albopictus (CoMO), and prevalence of the virus in saliva samples. (A) Midgut infection rates of MAYV TRVL in midguts of CoMO mosquitoes at 2 d (n = 10) and 4 d (n = 18) pibm. (B) Dissemination rates of the virus from infected midguts of CoMO mosquitoes at 2 d (n = 10) and 4 d (n = 18) pibm. (C) Virus titers in midguts and carcasses of Ae. albopictus CoMO females were analyzed at 2 d (n = 10) and 4 d (n = 18) pibm by plaque assays in Vero cells. Each data point represents the MAYV TRVL titer of an individual midgut or carcass. The virus titer in the artificial bloodmeal was 1 × 107 PFU/ml. Only infected mosquitoes were included in the analysis. Black bars indicate medians. Different letters indicate median titers that were significantly different from each other (Mann–Whitney U test). Significantly different comparisons: P values ranged from 0.0003 to 0.0013. (D) Prevalence of MAYV TRVL in head tissue of CoMO mosquitoes analyzed at 3 d post-intrathoracic injection of 700 PFU/mosquito by plaque assays in Vero cells. Prevalence of the virus in saliva samples was assessed at 3 d post-intrathoracic injection by CPE analysis of Vero cells, which had been inoculated with the individual saliva samples.
The Effects of Mixed Infections Between CHIKV 37997 and MAYV IQT on the Transmission Potential of Both Viruses
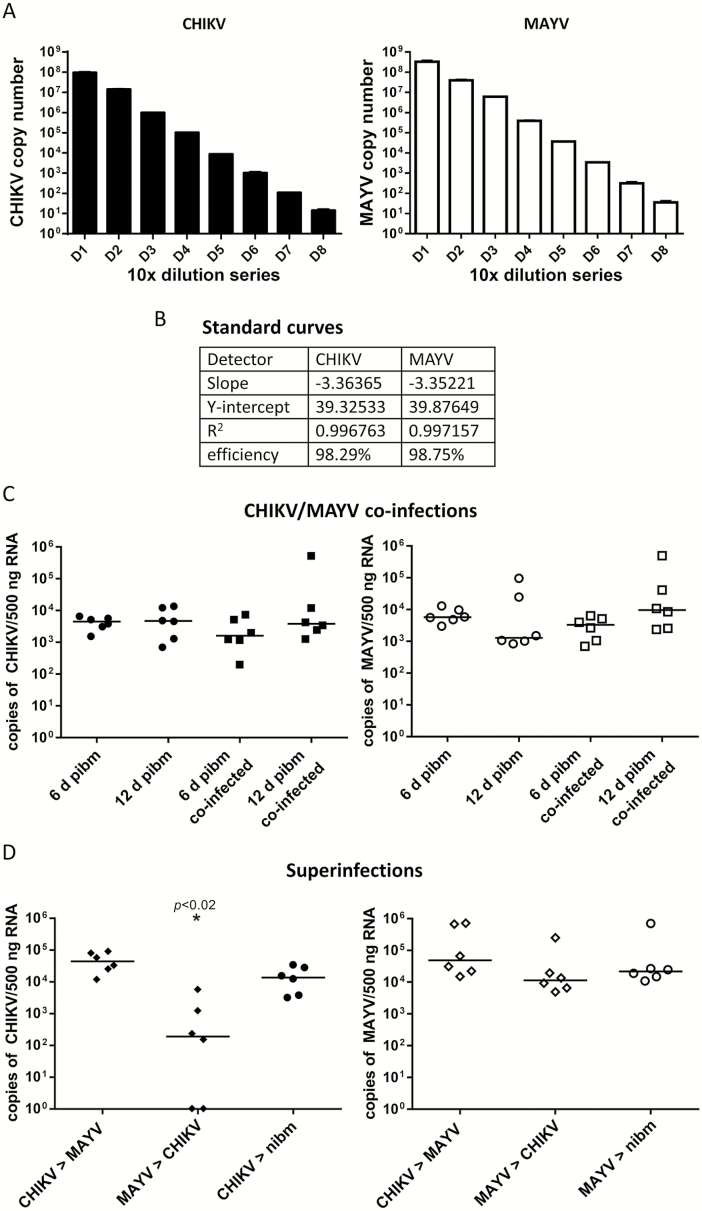
We conducted two experiments to reveal the effects of mixed infections between CHIKV 37997 and MAYV IQT on the transmission of both viruses by Ae. aegypti HWE. Initially, we established that our qRT–PCR assay would allow reliable, accurate virus-specific detection of viral genome copy number equivalents in mixed infections (Fig. 6A and B). Importantly, median bloodmeal (input) titers were 2.0 × 106 PFU/ml (CHIKV) and 5.3 × 106 PFU/ml (MAYV), allowing a direct comparison between the two viruses in the mixed infection experiments.
Fig. 6.
Effects of mixed infections between MAYV IQT and CHIKV 37997 on virus prevalence in mosquito saliva. (A) Tenfold dilution series values calculated from standard curves for CHIKV 37997 (black) and MAYV IQT (white) showing specific detection of viral genome copy number equivalents. Error bars represent standard deviations. (B) Standard curve quality parameters indicating high probability of virus-specific detection of viral RNA genome copy number equivalents. (C) Coinfection of Ae. aegypti HWE with CHIKV 37997 and MAYV IQT. Left panel (filled shapes): detection of CHIKV genome copy number equivalents/500 ng total RNA in six saliva samples each representing a pool of saliva from five females (n = 30). Saliva samples were collected at 6 or 12 d pibm. Artificial bloodmeals contained CHIKV only or a mixture of CHIKV and MAYV. Right panel (open shapes): detection of MAYV genome copy number equivalents/500 ng total RNA in six saliva samples each representing a pool of saliva from five females (n = 30). Saliva samples were collected at 6 or 12 d pibm. Artificial bloodmeals contained MAYV only or the same mixture of CHIKV and MAYV as shown in the left panel. Bars indicate median values. (D) Superinfections between CHIKV and MAYV. Left panel (filled shapes): CHIKV-specific detection. Females acquired CHIKV via an artificial bloodmeal followed by a second bloodmeal (at 6 d post–initial infectious bloodmeal) containing MAYV (CHIKV > MAYV). In another setup, females acquired MAYV via an artificial bloodmeal followed by a second bloodmeal (at 6 d post–initial infectious bloodmeal) containing CHIKV (MAYV > CHIKV). As a control, females acquired CHIKV via artificial bloodfeeding and 6 d later, females were given another, noninfectious bloodmeal (nibm). At 12 d post–initial infectious bloodmeal, six saliva samples each consisting of a pool of saliva from five females were collected and analyzed (n = 30). Right panel (open shapes): MAYV-specific detection of the same superinfection samples as shown in the left panel (filled shapes). As a control, females acquired MAYV via artificial bloodfeeding and 6 d later, females were given another, noninfectious bloodmeal (nibm). Samples were analyzed by qRT–PCR from a standard curve as described above. One-way ANOVA followed by Dunn’s multiple comparison test was used for statistical analysis to determine P values; * indicates P < 0.020. Bars indicate median values.
In the first experimental setup, mosquitoes simultaneously acquired MAYV and CHIKV via an artificial bloodmeal (coinfection). In this coinfection experiment, both viruses reached similar concentrations in mosquito saliva at 12 d pibm, which amounted to 5 × 103–1 × 104 median viral RNA copy number equivalents per pooled saliva sample (Fig. 6C). When CHIKV had been orally acquired before MAYV (MAYV superinfection), median viral RNA copy number equivalents in saliva were similar for both viruses and resembled the values obtained for mosquitoes that had acquired only one of the two viruses (Fig. 6D). Furthermore, when MAYV had been orally acquired before CHIKV (CHIKV superinfection), the median MAYV RNA copy number equivalent in saliva was similar to those obtained from mosquitoes that had acquired one of the two viruses only. However, the median CHIKV RNA copy number equivalent was significantly lower (P = 0.020; and in two out of six pooled saliva samples [=10/30 samples total] the virus was undetectable) in MAYV-infected mosquitoes at 12 d pibm, suggesting that a primary MAYV infection leads to a superinfection exclusion for CHIKV. In contrast, a primary CHIKV infection did not cause a superinfection exclusion for MAYV. In single infections (in absence of a second, noninfectious bloodmeal), CHIKV and MAYV produced similar median RNA copy number equivalents at 6 d pibm in saliva (Fig. 6C), suggesting that the low median CHIKV RNA copy number equivalent detected in saliva of superinfected mosquitoes was not based on principal differences in the two viruses’ replication efficiencies. We also observed that at 6 d post-nibm, mosquitoes had up to 1 log increased median viral RNA copy number equivalents in saliva in comparison to mosquitoes that had received only a single, virus-containing bloodmeal (Fig. 6C and D).
Discussion
Although MAYV has been known to circulate in Central and South America for over 60 yr, only a few vector competence studies in regard to the virus have been conducted to date (Smith and Francy 1991, Long et al. 2011, Brustolin et al. 2018, Wiggins et al. 2018). Our results show that the MAYV strains IQT and TRVL produced different infection patterns in two Ae. aegypti strains, HWE and ORL. Despite similar input titers, TRVL produced ~1 log lower median titers than IQT in midguts and carcasses of both Ae. aegypti strains throughout the time course study. Furthermore, MAYV IQT produced midgut infection and dissemination rates of >90% at 2 d pibm, whereas the TRVL strain required 7 d to generate similar infection rates in both mosquito strains. Similar to HWE and ORL mosquitoes in our study, field-collected Ae. aegypti from Maynas Province of Peru previously proved to be competent vectors for MAYV IQT following its oral acquisition from artificial bloodmeals or viremic mice. Wild-type mosquito infection rates and virus transmission rates were as high as 84% and 70%, respectively (Long et al. 2011). In Ae. albopictus CoMO, midgut infection and dissemination rates of MAYV TRVL were higher than those observed for the same virus strain in Ae. aegypti ORL at both time points and in HWE at 2 d pibm. Due to the initially observed low feeding rates of the CoMO females on artificial bloodmeals, we did not test the presence of bloodmeal-acquired virus in saliva of these mosquitoes. Regardless, at 72 h post-intrathoracic injection, 43% of the tested Ae. albopictus CoMO individuals contained the virus in saliva. The TRVL strain had been tested earlier in Ae. albopictus wild-types from Brazil and recently in Ae. albopictus and Ae. aegypti wild-types from Florida (Smith and Francy 1991, Wiggins et al. 2018). In both studies, infection rates of >80% were reported for Ae. albopictus, suggesting that, similar to our CoMO colony, these wild-types were highly susceptible to the virus. A striking difference was the observed low level of prevalence of MAYV TRVL in saliva of the Ae. albopictus and Ae. aegypti wild-types from Florida, not exceeding 10% before 9 d pibm, indicative of a relatively strong barrier to the virus at the salivary gland level (Wiggins et al. 2018). This is in contrast to the higher prevalence of the virus in saliva of HWE (32%) and ORL (50%) mosquitoes at 7 d pibm. However, it needs to be pointed out that different laboratory environments and techniques employed by the various work groups may contribute to the divergent results.
Previously, we conducted a similar vector competence analysis in HWE and ORL mosquitoes for CHIKV strain 37997 allowing us to directly compare the interactions of MAYV and CHIKV in these mosquitoes (Dong et al. 2016). Until 5 d pibm, median CHIKV titers were significantly (~1 log) lower in midguts and carcasses of ORL mosquitoes than in those of the HWE strain. Overall, MAYV IQT produced an infection pattern similar to that of CHIKV 37997 in HWE midgut and carcasses, whereas the MAYV TRVL infection pattern in midgut and carcasses of both mosquito strains resembled that of CHIKV in ORL mosquitoes. In both Ae. aegypti strains, head tissue infections with MAYV TRVL were delayed by 2 d when compared with MAYV IQT correlating with a 2-d delayed presence of the former in mosquito saliva. However, similar to CHIKV in our previous study, MAYV IQT was already detectable in head tissue and saliva at 2 d pibm. Regarding the presence of infectious virus in saliva, the barrier of infection at the salivary gland level for MAYV IQT was similar to that for CHIKV as well (Dubrulle et al. 2009, Long et al. 2011, Dong et al. 2016). EIPs as short as 2 d have been also reported for Venezuelan equine encephalitis virus (Togaviridae; Alphavirus; VEEV) in Ae. aegypti and Rift Valley fever virus (Bunyaviridae; Phlebovirus; RVFV) in Culex pipiens (Gaidamovich et al. 1973, Faran et al. 1988).
Our data suggest that the IQT strain was better adapted to Ae. aegypti than MAYV TRVL. This finding is supported by the fact that previously the IQT strain had been repeatedly passaged in Ae. albopictus C6/36 cells (Powers et al. 2006), whereas the TRVL strain had not been passaged in invertebrate cells. Thus, passage in C6/36 cells may have introduced adaptive mutations to the IQT strain resulting in a higher infectivity in Ae. aegypti in comparison to the TRVL strain, which had been maintained in mammalian cell lines (Anderson et al. 1957, Casals and Whitman 1957). Full-length sequencing and phylogenetic analysis of the TRVL and IQT strains confirmed that both strains belong to the D genotype with MAYV TRVL clustering together with another strain from Trinidad, TRVL 15537. These two strains are more ancestral to MAYV IQT (and other D genotype viruses). Both Trinidad strains have specific amino acid residues in five viral proteins, nsP1, nsP2, nsP3, E2, and E1, whereas the IQT strain had specific amino acid residues only in nsP1 and nsP2. Further studies will be needed to elucidate which mutations would be required to adapt a forest-dwelling mosquito–transmitted MAYV strain to efficient transmission by urban, anthropophilic mosquito species, such as Ae. aegypti and Ae. albopictus.
We showed that simultaneously acquired MAYV and CHIKV (coinfection) can both be cotransmitted by the same vector with similar efficiencies, at least under laboratory conditions. These findings are in accordance with other studies demonstrating that in mosquitoes, coinfecting arboviruses do not interfere with each other (Vazeille et al. 2010, Goertz et al. 2017, Rückert et al. 2017). In nature, coinfection of mosquitoes with several arboviruses is not an unlikely event since human individuals in endemic regions have been identified as being highly viremic for more than one mosquito-borne virus including DENV, CHIKV, and/or ZIKV (Myers and Carey 1967, Dupont-Rouzeyrol et al. 2015, Villamil-Gomez et al. 2016, Waggoner et al. 2016, Zambrano et al. 2016). Even more likely than coinfection is sequential arbovirus infection (superinfection) of mosquitoes in regions where different arboviruses cocirculate. Under this scenario, the same mosquito could sequentially acquire different viruses when biting several hosts who would be viremic for different viruses. We observed superinfection exclusion for CHIKV when it was orally acquired by MAYV-infected mosquitoes, as CHIKV, but not MAYV, was absent or significantly less detectable in saliva of those mosquitoes. Interestingly, there was no interference for either virus when MAYV had been acquired by CHIKV-infected mosquitoes.
Superinfection exclusion, causing a cell persistently infected with one virus to prevent infection with a second, closely related virus has been reported before for a range of arboviruses including alphaviruses (Condreay and Brown 1986, Karpf et al. 1997). Most of the studies addressing viral superinfection exclusion have been conducted in cell culture. For example, recently, it was shown that yellow fever virus (Flaviviridae; Flavivirus) and DENV2 exclude each other following sequential inoculation of C6/36 Ae. albopictus cells (Abrao and da Fonseca 2016). Fewer studies have been conducted to analyze superinfection exclusion of arboviruses in vivo (Sundin and Beaty 1988, Borucki et al. 1999, Pesko and Mores 2009, Campbell et al. 2014, Nasar et al. 2015, Nuckols et al. 2015). Investigating superinfection exclusion has gained additional attention when characterizing insect-specific flaviviruses such as Nhumirim virus, only found in Culex spp., or the insect-specific alphavirus, Eilat virus (Kent et al. 2010, Bolling, et al. 2012, Goenaga et al. 2015, Nasar et al. 2015). It has been suggested that these mosquito-specific viruses may interfere with the replication and transmission of human-infecting arboviruses in the mosquito vector. For example, the presence of Nhumirim virus antagonized West Nile virus (Flaviviridae; Flavivirus) transmission by Cx. quinquefasciatus, and the presence of Eilat virus delayed CHIKV dissemination from the midgut in Ae. aegypti by 3 d (Goenaga et al. 2015, Nasar et al. 2015). Our study demonstrates for the first time in vivo that two human-pathogenic alphaviruses can antagonize each other depending on their sequence of infection in the mosquito.
Mechanistically, viral superinfection exclusion is not well understood. It is possible that in our study, prior infection with MAYV caused diminishment of cellular resources required for optimal CHIKV replication and infection in the mosquito. Reduced virion binding to cell surface receptors, cell entry capabilities, inhibition of nucleocapsid uncoating, and/or inhibition of viral replication complexes all have been described to explain the phenomenon for alphaviruses (Singh et al. 1997, Nasar et al. 2015). It will require extensive molecular analyses to reveal the mechanism underlying CHIKV exclusion in MAYV-infected Ae. aegypti. We also observed that a second, noninfectious bloodmeal acquired by virus-infected mosquitoes leads to an enhanced virus concentration in their saliva in comparison to those mosquitoes that had received only a single, infectious bloodmeal. This observation is in line with our recent ultrastructural studies in which we showed that acquisition of a second (noninfectious) bloodmeal enhanced virus dissemination from the infected midgut (Kantor et al. 2018).
In conclusion, we confirmed that Ae. aegypti and Ae. albopictus can act as efficient vectors for MAYV, as they have shown before for CHIKV and that both, CHIKV and MAYV, can be efficiently cotransmitted by the same infected mosquito. We also showed that mosquito infection and transmission efficiencies can vary significantly between different MAYV strains.
Acknowledgments
The viruses used in this work were obtained through the UTMB-Galveston Arbovirus Reference Collection maintained at the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). All virus infection experiments have been carried out in the BSL3 virology suite of the Laboratory of Infectious Diseases Research (LIDR) at the University of Missouri. This work was funded by grants from the National Institutes of Health – National Institute of Allergy and Infectious Diseases (NIH–NIAID), R01AI091972 and R01AI134661. NIH did not have a role in study design, data collection and interpretation, or the decision to submit the work for publication.
References Cited
- Abrao E. P., and da Fonseca B. A.. 2016. Infection of mosquito cells (C6/36) by dengue-2 virus interferes with subsequent infection by yellow fever virus. Vector Borne Zoonotic Dis. 16: 124–130. [DOI] [PubMed] [Google Scholar]
- Aitken T. H., Downs W. G., Anderson C. R., Spence L., and Casals J.. 1960. Mayaro virus isolated from a Trinidadian mosquito, Mansonia venezuelensis. Science. 131: 986. [DOI] [PubMed] [Google Scholar]
- Anderson C. R., Downs W. G., Wattley G. H., Ahin N. W., and Reese A. A.. 1957. Mayaro virus: a new human disease agent. II. Isolation from blood of patients in Trinidad, B.W.I. Am. J. Trop. Med. Hyg. 6: 1012–1016. [DOI] [PubMed] [Google Scholar]
- Auguste A. J., Liria J., Forrester N. L., Giambalvo D., Moncada M., Long K. C., Morón D., de Manzione N., Tesh R. B., Halsey E. S., et al. 2015. Evolutionary and ecological characterization of mayaro virus strains isolated during an outbreak, Venezuela, 2010. Emerg. Infect. Dis. 21: 1742–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling B. G., Olea-Popelka F. J., Eisen L., Moore C. G., and Blair C. D.. 2012. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology. 427: 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki M. K., Chandler L. J., Parker B. M., Blair C. D., and Beaty B. J.. 1999. Bunyavirus superinfection and segment reassortment in transovarially infected mosquitoes. J. Gen. Virol. 80 (Pt 12): 3173–3179. [DOI] [PubMed] [Google Scholar]
- Brustolin M., Pujhari S., Henderson C. A., and Rasgon J. L.. 2018. Anopheles mosquitoes may drive invasion and transmission of Mayaro virus across geographically diverse regions. Plos Negl. Trop. Dis. 12: e0006895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C. H., Gutiérrez E., Maness K. S., and Lord R. D.. 1974. Isolation of mayaro virus from a migrating bird captured in Louisiana in 1967. Bull. Pan Am. Health Organ. 8: 243–248. [PubMed] [Google Scholar]
- Campbell C. L., Smith D. R., Sanchez-Vargas I., Zhang B., Shi P. Y., and Ebel G. D.. 2014. A positively selected mutation in the WNV 2K peptide confers resistance to superinfection exclusion in vivo. Virology. 464–465: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals J., and Whitman L.. 1957. Mayaro virus: a new human disease agent. I. Relationship to other arbor viruses. Am. J. Trop. Med. Hyg. 6: 1004–1011. [DOI] [PubMed] [Google Scholar]
- Causey O. R., and Maroja O. M.. 1957. Mayaro virus: a new human disease agent. III. Investigation of an epidemic of acute febrile illness on the river Guama in Pará, Brazil, and isolation of mayaro virus as causative agent. Am. J. Trop. Med. Hyg. 6: 1017–1023. [PubMed] [Google Scholar]
- Condreay L. D., and Brown D. T.. 1986. Exclusion of superinfecting homologous virus by sindbis virus-infected Aedes albopictus (mosquito) cells. J. Virol. 58: 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Kantor A. M., Lin J., Passarelli A. L., Clem R. J., and Franz A. W.. 2016. Infection pattern and transmission potential of chikungunya virus in two new world laboratory-adapted Aedes aegypti strains. Sci. Rep. 6: 24729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle M., Mousson L., Moutailler S., Vazeille M., and Failloux A. B.. 2009. Chikungunya virus and aedes mosquitoes: saliva is infectious as soon as two days after oral infection. PLoS One. 4: e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont-Rouzeyrol M., O’Connor O., Calvez E., Daurès M., John M., Grangeon J. P., and Gourinat A. C.. 2015. Co-infection with zika and dengue viruses in 2 patients, New Caledonia, 2014. Emerg. Infect. Dis. 21: 381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faran M. E., Romoser W. S., Routier R. G., and Bailey C. L.. 1988. The distribution of Rift Valley fever virus in the mosquito Culex pipiens as revealed by viral titration of dissected organs and tissues. Am. J. Trop. Med. Hyg. 39: 206–213. [DOI] [PubMed] [Google Scholar]
- Furuya-Kanamori L., Liang S., Milinovich G., Soares Magalhaes R. J., Clements A. C., Hu W., Brasil P., Frentiu F. D., Dunning R., and Yakob L.. 2016. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect. Dis. 16: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidamovich S. Y., Khutoretskaya N. V., Lvova A. I., and Sveshnikova N. A.. 1973. Immunofluorescent staining study of the salivary glands of mosquitoes infected with group A arboviruses. Intervirology. 1: 193–200. [DOI] [PubMed] [Google Scholar]
- Goenaga S., Kenney J. L., Duggal N. K., Delorey M., Ebel G. D., Zhang B., Levis S. C., Enria D. A., and Brault A. C.. 2015. Potential for co-infection of a mosquito-specific flavivirus, nhumirim virus, to block west nile virus transmission in mosquitoes. Viruses. 7: 5801–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göertz G. P., Vogels C. B. F., Geertsema C., Koenraadt C. J. M., and Pijlman G. P.. 2017. Mosquito co-infection with Zika and chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. Plos Negl. Trop. Dis. 11: e0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch A. L., Peterson N. E., LeDuc J. W., and Pinheiro F. P.. 1981. An outbreak of mayaro virus disease in Belterra, Brazil. III. Entomological and ecological studies. Am. J. Trop. Med. Hyg. 30: 689–698. [DOI] [PubMed] [Google Scholar]
- Izurieta R. O., Macaluso M., Watts D. M., Tesh R. B., Guerra B., Cruz L. M., Galwankar S., and Vermund S. H.. 2011. Hunting in the rainforest and mayaro virus infection: an emerging alphavirus in Ecuador. J. Glob. Infect. Dis. 3: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor A. M., Grant D. G., Balaraman V., White T. A., and Franz A. W. E.. 2018. Ultrastructural analysis of chikungunya virus dissemination from the midgut of the yellow fever mosquito, Aedes aegypti. Viruses 10: E571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf A. R., Lenches E., Strauss E. G., Strauss J. H., and Brown D. T.. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71: 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent R. J., Crabtree M. B., and Miller B. R.. 2010. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. Plos Negl. Trop. Dis. 4: e671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., and Nei M.. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10: 189–191. [DOI] [PubMed] [Google Scholar]
- Lednicky J., De Rochars V. M., Elbadry M., Loeb J., Telisma T., Chavannes S., Anilis G., Cella E., Ciccozzi M., Okech B., et al. 2016. Mayaro virus in child with acute febrile illness, Haiti, 2015. Emerg. Infect. Dis. 22: 2000–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K. C., Ziegler S. A., Thangamani S., Hausser N. L., Kochel T. J., Higgs S., and Tesh R. B.. 2011. Experimental transmission of mayaro virus by Aedes aegypti. Am. J. Trop. Med. Hyg. 85: 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavian C., Rife B. D., Dollar J. J., Cella E., Ciccozzi M., Prosperi M. C. F., Lednicky J., Morris J. G., Capua I., and Salemi M.. 2017. Emergence of recombinant mayaro virus strains from the Amazon basin. Sci. Rep. 7: 8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., and Carey D. E.. 1967. Concurrent isolation from patient of two arboviruses, chikungunya and dengue type 2. Science. 157: 1307–1308. [DOI] [PubMed] [Google Scholar]
- Nasar F., Erasmus J. H., Haddow A. D., Tesh R. B., and Weaver S. C.. 2015. Eilat virus induces both homologous and heterologous interference. Virology. 484: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuckols J. T., Huang Y. J., Higgs S., Miller A. L., Pyles R. B., Spratt H. M., Horne K. M., and Vanlandingham D. L.. 2015. Evaluation of simultaneous transmission of chikungunya virus and dengue virus type 2 in infected Aedes aegypti and Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 52: 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauvolid-Corrêa A., Juliano R. S., Campos Z., Velez J., Nogueira R. M., and Komar N.. 2015. Neutralising antibodies for mayaro virus in Pantanal, Brazil. Mem. Inst. Oswaldo Cruz. 110: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesko K., and Mores C. N.. 2009. Effect of sequential exposure on infection and dissemination rates for West Nile and St. Louis encephalitis viruses in Culex quinquefasciatus. Vector Borne Zoonotic Dis. 9: 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. M., Aguilar P. V., Chandler L. J., Brault A. C., Meakins T. A., Watts D., Russell K. L., Olson J., Vasconcelos P. F., Da Rosa A. T., et al. 2006. Genetic relationships among Mayaro and Una viruses suggest distinct patterns of transmission. Am. J. Trop. Med. Hyg. 75: 461–469. [PubMed] [Google Scholar]
- Rückert C., Weger-Lucarelli J., Garcia-Luna S. M., Young M. C., Byas A. D., Murrieta R. A., Fauver J. R., and Ebel G. D.. 2017. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat. Commun. 8: 15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I. R., Suomalainen M., Varadarajan S., Garoff H., and Helenius A.. 1997. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology. 231: 59–71. [DOI] [PubMed] [Google Scholar]
- Smith G. C., and Francy D. B.. 1991. Laboratory studies of a Brazilian strain of Aedes albopictus as a potential vector of mayaro and oropouche viruses. J. Am. Mosq. Control Assoc. 7: 89–93. [PubMed] [Google Scholar]
- Sundin D. R., and Beaty B. J.. 1988. Interference to oral superinfection of Aedes triseriatus infected with La Crosse virus. Am. J. Trop. Med. Hyg. 38: 428–432. [DOI] [PubMed] [Google Scholar]
- Tesh R. B., Watts D. M., Russell K. L., Damodaran C., Calampa C., Cabezas C., Ramirez G., Vasquez B., Hayes C. G., Rossi C. A., et al. 1999. Mayaro virus disease: an emerging mosquito-borne zoonosis in tropical South America. Clin. Infect. Dis. 28: 67–73. [DOI] [PubMed] [Google Scholar]
- de Thoisy B., Gardon J., Salas R. A., Morvan J., and Kazanji M.. 2003. Mayaro virus in wild mammals, French Guiana. Emerg. Infect. Dis. 9: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis B. V., Morton F. A., and Cochran J. H.. 1946. Insect repellents used as skin treatments by the Armed Forces. J. Econ. Entomol. 39: 627–630. [DOI] [PubMed] [Google Scholar]
- Vanlandingham D. L., Hong C., Klingler K., Tsetsarkin K., McElroy K. L., Powers A. M., Lehane M. J., and Higgs S.. 2005. Differential infectivities of o’nyong-nyong and chikungunya virus isolates in Anopheles gambiae and Aedes aegypti mosquitoes. Am. J. Trop. Med. Hyg. 72: 616–621. [PubMed] [Google Scholar]
- Vazeille M., Mousson L., Martin E., and Failloux A. B.. 2010. Orally co-Infected Aedes albopictus from La Reunion Island, Indian Ocean, can deliver both dengue and chikungunya infectious viral particles in their saliva. Plos Negl. Trop. Dis. 4: e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamil-Gómez W. E., Rodríguez-Morales A. J., Uribe-García A. M., González-Arismendy E., Castellanos J. E., Calvo E. P., Álvarez-Mon M., and Musso D.. 2016. Zika, dengue, and chikungunya co-infection in a pregnant woman from Colombia. Int. J. Infect. Dis. 51: 135–138. [DOI] [PubMed] [Google Scholar]
- Waggoner J. J., Gresh L., Vargas M. J., Ballesteros G., Tellez Y., Soda K. J., Sahoo M. K., Nuñez A., Balmaseda A., Harris E., et al. 2016. Viremia and clinical presentation in nicaraguan patients infected with zika virus, chikungunya virus, and dengue virus. Clin. Infect. Dis. 63: 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell M. D., Wilson T. G., Higgs S., and Black W. C.. 2000. Chemical and gamma-ray mutagenesis of the white gene in Aedes aegypti. Insect Mol. Biol. 9: 119–125. [DOI] [PubMed] [Google Scholar]
- Wiggins K., Eastmond B., and Alto B. W.. 2018. Transmission potential of Mayaro virus in Florida Aedes aegypti and Aedes albopictus mosquitoes. Med. Vet. Entomol. 32: 436–442. [DOI] [PubMed] [Google Scholar]
- Zambrano H., Waggoner J. J., Almeida C., Rivera L., Benjamin J. Q., and Pinsky B. A.. 2016. Zika virus and chikungunya virus coinfections: a series of three cases from a single center in Ecuador. Am. J. Trop. Med. Hyg. 95: 894–896. [DOI] [PMC free article] [PubMed] [Google Scholar]