Abstract
Current regimens for the detection and surveillance of bladder cancer (BLCA) are invasive and have suboptimal sensitivity. Here, we present a novel high-throughput sequencing (HTS) method for detection of urine tumor DNA (utDNA) called utDNA CAPP-Seq (uCAPP-Seq) and apply it to 67 healthy adults and 118 patients with early-stage BLCA who either had urine collected prior to treatment or during surveillance. Using this targeted sequencing approach, we detected a median of 6 mutations per BLCA patient and observed surprisingly frequent mutations of the PLEKHS1 promoter (46%), suggesting these mutations represent a useful biomarker for detection of BLCA. We detected utDNA pre-treatment in 93% of cases using a tumor mutation-informed approach and in 84% when blinded to tumor mutation status, with 96–100% specificity. In the surveillance setting, we detected utDNA in 91% of patients who ultimately recurred, with utDNA detection preceding clinical progression in 92% of cases. uCAPP-Seq outperformed a commonly used ancillary test (UroVysion, p=0.02) and cytology and cystoscopy combined (p≤0.006), detecting 100% of BLCA cases detected by cytology and 82% that cytology missed. Our results indicate that uCAPP-Seq is a promising approach for early detection and surveillance of BLCA.
Keywords: Urothelial carcinoma, minimal residual disease, cell free tumor DNA
Introduction:
BLCA is the sixth leading cause of cancer in the U.S., with an estimated 79,030 new cases in 2017 (1). After diagnosis and treatment for localized disease, the National Comprehensive Cancer Network guidelines recommend that patients undergo cystoscopy and urine cytology evaluation to monitor for recurrence every 3–6 months for two years and then at increasing intervals (1). Unfortunately, cystoscopy is invasive and cytology has a low sensitivity, ranging from 20 to 53% (2). As a result of the need for this procedure-based, long-term follow-up, BLCA management costs more per patient lifetime than any other cancer (3).
Many attempts have been made to overcome these challenges by developing biomarkers for BLCA surveillance. The Food and Drug Administration has approved six different urine-based tests for BLCA recurrence, but they achieve modest sensitivities (55–70%) and specificities (71–83%), and none of the available tests have achieved widespread adoption (4).
Recent work has demonstrated the promise of analyzing circulating tumor DNA (ctDNA) in plasma to detect minimal residual disease (MRD) in a variety of tumor types (5–7). For example, recent work from our group has shown that hybrid-capture based ctDNA analysis using CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) can achieve 94% sensitivity and 100% specificity for detecting MRD post-treatment in plasma from patients with localized lung cancer (6,8,9). In the case of localized BLCA, analysis of tumor DNA in urine has been explored as a potential approach for detection and surveillance using amplicon-based HTS approaches, with sensitivities and specificities ranging from 68–85% and 80–100%, respectively (10–13).
In this study, we developed a novel HTS-based hybrid capture method for detection of utDNA called uCAPP-Seq and apply it to urine supernatant specimens. We hypothesized that assessment of utDNA would have superior performance characteristics compared to cytology for detecting early-stage BLCA and post-treatment residual disease.
Results:
Development of a novel assay for utDNA detection:
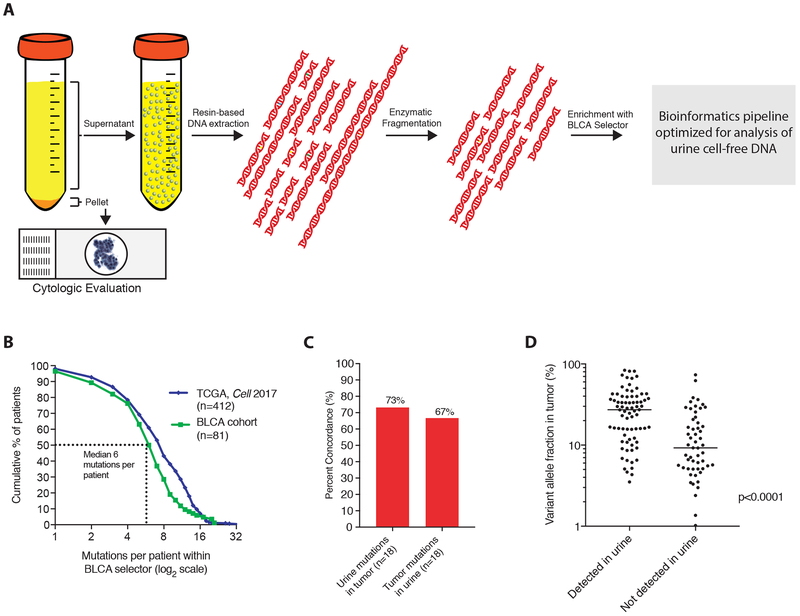
Given practical challenges posed by purifying nucleic acids from large volumes of fluid, we adapted and optimized a previously described resin-based cell-free DNA (cfDNA) extraction protocol for urine samples (Fig. 1A), which performed as well as commercially-available kits but allows analysis of larger volumes of urine (Supplemental Fig. 1A) (14). We next established that in the presence of EDTA, urine cfDNA concentrations remain stable for at least 7 days at 4 degrees Celsius but not at room temperature (Supplemental Figs. S1B,C). Applying our optimized protocol to 185 urine samples with a median urine volume of 50 mL, we observed a median urine cfDNA concentration of 7.7 ng/ml and yield of 348.1 ng per sample (Supplemental Tables S1, S2).
Figure 1: Schematic and validation of workflow for uCAPP-Seq.
(A) Workflow for uCAPP-Seq. Voided urine specimens were centrifuged and the cellular fraction was submitted for cytologic evaluation or other analyses. utDNA was extracted from the supernatant using a resin-based extraction protocol and then subjected to enzymatic fragmentation, library preparation, and hybrid capture with a panel optimized for bladder cancer. After next-generation sequencing, reads were processed through a bioinformatic pipeline consisting of adapter trimming, quality filtering, BWA-MEM based mapping, and variable read-length barcode-based deduplication. (B) A ~311kb hybrid capture panel was designed for bladder cancer targeting recurrently mutated regions identified in the literature, covering a median of 7 mutations per patient in the 2017 TCGA dataset on urothelial carcinoma (n=412) and 6 mutations per patient across 81 tumor and utDNA samples in this study. (C) Across samples with early-stage bladder cancer and paired tumor available for genotyping (n=18), a median of 73% of utDNA mutations were identified in paired tumor and 67% tumor mutations were identified in utDNA. (D) Tumor mutations also identified in urine had a higher median allele fraction (27% vs. 9%, p<0.0001) than those not identified in urine. The p-value was calculated by the Mann Whitney test. TCGA, The Cancer Genome Atlas; CAPP-Seq, Cancer Personalized Profiling by Deep Sequencing. BLCA, bladder cancer; BWA-MEM, Burrows-Wheeler Aligner with maximal exact matches.
We next designed a custom capture panel for BLCA targeting recurrent single-nucleotide variants (SNVs), insertions/deletions, and copy number alterations. We began by including genomic regions covering known driver mutations in BLCA (15–17). We then applied our previously described algorithm to maximize patient coverage in the smallest possible genomic space using data from 412 BLCA cases from The Cancer Genome Atlas (TCGA) (8,18). The final panel covered ~311 kb of genomic space, included regions from 460 genes, and was predicted to identify a median of 7 mutations per BLCA patient (18) (Supplemental Table S3).
Urine contains a wider range of cfDNA fragment sizes than plasma (Supplemental Figs 2A-C), necessitating modifications to the library preparation protocol and bioinformatic analyses that we had previously optimized for CAPP-Seq analysis of plasma. First, we tested if mutant DNA fragments from BLCAs were enriched in short (<500bp) or long (>500bp) fragments of urine cfDNA. We observed similar variant allele fractions for driver mutations in patients with known BLCA in both size ranges (Supplemental Figs. S3A-C), indicating that size-selection of DNA fragments prior to library preparation was unnecessary. Furthermore, we found that enzymatic fragmentation yielded significantly higher DNA recovery than acoustic shearing (Supplemental Fig S3D; p=0.0006) and increased the deduplicated sequencing depth over unfragmented urine cfDNA by approximately two-fold (Supplemental Figs. S3E,F). Finally, we modified our previously published bioinformatics approach for analysis of plasma cfDNA to efficiently recover shorter DNA molecules present in fragmented urine cfDNA (see Methods).
To explore the utility of uCAPP-Seq we applied our optimized protocol and the BLCA panel to 118 urine and 60 tumor samples from 130 patients with BLCA (Supplemental Tables S1,S2,S4). These samples were derived from two independent patient groups (Methods, Supplemental Fig. S4), including one where urine was collected at the time of diagnosis (“Early Stage BLCA Group”, n=54) and a second where urine was collected during surveillance after treatment for localized BLCA (“Surveillance Group”, n=64).
In line with expectations from in silico predictions, we observed a median of 6 mutations per patient (Fig 1B; Supplemental Tables S5-S6). Next, we tested the concordance of mutations detected in tumor tissue and urine using 18 patients for whom paired urine and tumor tissue were available. Across these cases, a median of 66.7% mutations that were found in tumor tissue were also identified in utDNA while a median of 73.2% of mutations that were found in utDNA were also detected in paired tumor (Fig. 1C), with higher concordance between putative driver versus passenger mutations (p=0.009). Within tumor tissue, mutations that were also found in urine had higher median allele fractions than those not found in urine (27.2% vs. 9.2%, p<0.0001; Fig. 1D). Taken together, these results suggest that overall concordance between mutations found in bladder tumors and utDNA is high and is likely higher for truncal mutations than subclonal variants.
Tumor and urine genotypes in BLCA:
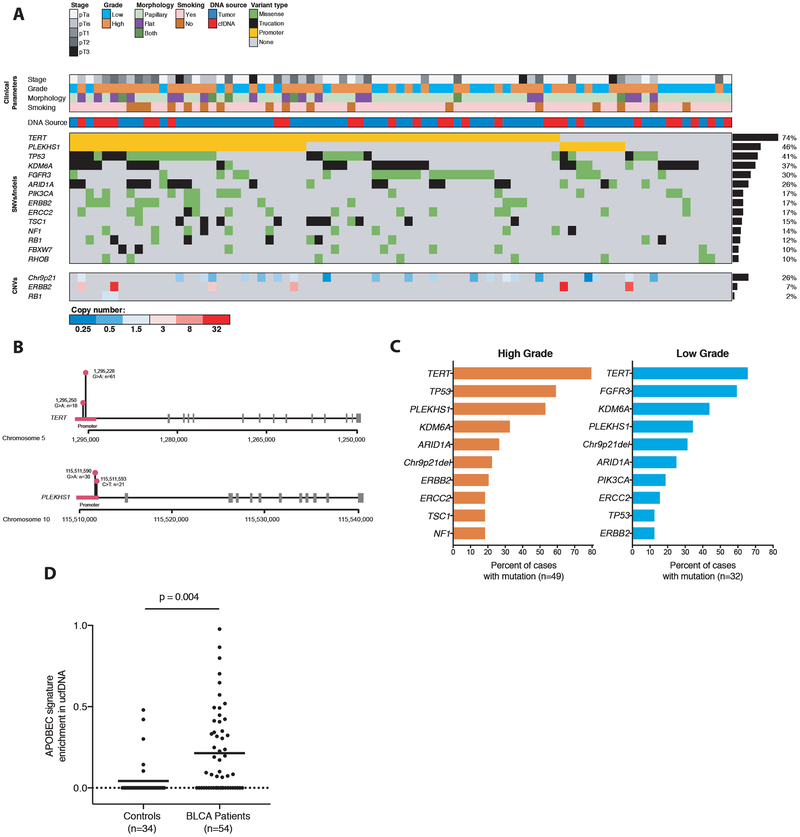
Across the 81 patients for whom we profiled either tumor tissue or urine at time of BLCA diagnosis, the two most commonly mutated regions were the TERT and PLEKHS1 promoters (Fig. 2A). TERT promoter mutations were present in 74% of cases, with all mutations occurring at the two previously described hotspots (Fig. 2B) (17). PLEKHS1 was originally described as a recurrent noncoding mutation in cancers by Weinhold and colleagues in an analysis of whole genome sequencing data, with mutations at one of two single-nucleotide hotspots (hg19 chr10:g.115511590 and 115511593) found in 20/863 (~2%) of cancers, including 8/20 of BLCAs (17). We identified PLEKHS1 promoter mutations in 37/81 (46%) of cases in our group, with all mutations clustered at these two previously described hotspots (Fig. 2B).
Figure 2: Genetic findings across bladder cancers profiled in study.
(A) Spectrum of genetic mutations and copy-number changes observed across 81 tumor and utDNA cases in this study, with clinicopathologic correlates. All tumor cases and all utDNA cases from patients with active cancer and at least one variant detected by genotyping were included in this analysis. All genes mutated in ≥ 10% of cases are shown, as well as all genes evaluated for copy-number variants. (B) Distribution of mutations in the TERT and PLEKHS1 promoters. (C) Comparison of mutations across high vs. low grade bladder cancers profiled in this study. (D) Enrichment of the APOBEC mutational signature in the cfDNA of patients with active bladder cancer versus healthy controls. P-values were calculated by multivariate regression controlling for total mutation count, median deduplicated sequencing depth, and the interaction between the two. APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like.
Nearly all other genes mutated in over 10% of the BLCA tumors we profiled were well-characterized driver genes, including TP53, FGFR3, ERBB2, and RB1. Consistent with prior studies, we observed a significantly higher frequency of TP53 mutations in high versus low grade tumors (59.2% vs. 12.5%; p<0.0001), and conversely, found disproportionately more FGFR3 mutations in low versus high grade cases (59.4% vs. 10.2%; p<0.0001) (Fig. 2C) (19). We also assessed copy number variants across genes known to be significantly altered in BLCA that were covered in our panel (16,18) and found deletions involving chromosome 9p21 in 27% of cases, amplifications of ERBB2 in 7%, and deletions of RB1 in 2% (Fig. 2A; Supplemental Tables S7,S8). We did not observe a correlation between the total mutation count and the stage of disease.
Since prior work had demonstrated enrichment of an APOBEC-related mutational signature in BLCA tumors by whole exome sequencing (18), we tested whether this signature was evident in utDNA by more targeted sequencing. Strikingly, we observed a significant enrichment of this APOBEC mutational signature in the urine cfDNA of patients with known BLCA compared to that of controls (p=0.004, Fig. 2D). The extent of APOBEC signature enrichment was significantly correlated in paired urine and tumor samples (r=0.64, p=0.0007; Supplemental Fig. S5) and, if validated, could suggest that cancer-related mutational signatures identified in cfDNA might serve as biomarkers for the presence of cancer.
Detection of early-stage BLCA using uCAPP-Seq:
We next tested the ability uCAPP-Seq to detect early-stage BLCA. We applied two different approaches to the detection of mutations in urine cfDNA, here designated “tumor-informed” and “tumor-naïve” profiling. The tumor-informed approach leverages prior knowledge by first sequencing a patient’s tumor specimen and germline tissue and then testing for the presence of these mutations in a urine sample using a Monte Carlo-based statistical approach, as previously described (8). In contrast, the tumor-naïve approach is designed to detect putative driver mutations without prior knowledge of the tumor genotype (see Supplemental Methods). We used 33 independent control subjects to establish threshold parameters for both the tumor-informed and tumor-naïve approaches, while 34 controls served to validate specificity (see Methods; Supplemental Fig. S6).
Our early-stage BLCA group consisted of pre-treatment urine from 54 individuals with biopsy-proven BLCA (74% pTa, 6% pTis, 9% pT1, 11% pT2) and 34 controls without BLCA. Although we attempted to match BLCA cases and non-BLCA controls for age and smoking status, controls were slightly younger (median 63.5 vs. 71) and contained fewer smokers (58% vs 85%; Supplemental Tables S9,S10). We observed a median urine cfDNA concentration of 12.0 ng/mL and yield of 523.3 ng among patients with active BLCA and a median urine cfDNA concentration of 6.88 ng/mL and yield of 254.5 ng among controls (p=ns for both comparisons).
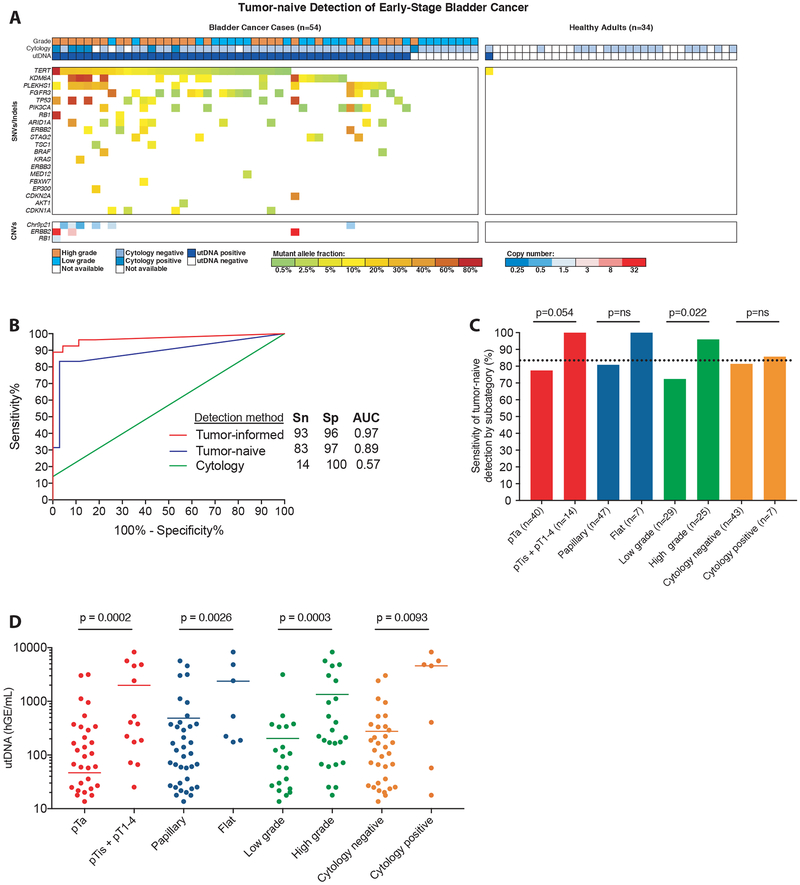
Using the tumor-naïve approach, we detected 134 putative driver mutations in the urine of cases and only 1 in the controls (Fig. 3A). The 1 mutation detected in the control cohort was a TERT promoter mutation at 11.4% variant allele fraction in patient CTR9500. A follow-up cystoscopy eight months later was negative for BLCA but identified a small raised lesion in the patient’s urethra that could potentially represent an early papilloma or other pre-neoplastic lesion. Among BLCA patients, cytology achieved a sensitivity of 14% while tumor-naïve utDNA profiling achieved a sensitivity of 83% (p<0.0001), both with high specificity (100% vs. 97%; p=ns) (Fig. 3B). Tumor-naïve profiling detected 77.5% of pTa and 100% of higher stage lesions (p=0.054) and a significantly greater fraction of high versus low grade cases (96% vs. 72.4%, p=0.022; Fig. 3C). In the subset of patients with available tumor tissue (n=27), tumor-informed profiling achieved an even higher sensitivity of 93% (significantly higher compared with cytology, p<0.0001), and a specificity of 96% (comparable to cytology, p=ns; Fig. 3B). Concentrations of utDNA were significantly correlated with clinical risk parameters, including T stage, lesion morphology, and grade (Fig. 3D). Urine samples that contained utDNA but which were negative by cytology tended to have lower concentrations of utDNA (median 94.2 hGE/mL vs 4592.8 hGE/mL, P=0.009), consistent with lower tumor burden.
Figure 3: Application of uCAPP-Seq to detect early-stage bladder cancer.
(A) Distribution of putative driver mutations identified in utDNA using tumor naïve profiling across patients with biopsy-proven bladder cancer (n=54) and controls (n=34), with associated tumor grade and cytology result. (B) Receiver operating characteristic analysis of tumor-informed profiling (n=27 cases, 34 controls), tumor-naive profiling (n=54 cases, 34 controls), and cytology (n=50 cases, 18 controls). (C) Correlates of detection by tumor-naive profiling among bladder cancer cases (n=54). P-values were calculated by the N-1 Chi Square test for comparing proportions. (D) Correlates of utDNA levels (haploid genome equivalents per mL, hGE/mL) among bladder cancer cases (n=54). P-values were calculated by the Mann Whitney test. SNVs, single-nucleotide variants; CNVs, copy-number variants; Sn, sensitivity; Sp, specificity; AUC, area under the curve; utDNA, urinary tumor DNA.
Surveillance of BLCA using uCAPP-Seq:
We next tested the performance of uCAPP-Seq for detecting recurrence in patients undergoing surveillance after local BLCA treatment. We prospectively collected urine samples from patients undergoing standard-of-care urine cytology testing for surveillance of BLCA and analyzed utDNA in the earliest available timepoint from patients who ultimately developed recurrent disease (n=37 patients). Recurrence was defined as biopsy-proven cancer (32/37 cases) or strong alternative evidence of recurrence such as positive urine cytology (5/37 cases; Supplemental Table S11, S12). We also analyzed the earliest timepoint available from patients who remained disease-free over at least 9 months of clinical follow-up (n=27 patients; median of 12.4 months; Supplemental Tables S11,S12).
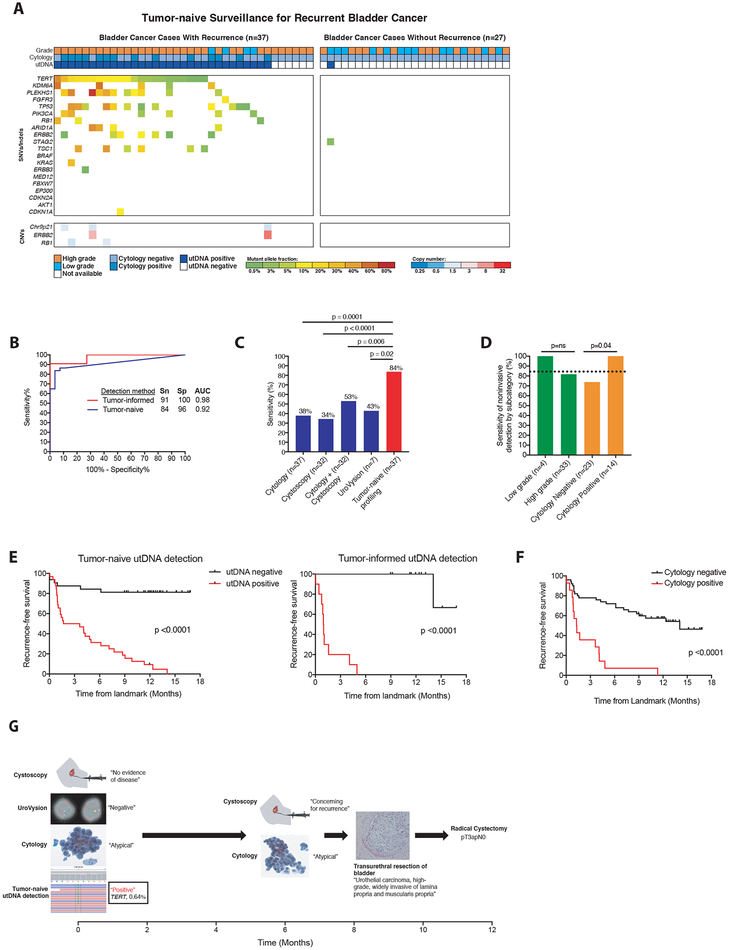
Using tumor-naïve utDNA profiling, we observed 88 putative driver mutations among patients experiencing disease recurrence and only 1 among those who did not (Fig. 4A). The 1 mutation detected in a patient who did not recur was a STAG2 nonsense mutation that was near the terminal end of the protein (residue 1029/1268) and therefore may not actually inactivate it. Of note, this variant was not present in paired tumor tissue from the same patient and thus most likely does not originate from tumor cells. Cytology was positive in 37.8% of patients who developed recurrence while utDNA was positive in 84% (p=0.0001) (Fig 4B, C). A concurrent multiplexed fluorescence in situ hybridization (FISH) test for aneuploidy (UroVysion) was available for 7 patients who developed recurrent disease and was positive for 3 (42.9%). This sensitivity was comparable to cytology (p=ns) but significantly lower than tumor-naïve utDNA profiling (p=0.019; Fig. 4C). Among 32 cases where cystoscopy was performed at the same time as the utDNA specimen, gross tumor recurrence was diagnosed in 11 patients (34.4%), again resulting in significantly lower sensitivity than uCAPP-Seq (p<0.0001). The sensitivity of cytology and cystoscopy combined (53.1%) was also significantly lower than tumor-naive utDNA profiling (p=0.0057). We detected mutant DNA in 1/27 patients who had at least nine months of negative follow-up, resulting in a specificity of 96% (Fig. 4B). The specificity of cytology could not be assessed in the control group as a positive cytology was an exclusion criterion.
Figure 4: Application of uCAPP-Seq to detect residual disease in the surveillance setting.
(A) Distribution of putative driver mutations identified in urine cfDNA using tumor naïve profiling across cases that developed recurrent cancer (n=37) and cases with at least 9 months of negative clinical follow-up (n=27), with recurrent cancer defined by biopsy (32 cases) or alternative clinical evidence (5 cases), as specified in Supplemental Table S12. (B) Receiver operating characteristic analysis of tumor-naive profiling (n=37 cases and 27 controls) and tumor-informed profiling (n=11 cases and 11 controls) across surveillance group. (C) Comparison of the sensitivities of cytology (n=37), cystoscopy (n=32), cytology plus cystoscopy (n=32), UroVysion (n=7), and tumor-naive profiling (n=37) in detecting residual BLCA. (D) Correlates of sensitivity for detecting disease by tumor-naive profiling. P-values for (C) and (D) were calculated by the N-1 Chi Square test for comparing proportions. (E) Kaplan-Meier analysis of recurrence-free survival stratified by utDNA detection by tumor-naive and tumor-informed profiling (HR 8.8 and 27.3), respectively and (F) by cytology (HR 4.6). P-values and HR were calculated by the log-rank test. (G) Example of patient detected by tumor-naive profiling but missed by cystoscopy, cytology, and UroVysion who later was diagnosed with muscle-invasive bladder cancer, requiring a radical cystectomy. SNVs, single-nucleotide variants; CNVs, copy-number variants; Sn, sensitivity; Sp, specificity; AUC, area under the curve; MRD, minimal residual disease; utDNA, urinary tumor DNA.
Tumor-naïve profiling detected 100% of recurrent cases that had positive cytology (n=14) and 73.9% that had negative cytology (n=23; p=0.04). We did not observe a significant difference in the detection rate between low- and high-grade tumors (Fig. 4D). In the subset of patients with available paired tumor tissue (n=22), tumor-informed profiling achieved a 91% sensitivity (significantly improving on cytology, p=0.002) with 100% specificity (Fig. 4B). Tumor-naive profiling, tumor-informed profiling, and cytology were all predictive of recurrence-free survival but utDNA achieved a wider separation of outcomes in patients with positive and negative findings (HR 8.8, 27.3, 4.6, respectively, all with p<0.0001; Figs. 4E and F). No significant differences were observed between patients who recurred and those who did not for age, sex, smoking history, prior tumor stage or morphology, or prior treatment type (surgery vs. intravesical therapy). Patients who did not recur, however, were more likely to have had prior low grade tumors and a longer interval between their last treatment and the specimen analyzed (Supplemental Tables S11,S12). Nevertheless, in multivariate logistic regression including these parameters, tumor-naïve utDNA profiling results remained highly significant (p=0.001), with an adjusted odds ratio for a “positive” classification of 128 (Supplemental Table S13).
Detection of utDNA preceded clinical disease recurrence in 92% of patients by a median of 2.7 months. This lead time is likely an underestimate, since many patients who recurred in our cohort had earlier surveillance time points that were negative by cytology but that we did not have access to for utDNA analysis. In addition to allowing noninvasive detection, our approach may therefore also facilitate earlier diagnosis of recurrence. The potential utility of this approach is highlighted by a case that was clinically classified as negative by cytology, UroVysion, and cystoscopy at the landmark timepoint but as positive by tumor-naive utDNA profiling (Fig. 4G). At the follow-up screening interval 6.1 months later, cytology was again negative but cystoscopy raised concern for recurrence. A transurethral resection of the bladder at 7.8 months revealed muscle-invasive BLCA, leading to a radical cystectomy at 11.7 months.
Discussion:
Here, we present a novel method for profiling utDNA called uCAPP-Seq and apply it to patients with BLCA. Our approach demonstrates high concordance for mutations between tumor and utDNA and enables genotyping of multiple somatic aberration types across a broad genomic space in a single integrated assay. Using this approach, we demonstrate that PLEKHS1 promoter mutations are among the most common somatic alterations in BLCA and are shed into the urine. We also highlight the potential of an APOBEC mutational signature as a utDNA biomarker for BLCA. In a group of patients with BLCA amenable to transurethral resection, we achieved a ~6-fold improvement in sensitivity over cytology while maintaining high specificity. Additionally, in a group of patients undergoing surveillance for recurrent BLCA after local therapy, we achieved ~2-fold improvements in sensitivity over cytology, the most commonly used ancillary test for BLCA (UroVysion), and gross evidence of disease by cystoscopy, detecting 92% of recurrences at a median of 2.7 months before clinical recurrence. Given the long lead time observed in some cases (Fig. 4G), it is tempting to speculate that intervention at the time of utDNA positivity might increase the likelihood of success of bladder-sparing interventions, though this will need to be tested in prospective trials. In summary, profiling of urine supernatants that are currently discarded in patients undergoing cytology could have significant value for disease detection and surveillance.
Across both groups, utDNA was detected in 21/21 (100%) of BLCA cases that were positive by cytology and 54/66 (82%) of BLCA cases that cytology missed. Although numerous studies have shown ancillary molecular testing can provide added sensitivity when used in conjunction with urine cytology, currently available tests typically miss a substantial fraction of cases cytology detects and fail to achieve cytology’s high specificity (2,4). Although uCAPP-Seq will need to be tested prospectively and in larger studies, our initial results suggest that it could offer a higher sensitivity alternative to cytology for the non-invasive detection of BLCA, for example, in the work-up of patients with microscopic hematuria. It could reduce the frequency of expensive and invasive cystoscopy procedures in a surveillance context. Although not explored in this study, uCAPP-Seq could potentially allow the monitoring of response to therapies such as intravesicular Bacillus Calmette-Guerin (BCG) or neoadjuvant chemotherapy through frequent monitoring of utDNA levels. Additionally, our approach could enable repeated monitoring of genome evolution during treatment (20).
Several recent studies have explored the use of cellular or cell-free urine DNA as a potential biomarker for BLCA (10–13,21). This includes two studies using amplicon-based HTS approaches (10,12). Specifically, Ward et al. and Springer et al. evaluated DNA in the cellular fraction of urine samples, achieving 70–85% sensitivity and 93–97% specificity in identifying bladder cancer at diagnosis, and in the case of Springer et al, 68% sensitivity and 80% specificity in identifying residual bladder cancer in a surveillance context (10,12). Our approach differs in using hybrid-capture target enrichment, interrogating a significantly larger genomic territory and number of mutations per case, and detecting somatic variants and copy number alterations in one assay. In contrast to Springer et al, we analyzed urine cfDNA instead of cellular DNA. Focusing on urine cfDNA in the supernatant has the advantage of utilizing material that is currently discarded and that, in some cases, may contain higher variant allele fractions than DNA isolated from the cellular pellet (13,21).
Limitations of our study include the case-control study design employed for our surveillance group, which enriched for clinically lower risk patients in the control subgroup compared to the cases. However, this imbalance has no impact on our comparison to cytology and UroVysion, as the same imbalances similarly affect those methods and would be expected to enhance their performance (2). Separately, the finding of utDNA positivity in the surveillance group remained highly significant in a multivariate analysis. Likewise, the cases and controls in our early-detection group were also slightly mismatched for median age and smoking status, raising the possibility that specificity might be lower in a perfectly matched control group. Finally, our cohort sizes are relatively small, and it will be important to test our method in larger patient cohorts and prospective trials.
In conclusion, we have developed a novel HTS-based approach to detecting utDNA and used it to explore genomic features and non-invasive diagnosis of BLCA. Our approach substantially improves on the performance of cytology while maintaining high specificity. Importantly, our approach will need to be assessed in additional groups and prospective clinical trials to establish its clinical utility.
Methods:
Patient selection and sample collection
Urine samples were collected from two groups, including (1) 54 patients with biopsy-proven early stage BLCA and controls at the Veterans Affairs Palo Alto Healthcare System prior to cystoscopy. Samples were also collected from (2) 410 patients undergoing surveillance for recurrent BLCA between June 2016 and August 2017 from the Stanford Cytopathology lab, and from 33 healthy volunteers. Characteristics of samples and patients from each group are listed in Supplemental Tables S5 and S7, respectively. Across both groups, tumor and germline tissue was collected where available from formalin fixed, paraffin embedded tissue blocks. This study was conducted with Institutional Review Board approval from both institutions in accordance with the Declaration of Helsinki. Written informed consent was obtained for samples acquired within the Veterans Affairs Palo Alto Healthcare System. Informed consent was waived by the Institutional Review Board for the use of discarded samples acquired at Stanford Hospital.
Sample processing
Samples were centrifuged to collect the pellet for clinical cytology, and the supernatant was combined with EDTA to a final concentration of 0.5 mM. Samples were stored at 4 degrees Celsius for up to one week, then at −80° Celsius. Among samples selected for further analysis, 10–50mL of urine supernatant was combined with Q-sepharose resin slurry (GE Healthcare, Chicago, IL) at a ratio of 100ul per 10mL urine and mixed for 30 minutes on a rolling drum. Mixtures were centrifuged at 1800g for 5 minutes to collect the resin and the supernatant was discarded. The resin was then washed on Econo-Pac Chromatography Columns (Biorad, Hercules, CA) with 10mL of 0.3M LiCl / 10mM sodium acetate (pH 5.5) and DNA was eluted with 1.675 mL of 2M LiCl / 10mM sodium acetate (pH 5.5) into 5mL 95% ethanol. The solution was then washed on a QIAquick column (Qiagen, Hilden, Germany) on a vacuum manifold with 1mL 2M LiCl in 70% ethanol, followed by 1 mL 75 mM potassium acetate (pH 5.0) in 80% ethanol. Residual fluid was removed by centrifugation for 3 minutes at 20,000g prior to elution in Buffer EB (Qiagen, Hilden, Germany). This protocol was adapted from Shekhtman et al. and scaled up for large fluid volumes (14). For each subject, a maximum of 42ng of urine cfDNA (Supplemental Tables S2 and S3) was subjected to enzymatic fragmentation with the Kapa Hyperplus Kit (Roche, Basel, Switzerland) for 30 minutes at 37 Celsius, after which library preparation proceeded as previously described (8).
Panel design and hybrid capture
Hybrid selection was performed with a custom SeqCap EZ Choice Library (Roche, Basel, Switzerland) designed through the NimbleDesign portal with support from the BioProd division, using genome build hg19/GRCh37. Hybrid capture and further processing were performed as described previously, prior to 151×2bp paired end sequencing on an Illumina HiSeq4000 (Illumina, San Diego, CA) with an 8 base indexing read.
Bioinformatics pipeline
Raw reads were demultiplexed, subjected to quality control and adapter content removal with the AfterQC package (22), and then mapped to human genome assembly hg19/GRCh37 using BWA-MEM (https://arxivorg/abs/13033997). Molecular barcoding, PCR duplicate removal, and adaptive variant calling were performed as previously described (8,24) with modifications to support variable fragment lengths. Raw variant calls were subjected to removal of stereotyped technical artifacts calibrated on 12 healthy control urine cfDNA samples (i.e. “polished”), as previously described (23). APOBEC mutational signature enrichment was identified by the deconstructSigs R package.
Statistical Analyses
Sensitivity and specificity were assessed in Receiver Operating Characteristic analyses, as described in the Supplemental Methods. The gold standard for a true positive was biopsy-proven cancer in all the cases in the early detection cohort and in 32/37 cases in the surveillance cohort. There were 5 cases in the surveillance cohort that did not have a supporting biopsy but did have strong alternative evidence of recurrent disease, including one or more “malignant” cytology diagnoses combined with a positive cystoscopy or imaging finding. Time-to-event analysis for recurrence-free survival was done using the log-rank test to estimate both p values and Hazard ratios and expressed as Kaplan-Meier plots. All statistical analyses were done using Prism 7 (GraphPad Software, La Jolla, CA), R v3.2.2 (http://www.r-project.org) through the RStudio environment, or medcalc.org. In calculating the sensitivity and specificity of cytology, ‘negative’ and ‘atypical’ results were considered clinically negative while ‘suspicious’ and ‘positive’ results were considered clinically positive (24). See Supplemental Methods for further details.
Supplementary Material
Significance:
This study shows that utDNA can be detected using high-throughput sequencing with high sensitivity and specificity in patients with early-stage BLCA and during post-treatment surveillance, significantly outperforming standard diagnostic modalities, and facilitating non-invasive detection, genotyping, and monitoring.
Acknowledgements:
We are grateful to the patients and families involved in this study and to the Stanford Cytopathology laboratory for preserving samples for this study. This work was supported with grants from the Joint Initiative for Metrology in Biology (J. Dudley; M. Diehn; 1186777–204-SBARL), the Stanford Pathology Department (J. Dudley; M. Diehn; 1194947–105-DHCRE), the Stanford Cancer Institute (J. Schroers-Martin), Albert Institute for Bladder Cancer Care and Research (J. Liao and M. Diehn), the National Cancer Institute (M. Diehn, A. Alizadeh; R01CA188298), the US National Institutes of Health Director’s New Innovator Award Program (M. Diehn; 1-DP2-CA186569), the Virginia and D.K. Ludwig Fund for Cancer Research (M. Diehn and A Alizadeh), and the CRK Faculty Scholar Fund (M. Diehn). This work used the Genome Sequencing Service Center by Stanford Center for Genomics and Personalized Medicine Sequencing Center, supported by the grant award NIH S10OD020141. Funding sources played no role in the writing of this manuscript or decision to submit it for publication.
Footnotes
Disclosure of Potential Conflicts of Interest:
J.D. has served as a consultant for Merck. M.L.Y.S. is an employee at Cepheid. A.A.A. and M.D. are co-inventors on patent applications related to CAPP-Seq. A.A.A. has equity in FortySeven and CiberMed and has served as a consultant for Roche, Genentech, Chugai, and Pharmacyclics. M.D. has equity in CiberMed has served as a consultant for Roche, Novartis, AstraZeneca, Varian Medical Systems, and Quanticel Pharmaceuticals.
References:
- 1.Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, Clark PE, et al. Bladder Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15(10):1240–67 doi 10.6004/jnccn.2017.0156. [DOI] [PubMed] [Google Scholar]
- 2.Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology 2003;61(1):109–18; discussion 18. [DOI] [PubMed] [Google Scholar]
- 3.Avritscher EB, Cooksley CD, Grossman HB, Sabichi AL, Hamblin L, Dinney CP, et al. Clinical model of lifetime cost of treating bladder cancer and associated complications. Urology 2006;68(3):549–53 doi 10.1016/j.urology.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 4.Chou R, Gore JL, Buckley D, Fu R, Gustafson K, Griffin JC, et al. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Ann Intern Med 2015;163(12):922–31 doi 10.7326/M15-0997. [DOI] [PubMed] [Google Scholar]
- 5.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8(346):346ra92 doi 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017. doi 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7(302):302ra133 doi 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 8.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20(5):548–54 doi 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34(5):547–55 doi 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer SU, Chen CH, Rodriguez Pena MDC, Li L, Douville C, Wang Y, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7 doi 10.7554/eLife.32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birkenkamp-Demtroder K, Nordentoft I, Christensen E, Hoyer S, Reinert T, Vang S, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70(1):75–82 doi 10.1016/j.eururo.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Ward DG, Baxter L, Gordon NS, Ott S, Savage RS, Beggs AD, et al. Multiplex PCR and Next Generation Sequencing for the Non-Invasive Detection of Bladder Cancer. PLoS One 2016;11(2):e0149756 doi 10.1371/journal.pone.0149756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel KM, van der Vos KE, Smith CG, Mouliere F, Tsui D, Morris J, et al. Association Of Plasma And Urinary Mutant DNA With Clinical Outcomes In Muscle Invasive Bladder Cancer. Sci Rep 2017;7(1):5554 doi 10.1038/s41598-017-05623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shekhtman EM, Anne K, Melkonyan HS, Robbins DJ, Warsof SL, Umansky SR. Optimization of transrenal DNA analysis: detection of fetal DNA in maternal urine. Clin Chem 2009;55(4):723–9 doi 10.1373/clinchem.2008.113050. [DOI] [PubMed] [Google Scholar]
- 15.Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol 2013;31(25):3133–40 doi 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507(7492):315–22 doi 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 2014;46(11):1160–5 doi 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171(3):540–56 e25 doi 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15(1):25–41 doi 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 20.Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov 2014;4(10):1140–53 doi 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Togneri FS, Ward DG, Foster JM, Devall AJ, Wojtowicz P, Alyas S, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet 2016;24(8):1167–74 doi 10.1038/ejhg.2015.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Huang T, Zhou Y, Han Y, Xu M, Gu J. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 2017;18(Suppl 3):80 doi 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nature Biotechnology 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brimo F, Vollmer RT, Case B, Aprikian A, Kassouf W, Auger M. Accuracy of urine cytology and the significance of an atypical category. Am J Clin Pathol 2009;132(5):785–93 doi 10.1309/AJCPPRZLG9KT9AXL. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.