Abstract
Engineering microbial communities in open environments remains challenging. Here, we describe a platform to identify and modify genetically tractable mammalian microbiota by engineering community-wide horizontal gene transfer events in situ. With this approach, we demonstrate that diverse taxa in the murine gut microbiome can be modified directly with a desired genetic payload. In situ microbiome engineering in living animals enables introduction of novel capabilities into established communities in their native milieu.
In nature, microbes live in open, dynamic and complex habitats that can be difficult, if not impossible, to recapitulate in a laboratory setting. Recent advances in deep sequencing have shed light on the vast microbial diversity that exists in many environments, including the mammalian gut. However, our ability to genetically access and alter these microbiomes remains limited despite advances in culturomics and synthetic biology1–4. While genetic intractability is often attributed to host immunity such as restriction-methylation5 or CRISPR-Cas processes6, a myriad of other factors (e.g., DNA transformation, growth state, fitness burden) may also influence gene transfer potential7. The inability to genetically alter a bacterium greatly limits our basic understanding of the organism and its biotechnological potential. To overcome these challenges, we devised an approach, Metagenomic Alteration of Gut microbiome by In situ Conjugation (MAGIC), to genetically modify gut microbiota in their native habitat by engineering the mobilome—the repertoire of mobile genetic elements that permeate the gut microbiome. Implementing MAGIC directly on a complex gut microbiome in its natural habitat enables genetic modification of diverse microbes and development of new modifiable microbial chasses for synthetic applications (Figure 1a).
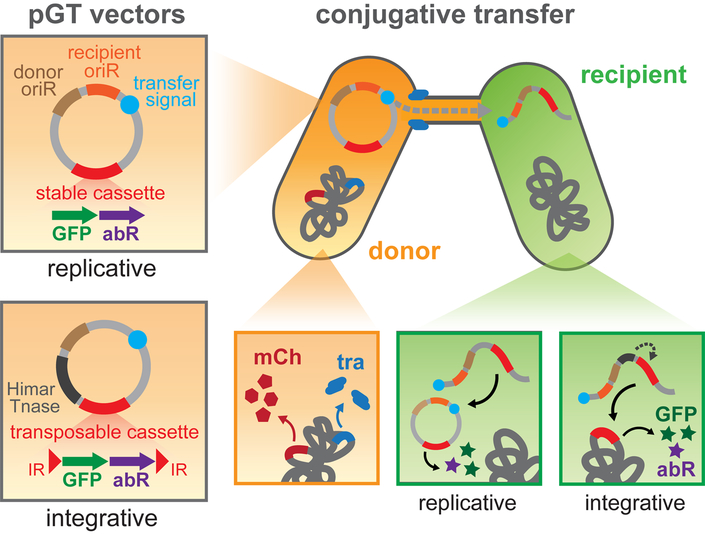
Figure 1. Overview of Metagenomic Alteration of Gut microbiome by In situ Conjugation (MAGIC).
(a) In contrast to traditional approaches to cultivate microbes first and then test for genetic accessibility, MAGIC harnesses horizontal gene transfer in the native environment to genetically modify bacteria in situ. Transconjugant bacteria can be detected by FACS or antibiotic selection and further manipulated. (b) MAGIC implementation to transfer replicative or integrative pGT vectors from an engineered donor strain into amenable recipients in a complex microbiome. Replicative vectors feature a broad-host range origin of replication (oriR), while integrative vectors contain a transposable Himar cassette and transposase. The donor E. coli strain contains genomically integrated conjugative transfer genes (tra) and a mCherry gene. Transconjugant bacteria are detectable based on expression of an engineered payload that includes GFP and an antibiotic resistance gene (abR).
We sought to utilize the mammalian gut as a testbed for MAGIC because it harbors a diverse microbial community that also plays a key functional role in host physiology8. We constructed a laboratory Escherichia coli donor strain that can deliver a genetic payload into target recipients by broad host-range bacterial conjugation (Figure 1b). The IncPα-family RP4 conjugation system9, which can efficiently conjugate into both Gram-positive and Gram-negative cells, was integrated into the EcGT1 donor genome along with a constitutively expressing mCherry-specR cassette (ΔgalK::mCherry-specR). To strengthen biocontainment of the donor and to facilitate in vitro selection of recipients, an alternative strain EcGT2 (Δasd::mCherry-specR) was generated to be auxotrophic for the essential cell wall component diaminopimelic acid (DAP), thus requiring DAP supplementation in the growth media10.
We developed a modular suite of mobile plasmids (pGT) that featured replicative origins with narrow to broad host-ranges, a RP4 transfer origin, a selectable marker, and the desired genetic payload (Suppl. Tables 1–3, Suppl. Figure S1). A broad host-range Himar transposon system was also utilized for delivering integrative payloads. As a demonstration of the system, we used a dual-reporter payload harboring a green fluorescent protein (GFP) and an antibiotic resistance gene (AbR). The use of Fluorescence Activated Cell Sorting (FACS) combined with 16S metagenomic analysis enables identification of successfully modified recipients or transconjugants, which can then be readily isolated on antibiotic selective plates. This multi-pronged strategy can increase the diversity of genetically tractable microbiota that can be captured. We first validated and optimized MAGIC protocols in vitro by assessing the gating stringency of FACS with control spike-ins of GFP-tagged bacteria into a complex sample community (Suppl. Figure S2). Subsequently, in vitro conjugations with defined recipient species (Suppl. Figure S3) and live bacterial communities extracted from mouse feces (Suppl. Figure S4) demonstrated the transfer of the payload from donors to recipients to yield GFP+ transconjugants that could be enriched by FACS (Suppl. Figure S5), which were confirmed by fluorescence microscopy (Suppl. Figure S6). 16S rRNA sequencing of FACS-enriched transconjugant populations revealed a diverse range of recipient bacteria (Suppl. Figure S7).
Since the gut microbiome can vary widely between hosts and can change due to dietary or other environmental variables, we explored the possibility of implementing MAGIC in vivo, directly in the native gut microbiome of an animal. We hypothesized that different groups of microbiota may be modified by using a library of pGT vectors that exhibit a range of gene expression levels and plasmid replication elements suitable for different gut bacteria. Libraries of pGT vectors (pGT-L1 to pGT-L6) were generated by modularly permuting pGT parts, including regulatory sequences of varying activity, payload selectable genes (bla, catP, tetQ), transposon elements (Himar), and plasmid origins (RSF1010, pBBR1, p15A) (Suppl. Table S1–S2). We carried out 4 separate in vivo studies where EcGT2 donors containing pGT libraries were orally gavaged into conventionally raised C57BL6/J mice obtained from commercial vendors. To assess the transfer capacity of individual pGT replicative or integrative designs (pBBR1, p15A-Himar, and RSF1010), we introduced each pGT library (pGT-L1, pGT-L2, or pGT-L3, respectively) separately into a mice cohort from Taconic (Suppl. Figure S8). We tested larger combinatorial libraries (pGT-L3 to pGT-L6) in two independent mice cohorts to assess variability across cohorts (Figure 2, Suppl. Figure S9). To compare in situ transfer in different gut communities, we tested the pGT-L6 library in mice from a different source (Charles River) (Suppl. Figure S10).
Figure 2. Identification and isolation of genetically tractable bacteria from the murine gut using MAGIC.
(a) Implementation of MAGIC in a murine model with fecal bacterial analysis by FACS, antibiotic selection, and sequencing. (b) FACS dot plots of fecal bacteria, pre- and post-gavage of EcGT2 donors containing pGT-L3 or pGT-L6 vector libraries. Green boxes define the sorted GFP+/mCherry- transconjugant populations. For each vector library, fecal samples from 3 co-housed mice were independently evaluated by flow cytometry with similar results. (c) Longitudinal analysis of fecal microbiome by flow cytometry for presence of EcGT2 pGT-NT donor cells (red triangles, n= 4 mice) and transconjugants of vector libraries pGT-L3 (purple circles, n=3 mice), pGT-L6 (maroon circles, n=3 mice), pGT-NT control (green circles, n= 4 mice), or PBS (no donor) control (orange circles, n=2 mice). Donor cells and transconjugants were lost within 48 hours. The dotted line shows the detection limit. (d) 16S taxonomic classification of transconjugants (GFP+/mCh−) enriched by FACS of pGT-L3 and pGT-L6 recipient groups. Each column represents transconjugants from one mouse. Each OTU’s relative abundance in the total bacterial population is shown in the grayscale heat-map, while each OTU’s fold enrichment among transconjugants is shown in the orange heat-map.. Bracketed values indicate confidence of taxonomic assignment by RDP classifier. Genera with successfully cultivated isolates are denoted by white stars. (e) PCR confirmed the presence of the antibiotic resistance/GFP payload cassette from pGT-L3 and pGT-L6 vectors in diverse isolates that were engineered in the murine gut and isolated by selective plating with carbenicillin or tetracycline. NA indicates 16S sequences that were not available.
We performed FACS enrichment and 16S metagenomic analysis on fecal material from all mice studies collected over time after oral gavage of pGT libraries (Figure 2a). Across in situ studies, up to 5% of resulting bacteria appeared to be successful transconjugants (i.e., GFP+/mCherry−) six hours post-gavage, compared to control groups (mice gavaged with PBS or EcGT2 carrying a non-transferrable vector pGT-NT) (Figure 2b, Suppl. Figures S8a, S9a, S10a). These GFP+/mCherry− transconjugants persisted for up to 72 hours post-gavage (Figure 2c, Supp. Fig S9b). 16S metagenomic sequencing of these transconjugant populations revealed a wide phylogenetic breadth (Figure 2d, Suppl. Figures S8b, S9c, 10b). Importantly, we observed significant reproducible enrichment of Proteobacteria and Firmicutes, especially Clostridiales and Bacillales, amongst successful transconjugants across multiple independent experiments. Using the same pGT-L6 library in mice from different vendors, which harbored distinct microbiomes (Suppl. Figure S10c), yielded shared and distinct transconjugants (Suppl. Figure S10d). In parallel to FACS-metagenomic studies, we isolated individual transconjugants from these fecal samples by selective plating for the payload antibiotic resistance gene and confirmed the presence of the GFP-AbR payload by PCR (Figure 2e). Across all experiments, we isolated and validated over 297 transconjugants belonging to 19 genera across 4 phyla (Suppl. Figure S11, Suppl. Table 4), validating the capacity of MAGIC to broadly transfer genetic material in situ into diverse recipients in the mammalian gut. In contrast, only 7 genera could be isolated from in vitro conjugation experiments using the same pGT vectors despite comparable diversity of transconjugants detected by FACS-metagenomics (Suppl. Figure S7). This difference may be due to in vitro conditions that sub-optimally support growth of diverse species during conjugation reactions, which underscores the value of implementing MAGIC in situ in an established complex microbiome.
Since transconjugants were no longer detected by 72 hours in situ (Figure 2c, Suppl. Figure S9b), we speculated that the genetic payload (GFP-AbR) on pGT vectors might be unstable or toxic, thus causing its negative selection in transconjugants. This hypothesis was tested in vitro by 20–30 serial passages of two transconjugant isolates of Escherichia fergusonii that contained the GFP-carbR payload either on a pGT-B1 (replicative pBBR1 origin) or a pGT-Ah1 (integrative Himar transposon) plasmid (Suppl. Figure S12). For the pGT-B1 population, we observed a significant increase in the fraction of GFP(−) cells (Suppl. Figure S12a–c). PCR assay of the origin of replication indicated that the pGT-B1 plasmid was no longer present in these GFP(−) cells (Suppl. Figure S12d). In contrast, cells in the pGT-Ah1 population remained GFP(+) despite a detectable loss of the plasmid in parts of the population over time (Suppl. Figure S12 e–g), which suggests a more stable maintenance of the GFP-CarbR payload as a integrative transposon within the host genome. Together, these results highlight the challenges of maintaining long-term in vivo stability of engineered genetic constructs in complex microbial communities, and suggest design considerations for more precise tuning of payload life-span and for improving payload biocontainment.
Microbes in the wild are known to possess a variety of natural mobile DNA elements and conjugation systems. Whole genome sequencing of three transconjugant strains of Proteus mirabilis and Escherichia fergusonii from our studies (designated as Modifiable Gut Bacteria MGB3, MGB4, and MBG9) revealed the presence of putative endogenous DNA mobilization systems (Suppl. Figure S13a–c). We wondered whether these native mobilization systems could interface with our engineered pGT vectors and thus performed in vitro conjugations of the MGB strains with laboratory E. coli recipients. Surprisingly, we discovered that MGB4 and MGB9 (both E. fergusonii) were able to mobilize pGT vectors into recipients, although at a lower efficiency than our engineered EcGT2 donor (Figure 3a, Suppl. Figure S13d). These results suggest that some native gut bacteria can promote secondary transfer of engineered payloads using their endogenous conjugation machinery, which may improve payload transfer in situ.
Figure 3. Transconjugant native gut bacteria recolonize the gut and mediate secondary transfer of engineered genetic payloads.
(a) Left panel: GFP expression profiles of three isolates (MGB3, MGB4, MGB9; n=5 for each) versus control strain (E. coli MG1655, n=5). MGB isolates were P. mirabilis (orange bar) and E. fergusonii (blue bars) containing either vector pGT-Ah1 (red border) or vector pGT-B1 (purple border). E. fergusonii strains were genetically identical, but received two different vectors. Right panel: efficiency of in vitro conjugation of pGT vectors from MGB strains to E. coli MG1655 recipients. EcGT2 donors were used as positive controls (gray bars). Sample sizes are n=2–4. Bars indicate means; error bars indicate standard deviation. (b) Schematic diagram of experiment: genetically tractable gut microbiota were isolated from the murine microbiome in vitro and then orally gavaged to recolonize the gut. (c) Colonization of MGB strains and EcGT2 lab strain in mice (n=6, n=4 respectively) over time, after initial oral gavage. Cell densities were determined by both plating (light green) and flow cytometry (dark green) of fecal bacteria, and by flow cytometry for E. coli (orange). Error bars indicate standard deviation. (d) FACS enrichment and 16S taxonomic classification of top in vivo transconjugants at 6 hours post-gavage with MGB strains. Fecal samples from 6 mice were combined for analysis. Each OTU’s relative abundance in the total bacterial population is shown in the grayscale heat-map, while each OTU’s fold enrichment among transconjugants is shown in the orange heat-map. Bracketed values indicate confidence of taxonomic assignment by RDP classifier. Red asterisks denote OTUs that share the same genus as MGB donors.
In general, non-gut adapted bacteria, including common probiotics, do not colonize an established gut microbiome. Infiltration of foreign species usually requires drastic perturbations, such as use of broad-spectrum antibiotics to suppress the natural flora. Even then, exogenous species do not persist upon discontinuation of antibiotic suppression11. Since our donor strains did not readily colonize the murine gut and transconjugants were lost soon after (Figure 2c, Suppl. Figures S9b, S14a), we reasoned that using a colonizing donor strain may extend the persistence of payload constructs in situ. To explore this possibility, we first tested whether a mixed population of MGB strains (MGB3, MGB4, MGB9) could stably recolonize the native murine gut after a single oral dose without any antibiotic co-administration (Figure 3b). In sharp contrast to the rapid loss of a non-gut-adapted strain (EcGT1) within 48 hours, MGB strains (especially MGB4) recolonized the murine gut and stably persisted for at least 15 days (Figure 3c, Suppl. Figure S15a), populating along the entire gastrointestinal tract (Suppl. Figure S15b). FACS enrichment and 16S sequencing of GFP-expressing bacteria in feces from these mice revealed transconjugants resulting from in situ transfer of the pGT payload from MGB strains to the native microbiome after 6 hours (Figure 3d) and even 11 days post-gavage (Suppl. Figure S15c). These transconjugant populations had similar phylogeny although less diversity than those from prior in situ experiments using the non-colonizing EcGT2 donor (Figure 2d, Suppl. Figure 9c). Together these results highlight the utility of MAGIC to isolate host-derived engineerable strains that can be modified and then used to stably recolonize the native community and mediate further transfer of engineered functions in situ.
In summary, MAGIC enables metagenomic infiltration of genetic payloads into a native microbiome and isolation of genetically modifiable strains from diverse communities. These modifiable native strains can then be reintroduced into their original community to maintain engineered functions via sustained vertical and horizontal transmission in situ. Future improvements to the system, such as optimization of vector stability and donor strain dosage (Suppl. Figure S14b), could enable better quantitative and temporal control of retention of genetic payloads in situ, which may be useful in applications requiring short-term or long-term actuation of engineered functions12–14. Designing genetic programs based on recipient-specific properties should enhance targeted execution of desired functions only in a defined subset of species in a community15, 16. Beyond the gut microbiome, MAGIC and complementary strategies to engineer the horizontal gene pool can facilitate programmable execution of genetic circuits in other microbial communities17–20. Isolation of genetically tractable representatives from diverse microbiomes will expand the repertoire of new microbial chasses for emerging applications in synthetic biology and microbial ecology.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Media, chemicals and reagents.
E. coli, S. enterica, V. cholera, and P. aeruginosa strains were grown in rich LB-Lennox media (BD) buffered to pH 7.45 with NaOH in aerobic conditions at 37°C, while L. reuteri was grown in MRS media (BD). B. thetaiotaomicron and E. faecalis were grown anaerobically at 37°C in Gifu Anaerobic Modified Medium (GAM) (Nissui Pharmaceutical) or BHI media (BD) supplemented with cysteine (1 g/L), hemin (5 mg/L), resazurin (1 mg/L), and Vitamin K (1 μL/L). All gut bacteria used in the study were grown in LB-Lennox or Gifu Anaerobic Modified Medium (GAM). Antibiotics were used at the following concentrations to select for E. coli: chloramphenicol (chlor) at 20 μg/ml, carbenicillin (carb) at 50 μg/ml, spectinomycin (spec) at 250 μg/ml, kanamycin (kan) at 50 μg/ml, tetracycline (tet) at 25 μg/ml, and erythromycin (erm) at 25 μg/ml. Antibiotics were used at the following ranges of concentrations to select for transconjugant gut bacteria: chloramphenicol (chlor) at 5–20 μg/ml, carbenicillin (carb) at 10–50 μg/ml, tetracycline (tet) at 5–25 μg/ml. Diaminopimelic acid (DAP) was supplemented at 50 μM as needed.
Isolation of live murine gut bacteria.
Fresh fecal pellets were harvested from mice, and live gut bacteria were isolated by mechanical homogenization. Briefly, 250 μL of PBS was added to previously weighed pellets in a microcentrifuge tube. Pellets were thoroughly mechanically disrupted using a motorized pellet pestle before adding 750 μL of PBS. The disrupted pellets in PBS were then subjected to four iterations of vortex mixing for 15 sec at medium speed, centrifugation at 1,000 rpm for 30 sec at room temperature, recovery of 750 μL of supernatant into a new tube, and replacement of that volume of PBS before the next iteration. The resulting 3 ml of isolated cells were pelleted by centrifugation at 4,000×g for 5 min at room temperature, the supernatant was discarded, and cells were re-suspended in 0.5–1.0 ml of PBS. All gut bacteria isolations were performed in an anaerobic chamber (Coy Labs).
Donor strain construction.
Donor strains EcGT1 and EcGT2 were derived from the S17 λpir E. coli strain21 by generating modifications ΔgalK::mCherry-specR and Δasd::mCherry-specR, respectively, with λ-red recombineering using the pKD46 system22. Synthetic cassettes containing constitutively active mCherry and spectinomycin resistance genes were constructed with ~40 bp of homology on both ends to galK or asd flanking regions on the E. coli genome. 100 ng of mCherry-specR cassette DNA were electroporated into recombineering-competent S17-pKD46 cells. Cells were allowed to recover in 3 mL LB+carb at 30°C for 3 hours prior to plating on LB+spec. Spectinomycin-resistant colonies were genotyped by PCR for validation of mutations. The pKD46 recombineering plasmid was cured out of validated recombinants by growth at 37°C in the absence of carbenicillin to yield the EcGT1 and EcGT2 strains used throughout the study. When generating the EcGT2 strain, the growth media was supplemented with DAP at all stages of the protocol.
Plasmid construction.
pGT vectors were designed to have modular components (e.g., selectable markers, regulatory elements, replication origins) that are interchangeable by isothermal assembly (ITA) or Golden Gate Assembly. Vector selection markers for E. coli were constitutively expressed, while the deliverable cargo or transposase cassettes were expressed using different regulatory elements to enable broad-host or narrow-host range gene expression. Regulatory elements used in this study exhibit a range of activity (Suppl. Table 1). Vector libraries used in this study are detailed in Suppl. Table 2. Full vector component sequences are listed in Suppl. Table 3. The non-transferrable vector pGT-NT used as a negative control was a minimal p15A cloning vector with no origin of transfer, containing a constitutively expressed sfGFP gene.
All plasmids were constructed by isothermal assembly (ITA) with NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). Component parts were made by high-fidelity PCR with Q5 (NEB) or KAPA Hifi (Kapa Biosystems) polymerases, using existing vectors or gBlocks (Integrated DNA Technologies) as PCR templates. PCR products were digested with DpnI (NEB) and purified with the QIAquick PCR purification kit (Qiagen) prior to ITA and transformation into E. coli. All assembled plasmids were Sanger sequence-verified.
In vitro MAGIC studies on synthetic recipient community.
Donor strains harboring pGT vectors and representative recipients (E. coli MG1655, S. enterica ATCC 700931, V. cholera C9503, P. aeruginosa PA01, E. faecalis ATCC 29200, L. reuteri ATCC 23272, B. thetaiotaomicron ATCC 29148) were grown overnight in appropriate media and cultivation conditions, and a 1:1000 dilution culture was re-grown for 14 hours at 37°C prior to conjugation studies. To prepare cells for in vitro conjugation, donor and recipient populations were washed twice in PBS and cells were quantified by OD600 or flow cytometry using SYTO9 staining (Thermo Fisher). 108 donor cells and 108 recipient cells were mixed together, pelleted by centrifugation, and re-suspended in 10μL PBS. Donor and recipient mixes were spotted on an agar plate and incubated for 5 hours at 30°C or 37°C for conjugation. In vitro conjugations were performed on LB-Lennox (E. coli, S. enterica, V. cholera, P. aeruginosa, E. faecalis), MRS (L. reuteri), or supplemented BHI agar (B. thetaiotaomicron). Post-conjugation, cells were scraped from the plate into 1 mL PBS, and 100 uL was plated on appropriate antibiotics and incubated overnight at 30°C or 37°C to determine the number of colony forming units (CFU) of transconjugants.
In vitro MAGIC studies on natural recipient community.
Donor strains harboring pGT vectors were streaked onto LB-Lennox agar plates with appropriate antibiotics and supplements, grown at 37°C overnight, and then grown from a single colony in 2 mL liquid media for 10 hours at 37°C prior to conjugation. The recipient community was isolated anaerobically from fresh murine feces as described above, immediately before conjugation. Donor cells were washed twice in PBS and quantified by OD600, while recipient cells were quantified by flow cytometry using SYTO9 staining. 108 donor cells and 109 recipient cells were mixed, pelleted by centrifugation at 5000 × g, and resuspended in 25 μL PBS. The mixes were spotted on PBS + 1.5% agar plates and incubated at 37°C either aerobically or anaerobically overnight (9–10 hours). Post-conjugation, cells were scraped from the plate into 1 mL of PBS and subjected to antibiotic selection on GAM media, FACS enrichment, and metagenomic 16S analysis (see below).
In vitro assessment of pGT vectors horizontal gene transfer mediated by natural isolates.
MGB natural isolates harboring pGT vectors (MGB3, MGB9, MBG4) were conjugated with a recipient E. coli strain harboring a kanamycin resistance plasmid compatible with pGT vectors. Prior to conjugations, all strains were streaked onto GAM agar plates with appropriate antibiotics, grown at 37°C overnight, and then grown from a single colony in 5 mL liquid GAM for 10 hours at 37°C prior to conjugation. MGB donor and recipient cells were washed twice in PBS and quantified by OD600. 109 cells each of MGB and recipient strains were mixed, pelleted by centrifugation at 5000 × g, and resuspended in 15 μL PBS. The mixtures were spotted on GAM agar plates and incubated at 37°C aerobically for 6 hours. Post-conjugation, cells were scraped from the plate into 1 mL of PBS and plated on selective and non-selective GAM media. Conjugation efficiency was calculated as , where t is the number of E. coli transconjugant CFUs and n is the total number of E. coli CFUs.
Measurement of GFP expression in MGB strains.
MGB isolates harboring pGT vectors (MGB3, MGB9, MBG4) were streaked onto GAM agar plates with appropriate antibiotics, grown at 37°C overnight, and then diluted to OD600 0.001 in liquid GAM into a 96 well plate. The plate was incubated in a Synergy H1 (BioTek) microplate reader for 24 hours at 37°C with orbital shaking. Measurements of OD600 and GFP expression (excitation 488 nm, emission 510 nm) were taken using Gen5 software (BioTek) at the end of 24 hours.
In vivo MAGIC studies in mice.
Conventionally raised C57BL/6 female mice (Taconic Biosciences or Charles River Laboratories) were used throughout the study. Two control groups of 4 mice each were gavaged with PBS and EcGT2 containing a non-transferable GFP vector (pGT-NT). Three to four mice were used in each group gavaged with a pGT donor mix or with MGB strains. To equilibrate the murine gut microbiome ahead of time, mice from multiple litters were mixed, co-housed for at least 1 week prior to all experiments, and randomly allocated into groups. Mice were gavaged with 109 donor cells (EcGT2 or MGB strains) in 300 uL of PBS at 8–10 weeks old. Control mice were gavaged with 300 uL of PBS. Fecal matter was collected immediately before gavage and periodically after gavage to analyze the resulting microbiome populations by FACS, metagenomic 16S sequencing, and plating. Upon completion of the study, mice were euthanized and small and large intestinal tissues were extracted. Luminal contents were washed from each tissue sample with PBS and bacteria were extracted by homogenization of the luminal contents for plating and final CFU determination.
Flow cytometry and fluorescence-activated cell sorting (FACS) measurements.
Gut bacteria isolated from fresh fecal pellets were analyzed for evidence of successful conjugation on a flow cytometer (Guava easyCyte HT) using red (642 nm) and blue (488 nm) lasers with Red2 and Green photodiodes to detect mCherry (587/610 nm) and sfGFP (485/510 nm) fluorescence, respectively. Bacteria at 100x and 1,000x dilutions in PBS were used for optimal detection of donor (GFP+/mCh+), gut microbes without a transferred vector (GFP−/mCh−), and transconjugants (GFP+/mCh−). Data were collected and analyzed usingInCyte 3.1 software. For FACS enrichment studies, a BD FACS Aria II cell sorter operated with BD FACSDiva software was used to gate for sfGFP (FITC filter 515/10nm) and mCherry (mCherry filter 616/26nm). A double gating on GFP and mCherry channels was used to select for cells with GFP+/mCh− fluorescence. In addition, background events were also taken into account by using the GFP+/mCh− fluorescence detected in the fecal sample prior to gavage as baseline signal. An increase over the baseline signifies an enrichment of transconjugants. Population density (cells/gram fecal matter) was calculated based on number of cells sorted over the mass of the sorted fecal sample. Additional plating and direct colony counting were used to validate flow cytometric measurements. FACS plots were formatted using FCS Express 6.
Fluorescence microscopy of fecal bacteria.
Bacteria were suspended in PBS and centrifuged at 5000×g to concentrate into a smaller volume, which varied depending on the concentration of bacteria. The bacteria were resuspended by pipetting, and a volume of 15 uL was dropped onto a Superfrost Plus microscope slide (Thermo Shandon) and covered with a glass cover slip. Slides were air-dried until the PBS receded from the edges of the cover slip and then sealed with clear nail polish. Bacteria were imaged at 40x magnification on a Nikon Eclipse Ti2 microscope on bright field, RFP, and GFP channels using NIS-Elements-AR software.
Validation of pGT vectors in transconjugants.
Transconjugant validation was performed by colony PCR of the GFP-antibiotic resistance payload and/or the pGT vector backbone. PCR products with the expected size were further verified by Sanger sequencing. Taxonomy assignment of isolated colonies was based on 16S rRNA PCR amplification and Sanger sequencing. All transconjugant strains validated in the study are listed in Suppl. Table 4.
In vitro evolution of transconjugant gut bacteria.
Escherichia fergusonii transconjugants MGB4 and MGB9 were serially passaged in LB media for 11–15 days. Starting from a single colony, the strains were inoculated into LB and grown at 37C with shaking. Every 12 hours the liquid culture was diluted 1:1000 into fresh LB media. At selected time points an aliquot of the saturated culture was plated on selective (50 μg/mL carbenicillin) and non-selective plates to quantify the percentage of cells expressing the payload antibiotic resistance and GFP genes. MGB9 cultures were also plated on selective plates with 20 μg/mL chloramphenicol to check for maintenance of the plasmid backbone.
Metagenomic 16S sequencing.
Genomic DNA was extracted from isolated bacteria populations using the MasterPure Gram Positive DNA Purification Kit (Epicentre). PCR amplification of the 16S rRNA V4 region and multiplexed barcoding of samples were performed based on previous protocols3. The V4 region of the 16S rRNA gene was amplified using customized primers based on the method described in Kozich et al.23 with the following modifications: (i) alteration of 16S primers to match updated EMP 505f and 806rB primers24–26 and (ii) use of NexteraXT indices such that each index pair is separated by a Hamming distance of >2 and Illumina low-plex pooling guidelines can be used. Sequencing was performed using the Illumina MiSeq system (500V2 kit).
Analysis of 16S next-generation sequencing (NGS) data.
Bacteria from fecal samples taken right before gavage (T0) and 6 hours post-gavage (T6) and were sorted by FACS to enrich for transconjugants. The compositions of the sorted transconjugant and total populations for each sample were determined from 16S sequencing data using the UPARSE pipeline27 (USEARCH version 10.0.240) to generate Operational Taxonomic Unit (OTU) tables and abundances and the RDP classifier28 to assign the taxonomy. Phylogenetic associations were analyzed at the genus level with at least 90% confidence for 16S assignment. In all MiSeq runs, two blank controls with sterile water as input material were included to check for contaminants in the reagents and to filter out contaminant OTUs if present. Reads mapping to non-bacterial DNA (e.g., mitochondria, plastids, or other eukaryotic DNA) were also excluded from analysis. Only OTUs with more than 10 reads were considered in downstream analysis.
Relative abundances of OTUs in unsorted total fecal populations were calculated as the normalized number of reads in a sample. Relative abundances of OTUs in T0 FACS-enriched populations were used to measure false positive background fluorescence, which was subtracted from the T6 transconjugant populations. The corrected relative abundance of each OTU in a T6 FACS-enriched population is given by the formula:
where RAt,i,sorted is the corrected relative abundance of OTU at time t, At,i is the normalized number of reads of OTU i at time t in the FACS-sorted sample, and Nt is the fraction of mCherry-/GFP+ FACS-sorted events at time t. OTUs for which RA6,i,sorted is negative are eliminated from subsequent analysis, and all remaining RA6,i,sorted values are renormalized.
The fold enrichment of each OTU in the FACS-sorted population is defined as its relative abundance in the FACS-sorted population divided by its relative abundance in the unsorted total population at T6. To overcome the problem of detection limits (i.e., OTU i appears in the sorted population but is below the detection limit in the total population), we added a pseudo-count of p to all relative abundances when calculating fold enrichments. p is given by
where n is the total number of reads in the FACS-sorted sample and [−log10 n]is the floor function - the greatest integer less than or equal to - of −log10 n. The fold enrichment of OTU i with the pseudo-count correction is calculated as
If the relative abundance of OTU i in the unsorted population is below the detection limit, then the fold enrichment is calculable as , instead of.
The pseudo-count-corrected fold enrichment Fi overestimates the true fold enrichment () by at most 10%, while possibly underestimating it. Because and,
In all heat maps showing fold enrichment versus relative abundance, only OTUs with Fi > 10 are displayed to show more stringent and high confidence results. R code for this analysis is available upon request.
Whole genome sequencing of engineered mouse gut bacteria (MGB) isolates.
To sequence MGB isolates, we prepared a sequencing library using the Nextera kit (Illumina) and utilized the Illumina HiSeq 2500 platform for 100 bp single-end reads. The SPAdes single cell assembler pipeline (version 3.9.1)29 was employed to generate whole genome contigs. BLAST and PlasmidFinder (version 1.3)30 were used to analyze the sequences and identify native mobilization systems. Geneious (version 7.1.5) was used to visualize contig alignments to genomes and plasmids.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Wang lab for helpful scientific discussions and feedback, especially Ravi Sheth for helpful insights on NGS library preparation. H.H.W. acknowledges funding from DARPA (W911NF-15-2-0065), NIH (1DP5OD009172, 5R01AI132403, 1R01DK118044), NSF (MCB-1453219), ONR (N00014–15-1-2704), and Burroughs Wellcome PATH (1016691). C.R. is supported by a Junior Fellowship of the Simons Society of Fellows (#527896). S.P.C. is supported by a NIDDK F30 fellowship (F30 DK111145–01A1) and a NIH MSTP training grant (NIH T32GM007367). We also thank Amir Figueroa at the Columbia University Medical Center Flow Cytometry Core for assistance with FACS studies.
Footnotes
DATA AVAILABILITY
The raw data from this study are available from the corresponding author upon request.
MATERIALS AVAILABILITY
All modular vector part sequences are listed in Supplementary Table 3. Full plasmid maps, vectors and strains used in this study are available from the corresponding author upon request.
ANIMAL ETHICS STATEMENT
All animal experiments were performed in compliance with Columbia University Medical Center IACUC protocols AC-AAAU6464 and AC-AAAL2503.
COMPETING FINANCIAL INTERESTS
A provisional patent application has been filed by The Trustees of Columbia University in the City of New York based on this work. The authors declare no additional competing financial interests.
REFERENCES
- 1.Lagier JC et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 1, 16203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Yaung SJ, Church GM & Wang HH Recent progress in engineering human-associated microbiomes. Methods in molecular biology 1151, 3–25 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Cuiv PO et al. Isolation of Genetically Tractable Most-Wanted Bacteria by Metaparental Mating. Sci Rep 5, 13282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mimee M, Tucker AC, Voigt CA & Lu TK Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tock MR & Dryden DT The biology of restriction and anti-restriction. Curr Opin Microbiol 8, 466–472 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Marraffini LA CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Thomas CM & Nielsen KM Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3, 711–721 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pansegrau W et al. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J Mol Biol 239, 623–663 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Hapfelmeier S et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myhal ML, Laux DC & Cohen PS Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol 1, 186–192 (1982). [DOI] [PubMed] [Google Scholar]
- 12.Kommineni S et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saeidi N et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol 7, 521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steidler L et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Wegmann U, Horn N & Carding SR Defining the bacteroides ribosomal binding site. Applied and environmental microbiology 79, 1980–1989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheth RU, Cabral V, Chen SP & Wang HH Manipulating Bacterial Communities by in situ Microbiome Engineering. Trends Genet 32, 189–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klumper U et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. The ISME journal 9, 934–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlberg C, Bergstrom M & Hermansson M In Situ Detection of High Levels of Horizontal Plasmid Transfer in Marine Bacterial Communities. Appl Environ Microbiol 64, 2670–2675 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bikard D et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature biotechnology 32, 1146–1150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brophy JAN et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol 3, 1043–1053 (2018). [DOI] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES
- 21.Simon R, Priefer U & Puhler A A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech 1, 784–791 (1983). [Google Scholar]
- 22.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. P Natl Acad Sci USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parada AE, Needham DM & Fuhrman JA Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Apprill A, McNally S, Parsons R & Weber L Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology 75, 129–137 (2015). [Google Scholar]
- 27.Edgar RC UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM & Cole JR Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurk S et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20, 714–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carattoli A et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58, 3895–3903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stalker DM, Kolter R & Helinski DR Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proceedings of the National Academy of Sciences of the United States of America 76, 1150–1154 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiszczynska-Sawicka E & Kur J Effect of Escherichia coli IHF mutations on plasmid p15A copy number. Plasmid 38, 174–179 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Kues U & Stahl U Replication of plasmids in gram-negative bacteria. Microbiol Rev 53, 491–516 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antoine R & Locht C Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol Microbiol 6, 1785–1799 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Frey J, Bagdasarian MM & Bagdasarian M Replication and copy number control of the broad-host-range plasmid RSF1010. Gene 113, 101–106 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Bryksin AV & Matsumura I Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PloS one 5, e13244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ & Robertson HM Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci U S A 96, 11428–11433 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerry P, van Embden J & Falkow S Molecular nature of two nonconjugative plasmids carrying drug resistance genes. Journal of bacteriology 117, 619–630 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholz P et al. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75, 271–288 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.