Abstract
Otoancorin (OTOA), encoded by OTOA, is required for the development of the tectorial membrane (TM) in the inner ear. Mutations in this gene cause non-syndromic hearing loss (DFNB22). The molecular mechanisms underlying most of DFNB22 remains poorly understood. Disruption of glycosylphosphatidylinositol (GPI) anchorage has been assumed to be the pathophysiology, mandating experimental validation. From a Korean deaf family, we identified two trans OTOA variants (c.1320+5G>C and p.Gln589ArgfsX55 (NM_144672.3)). Pathogenic potential of c.1320+5G>C was confirmed by a minigene splicing assay. To experimentally determine the GPI anchorage, wildtype and the mutant OTOA harboring p.Gln589ArgfsX55 was expressed in the HEK293T cells. The mutant OTOA with p.Gln589ArgfsX55 resulted in an uncontrolled release of OTOA into the medium in contrast with phosphatidylinositol-specific phospholipase C-induced controlled release of wildtype OTOA from the cell surface. Together, this reverse translational study confirmed GPI-anchorage of OTOA and showed that downstream sequences from the 589th aa is critical for GPI-anchorage.
Keywords: OTOA, Deafness, DFNB22, Tectorial membrane, Glycosylphosphatidylinositol (GPI)-anchorage
OTOA encodes otoancorin (OTOA), a protein required for limbal attachment of the tectorial membrane (TM), which is necessary for conditioning the proper stimulation of the inner hair cells (Lukashkin et al., 2012). Association of OTOA and hearing loss (DFNB22) was first reported in 2002 (Zwaenepoel et al., 2002). Until recently, most alterations of OTOA associated with deafness have been detected in North African or Middle Eastern nationals, including Palestinians, Pakistanis, Qataris, Turkish, and Algerians (Alkowari et al., 2017; Bademci et al., 2014; K. Lee et al., 2013; Shahin et al., 2010; Walsh et al., 2006; Zwaenepoel et al., 2002).
Copy number variations (CNVs) including large deletions or segmental duplications of OTOA, which could have evaded conventional NGS-based sequencing, have been reported in DFNB22. Comprehensive analysis of the CNVs of OTOA (Shearer et al., 2014) first reported DFNB 22 in patients of non-Middle Eastern ethnicity. DFNB22 was later reported in other ethnicities including Han Chinese and European populations etc. (He et al., 2018; Sloan-Heggen et al., 2016; Sommen et al., 2016). Currently, eight missense variants, two frameshift variants, one nonsense and one splice site variant of OTOA have been known to be associated with DFNB22 (Supp. Table S1), in addition to large/ whole deletions or duplications of this gene (Ammar-Khodja et al., 2015; He et al., 2018; K. Lee et al., 2013; Sloan-Heggen et al., 2016; Sommen et al., 2016; Walsh et al., 2006; Zwaenepoel et al., 2002). However, no point mutation or CNV of OTOA has yet been reported in the Korean deaf population.
OTOA, which contains an N-terminal secretory signal peptide (SP) and a C-terminal hydrophobic patch, was previously predicted as a glycosylphosphatidylinositol (GPI)-anchored protein (AP) using a computational GPI-anchorage prediction tool (Zwaenepoel et al., 2002). Since this report, disruption of GPI-anchorage has been assumed to play a role in the pathophysiology of OTOA-related deafness. However, it is critical to experimentally validate the GPI-anchorage of predicted proteins, since the efficiency of a GPI-anchorage varies depending on GPI-attachment sequences. Indeed, some proteins that pass the prediction algorism are not GPI-anchored (Galian, Bjorkholm, Bulleid, & von Heijne, 2012). Conversely, other proteins, which were regarded as a type I transmembrane protein, were proven to be GPI-anchored, contrary to prediction (Davies et al., 2010). GPI-anchorage of OTOA itself has not yet been experimentally verified, and the mechanism involving alterations in the OTOA gene leading to hearing loss is still not known.
Here, we report a novel causative variant of OTOA in the Korean deaf population. Further, through reverse translational research, we experimentally confirmed that OTOA is indeed GPI anchored and showed at least some downstream sequences from the 589th amino acid is critical for its function, suggesting that disruption of GPI-anchorage could be involved in the pathophysiology of DFNB22.
We identified two potential OTOA pathogenic variants, c.1320+5G>C and c.1765delC (p.Gln589ArgfsX55) in a Korean hearing-impaired pedigree (SB285) through the whole-exome sequencing (WES) and subsequent bioinformatics analysis (Chang et al., 2018; Han et al., 2017; Kim et al., 2018) (see the Supplementary Materials and Methods). We obtained a written informed consent from SB285–618 and SB285–586. For SB285–576, the written informed consent was obtained from his parents (Supp. Figure S1).
Because the patient (a 21-month-old Korean male, designated here as SB285–576) failed to pass universal newborn screening and displayed moderate to severe sensorineural hearing loss (Supp. Figure S1A and S1B), we analyzed the SB285, to identify potentially pathogenic genes. A stepwise filtering process including in silico analysis and a segregation study revealed two variants possibly responsible for the hearing loss: c.1320+5G>C in the splice region site just downstream of exon 13 and c.1765delC (p.Gln589ArgfsX55) in exon 17 of OTOA (NM_144672.3) (Supp. Figure S1C and Supp. Table S2). The c.1320+5G>C variant was identified as a novel splice region variant while c.1765delC (p.Gln589ArgfsX55) was previously detected as a single heterozygous state in a profoundly deaf patient, where it was assumed to be fortuitously detected irrespective of its pathogenic potential and causality were doubted (Park et al., 2014). In contrast, c.1765delC (p.Gln589ArgfsX55) was confirmed to be in a trans configuration with another potentially pathogenic variant in our present study, increasing the likelihood of the pathogenic contribution of this variant to deafness of SB285.
Disruption of the splicing region by c.1320+5G>C was initially suggested by ESEfinder, NNSplice, MaxEntScan::score5ss, and NetGene2. The c.1320+5G>C and p.Gln589ArgfsX55 alleles were rarely or never detected in the ExAC database, gnomAD and among 1,722 ethnicity-matched Korean control subjects (KRGDB database), suggesting its potential pathogenicity (Supp. Table S2). It was also classified as likely pathogenic in Deafness Variation Database (http://deafnessvariationdatabase.org/) (Accessed: December 31, 2018) (Azaiez et al., 2018). In addition, the c.1320+5G and p.Gln589 residues were well conserved across several different species in vertebrates, as supported by high GERP++ scores of 5.21 and 5.22, respectively, and showed deleterious effects according to the Mutation taster and CADD score (Supp. Table S2). Taken together, the two variants of OTOA found in SB285 were predicted to be likely pathogenic (c.1320+5G>C) or pathogenic (c.1765delC), according to the guidelines for the interpretation of variants (Oza et al., 2018; Richards et al., 2015).
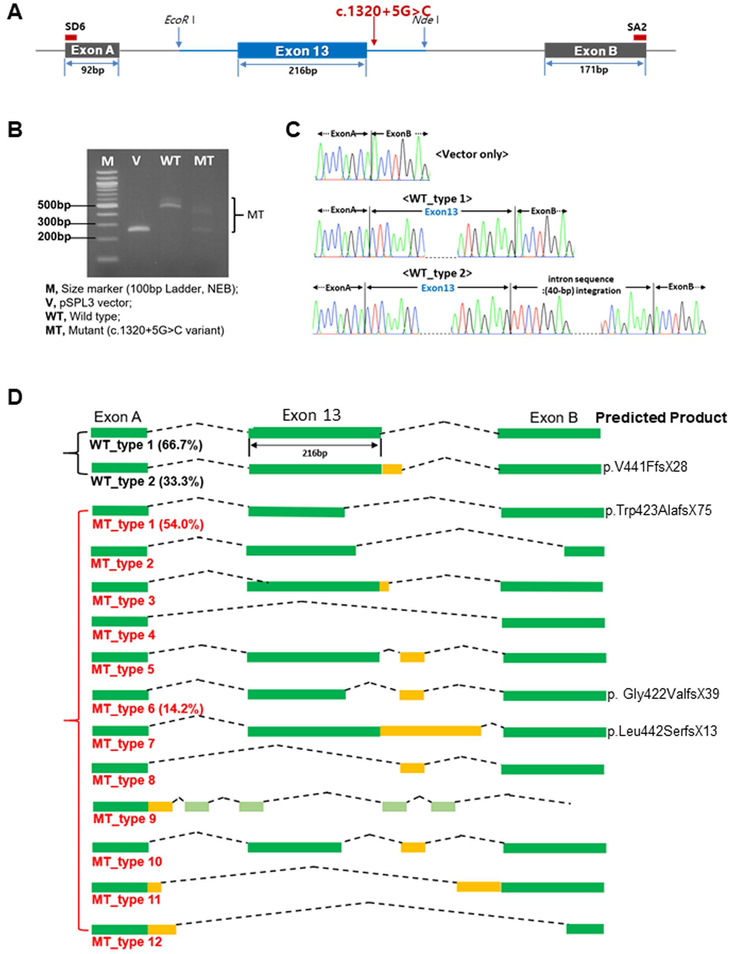
The altered splice region site by c.1320+5G>C may have generated alternative transcripts, resulting in production of pathogenic OTOA mutants with variable C-terminal domains. To test this hypothesis, we carried out a minigene splicing assay using plasmids harboring downstream of intron 12 and exon 13, and upstream of intron 13, of either the wild-type (WT) OTOA or c.1320+5G>C (Figure 1A). We transfected the pSPL3 vector only, the WT, and c.1320+5G>C into Cos-7 cells independently, and collected cells to extract transcripts. Using the reverse transcription-polymerase chain reaction (RT-PCR), the quantity and size of transcripts from each construct were visualized on an agarose gel (Figure 1B). While the WT-transfected cells expressed two major transcripts (479 bp and 519 bp), the mutant-transfected cells expressed shorter (200 bp) or longer (643 bp) transcripts, with a smeared banding pattern. The splicing patterns of the transgenes were further confirmed by Sanger sequencing (Figure 1C). The WT construct generated two forms of transcripts: 1) [(Exon A-Exon 13-Exon B) (6/9 colony pick, 66.7%)] and 2) [(Exon A-Exon 13–40 bp of upstream sequences of Intron 13) (3/9 colony pick, 33.3%)], indicating (Exon A-Exon 13-Exon B) was a dominant form, consistent with the RT-PCR data shown in Figure 1B. However, the mutant construct generated 12 distinctive types of transcripts [mutant (MT) type 1, 61/113 colony pick, 54.0%; MT type 6, 16/113 colony pick, 14.2%], each featuring different splicing patterns (Figure 1D and Supp. Table S3). Notably, no normal splicing pattern was detected in the c.1320+5G>C mutant. Statistical analysis showed a significantly higher portion of normal splicing patterns in the WT than in the mutant (P < 0.00001), indicating that c.1320+5G>C resulted in the production of multiple mutant forms of OTOA (Supp. Table S3). A minigene splicing assay performed on the previously proven pathogenic variant: c.1320+2T>C (Zwaenepoel et al., 2002) for doublechecking the integrity of this system showed a similar pattern on an agarose gel with that of c.1320+5G>C and the most prominent band turned out to be identical with the MT type 7 from c.1320+5G>C (Supp. Figure S2).
Figure 1. Mutation in the OTOA splicing region site by c.1320+5G>C generated multiple splicing isoforms and affected transcript stability.
(A) Generation of pSPL3 plasmid incorporating the c.1320+5G>C splicing variant.
(B) Reverse transcription-polymerase chain reaction products shown on an agarose gel with multiple bands for the wild-type and mutant samples.
(C) Sanger sequencing chromatograms of two types of spliced transcripts from the wild-type and one type of transcript from the vector only, directly quantitated using the polymerase chain reaction, TA cloning, and Sanger sequencing results.
(D) Schematic illustration of the two types of splicing products from the wild-type and 12 types of those from the mutants, with the two most prevalent ones showing percentages, directly quantitated using the polymerase chain reaction, TA cloning, and Sanger sequencing.
Regarding a GPI-anchorage of OTOA, we performed an in vitro assay to test whether WT OTOA contains a GPI-anchor for the surface-tethering of the protein. In detail, we transfected HEK293T cells with a full-length OTOA construct, which was myc-tagged just downstream to the N-terminal SP (myc-OTOA), and the levels of OTOA in the cell lysate and the medium were then determined by western blotting (Figure 2A). While OTOA was not initially detected in the medium, treatment of transfected cells with a bacterial phosphatidylinositol-specific phospholipase C (PI-PLC) induced the release of OTOA into the medium. Migration of the released OTOA by PI-PLC was slower than that in the cell lysate, and additional peptide:N-glycosidase (PNGase) F treatment produced the same size band from the cell lysate and the medium, indicating that hyper-glycosylated OTOA was expressed on the cell surface, while hypo-glycosylated proteins were retained inside the cells (Figure 2A). Given that PI-PLC is expected to cleave the GPI-anchor on the cell surface and thus release OTOA into the medium, our results strongly indicate that mature OTOA protein was tethered on the plasma membrane via a GPI-anchor. To determine the degree of the GPI-anchorage of OTOA on the cell surface, we performed surface staining of OTOA in live cells, followed by total staining in permeabilized cells. OTOA was normally expressed on the cell surface and was completely removed from the cell surface by the PI-PLC treatment, showing that surface OTOA was predominantly GPI-anchored on the cell surface (Figure 2B). Schematic depiction of the anchoring of WT OTOA to the cell membrane by GPI anchor is illustrated (Figure 2C and 2D).
Figure 2. Wild-type (WT) OTOA is localized on the cell surface via a GPI-anchor, while truncated OTOA (p.Gln589ArgfsX55) is constitutively secreted from cells.
(A) HEK293T cells were transfected with myc-tagged WT OTOA. Western blot of cell lysate and medium samples with α-myc antibody showed that OTOA is not released into the medium. Treatment of cells with bacterial phosphatidylinositol- phospholipase C (PI-PLC), which cleaves a GPI-anchor on the cell surface, facilitates the release of OTOA into the medium. The released OTOA by PI-PLC migrated slower than that in the cell lysate. Deglycosylation by PNGase F treatment produced the same smaller size bands in the cell lysate and the medium, indicating hyper-glycosylated OTOA is localized on the cell surface via a GPI-anchorage.
(B) Live cell surface labeling. HEK293T cells were transfected with WT OTOA and subsequently immunostained with an α-myc antibody (raised in rabbit) in the absence of a detergent for surface labeling. Cells were then treated with blocking solution containing 1% Triton X-100, and incubated with an α-myc antibody (raised in mouse) for total staining. Surface and total Otoa is shown as red and green, respectively. OTOA is observed on the cell surface, which is completely removed by PI-PLC treatment.
(C and D) Schematic cartoons illustrating the shape and function of WT OTOA. OTOA is tethered to the plasma membrane via a GPI-anchorage, which is attached to the C-terminus of the mature protein. Red and green color of the mature protein indicated the preserved and truncated domain of Gln589ArgfsX55 mutant, respectively.
(E) HEK293T cells were transfected with myc-tagged truncated OTOA (p.Gln589ArgfsX55). The truncated OTOA was constitutively secreted into the medium without PI-PLC treatment. The released protein migrated slower than that in the cell lysate. Deglycosylation by PNGase F treatment produced the same smaller size bands in the cell lysate and the medium, indicating hyper-glycosylated is secreted into the medium.
(F) Live cell staining of HEK293T cells transfected with myc-tagged truncated OTOA (p.Gln589ArgfsX55) showed that the truncated OTOA is not tethered to the cell surface.
(G and H) The truncated OTOA lacks a C-terminal GPI-anchorage and is constitutively secreted from the producing cells.
Since a substantial portion of the human DFNB22 phenotypes resulted from point mutations of OTOA rather than from total absence of the protein (He et al., 2018; K. Lee et al., 2013; Sloan-Heggen et al., 2016; Sommen et al., 2016; Walsh et al., 2006; Zwaenepoel et al., 2002), Otoa knockout mice could not faithfully model DFNB22. Noting that cell surface WT OTOA is exclusively GPI-anchored (Figure 2B), we characterized the pathogenic potential of our variants, focusing on the GPI-anchorage and surface expression. p.Gln589ArgfsX55 was chosen for this experiment because it was predicted to generate the longer translation product than c.1320+5G>C, if it was not subject to nonsense medicated decay (NMD). Using this second variant with the longer translation product, we expected to be able to better reduce the portion of the C-terminal part with unknown functions than using c.1320+5G>C.
First, we monitored the cellular localization of OTOA (p.Gln589ArgfsX55), which lacked the C-terminal domain, presumably with the GPI-anchorage signal. OTOA (p.Gln589ArgfsX55) was constitutively secreted into the medium and PI-PLC treatment did not facilitate its release (Figure 2E). Consistent with the western blotting data, we could not detect the surface expression of the mutant by surface staining (Figure 2F). Schematic depiction of the uncontrolled secretion of the OTOA (p.Gln589ArgfsX55) is illustrated (Figure 2G and 2H).
TECTA is another GPI-anchored protein that plays critical roles in the formation of the TM via crosslinking TM components (Andrade, Salles, Grati, Manor, & Kachar, 2016). Both Otoa and Tecta are expressed in the spiral limbus during TM development (Lukashkin et al., 2012; Zwaenepoel et al., 2002). Thus, we tested whether TECTA sequestered OTOA (p.Gln589ArgfsX55) on the cell surface. Expression of TECTA did not block the constitutive release of OTOA (p.Gln589ArgfsX55) (Supp. Figure S3).
Overall, the data suggested that OTOA is predominantly GPI-anchored on the cell surface and also that the loss of surface tethering followed by uncontrolled release of OTOA (p.Gln589ArgfsX55) may have been the cause of hearing deficits observed in the proband. However, we could not exclude the possibility that another mechanism yet to be identified from the C-terminal domain downstream to the 589th amino acid (Gln) may have exerted more deleterious pathogenic effects alone or in combination with disruption of the GPI-anchorage. Nonetheless, our present study of the frameshift variant provides the first reported evidence that OTOA is a GPI-anchored protein. The pathogenic potential of the splice region variant, c.1320+5G>C was further supported by the similar minigene splicing assay results with those obtained from the previously reported pathogenic variant, c.1320+2T>C, which is only 3 bp ahead of our variant.
To exclude the possibility that our two variants (c.1320+5G>C and p.Gln589ArgfsX55 located between exon 13 and exon17) reside in the pseudogene, OTOAP1, we checked the sequence similarity between OTOA (NM_144672.3) and OTOAP1 (NR_003676) using the BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and MAFFT (https://mafft.cbrc.jp/alignment/software/), which showed 28.29% sequence homology on the region between the exon 1 and exon 20, while 99.57% homology was observed on the remaining region (between exon 21 and C-terminal), further supporting the presence of our two variants in the real OTOA gene.
The contribution of OTOA variants, including CNVs encompassing this gene, to deafness in Koreans was indirectly verified through the analysis of normal control whole-exome sequencing (WES) data. CNVs were analyzed in 944 cases of normal control data from the Korean Variant Archive (KOVA) (S. Lee et al., 2017) with the detection of only two CNVs of OTOA in contrast to the significantly higher detection rate (18/686) of OTOA CNVs in the non-syndromic hearing loss (NSHL) population (Shearer et al., 2014), indicating significantly more CNVs in the NSHL group than in the normal control population (p < 0.001), although the difference in ethnicity might have biased the interpretation. Indels were also investigated in 1,944 cases of combined normal control data from KOVA and Samsung Genome Institute (SGI), with only three indels in the intron detected (unpublished data), supporting the pathogenic potential of variants of OTOA in the deaf population.
Although OTOA was reported to be associated with DFNB22 in 2002, the molecular mechanism underlying the OTOA-related hearing loss (DFNB22) in humans has not been clearly elucidated. Initially, the association of TM and hearing loss was proposed for OTOA-related DFNB22 (Zwaenepoel et al., 2002). The TM of the mammalian cochlea is a complex structure composed of collagen fibrils embedded in a non-collagenous matrix, where a number of different glycoproteins, including Tecta, Tectb, otogelin, otolin, and Ceacam16 are located (Cohen-Salmon, El-Amraoui, Leibovici, & Petit, 1997; Deans, Peterson, & Wong, 2010; Hasko & Richardson, 1988; Legan, Rau, Keen, & Richardson, 1997; Zheng et al., 2011). Alterations in these glycoproteins have been reported to be associated with both multiple autosomal dominant and recessive hereditary deafness (Alasti et al., 2008; Mustapha et al., 1999; Verhoeven et al., 1998). A recent study using a null function mouse model of Otoa revealed that detachment of TM from the spiral limbus and subsequent disruption of inner hair cell stimulation might play an important role in the pathogenesis of DFNB22 (Lukashkin et al., 2012). However, mice with complete loss of Otoa function did not always faithfully model the human phenotype (DFNB22), with a varying range of OTOA functional losses. A delicate and sophisticated subtle pathogenic mechanism would be masked with an overwhelming null function, and may not have been elucidated with this null mutant model.
In the study, we could not properly evaluate the possibility of NMD for two mutant truncating transcripts from c.1320+5G>C and p.Gln589ArgfsX55, because OTOA transcript was not detected even from wild-type lymphoblastoid cell line (unpublished data). Therefore, there still remains the possibility of significant reduction of transcripts and proteins due to NMD. However, failure in GPI-anchorage of OTOA can still hold true even for null alleles of OTOA
In the present study, we experimentally proved GPI-anchorage of OTOA and also suggest the failure of GPI-anchorage as an important mechanism underlying the OTOA-associated, TM-related hearing loss, which was confirmed by an actual human OTOA variant from hearing-impaired patients and by subsequent, reverse translational molecular analysis.
Supplementary Material
Acknowledgments:
The authors acknowledge Dr. Jung-Wook Kim from Seoul National University, Republic of Korea for kindly providing pSPL3 exon trapping vector.
Funding information
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1A2B2001054 to B.Y.C., 2017R1D1A1B03034401 to D.Y.O. and 2018R1D1A1B07046159 to B.J.K.), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI15C1632, and HI17C0952 to B.Y.C.), and the National Institutes of Health (R21DC016750 to S.P.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest
The authors declare that no competing interests exist.
References
- Alasti F, Sanati MH, Behrouzifard AH, Sadeghi A, de Brouwer AP, Kremer H, Smith RJ, & Van Camp G (2008). A novel TECTA mutation confirms the recognizable phenotype among autosomal recessive hearing impairment families. Int J Pediatr Otorhinolaryngol, 72(2), 249–255. doi: 10.1016/j.ijporl.2007.09.023 [DOI] [PubMed] [Google Scholar]
- Alkowari MK, Vozzi D, Bhagat S, Krishnamoorthy N, Morgan A, Hayder Y, Logendra B, Najjar N, Gandin I, Gasparini P, Badii R, Girotto G, & Abdulhadi K (2017). Targeted sequencing identifies novel variants involved in autosomal recessive hereditary hearing loss in Qatari families. Mutat Res, 800–802, 29–36. doi: 10.1016/j.mrfmmm.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Ammar-Khodja F, Bonnet C, Dahmani M, Ouhab S, Lefevre GM, Ibrahim H, Hardelin JP, Weil D, Louha M, & Petit C (2015). Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing. Mol Genet Genomic Med, 3(3), 189–196. doi: 10.1002/mgg3.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LR, Salles FT, Grati M, Manor U, & Kachar B (2016). Tectorins crosslink type II collagen fibrils and connect the tectorial membrane to the spiral limbus. J Struct Biol, 194(2), 139–146. doi: 10.1016/j.jsb.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, Shearer AE, Sloan-Heggen CM, Kolbe D, Casavant T, Schnieders MJ, Nishimura C, Braun T, & Smith RJH (2018). Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am J Hum Genet, 103(4), 484–497. doi: 10.1016/j.ajhg.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Diaz-Horta O, Guo S, Duman D, Van Booven D, Foster J 2nd, Cengiz FB, Blanton S, & Tekin M (2014). Identification of copy number variants through whole-exome sequencing in autosomal recessive nonsyndromic hearing loss. Genet Test Mol Biomarkers, 18(9), 658–661. doi: 10.1089/gtmb.2014.0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MY, Lee C, Han JH, Kim MY, Park HR, Kim N, Park WY, Oh DY, & Choi BY (2018). Expansion of phenotypic spectrum of MYO15A pathogenic variants to include postlingual onset of progressive partial deafness. BMC Med Genet, 19(1), 29. doi: 10.1186/s12881-018-0541-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Salmon M, El-Amraoui A, Leibovici M, & Petit C (1997). Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc Natl Acad Sci U S A, 94(26), 14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, Kadurin I, Alvarez-Laviada A, Douglas L, Nieto-Rostro M, Bauer CS, Pratt WS, & Dolphin AC (2010). The alpha2delta subunits of voltage-gated calcium channels form GPI-anchored proteins, a posttranslational modification essential for function. Proc Natl Acad Sci U S A, 107(4), 1654–1659. doi: 10.1073/pnas.0908735107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR, Peterson JM, & Wong GW (2010). Mammalian Otolin: a multimeric glycoprotein specific to the inner ear that interacts with otoconial matrix protein Otoconin-90 and Cerebellin-1. PLoS One, 5(9), e12765. doi: 10.1371/journal.pone.0012765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galian C, Bjorkholm P, Bulleid N, & von Heijne G (2012). Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J Biol Chem, 287(20), 16399–16409. doi: 10.1074/jbc.M112.350009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KH, Oh DY, Lee S, Lee C, Han JH, Kim MY, Park HR, Park MK, Kim NKD, Lee J, Yi E, Kim JM, Kim JW, Chae JH, Oh SH, Park WY, & Choi BY (2017). ATP1A3 mutations can cause progressive auditory neuropathy: a new gene of auditory synaptopathy. Sci Rep, 7(1), 16504. doi: 10.1038/s41598-017-16676-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasko JA, & Richardson GP (1988). The ultrastructural organization and properties of the mouse tectorial membrane matrix. Hear Res, 35(1), 21–38. [DOI] [PubMed] [Google Scholar]
- He L, Pang X, Liu H, Chai Y, Wu H, & Yang T (2018). Targeted next-generation sequencing and parental genotyping in sporadic Chinese Han deaf patients. Clin Genet, 93(4), 899–904. doi: 10.1111/cge.13182 [DOI] [PubMed] [Google Scholar]
- Kim BJ, Han JH, Park HR, Kim MY, Kim AR, Oh SH, Park WY, Oh DY, Lee S, & Choi BY (2018). A clinical guidance to DFNA22 drawn from a Korean cohort study with an autosomal dominant deaf population: A retrospective cohort study. J Gene Med, e3019. doi: 10.1002/jgm.3019 [DOI] [PubMed] [Google Scholar]
- Lee K, Chiu I, Santos-Cortez RL, Basit S, Khan S, Azeem Z, Andrade PB, Kim SS, Ahmad W, & Leal SM (2013). Novel OTOA mutations cause autosomal recessive non-syndromic hearing impairment in Pakistani families. Clin Genet, 84(3), 294–296. doi: 10.1111/cge.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, Bjornson RD, Chae JH, Jang IJ, Lee S, Park WY, Baek D, & Choi M (2017). Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep, 7(1), 4287. doi: 10.1038/s41598-017-04642-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legan PK, Rau A, Keen JN, & Richardson GP (1997). The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J Biol Chem, 272(13), 8791–8801. [DOI] [PubMed] [Google Scholar]
- Lukashkin AN, Legan PK, Weddell TD, Lukashkina VA, Goodyear RJ, Welstead LJ, Petit C, Russell IJ, & Richardson GP (2012). A mouse model for human deafness DFNB22 reveals that hearing impairment is due to a loss of inner hair cell stimulation. Proc Natl Acad Sci U S A, 109(47), 19351–19356. doi: 10.1073/pnas.1210159109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, & Petit C (1999). An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet, 8(3), 409–412. [DOI] [PubMed] [Google Scholar]
- Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, Shen J, Chapin A, Boczek NJ, Schimmenti LA, Murry JB, Hasadsri L, Nara K, Kenna M, Booth KT, Azaiez H, Griffith A, Avraham KB, Kremer H, Rehm HL, et al. (2018). Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat, 39(11), 1593–1613. doi: 10.1002/humu.23630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim NK, Kim AR, Rhee J, Oh SH, Koo JW, Nam JY, Park WY, & Choi BY (2014). Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis, 9, 167. doi: 10.1186/s13023-014-0167-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, & Committee, Acmg Laboratory Quality Assurance. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Rayyan AA, Lee MK, Higgins J, Dickel D, Lewis K, Thompson J, Baker C, Nord AS, Stray S, Gurwitz D, Avraham KB, King MC, & Kanaan M (2010). Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet, 18(4), 407–413. doi: 10.1038/ejhg.2009.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Kolbe DL, Azaiez H, Sloan CM, Frees KL, Weaver AE, Clark ET, Nishimura CJ, Black-Ziegelbein EA, & Smith RJ (2014). Copy number variants are a common cause of non-syndromic hearing loss. Genome Med, 6(5), 37. doi: 10.1186/gm554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, Ephraim SS, Shibata SB, Booth KT, Campbell CA, Ranum PT, Weaver AE, Black-Ziegelbein EA, Wang D, Azaiez H, & Smith RJ (2016). Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet, 135(4), 441–450. doi: 10.1007/s00439-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommen M, Schrauwen I, Vandeweyer G, Boeckx N, Corneveaux JJ, van den Ende J, Boudewyns A, De Leenheer E, Janssens S, Claes K, Verstreken M, Strenzke N, Predohl F, Wuyts W, Mortier G, Bitner-Glindzicz M, Moser T, Coucke P, Huentelman MJ, & Van Camp G (2016). DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum Mutat, 37(8), 812–819. doi: 10.1002/humu.22999 [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, Smith RJ, Somers T, Offeciers FE, Van de Heyning P, Richardson GP, Wachtler F, Kimberling WJ, Willems PJ, Govaerts PJ, & Van Camp G (1998). Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet, 19(1), 60–62. doi: 10.1038/ng0598-60 [DOI] [PubMed] [Google Scholar]
- Walsh T, Abu Rayan A, Abu Sa’ed J, Shahin H, Shepshelovich J, Lee MK, Hirschberg K, Tekin M, Salhab W, Avraham KB, King MC, & Kanaan M (2006). Genomic analysis of a heterogeneous Mendelian phenotype: multiple novel alleles for inherited hearing loss in the Palestinian population. Hum Genomics, 2(4), 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Miller KK, Yang T, Hildebrand MS, Shearer AE, DeLuca AP, Scheetz TE, Drummond J, Scherer SE, Legan PK, Goodyear RJ, Richardson GP, Cheatham MA, Smith RJ, & Dallos P (2011). Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4). Proc Natl Acad Sci U S A, 108(10), 4218–4223. doi: 10.1073/pnas.1005842108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel I, Mustapha M, Leibovici M, Verpy E, Goodyear R, Liu XZ, Nouaille S, Nance WE, Kanaan M, Avraham KB, Tekaia F, Loiselet J, Lathrop M, Richardson G, & Petit C (2002). Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci U S A, 99(9), 6240–6245. doi: 10.1073/pnas.082515999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.