Abstract
Δ5 and Δ6 fatty acid desaturases are critical enzymes in the pathways for the biosynthesis of the polyunsaturated fatty acids arachidonic, eicosapentaenoic, and docosahexaenoic acids. They are encoded by distinct genes in mammals and Caenorhabditis elegans. This paper describes a cDNA isolated from zebrafish (Danio rerio) with high similarity to mammalian Δ6 desaturase genes. The 1,590-bp sequence specifies a protein that, in common with other fatty acid desaturases, contains an N-terminal cytochrome b5 domain and three histidine boxes, believed to be involved in catalysis. When the zebrafish cDNA was expressed in Saccharomyces cerevisiae it conferred the ability to convert linoleic acid (18:2n-6) and α-linolenic acid (18:3n-3) to their corresponding Δ6 desaturated products, 18:3n-6 and 18:4n-3. However, in addition it conferred on the yeast the ability to convert di-homo-γ-linoleic acid (20:3n-6) and eicosatetraenoic acid (20:4n-3) to arachidonic acid (20:4n-6) and eicosapentaenoic acid (20:5n-3), respectively, indicating that the zebrafish gene encodes an enzyme having both Δ5 and Δ6 desaturase activity. The zebrafish Δ5/Δ6 desaturase may represent a component of a prototypic vertebrate polyunsaturated fatty acids biosynthesis pathway.
Vertebrates lack the Δ12 and Δ15 fatty acid desaturases responsible for converting oleic acid (18:1n-9) into linoleic acid (18:2n-6) and α-linolenic acid (18:3n-3) and thus are unable to biosynthesize polyunsaturated fatty acids (PUFA) de novo (1, 2). PUFAs therefore are essential dietary nutrients for vertebrates (1, 2). The physiologically active PUFAs are arachidonic acid (20:4n-6), eicosapentaenoic acid (20:5n-3), and docosahexaenoic acid (22:6n-3) and are required for optimal health and normal development of vertebrates (3–6). The pathway from 18:2n-6 to arachidonic acid and from 18:3n-3 to eicosapentaenoic acid involves desaturations at the Δ6 and Δ5 positions in the carbon backbone, and an intermediate 2-carbon chain elongation step (7). Production of docosahexaenoic acid from eicosapentaenoic acid requires an additional desaturation and 2-carbon chain elongation, although the mechanism may be more complicated (8). However, there is considerable variation between animal species in their abilities to synthesize the C20 and C22 PUFAs from the plant-derived C18 precursors 18:2n-6 and 18:3n-3. Some animals, notably extreme carnivores, have a very limited ability to synthesize C20 and C22 PUFAs and consequently have a strict requirement for a dietary source of preformed C20 and C22 PUFAs (9–12). Although humans generally possess the ability to synthesize C20 and C22 PUFAs from linoleic and α-linolenic acids, dietary changes as a consequence of intensification of agriculture have resulted in an increase in the 18:2n-6/18:3n-3 ratio in foods (13). There is considerable evidence that these changes have had, and continue to have, negative impacts on health and development in affected populations (4, 13). The dietary 18:2n-6/18:3n-3 ratio seems to have increased because of a steady decline in dietary n-3 fatty acids over a period of several hundred years, and this increase has been compounded by an increased dietary intake of 18:2n-6 in recent decades, particularly in Western societies (13–16).
Meeting the dietary demands of a burgeoning human population for a correct dietary balance of PUFAs, and at levels required for normal health and development, is a major challenge. Clearly, an understanding of the molecular basis of PUFA biosynthesis could underpin efforts to meet this challenge. However, until recently, and although the biochemical pathways involved in PUFA synthesis were described, little was known of the enzymes involved and of the factors affecting their function(s). Some progress has been made recently in characterizing the elongase and desaturases involved in PUFA synthesis (17). Full-length cDNAs for Δ6 desaturases have been isolated from the nematode Caenorhabditis elegans (18), rat (19), mouse, and human (20). Fatty acid Δ5 desaturase genes have been isolated from C. elegans (21, 22) and humans (23, 24).
We have undertaken to study the PUFA synthesis pathway in fish for two reasons. First, fish are an important source of PUFA, especially of the long chain C20 and C22 n-3 PUFAs that often are deficient in human diets (14–16). Second, there is wide variation between fish species in their ability to synthesize PUFA (25–27). Many freshwater species such as trout, tilapia, and carp are able to convert dietary C18 precursor fatty acids to arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid. However, marine species such as turbot and sea bream, which are inherently piscivorous, have very limited abilities to perform these conversions (26–31). A comparison of the genes encoding key elements in the fatty acid desaturation pathway between freshwater and marine species may, therefore, increase knowledge of the molecular components of the pathway itself and of the molecular genetic basis of phenotypic variation in PUFA biosynthesis.
We have demonstrated recently that zebrafish (Danio rerio) share the capacity of other freshwater fish species to synthesize C20 and C22 PUFAs from vegetable oil-derived C18 dietary precursors (32). Therefore, because Δ6 desaturation generally is considered to be the rate-limiting step in PUFA synthesis (33), we have initially targeted the Δ6 desaturase gene(s) of zebrafish. Here we describe the cloning and functional characterization of a fatty acid desaturase gene obtained from zebrafish, which, uniquely among such genes described to date, has both Δ5 and Δ6 desaturation activities.
Materials and Methods
Isolation of a Zebrafish Desaturase cDNA and Sequence Analysis.
A zebrafish expressed sequence tag sequence (GenBank accession no. AI497337) was identified that displayed high homology to mammalian Δ5 and Δ6 desaturase genes. cDNA was synthesized from zebrafish liver total RNA by using Moloney murine leukemia virus reverse transcriptase primed by the oligonucleotide 5′-GATAGCGGCCGCGTTTTTTTTTTTTTTTT(AGC)-3′. Then a portion of this cDNA was subjected to PCR amplification (Ready-to-Go PCR beads, Amersham Pharmacia) with the primer described above and an oligonucleotide (5′-ATGGGTGGCGGAGGACAGC-3′) predicted from the zebrafish expressed sequence tag sequence to contain the protein initiation codon. Amplification involved an initial denaturation step at 94°C for 1 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 3 min. The products were cloned into the pYES2 plasmid (Invitrogen) by using standard methods, and nucleotide sequences were determined by using a Perkin–Elmer ABI-377 DNA sequencer. Deduced amino acid sequences were aligned by using CLUSTALX, and sequence phylogenies were predicted by using the neighbor-joining method of Saitou and Nei (34). Confidence in the resulting phylogenetic tree branch topology was measured by bootstrapping the data through 1,000 iterations.
Expression of the Zebrafish Desaturase cDNA.
The coding sequence of the zebrafish cDNA was amplified by using the forward primer 5′-CCCAAGCTTACTATGGGTGGCGGAGGACAGC-3′ and reverse primer 5′-CCGCTGGAGTTATTTGTTGAGATACGC-3′ containing HindIII and XhoI sites, respectively. The amplified product was ligated into the HindIII and XhoI sites of the pYES yeast expression vector (Invitrogen). The resulting plasmid construct, pYESZFB10, was transformed into Saccharomyces cerevisiae (strain INVSc1) by using the EasyComp transformation kit (Invitrogen). Yeast transformed with either the pYES vector or pYESZFB10 were cultured overnight in 2% raffinose, 0.67% nitrogen base, 1% tergitol type Nonidet P-40, and 0.19% uracil dropout medium at 30°C. The cultures then were diluted to an OD600 of 0.4 and grown until they reached an OD600 of 1, after which expression of the transgene was induced by the addition of galactose to 2% (wt/vol). At this point the cultures were supplemented with one of 0.5 mM 18:2(n-6), 18:3(n-3), 20:3(n-6), or 20:4(n-3) and then maintained at 30°C in a shaking incubator. Samples were taken for analysis 48 h after galactose induction.
Fatty Acid Analysis.
Approximately equal amounts of yeast cells were transferred into glass conical test tubes after determination of culture densities at OD600. The cells were collected by centrifugation at 500 × g for 2 min, and the pellets were washed twice with 5 ml of ice-cold Hanks' balance salt solution and dried under a stream of oxygen-free nitrogen. Fatty acid methyl esters (FAMEs) were prepared by incubating the dried yeast cells directly with 1 ml of methylation reagent containing 10% (vol/vol) concentrated HCl, 5% (vol/vol) 2,2-dimethoxypropane, and 85% (vol/vol) dry methanol for 1 h at 85°C. After incubation, FAMEs were extracted by the addition of 1 ml of 1% NaCl solution and 0.5 ml of hexane containing 0.01% butylated hydroxytoluene as antioxidant. The mixture was mixed vigorously and centrifuged at 600 × g for 5 min to promote phase separation. The top phase was removed carefully and filtered through Whatman No. 1 filter paper into a clean glass test tube, and the solvent evaporated under a stream of oxygen-free nitrogen. The FAMEs were purified by TLC and then resuspended in hexane, all as described previously (35). FAMEs were separated in a Fisons GC8160 gas chromatograph equipped with a chemically bonded CP Wax 52CB fused silica wall coated capillary column (30 m × 0.32 mm i.d., Chrompack U.K., London) with an on-column injection system and flame ionization detection. Hydrogen was used as carrier gas with an oven thermal gradient from an initial 50 to 180°C at 40°C/min and then to a final temperature of 235°C at 2°C/min. Individual FAMEs were identified by comparison with known standards, with a well characterized fish oil, and by reference to published data as described previously (35). FAMEs were quantified by using a directly linked PC operating CHROM-CARD software (Thermo-Quest Italia S.P.A., Milan, Italy). All solvents contained 0.01% butylated hydroxytoluene as an antioxidant.
GC-MS.
The identities of fatty acids and positions of their double bonds were confirmed by subjecting the picolinyl esters to electron ionization (EI) GC-MS. Free fatty acids were prepared from FAMEs by alkaline hydrolysis as described by Christie (36). Picolinyl esters were prepared by the method of Balazy and Nies (37). This method involves activating the free fatty acid by reaction with 1,1′-carbonyldiimidazole to form the imidazolide, which then reacts with 3-(hydroxymethyl)pyridine under basic conditions to form the picolinyl ester. GC-MS of the picolinyl esters was performed by using a Fisons GC8000 gas chromatograph coupled to an MD800 mass spectrometer (Fisons Instruments, Crawley, U.K.). The gas chromatograph was equipped with a fused silica capillary column (60 m × 0.32 mm i.d., 0.25-mm internal film thickness) coated with Zebron ZB-Wax (Phenomenex, Macclesfield, U.K.) and used helium as carrier gas. Samples were applied by using on-column injection with the oven temperature programmed to rise from 80 to 250°C at 40°C/min.
Materials.
Eicosatetraenoic acid (20:4n-3, >98% pure) was purchased from Cayman Chemicals (Ann Arbor, MI). Linoleic (18:2n-6), α-linolenic (18:3n-3), and eicosatrienoic (20:3n-6) acids (all >99% pure), BHT, 1,1′-carbonyldiimidazole, 2,2-dimethoxypropane, fatty acid-free BSA, galactose, 3-(hydroxymethyl)pyridine, and Hanks' balanced salt solution, nitrogen base, raffinose, tergitol Nonidet P-40, and uracil dropout medium were obtained from Sigma. TLC (20 × 20 cm × 0.25 mm) plates precoated with silica gel 60 (without fluorescent indicator) were purchased from Merck. All solvents were HPLC grade and obtained from Rathburn Chemicals (Peebleshire, U.K.).
Results
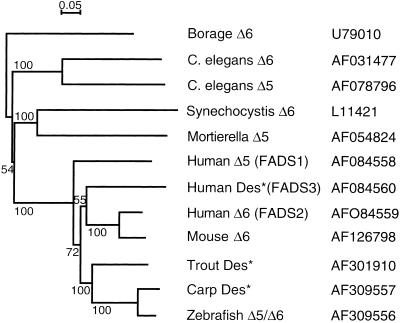
Sequencing revealed that the zebrafish cDNA (GenBank accession no. AF309556) comprised 1,590 bp, which included an ORF specifying a protein of 444 amino acids. The protein sequence included a number of characteristic features of microsomal fatty acid desaturases including three histidine boxes (Fig. 1). The protein sequence also contained an N-terminal cytochrome b5 domain containing the heme-binding motif, H-P-G-G, similar to that of other fatty acid desaturases. Further, the third histidine box contained a glutamine residue substituted for the first histidine. The amino acid sequence predicted by the zebrafish ORF indicated that the desaturase candidate possessed 64% identity and 78% similarity to human Δ6 desaturase (GenBank accession no. AF126799) and possessed 58% identity and 75% similarity to human Δ5 desaturase (GenBank accession no. AF199596). Phylogenetic analysis, comparing a variety of Δ5 and Δ6 desaturases, clustered the zebrafish sequence with mammalian Δ6 desaturase sequences and with an uncharacterized, putative fish Δ6 desaturase sequence (Fig. 2).
Figure 1.
Alignment of the predicted amino acid sequences of the zebrafish desaturase (DRD5/6) and human Δ5 (HSFADS1) and Δ6 desaturases (HSFADS2). The three ORFs encode 444 amino acid residues. Identical amino acids are in black, the cytochrome b5-like domain is underlined by asterisks, and the three histidine-rich domains are underlined.
Figure 2.
Phylogeny of desaturase deduced amino acid sequences. Sequences marked with an asterisk are not functionally characterized. Database accession numbers for the nucleic acid sequences are indicated. The conditions used for neighbor-joining tree construction are described in Materials and Methods and were applied by using CLUSTALW and NJPLOT. Horizontal branch lengths are proportional to the number of amino acid replacements per position, and the scale bar indicates this value. Numbers represent the percentage frequencies with which the tree topology presented here was replicated after 1,000 bootstrap iterations.
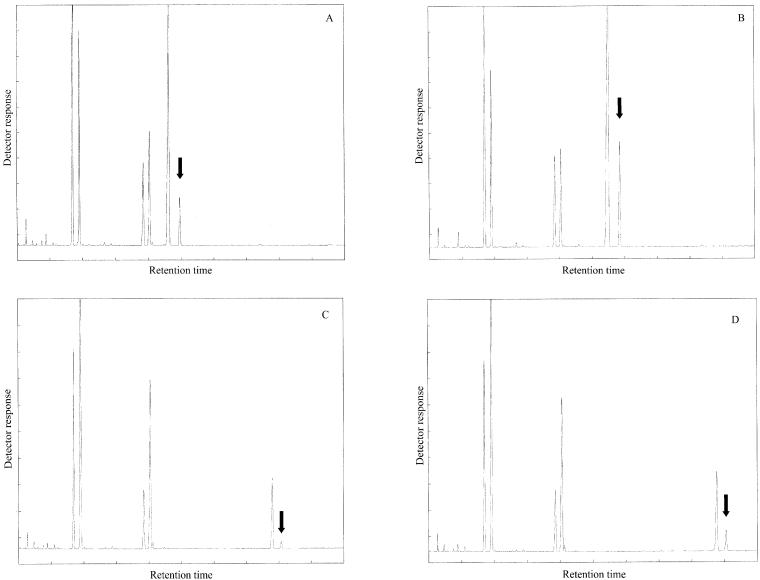
The zebrafish desaturase cDNA was functionally characterized by determining the fatty acid profiles of transformed S. cerevisiae containing either the pYES vector alone or the vector with the zebrafish cDNA insert (pYESZFB10) grown in the presence of 18:2n-6, 18:3n-3, 20:3n-6, or 20:4n-3. The fatty acid composition of the yeast transformed with the vector alone showed the four main fatty acids normally found in S. cerevisiae, namely 16:0, 16:1n-7, 18:0, and 18:1n-9, together with the four exogenously derived fatty acids (data not shown). This result is consistent with the fact that S. cerevisiae does not possess Δ5 or Δ6 fatty acid desaturase activities. Additional peaks were observed in the profiles of pYESZFB10-transformed yeast grown in the presence of the Δ6 desaturase substrate fatty acids, 18:2n-6 and 18:3n-3, and also in the profiles of pYESZFB10-transformed yeast grown in the presence of the Δ5 desaturase substrate fatty acids, 20:3n-6 and 20:4n-3 (Fig. 3 A–D). Based on GC retention times, the additional peaks associated with the presence of the zebrafish cDNA, indicated in Fig. 3 A–D, were identified as 18:3n-6, 18:4n-3, 20:4n-6, and 20:5n-3, respectively.
Figure 3.
Identification of fatty acid desaturation products in transgenic yeast. FAMEs were extracted from yeast transformed with pYESZFB10 grown in the presence of 18:2n-6 (A), 18:3n-3 (B), 20:3n-6 (C), or 20:4n-3 (D). The first four peaks in A–D are 16:0, 16:1n-7, 18:0, and 18:1n-9, respectively. The fifth peaks in each panel are the exogenously added fatty acids 18:2n-6 (A), 18:3n-3 (B), 20:3n-6 (C), and 20:4n-3 (D), respectively. The sixth peaks in each panel (arrowed) were identified tentatively (based on retention times) as 18:3n-6 (A), 18:4n-3 (B), 20:4n-6 (C), and 20:5n-3 (D), respectively.
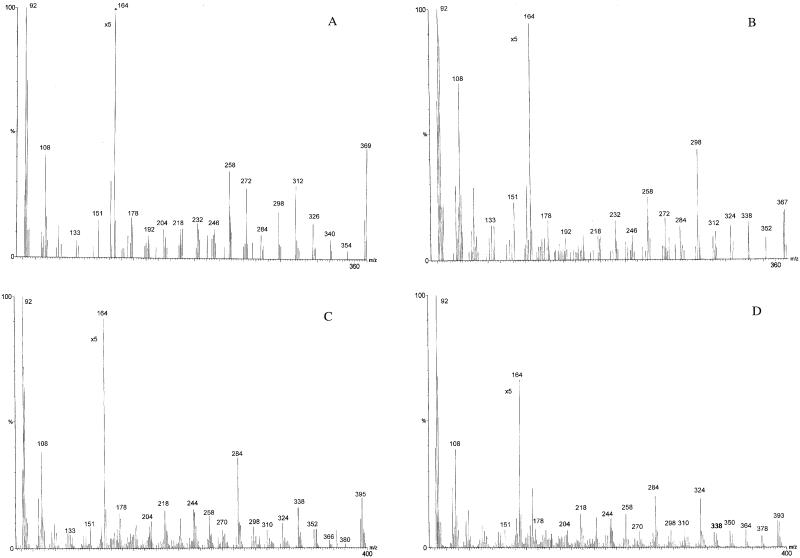
The FAME samples from the transformed yeast incubated with the exogenous PUFAs were converted to picolinyl esters and subjected to EI GC-MS to positively identify the structures represented by the additional PUFA peaks produced in cultures containing pYESZFB10. The samples all showed prominent ions at m/z = 92, 108, 151, and 164, which are characteristic of picolinyl esters representing fragments about the pyridine ring (Fig. 4; ref. 38). The EI spectra of the additional fatty acid in pYESZFB10p-transformed yeast incubated with 18:2n-6 showed a fragmentation pattern with a mass ion of 369 m/z and prominent peaks at 354, 340, 326, 312, 298, 272, 258, 232, 218, and 192 m/z (Fig. 4A). The initial interval of 15 (369–354) represented the terminal methyl and was followed by four intervals of 14, indicating four methylene groups. The intervals of 26 (298–272, 258–232, and 218–192) denoted the positions of three double bonds, indicating that this fatty acid is Δ12,9,618:3 = 18:3n-6. The EI spectra of the additional fatty acid from cells incubated with 18:3n-3 showed a mass ion of 367 m/z and fragments at 338, 312, 298, 272, 258, 232, 218, and 192 m/z, confirming this fatty acid as Δ15,12,9,618:4 = 18:4n-3 (Fig. 4B). The EI spectra of the additional fatty acid produced in cells incubated with 20:3n-6 showed a fragmentation pattern with a mass ion of 395 m/z and prominent ions at intervals of 26 (324–298, 284–258, 244–218, and 204–178 m/z), denoting the position of the double bonds and indicating that this fatty acid is Δ14,11,8,520:4 = 20:4n-6 (Fig. 4C). Similarly, the EI spectra of the additional fatty acid in cells incubated with 20:4n-3 showed a fragmentation pattern with a mass ion of 393 m/z with prominent ions at intervals of 26 (364–338, 324–298, 284–258, 244–218, and 204–178 m/z), confirming that this fatty acid is Δ17,14,11,8,520:5 = 20:5n-3 (Fig. 4D). The GC-MS data confirmed that the zebrafish clone is a fatty acid desaturase that introduces double bonds into 18:2n-6 and 18:3n-3 at the Δ6 position and also into 20:3n-6 and 20:4n-3 at the Δ5 position.
Figure 4.
Mass spectra of the arrowed peaks in Fig. 3. Picolinyl esters were prepared from FAMEs extracted from yeast transformed with pYESZFB10 grown in the presence of 18:2n-6 (A), 18:3n-3 (B), 20:3n-6 (C), and 20:4n-3 (D) and analyzed by GC-MS as described in Materials and Methods. The identities of the peaks were confirmed as 18:3n-6 (A), 18:5n-3 (B), 20:4n-6 (C), 20:5n-3 (D).
Thus, the analyses revealed that the cells transformed with pYESZFB10 had acquired functional Δ6 and Δ5 fatty acid desaturase activity. On the basis of the percentages of substrate fatty acids converted to product, the zebrafish gene is more active with Δ6 desaturase substrates than with Δ5 substrates and preferentially converts n-3 fatty acids rather than n-6 fatty acids (Table 1).
Table 1.
pYESZFB10 substrate conversion and activity type
| Substrate fatty acid | Substrate conversion, % | Desaturase activity |
|---|---|---|
| 18:2n-6 | 11.7 | Δ6 |
| 18:3n-3 | 29.4 | Δ6 |
| 20:3n-6 | 8.3 | Δ5 |
| 20:4n-3 | 20.4 | Δ5 |
Discussion
The 1,590-bp ORF of the zebrafish cDNA encodes a protein with substantial similarity to vertebrate Δ6 desaturases. Overall amino acid identities are 64% to human Δ6 desaturase and 58% to human Δ5 desaturase. In addition, the zebrafish protein contains a similar N-terminal cytochrome b5-like domain and the three catalytically important histidine boxes conserved in all members of the desaturase gene family. It also includes the variant third histidine box that seems typical of Δ5 and Δ6 desaturase genes described to date.
The analyses demonstrated unequivocally that the zebrafish cDNA encodes a polypeptide with both Δ5 and Δ6 fatty acid desaturase activities with slight biases toward n-3 substrates and Δ6 function.
Phylogenetic analysis indicates that with respect to functionally characterized genes, the zebrafish sequence has highest homology with mammalian Δ6 desaturases, with human Δ5 desaturase appearing to be distinct from the Δ6 desaturase sequences. Human Δ5 and Δ6 desaturase genes (FADS1 and FADS2, respectively) are clustered together with a related gene (FADS3) of unknown function on chromosome 11 and have arisen presumably from a gene-duplication event (39). The fact that the Δ5 and FADS3 sequences fall on distinct branches of the phylogenetic tree from that occupied by the Δ6 sequences suggests that this gene-duplication event(s) predates the divergence of mammalian and fish evolutionary lines. This interpretation suggests that a homologue of the human Δ5 gene should exist in fish species. Interestingly, the nematode C. elegans also possesses a cluster of Δ5 and Δ6 desaturase genes that seem to have arisen by gene duplication. However, this duplication clearly occurred after the divergence of nematodes and vertebrates (18, 21). Both the mammalian and nematode Δ5 and Δ6 desaturase enzymes have distinct, nonoverlapping substrate specificities. It therefore is remarkable that the zebrafish desaturase, when expressed in S. cerevisiae, exhibits both Δ5 and Δ6 fatty acid desaturase activities with a distinct preference for n-3 compared with n-6 substrates.
Although more fatty acid desaturase genes may be found in both zebrafish and mammals, it is conceivable that the bifunctional desaturase described here is a component of a prototypic vertebrate PUFA biosynthetic pathway that has persisted in a freshwater fish species. That humans and other mammals have two distinct enzymes for Δ5 and Δ6 desaturation may be an adaptation to a terrestrial diet providing relatively lower amounts of preformed C20 and C22 PUFAs than the diets of a vertebrate ancestor that they share with freshwater fish. Functional divergence of the products of a putative ancient gene-duplication event is a possible mechanism underlying adaptation to such a dietary change.
Conversely, the apparent deficiencies in the fatty acid desaturation pathway in some marine fish species, many of which are strictly piscivorous, may be a result of relaxation of constraints on a prototypic pathway in an environment providing a diet that is naturally rich in C20 and C22 PUFAs. Certainly the discovery of a bifunctional enzyme in zebrafish is consistent with findings in the piscivorous marine species turbot (Scophthalmus maximus). In turbot, a deficiency in the fatty acid desaturation/elongation pathway has been ascribed to a lack of C18–20 elongase function, while both Δ6 and Δ5 desaturase capabilities have been retained (40). However, in another marine species, gilthead sea bream (Sparus aurata), a block in the fatty acid desaturation/elongation pathway has been identified as a deficiency in Δ5 desaturase activity (41). In this case it is possible that a homologous desaturase enzyme has lost significant Δ5 desaturase activity, thus limiting the efficiency of the PUFA biosynthetic pathway in gilthead sea bream.
The ability of fish to provide 20:5n-3 and 22:6n-3 for human and other animal diets has become an increasingly important subject in recent years after the global stagnation and decline of capture marine fisheries (42). These fisheries hitherto have been the major source of 20:5n-3 and 22:6n-3 in human and animal diets (13). Increased fish farming potentially can offset the effects of this decline in a key marine resource. However, where farmed marine species are concerned, a dietary source of 20:5n-3 and 22:6n-3 is required, and this hitherto has been provided by fish oils from captured marine “feed species” such as sand eels, capelin, and anchovies (42). Such feed species are themselves finite resources that, in the case of anchovies, is reduced markedly by climatic phenomena such as El Niño. Understanding the molecular basis of the differences between marine and freshwater fish in their abilities to synthesize 20:5n-3 and 22:6n-3 may suggest ways in which the reliance of farmed marine species on marine fish oils could be reduced.
Here we report a fatty acid desaturase with both Δ6 and Δ5 activities. Such an enzyme could contribute to a biotechnological solution for 20:5n-3 and 22:6n-3 production, ultimately relieving pressure for unsustainable extraction of these key nutrients from natural marine sources.
Acknowledgments
We are grateful to Jonathan Napier and colleagues (University of Bristol, Bristol, U.K.) for helpful discussions and advice regarding functional analysis in yeast. N.H. received a postgraduate studentship provided by the Natural Environment Research Council (U.K.).
Abbreviations
- PUFA
polyunsaturated fatty acid
- FAME
fatty acid methyl ester
- EI
electron ionization
Footnotes
Data deposition: The sequence reported in this paper for zebrafish Δ5/Δ6 desaturase cDNA has been deposited in the GenBank database (accession no. AF309556).
References
- 1.Tinoco J. Prog Lipid Res. 1982;21:1–45. doi: 10.1016/0163-7827(82)90015-7. [DOI] [PubMed] [Google Scholar]
- 2.Holman R T. J Am Coll Nutr. 1986;5:183–211. doi: 10.1080/07315724.1986.10720125. [DOI] [PubMed] [Google Scholar]
- 3.Simopoulos A P. J Nutr. 1989;119:521–528. doi: 10.1093/jn/119.4.521. [DOI] [PubMed] [Google Scholar]
- 4.Simopoulos A P. Am J Clin Nutr. 1991;54:438–463. doi: 10.1093/ajcn/54.3.438. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. The Report of the British Nutrition Foundation's Taskforce. London: Chapman & Hall; 1992. [Google Scholar]
- 6.Lands W E M. FASEB J. 1992;6:2530–2536. doi: 10.1096/fasebj.6.8.1592205. [DOI] [PubMed] [Google Scholar]
- 7.Sprecher H. Prog Lipid Res. 1981;20:13–22. doi: 10.1016/0163-7827(81)90009-6. [DOI] [PubMed] [Google Scholar]
- 8.Sprecher H, Luthria D L, Mohammed B S, Baykousheva S P. J Lipid Res. 1995;36:2471–2477. [PubMed] [Google Scholar]
- 9.Rivers J P W, Sinclair A J, Crawford M A. Nature (London) 1975;258:171–173. doi: 10.1038/258171a0. [DOI] [PubMed] [Google Scholar]
- 10.Rivers J P W, Hassam A G, Crawford M A, Brambell M R. FEBS Lett. 1976;67:269–270. doi: 10.1016/0014-5793(76)80544-3. [DOI] [PubMed] [Google Scholar]
- 11.Hassam A G, Rivers J P W, Crawford M A. Nutr Metab. 1977;21:321–328. doi: 10.1159/000176079. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair A J, McLean J G, Monger E A. Lipids. 1979;14:932–936. doi: 10.1007/BF02533508. [DOI] [PubMed] [Google Scholar]
- 13.Simopoulos A P. Prostaglandins Leukotrienes Essent Fatty Acids. 1999;60:421–429. doi: 10.1016/s0952-3278(99)80023-4. [DOI] [PubMed] [Google Scholar]
- 14.Kris-Etherton P M, Shaffer Taylor D, Yu-Poth S, Huth P, Moriarty K, Fishell V, Hargrove R L, Zhao G, Etherton T D. Am J Clin Nutr. 2000;71:179S–188S. doi: 10.1093/ajcn/71.1.179S. [DOI] [PubMed] [Google Scholar]
- 15.Sanders T A B. Am J Clin Nutr. 2000;71:176S–178S. doi: 10.1093/ajcn/71.1.176s. [DOI] [PubMed] [Google Scholar]
- 16.Sugano M, Hirahara F. Am J Clin Nutr. 2000;71:189S–196S. doi: 10.1093/ajcn/71.1.189S. [DOI] [PubMed] [Google Scholar]
- 17.Tocher D R, Leaver M J, Hodgson P A. Prog Lipid Res. 1998;37:73–117. doi: 10.1016/s0163-7827(98)00005-8. [DOI] [PubMed] [Google Scholar]
- 18.Napier J A, Hey S J, Lacey D J, Shewry P R. Biochem J. 1998;330:611–614. doi: 10.1042/bj3300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aki T, Shimada Y, Inagaki K, Higashimoto H, Kawamoto S, Shigeta S, Ono K, Suzuki O. Biochem Biophys Res Commun. 1999;255:575–579. doi: 10.1006/bbrc.1999.0235. [DOI] [PubMed] [Google Scholar]
- 20.Cho H P, Nakamura M T, Clarke S D. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 21.Michaelson L V, Lazarus C M, Griffiths G, Napier J A, Stobart A K. J Biol Chem. 1998;273:19055–19059. doi: 10.1074/jbc.273.30.19055. [DOI] [PubMed] [Google Scholar]
- 22.Watts J L, Browse J. Arch Biochem Biophys. 1999;362:175–182. doi: 10.1006/abbi.1998.1024. [DOI] [PubMed] [Google Scholar]
- 23.Cho H P, Nakamura M T, Clarke S D. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 24.Leonard A E, Kelder B, Bobik E G, Chuang L-T, Parker-Barnes J M, Thurmond J M, Kroeger P E, Kopchick J J, Huang Y-S, Mukerji P. Biochem J. 2000;347:719–724. [PMC free article] [PubMed] [Google Scholar]
- 25.Sargent J R, Henderson R J, Tocher D R. In: Fish Nutrition. 2nd Ed. Halver J E, editor. San Diego: Academic; 1989. pp. 153–218. [Google Scholar]
- 26.Sargent J R, Bell J G, Henderson R J, Tocher D R. J Appl Ichthyol. 1995;11:183–198. [Google Scholar]
- 27.Sargent J R, Bell J G, McEvoy L, Tocher D R, Estevez A. Aquaculture. 1999;177:191–199. [Google Scholar]
- 28.Owen J M, Adron J A, Middleton C, Cowey C B. Lipids. 1975;10:528–531. doi: 10.1007/BF02532354. [DOI] [PubMed] [Google Scholar]
- 29.Henderson R J, Tocher D R. Prog Lipid Res. 1987;26:281–347. doi: 10.1016/0163-7827(87)90002-6. [DOI] [PubMed] [Google Scholar]
- 30.Mourente G, Tocher D R. Biochim Biophys Acta. 1994;1212:109–118. doi: 10.1016/0005-2760(94)90195-3. [DOI] [PubMed] [Google Scholar]
- 31.Buzzi M, Henderson R J, Sargent J R. Biochim Biophys Acta. 1996;1299:235–244. doi: 10.1016/0005-2760(95)00211-1. [DOI] [PubMed] [Google Scholar]
- 32.Tocher, D. R., Agaba, M., Hastings, N., Dick, J. R. & Teale, A. J. (2001) Fish Physiol. Biochem., in press.
- 33.Brenner R R. Prog Lipid Res. 1981;20:41–47. doi: 10.1016/0163-7827(81)90012-6. [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Tocher D R, Harvie D G. Fish Physiol Biochem. 1988;5:229–239. doi: 10.1007/BF01874800. [DOI] [PubMed] [Google Scholar]
- 36.Christie W W. Lipid Analysis. 2nd Ed. Oxford: Pergamon; 1982. [Google Scholar]
- 37.Balazy M, Nies A S. Biomed Environ Mass Spectrom. 1989;18:328–336. doi: 10.1002/bms.1200180508. [DOI] [PubMed] [Google Scholar]
- 38.Christie W W. Lipids. 1998;33:343–353. doi: 10.1007/s11745-998-0214-x. [DOI] [PubMed] [Google Scholar]
- 39.Marquardt A, Stohr H, White K, Weber B H F. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 40.Ghioni C, Tocher D R, Bell M V, Dick J R, Sargent J R. Biochim Biophys Acta. 1999;1437:170–181. doi: 10.1016/s1388-1981(99)00010-4. [DOI] [PubMed] [Google Scholar]
- 41.Tocher D R, Ghioni C. Lipids. 1999;34:433–440. doi: 10.1007/s11745-999-0382-8. [DOI] [PubMed] [Google Scholar]
- 42.Sargent J R, Tacon A. Proc Nutr Soc. 1999;8:377–383. doi: 10.1017/s0029665199001366. [DOI] [PubMed] [Google Scholar]