Abstract
The specific targeting of differentially expressed glycans in malignant cells has emerged as an attractive anti-cancer strategy. One such target is the oncodevelopmental antigen polysialic acid (polySia), a polymer of α2,8-linked sialic acid residues that is largely absent during postnatal development but is re-expressed during progression of several malignant human tumors including small cell and non-small cell lung carcinomas, glioma, neuroblastoma, and pancreatic carcinoma. In these cancers, expression of polySia correlates with tumor progression and poor prognosis and appears to modulate cancer cell adhesion, invasiveness, and metastasis. To evaluate the potential of PolySia as a target for anti-cancer therapy, we developed a chimeric human polySia-specific monoclonal antibody (mAb) that retained low nanomolar (nM) target affinity and exhibited exquisite selectivity for polySia structures. The engineered chimeric mAb recognized several polySia-positive tumor cell lines in vitro and induced rapid endocytosis of polySia antigens. To determine if this internalization could be exploited for delivery of conjugated cytotoxic drugs, we generated an antibody-drug conjugate (ADC) by covalently linking the chimeric human mAb to the tubulin-binding maytansinoid DM1 using a bioorthogonal chemical reaction scheme. The resulting polySia-directed ADC demonstrated potent target-dependent cytotoxicity against polySia-positive tumor cells in vitro. Collectively, these results establish polySia as a valid cell-surface, cancer-specific target for glycan-directed ADC and contribute to a growing body of evidence that the tumor glycocalyx is a promising target for synthetic immunotherapies.
Keywords: Antibody-drug conjugate, cancer, glycocalyx, glycosylation, immunotherapy, metastasis, monoclonal antibody, NCAM, polysialyltransferase, polysialic acid
Introduction
Glycosylation is the site-specific attachment of sugar assemblies known as glycans to a functional group of another molecule, most commonly proteins or lipids, resulting in the formation of a glycoconjugate. It is a tightly controlled cell- and microenvironment-specific mechanism that involves the coordinated expression and activity of numerous enzymes such as glycosyltransferases and glycosidases. Cellular glycosylation and its products are fundamental to a diverse range of biological processes involved in cancer progression including cell growth and proliferation, cell signaling and communication, cell-cell and cell-extracellular matrix (ECM) interactions, and immune recognition/response (1–4). Thus, it is not surprising that nearly all types of human cancers exhibit changes in glycosylation, a phenomenon that was first reported more than six decades ago (5,6). The glycosylation changes associated with oncogenic transformation typically involve either incomplete synthesis or neo-synthesis processes, both of which may arise from under- or overexpression of glycosyltransferases and glycosidases leading to the exposure of aberrant cell-surface glycans. The most common cancer-associated structural changes include N- and O-glycan branching, O-glycan truncation, increased sialylation, and increased “core” fucosylation, with these motifs occurring on all classes of glycoconjugates including glycoproteins, glycosphingolipids, and proteoglycans (7,8).
Many of these abnormal glycan epitopes are differentially expressed on malignant cells, thereby providing novel diagnostic and even therapeutic targets that are motivating the development of affinity reagents that recognize these distinct features. However, whereas a rich and diverse collection of antibodies and antibody-derived molecules have been developed for protein antigens, reliable binders that specifically recognize carbohydrates are much less common. Indeed, the paucity of glycan-specific binding reagents was noted by the National Academy of Sciences as a key barrier for advancing glycobiology (9). This shortage was also highlighted in the recently assembled Database for Anti-Glycan Reagents (DAGR), which indicates that while there are ~100 entries for antibodies against N- and O-linked carbohydrates, collectively these target an extremely small set of unique epitopes (10). Specifically, 55 of the 77 total antibodies to O-linked glycans target Tn, sialyl Tn, or TF antigens while 15 of the 25 total antibodies to N-linked glycans are derived from HIV patients. There is clearly a technological deficit when one considers that glycoproteins and glycolipids are estimated to contain approximately 3,000 glycan determinants (11).
Even when anti-glycan antibodies are available, information about their specificity is often limited and, in a surprising number of cases, antibodies reported to be specific for a designated antigen were found to cross-react with other glycans (12). Moreover, for many of the glycans that differentially occur in malignant cells, it remains to be determined whether they are druggable using “synthetic” immunotherapies (13) such as monoclonal antibodies (mAbs), antibody-drug conjugates (ADCs), bispecific antibodies (BsAbs), and chimeric antigen receptors (CARs), which all have the potential to initiate new immune or immune-like responses directed toward their tumor-expressed targets. One notable example along these lines is the synthetic immunotherapy dinutuximab, a first-in-class monoclonal antibody (mAb) that recognizes the disialoganglioside GD2 found on the surface of neuroblastic tumor cells and is administered as part of a multi-agent, multimodality therapy to pediatric patients with high-risk neuroblastoma (14). There is similar potential to develop other glycan-directed antibodies and antibody derivatives; however, this will require overcoming a number of key obstacles related to (i) the current lack of antibodies against structurally diverse glycan antigens beyond the small subset discussed above and (ii) the incomplete knowledgebase surrounding known antibodies in terms of their performance characteristics such as target specificity and therapeutic function (e.g., cytotoxicity).
Here, we addressed this latter gap by systematically characterizing the well-known mouse-derived mAb 735 (mo735) (15) and a newly created chimerized human derivative (ch735), both of which target the oncodevelopmental carbohydrate antigen polysialic acid (polySia). PolySia is a unique glycan homopolymer of α2,8-linked N-acetyl neuraminic acid (NeuNAc) that occurs as a terminating structure on the N-linked glycan associated with the neural cell adhesion molecule (NCAM) and also as a capsular polysaccharide (CPS) on the surface of bacterial pathogens causing meningitis (16). In vertebrates, the expression of polySia is abundant during early stages of development of the brain, heart, kidney, liver, pancreas, respiratory and digestive tracts, but becomes significantly reduced in adults with expression largely restricted to certain regions of the brain (16). Importantly, it is aberrantly re-expressed in many cancers, appearing as part of the tumor glycocalyx in small cell lung cancer (SCLC) (17), non-small cell lung cancer (NSCLC) (18), pancreatic cancer (19), Wilm’s tumor (20), neuroblastoma (21), and glioma (22) among others. PolySia expression, which is catalyzed by two polysialyltransferases, ST8SiaIV (PST) and particularly ST8SiaII (STX) in cancer cells (18), is known to promote cancer cell adhesion, migration and invasion (22–24) and is strongly correlated with aggressive and metastatic disease as well as poor prognosis in the clinic (25). For many of the aforementioned reasons, polySia was ranked as the second highest priority glycan antigen (after GD2) in a National Cancer Institute pilot project (26).
To investigate polySia targeting and its clinical potential, the mo735 and ch735 antibodies were subjected to a spectrum of biochemical and cell biological assays to characterize their polySia binding properties. Importantly, both antibodies were observed to bind polySia with high affinity and exquisite selectivity. We also confirmed that both antibodies recognized several polySia-positive tumor cell lines in vitro and induced rapid internalization of polySia into endosomal and lysosomal compartments. In light of these findings, we hypothesized that the antibody-induced endocytosis of polySia-receptors could be efficiently harnessed as part of an antitumor therapeutic strategy. To test this notion, we engineered an ADC using a bioorthogonal reaction scheme for stably linking the chimeric human ch735 mAb to the microtubule-inhibitory agent maytansinoid DM1, which has previously been developed as the cytotoxic payload in trastuzumab emtansine (T-DM1) for HER2-positive breast cancer (27). The resulting conjugate was found to exert potent target-dependent cytotoxicity against polySia-positive tumor cells in vitro, providing compelling proof-of-concept for the use of polySia-receptor internalization as a carrier for delivery of cytotoxic payloads to cancer cells. Taken together, our findings add to the growing body of literature implicating aberrant glycans in the tumor glycocalyx as an attractive collection of targets for the development of glycan-directed synthetic immunotherapies.
Materials and Methods
Construction of chimeric human mAb ch735.
The DNA sequences for the VH and VL domains of mAb mo735 (28) were obtained from the GenBank™/EBI Data Bank (accession number AB821355) and ordered from GeneArt Gene Synthesis (Thermo Fisher Scientific). The variable regions of mAb 735 were then swapped with the existing variable regions in pVITRO1-Trastuzumab-IgG1/k (Addgene plasmid #61883) as previously described to generate the vector pVITRO-735-IgG1/k (29). Briefly, polymerase incomplete primer extension (PIPE) PCR was performed using sets of primers (Supplementary Table 3) to generate four linear fragments of the construct with 5’ PIPE overhangs. All cloned plasmids were confirmed by DNA sequencing.
Cell culture and reagents.
Production of recombinant mAbs was performed using FreeStyle™ 293-F cells (ThermoFisher Scientific). FreeStyle™ 293-F cells were maintained in FreeStyle 293 expression medium (ThermoFisher Scientific). Cancer cell lines SH-SY5Y, H82, H69, K562, MCF7, and SKOV3 were obtained from American Type Culture Collection (ATCC) while cell lines SW2 and A549 were kindly provided by Dr. Karen Colley (University of Illinois at Chicago). SH-SY5Y cells were maintained in high glucose DMEM/F12 medium supplemented with 10% Hyclone FetalClone I serum (VWR), 1% MEM non-essential amino acids solution (ThermoFisher Scientific), penicillin (100 U/mL) and streptomycin (100 μg/mL) (ThermoFisher Scientific). H82, H69, K562, and A549 cells were maintained in RPMI 1640 with L-glutamine (ThermoFisher Scientific) supplemented with 10% Hyclone FetalClone I serum, penicillin (100 U/mL) and streptomycin (100 μg/mL). MCF7 cells were maintained in high glucose DMEM supplemented with 10% Hyclone FetalClone I serum, insulin (10 μg/mL, Sigma), penicillin (100 U/mL) and streptomycin (100 μg/mL). SKOV3 and SW2 cells were maintained in high glucose DMEM supplemented with 10% Hyclone FetalClone I serum, penicillin (100 U/mL) and streptomycin (100 μg/mL). All cell lines were maintained at low passage numbers and routinely checked for mycoplasma by PCR according to standard procedures.
Expression and purification of ch735 and trastuzumab.
293-F cells cultured in FreeStyle™ 293 Expression Medium (ThermoFisher Scientific) were transfected with pVITRO-735-IgG1/k or pVITRO1-Trastuzumab-IgG1/k using FreeStyle™MAX transfection reagent (ThermoFisher Scientific) according to the manufacturer’s instructions and selected under hygromycin B as previously described (29). Purified plasmid DNA was precipitated by mixing 1/10 the volume of 3 M sodium acetate pH 5.2 and 2–3 volumes of 100% ethanol and freezing at −80°C for 2 h. The DNA was collected by centrifugation at 13,000 x g at 4°C for 30 min and resuspended in 100 μL of sterile tissue culture grade water (Thermo Fisher). After selection, cultures were expanded to 1 L culture volume and maintained with 50% hygromycin B (25 μg/mL). Supernatants were harvested every 48 h, centrifuged at 1000 x g for 15 min, passed over 0.2 μm filters (VWR) and stored at 4°C until use.
Protein A/G agarose (Thermo Fisher) was used to purify antibodies from the supernatant according to the manufacturer’s recommendations. The agarose equilibrated with 10 mL phosphate-buffered saline (PBS) in a polypropylene gravity column. The supernatant was then allowed to completely pass through the column. The column was then washed with PBS until there was no signal in the flow through at an absorbance of 280 nm (Abs280). Antibodies were eluted from the column with 0.1 M glycine-HCl (pH 2.0) in 1-mL fractions and neutralized with 100 μL 1 M Tris (pH 8.0). Antibody purity was evaluated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing and non-reducing conditions and visualized by staining with Coomassie Blue G-250. Protein A/G-purified antibodies were analyzed by size exclusion chromatography (SEC) on a 4.6 mm ID x 30 cm TSKgel SuperSW3000 SEC column with 4-µm particles. Pure antibodies were eluted from the column at 8.66 min in 0.1 M phosphate buffer containing 0.1 M Na2SO4, pH 6.7.
ELISA.
Costar 96-well ELISA plates (Corning) were coated overnight at 4°C with 50 μl of 1 μg/mL chicken brain NCAM (Millipore) or endoN-treated NCAM in PBS. Chicken NCAM (Millipore, AG265) was digested with 1.5 μg of endoN per 50 μg of NCAM overnight at 37°C. After blocking with 5% (w/v) milk in PBS for 1–3 h at room temperature or overnight at 4°C, ELISA plates were washed three times with wash buffer (PBST with 0.3% BSA) and incubated with serially diluted purified ch735, mo735 (Absolute Antibody), or ab5032 (Millipore) for 1 h at room temperature. Antibody samples were quantified with a Nanodrop. After washing three times with wash buffer, 100 μl of 1:5,000-diluted rabbit anti-human IgG (Fc) antibody-HRP conjugate (Thermo Fisher), goat anti-mouse IgG-HRP (Abcam), or goat anti-rabbit IgG-HRP (Abcam) in wash buffer was added to each well for 1 h. Plates were washed and developed with TMB (ThermoFisher) using standard protocols.
Specificity profiling using glycan and glycoprotein arrays.
Specificity of mo735 and ch735 was determined using printed glycan arrays 5.3 and 5.4 at the CFG (http://www.functional-glycomics.org/static/consortium/resources/resourcecoreh.shtml). Both antibodies were analyzed at 10 μg/mL with 5 μg/mL of anti-mouse or anti-human Alexa-Fluor 647 (AF647)-conjugated secondary, respectively. Specificity of mo735 (1 μg/mL) and ch735 (10 μg/mL) was also assessed on a custom glycoprotein array that contained ~40–50 glycoproteins including chicken brain NCAM and endoN-treated chicken brain NCAM.
SPR.
Equilibrium binding-affinity measurements were made by SPR analysis on a Biacore 3000 system. Antibodies mo735 and ch735 were bound to the surface of a Protein A sensor chip with a target level of 1700 response units (RUs). Serial dilutions of the antigen, chicken NCAM, prepared in 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate-20 (HBS-EP buffer, GE Healthcare) at concentrations ranging from 0.25 to 250 nM were injected over the chip using the same buffer at a flow rate of 20 μl/min (10 min injection time, 2 min stabilization time, 20 min dissociation). The surface of the chip was regenerated between the injections of each serial dilution with 10 mM glycine, pH 1.5 (30 sec injection time, 3 min stabilization time). Kinetic parameters were determined by fitting the maximum response values for each concentration using the Hill slope non-linear regression analysis in Prism software.
CRISPR-Cas9 genome editing.
CRISPR guide RNAs targeting ST8SiaII (crRNA1: ATGCAGTGCGCACGTTGACG) and ST8Sia4 (crRNA1: ACCCGATGAGTTGCGTCTCC) were purchased from Genscript in the pLentiCRISPR v2 vector. Knockout cell lines were generated in SW2 cells using protocols described by Zhang and colleagues (30). Briefly, after lentiviral transduction, cells were maintained under selection with 1 μg/mL puromycin for at least 14 days prior to analysis by flow cytometry.
Flow cytometric analysis.
Cancer cells were trypsinized and collected with media, followed by three washes in PBS. To remove polySia, cells were treated with endoN at 3 μg/mL in PBS for 1 h at room temperature. The cells are resuspended in 4% paraformaldehyde (PFA), fixed at room temperature with constant agitation for 10 min, and then washed two times with PBS, and two times with 0.5% BSA in PBS. The cells were collected and resuspended to 1 million cells/100 μL and pipetted into a round bottom 96-well plate. The cells were pelleted in the 96-well plate and resuspended in 0.5% BSA in PBS containing mo735 or ch735 (5 μg/mL) and incubated for 30 min at room temperature with constant agitation. Cells were washed three times with 0.5% BSA in PBS and resuspended in anti-mouse IgG-Alexa-Fluor 488 (AF488) secondary or anti-human IgG-AF488 secondary (ThermoFisher Scientific) at a 1:200 dilution for 30 min at room temperature in the dark with constant agitation. Cells were washed three times, resuspended in 500 μL of 0.5% BSA in PBS, and analyzed on a BD FACSCalibur flow cytometer using Cell Quest Pro software (BD Biosciences).
Confocal microscopy.
Adherent cells were plated at 20,000 cells/cm2 on poly-L-lysine coated 35-mm glass bottom dishes and adhered overnight. To remove polySia, cells were then treated with endoN at 3 μg/mL in cell culture media overnight. Suspension cells were collected on the day of the experiment and labeled in suspension with the same protocol as the adherent cells. Cells were fixed with 4% paraformaldehyde and subsequently blocked with 5% normal goat serum PBS (NPBS) for 1 h at room temperature. Antibodies mo735 and ch735 were diluted to 5 μg/mL in 5% NPBS and incubated overnight at 4°C. Anti-mouse and anti-human AF488-conjugated secondary antibodies A32723 and A11013 (ThermoFisher Scientific) were diluted 1:200 in NPBS and incubated for 2 h at room temperature. Wheat germ agglutinin-AF647 (WGA-647) was diluted to 1 μg/mL in NPBS and incubated for 10 min at room temperature. Hoescht dye was used at 1 μg/mL in PBS for 5 min at room temperature. Samples were imaged on a Zeiss LSM inverted 880 confocal microscope using a 40x water immersion objective.
IHC.
The avidin-biotin complex (ABC) immunoperoxidase method was performed essentially as previously described (31). Briefly, the sections were quenched with 0.1% H2O2 in PBS for 15 min, blocked with avidin and biotin reagents (Vector, Burlingame, CA) for 10 min each, incubated in 10% serum from which the second antibody was raised and incubated with mAb 735 at 1 μg/ml for 1 h. This concentration was selected based on strong reactivity against known positive target cells and little or no background against stroma. The sections were subsequently incubated with biotinylated secondary antibodies for 30 min, and then incubated in ABC reagent per manufacturer’s protocols (ABC Kit, Vector Laboratories, PK-6102) for 30 min. Reactions were developed with liquid DAB+ Substrate Chromogen System (Dako, cat # K3468) for 3 min at room temperature. Slides were then counterstained with Mayer’s Hematoxylin (Dako Cyomation, cat # S3309) for 1 min at room temperature. The immunoreactivities were graded based on the percentage of positive cells and staining intensity above that seen on the negative control. Known positive and negative control slides were used in each experiment.
Internalization assays.
To calculate percent internalization, pre-chilled cells were incubated with 50 nM ch735 on ice for 1 h and then washed to remove unbound antibodies. For each time point, one aliquot of cells remained on ice and one was incubated at 37°C for 15, 30, or 60 min. Cells were fixed in 2% paraformaldehyde for 20 min and then stained with AF488-labeled antibody against human IgG and analyzed by flow cytometry and FlowJo software. Receptor-antibody complex internalization was calculated using the geometric mean as percent fluorescent intensity loss at 37°C relative to that on ice. For each sample, the geometric mean fluorescence intensity (MFI) of 10,000 cells was measured in triplicate.
Colocalization microscopy.
SH-SY5Y or SW2 cells were plated at 20,000 cells/cm2 on poly-L-lysine coated 35 mm glass bottom dishes and adhered overnight. To remove polySia, cells were treated as described above. To measure receptor-antibody internalization, cells were incubated with 150 nM AF488 labeled ch735, human IgG isotype control (ThermoFisher Scientific), mo735, or mouse IgG isotype control (anti-MBP mAb, NEB) and 100 nM AF647 transferrin or AF647 anti-LAMP-3 antibody (Santa Cruz) for 1 h. Cells were washed and then fixed as described above. To examine lysosomal trafficking, cells were incubated with 150 nM of ch735 or isotype at 37°C for 2 h, washed, fixed, and then permeabilized using 0.1% Triton X-100 NPBS. Cells were stained with AF488-labeled antibody against human IgG to visualize antigen-antibody complex (ThermoFisher Scientific) and mouse anti-human LAMP-1 clone D2D11 (Cell Signaling) followed by AF647-labeled anti-rabbit IgG to visualize the lysosomes (ThermoFisher Scientific). Hoescht dye was used at 1 μg/mL in PBS for 5 min at room temperature. Samples were imaged on a Zeiss LSM inverted 880 confocal microscope using a 40x water immersion objective. For colocalization analysis, a 5-μM line was drawn across the apparent vesicles. The fluorescence intensity of the plot profile was analyzed using FIJI software. Fluorescence intensity was normalized to the maximum value for each channel.
Drug linker synthesis.
To synthesize the drug linker, 650 µg of DM1 was incubated at 1.1 mM with 3 molar equivalents of maleimide-PEG3-TCO (Click Chemistry Tools) in 3:1 DMSO:PBS overnight at room temperature. The reaction mixture was purified on a C18 analytical RP-HPLC column on a gradient of 5–95% acetonitrile in water over 30 min. The product was dried, resuspended in DMSO, and quantified via Abs252 measurements (ext. coeff252nm = 26,790 M cm−1). Product mass was verified via LCMS (expected mass = 1,260.56).
ADC conjugation.
Purified ch735, purified trastuzumab, or human IgG1 isotype control (ThermoFisher Scientific) was reacted with 10 molar equivalents of methyltetrazine-PEG4-NHS ester overnight at 37°C. Excess reagent was removed by centrifugation dialysis. The Tz-conjugated antibody was then incubated with 3 molar equivalents of TCO-maleimide-DM1 drug linker for 5 h at 37°C. Excess reagent was removed by centrifugation dialysis. Average DAR was determined using absorbance spectroscopy to calculate the concentrations of antibody and drug (32). The following previously established extinction coefficients were used for each component: ε280 DM1 = 5,700 M−1 cm−1, ε252 DM1 = 26,790 M−1 cm−1 and ε280 Antibody = 218,134 M−1 cm−1, ε252 Antibody = 76,565 M−1 cm−1 (33).
Cell viability assay.
SKOV3, MCF7, and SH-SY5Y cells were plated at 5,000, 2,500, and 2,500 cells/well, respectively, and allowed to rest for 24 h. Five-fold serial dilutions of the antibodies were added starting at 150 nM and incubated for 72–144 h. The viability assays were then developed using Alamar blue according to manufacturer’s protocol (Bio-Rad). Percent viability is calculated by first subtracting the value of media alone from all samples. Subsequently, the resulting values are divided by the values measured from an un-treated control representing maximum viability.
Results
Construction and characterization of a chimeric human IgG targeting polySia.
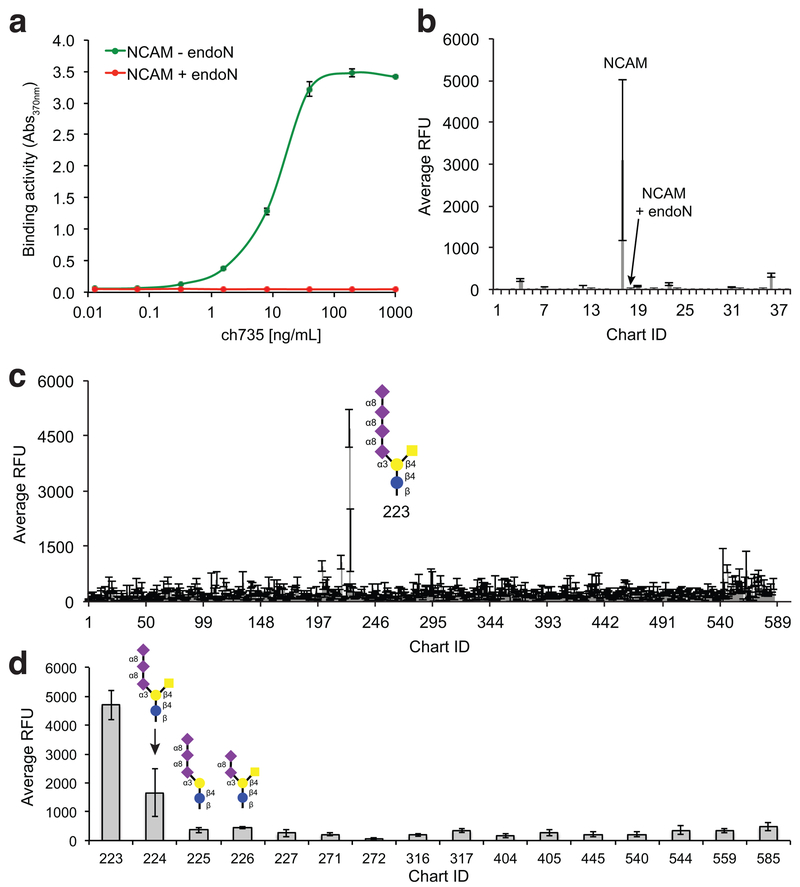
To generate a monoclonal antibody (mAb) that is more compatible with targeting human cancers, we converted the fully mouse IgG2a mAb 735 (mo735) into a chimeric human IgG1 (ch735) by swapping the variable regions according to an antibody cloning and expression method described by Beavil and coworkers (29). Using this approach, a stable cell line was generated that was capable of producing fully assembled ch735, which could be purified to near homogeneity at yields up to 6 mg/L (Supplementary Fig. 1a-c). Subsequent enzyme-linked immunosorbent assay (ELISA) analysis confirmed that both antibodies bound chicken brain-derived polysialylated neural cell adhesion molecule (NCAM) but not NCAM that was treated with endoneuraminidase N (endoN) that selectively removes polySia (Fig. 1a and Supplementary Fig. 2a). Probing of similarly prepared ELISA plates with an NCAM-specific antibody confirmed that both NCAM and endoN-treated NCAM were equally coated on ELISA plates (Supplementary Fig. 2b). Given the strict specificity of endoN for α2,8-linkages in sources as disparate as bacterial and neural membrane glycoconjugates, we conclude that both mo735 and ch735 specifically recognize polySia.
Figure 1. Binding specificity of the ch735 antibody.
(a) Antigen binding activity for recombinant purified ch735 determined by ELISA with either NCAM or endoN-treated NCAM immobilized as antigens. ELISA signals (Abs370) were obtained with anti-human IgG-HRP conjugate. (b) Glycoprotein-binding specificity of ch735 was probed using an array of ~50 glycoproteins. Antibodies were assayed at 1 μg/mL and detected with anti-human IgG antibodies. (c) Glycan-binding specificity of ch735 was measured against CFG glycan microarray (version 5.4) that contained ~585 natural and synthetic mammalian glycans (http://www.functionalglycomics.org). Antibodies were assayed at 10 μg/mL and detected with anti-human IgG antibodies. All data are the average of three replicate experiments and error bars are the standard deviation of the mean. (d) Same as in (c) but only showing data for glycan structures containing α2,8-linked sialic acid (see Supplementary Table 1 for a list of the corresponding structures).
To investigate glycan specificity, we also analyzed both antibodies on a glycoprotein array that contained ~50 glycoproteins and the current glycan array (version 5.3 for mo735, version 5.4 for ch735) of the Consortium for Functional Glycomics (CFG) that contained ~600 natural and synthetic mammalian glycans (http://www.functionalglycomics.org). The chimeric human IgG1 ch735 showed a strong preference for polysialylated NCAM in the glycoprotein array (Fig. 1b; Chart ID #17) and the tetra-sialic acid containing glycan GalNAcβ1–4(Neu5Acα2–8Neu5Acα2–8Neu5Acα2–8Neu5Acα2–3)Galβ1–4Glcβ-Sp0 in the glycan array (Fig. 1c and d; Chart ID #223 in microarray version 5.4). A lesser but still significant level of binding above background was detected for GalNAcβ1–4(Neu5Acα2–8Neu5Acα2–8Neu5Acα2–3)Galβ1–4Glcβ-Sp0 (Chart ID #224), but not for the closely related glycans #225 and 226 that differed from 224 by a single branched GalNac and Neu5Ac, respectively (Fig. 1d). In total, the glycan microarray included 145 glycans containing some form of sialic acid, often as the terminal sugar, of which 16 were α2,8-linked (Supplementary Table 1); hence, we conclude unequivocally that the specificity of ch735 is for α2,8-linked sialic acid with a degree of polymerization (DP) of three or greater. In line with this conclusion, there was no significant signal towards any other glycomolecules including endoN-treated NCAM that was spotted on the glycoprotein array. Nearly identical results were observed when the same microarrays were probed with mo735 (Supplementary Fig. 3a-c).
To determine affinity, we measured the equilibrium binding of both antibodies to polysialylated NCAM using surface plasmon resonance (SPR). Binding values were fit using the specific binding with Hill slope analysis in Prism software and the calculated Kd values for mo735 and ch735 were determined to be 10.22 and 4.79 nM, respectively (Supplementary Fig. 4a-b). These values were in close agreement with the previously reported Kd of ~5 nM for the mouse IgG against embryonic brain glycopeptides (34).
Antibody ch735 binds surface polySia and is internalized in cancer cells.
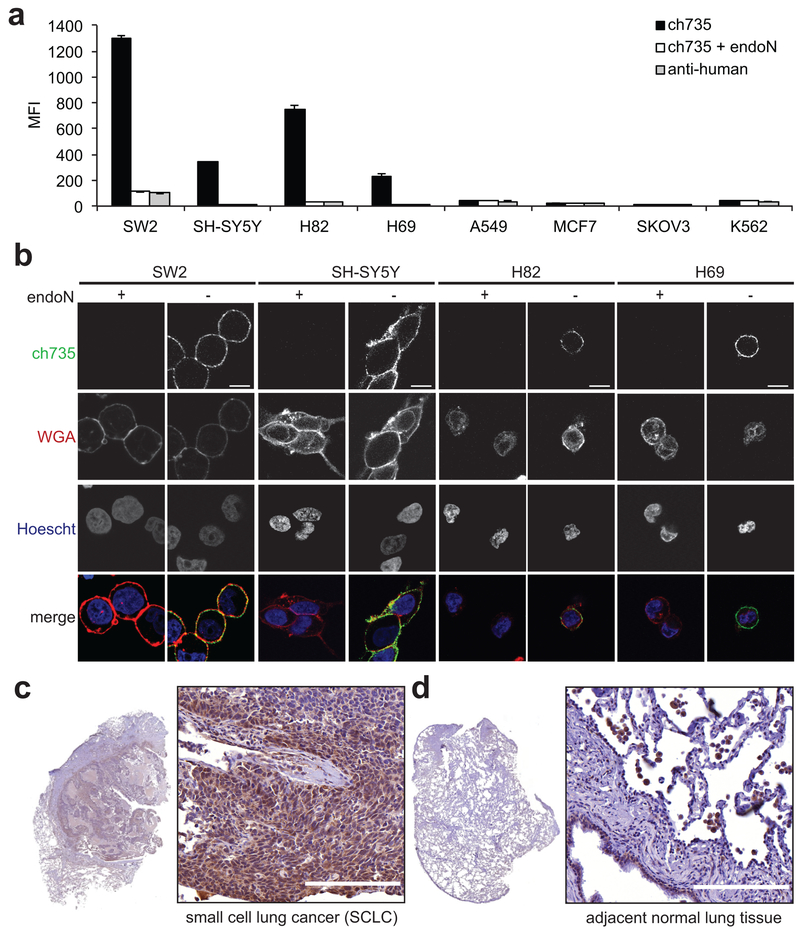
To demonstrate the relevance of this antibody in the context of human cancers, we used flow cytometric analysis to assess ch735 binding to polySia expressed on the surface of different cancer cell lines including the small cell lung cancer (SCLC) cell lines SW2, NCI-H69, and NCI-H82, as well as neuroblastoma cell line SH-SY5Y, non-small cell lung cancer (NSCLC) cell line A549, breast cancer cell line MCF7, ovarian cancer cell line SKOV3, and chronic myeloid leukemia (CML) cell line K562. Antibody ch735 bound most avidly to SW2 cells, and also recognized NCI-H69, NCI-H82, and SH-SY5Y cancer cells although with a lower intensity (Fig. 2a), in agreement with previous cell line characterization studies (35–37). Inspection of the representative histograms revealed not only differing levels of surface expression between the different polySia-positive cancer cells but also within each population especially for H69 cells (Supplementary Fig. 5a). The ability of ch735 to recognize polySia on the surfaces of these cells was corroborated by immunofluorescence microscopic images of the polySia-positive cell lines (Fig. 2b). When A549 and MCF7 cancer cells were similarly probed with ch735, we observed no significant binding above background, in agreement with previous studies (24,35). Likewise, SKOV3 and K562 cells were not recognized by ch735, establishing these as polySia-negative cell lines. It should also be noted that staining of each of these cell lines with mo735 yielded virtually identical polySia binding results (Supplementary Figs. 5b and 6a-b).
Figure 2. Immunostaining of antibody ch735 to polySia-expressing cancer cells.
(a) External levels of polySia on a panel of cancer cell lines with or without endoN treatment measured by flow cytometry using ch735 and fluorescent anti-human secondary. Data are the geometric mean fluorescence intensity (MFI), with the values reported as the average of three replicates and the error represented as the standard deviation of the mean. (b) Confocal microscopic images of endoN-treated and non-treated polySia expressing cancer cell lines to assess ch735 binding on the cell surface. Cells were stained with ch735 (green), wheat germ agglutinin (WGA, red) to stain the cell membrane, and Hoescht (blue) to stain nuclei. Scale bars, 10 μm. (c) Formalin-fixed, paraffin-embedded (FFPE) human tissue sections of SCLC and (d) adjacent normal tissue stained for polySia with mo735. Scale bars, 200 μm.
Importantly, surface binding was completely abrogated following endoN treatment, confirming that binding to these cell lines was specific to polySia. Further evidence of polySia-specific binding was demonstrated by a significant decrease in antibody labeling of SW2 cells in which the polysialyltransferases ST8SiaII and ST8SiaIV were knocked out by CRISPR-Cas9 gene editing (Supplementary Fig. 7a-c). In addition, immunohistochemistry (IHC) revealed strong staining of polySia in formalin-fixed, paraffin-embedded (FFPE) human tissue sections of SCLC (Fig. 2c), but little to no staining of the adjacent normal tissue except for the bronchial epithelial cells and alveolar macrophages (Fig. 2d), in close agreement with previous findings (31).
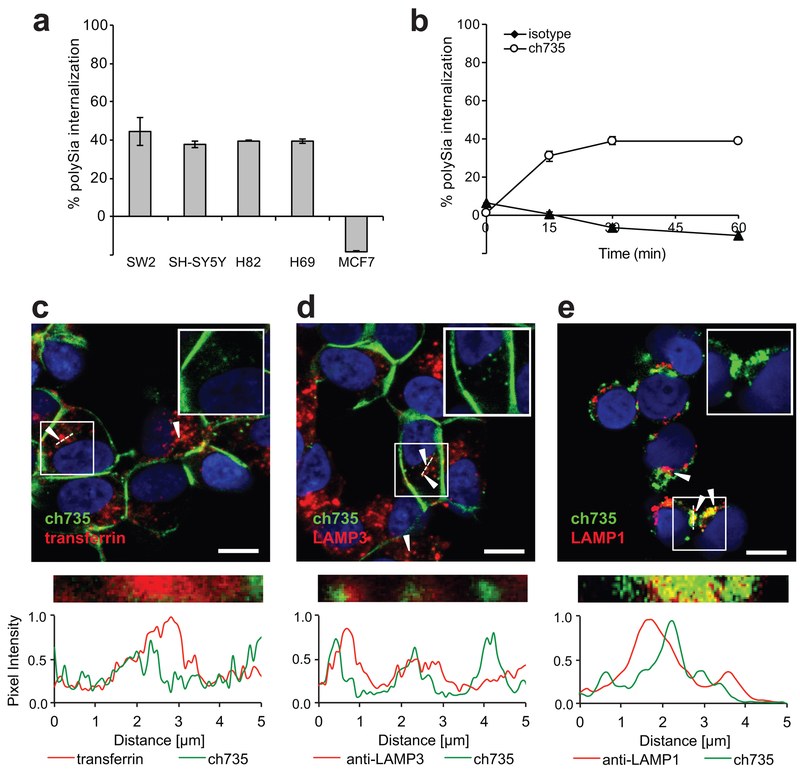
Given that clathrin‐mediated endocytosis is an essential pathway by which many glycoproteins are recycled or down‐regulated (38), we next investigated whether polySia undergoes a similar internalization process. Previous studies demonstrated that NCAM, one of the major carriers of polySia, was recycled by a clathrin-dependent endocytosis process (39,40), whereas polySia was only detectable at the cell surface (35,41) unless internalization was activated by the extracellular matrix (ECM) (42). To further investigate this issue here, each cell line that expressed cell surface polySia was evaluated for the ability to internalize polySia. This involved first binding ch735 to the surface of tumor cells at 4°C, after which an aliquot of cells remained at 4°C while the rest were incubated at 37°C and analyzed by flow cytometry at different time points. For each of the polySia-positive cell lines, we observed that ~40% of the antibody was internalized after 1 h while no internalization was observed for the MCF7 cell line (Fig. 3a), which were previously found to lack polySia at the cell surface (35). A time course of ch735 binding to SH-SY5Y cells revealed that antibody internalization occurred rapidly, with ~30% of the antibody internalized as early as 15 min and maximum internalization of 40% reached by 30 min (Fig. 3b). In contrast, an isotype control antibody showed no measurable internalization over the same time period. It is noteworthy that the internalization percentage and rate observed here with ch735 was on par with that reported previously with trastuzumab against HER-2-positive cancer cells (43). It should also be noted that comparable internalization of mAb mo735 into SW2 cells was observed, with intracellular fluorescence increasing as a function of polySia-specific antibody concentration and as a function of time (Supplementary Fig. 8a-d).
Figure 3. Internalization of ch735 into polySia-positive cancer cells.
(a) Internalization of ch735 in polySia-positive cell lines SH-SY5Y, SW2, H69, and H82, and in polySia-negative MCF7 cells after 1 h. Data are reported as the mean percent internalization and error bars are the standard deviation of the mean (n = 3). (b) Time course of antibody internalization in polySia-positive cell line SH-SY5Y treated with ch735 or isotype control. Data reported as the mean percent internalization and error bars are the standard deviation of the mean (n = 3). (c) Confocal microscopy images of SH-SY5Y cells incubated for 1 h with ch735 labeled with AF488 and transferrin labeled with AF647. Nuclei were stained by Hoescht (blue). Scale bar, 10 μm. Fluorescence intensity was measured across the dotted white line and normalized to the maximum value in each channel. White arrows indicate regions of colocalization. The inset shows only the ch735 (green) and DNA (blue) channels of the boxed region. (d) Confocal microscopy images of SH-SY5Y cells incubated for 1 h with ch735 labeled with AF488 and anti-LAMP-3 labeled with AF647. Nuclei were stained by Hoescht (blue). Scale bar, 10 μm. Fluorescence intensity was measured across the dotted white line and normalized to the maximum value in each channel. White arrows indicate regions of colocalization. The inset show only the ch735 (green) and DNA (blue) channels of the boxed region. (e) Confocal microscopy images of SH-SY5Y cells incubated for 120 min with ch735. Lysosomes were stained with anti-LAMP-1 and A647-labeled anti-rabbit antibody (red), ch735 was stained with AF488-labeled anti-human antibody (green), and nuclei were stained by Hoescht (blue). Scale bar, 10 μm. Fluorescence intensity was measured across the dotted white line and normalized to the maximum value in each channel. White arrows indicate regions of colocalization. The top right inset shows only the ch735 (green) and DNA (blue) channels of the boxed region.
Confocal microscopy was used to investigate the compartments where the ch735 mAb accumulated after internalization using markers of early endosomes, recycling endosomes or late endosome/lysosomes. Consistent with flow cytometry, ch735 initially labeled the plasma membrane of SH-SY5Y cells and after 1 h at 37°C was internalized, where it clearly colocalized with early endosomal and recycling endosomal marker transferrin (Fig. 3c) and late endosomal marker LAMP-3 (Fig. 3d). Accumulation of the ch735 mAb was also observed in late endosomal/lysosomal LAMP-1-positive compartments (Fig. 3e). As expected, no detectable binding, internalization or colocalization was observed for the isotype control (Supplementary Fig. 9a). Similar to ch735, the mo735 mAb compartmentalized in early and recycling endosomes as confirmed by colocalization with transferrin and LAMP-3 (Supplementary Fig. 9b and c). Based on these data, we conclude that mAb ch735 binds to tumor cell membranes in a target-specific manner, thereby inducing a subpopulation of bound antibodies to become rapidly internalized in endosomal/lysosomal compartments.
Glycan-directed ADC is cytotoxic against tumor cells expressing polySia.
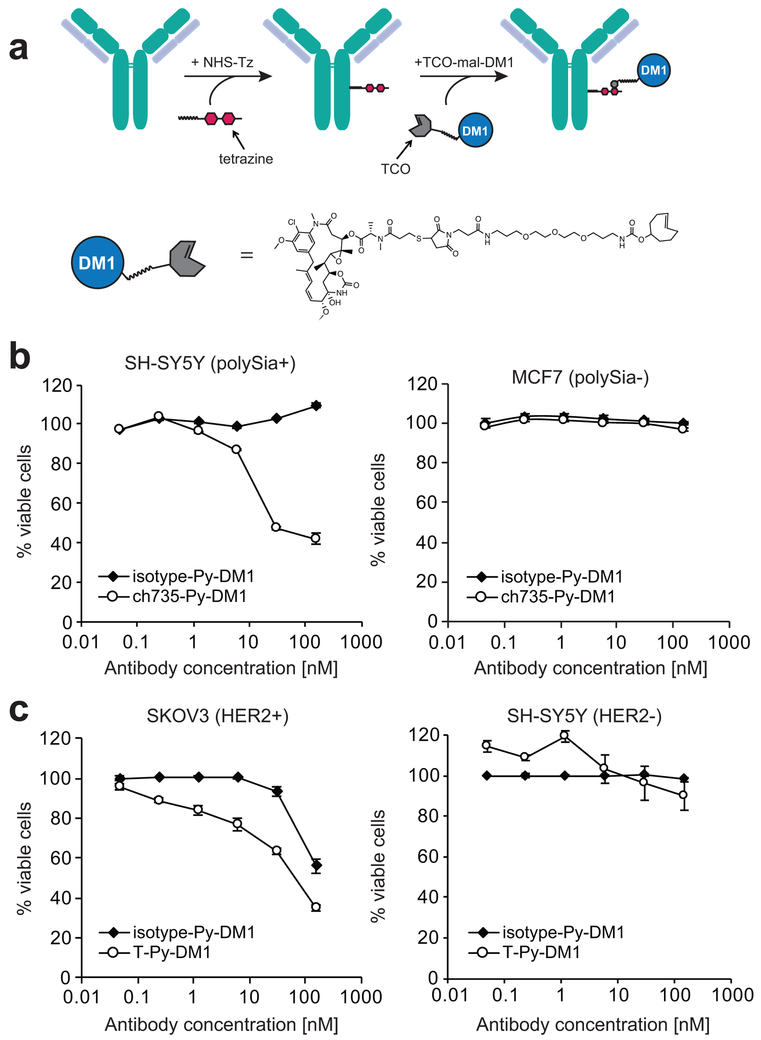
Given that ch735 induced internalization of polySia receptors in cancer cells, we next evaluated whether drug conjugation could be used to confer target-specific in vitro cytotoxicity to mAb ch735. To this end, we proposed a covalent, bioorthogonal reaction scheme between a tetrazine (Tz) and a trans-cyclooctene (TCO) as a means of linking ch735 to the cytotoxic maytansinoid DM1 that inhibits the assembly of microtubules (Fig 4a). We chose DM1 because it has been used successfully in other ADCs including T-DM1, an FDA-approved ADC for HER2-positive breast cancer (44). DM1 is membrane permeable and travels to the cytosol, its site of action, following lysosomal degradation of the ADC. Here, a trans-cyclooctene (TCO)-maleimide-DM1 non-cleavable drug linker was chemically synthesized (Supplementary Fig. 10a-c). Following synthesis, TCO-maleimide-DM1 was conjugated to Tz-modified ch735, forming a 1,4-dihydropyrazine (Py) linkage between the two and typically resulting in drug-to-antibody ratios (DARs) of ~2–3 (Supplementary Table 2). To evaluate in vitro cytotoxicity, we treated SH-SY5Y cells with ch735-Py-DM1 and then examined cell viability. The ch735-Py-DM1 conjugate, but not the isotype-Py-DM1 control, showed polySia-specific cell killing of SH-SY5Y cells, and neither showed any cytotoxicity towards MCF7 cells (Fig. 4b). For comparison, we similarly prepared a conjugate between Tz-modified trastuzumab and DM1 (T-Py-DM1) and found that it killed HER2-positive SKOV3 cells to an extent that was similar to ch735-Py-DM1 against SH-SY5Y cells (Fig 4c). Importantly, the comparable target-specific potency that we measured for ch735-Py-DM1 relative to T-Py-DM1 (IC50 values of 17 and 23 nM, respectively; Supplementary Table 2) reveals the therapeutic potential of this glycan-directed ADC against polySia-positive cancers including neuroblastoma, small cell and non-small cell lung carcinomas, multiple myeloma, and Wilms’ tumor.
Figure 4. Target-mediated in vitro cytotoxicity of glycan-directed ADC.
(a) Overview of the two-step ADC synthesis strategy used to generate ch735-Py-DM1. The first step involved conjugation of NHS-PEG4-tetrazine (NHS-Tz) to free lysines and the second step involved the reaction of the trans-cyclooctene (TCO) group on the TCO-maleimide-DM1 drug linker (TCO-mal-DM1) with the Tz on the antibody. (b) Chemical structure of the non-cleavable drug linker with DM1. (c) Viability of SH-SY5Y (polySia+) and MCF7 (polySia−) cells following treatment with different concentrations of ch735-Py-DM1 or isotype-Py-DM1. Percent viability is calculated based on the signal relative to untreated control cells. Representative data depicts mean percent viability and error bars are the standard deviation of the mean (n = 3). (d) Viability of SKOV3 (HER2+) and SH-SY5Y (HER2-) cells following treatment with different concentrations of T-Py-DM1 and isotype-Py-DM1. Representative data depicts mean percent viability and error bars are the standard deviation of the mean (n = 3).
Discussion
Here, we sought to expand the knowledge base surrounding cell surface polySia and affirm its potential as a target for antibody-based cancer therapy. PolySia is a rare posttranslational modification that is found on a select group of identified carrier proteins including NCAM, SynCAM-1, Neuropilin-2, and the voltage sensitive sodium channel α subunit (16). To create a more therapeutically relevant polySia-directed antibody, we engineered a chimerized human mAb, ch735, that was based on mouse-derived mo735, and determined that it recognized polySia with low nanomolar affinity and exquisite selectivity, binding α2,8-linked polySia structures with a DP of three sugar units or greater. It should be pointed out that previous experiments using SPR and ITC showed that mo735 prefers polySia chains of at least 8–11 sialic acid residues with increasing affinity as length increases (34), in line with our glycoprotein microarray results with polySia-NCAM that has a DP of ~50 (34). The binding to much shorter polySia chains that we observed with the glycan microarray could be due to differences in sensitivity and/or in how the immobilized glycans were presented to the antibody for binding (i.e., clustered). It is worth noting that a single-chain Fv (scFv) antibody derived from mo735 was observed to bind shorter α2–8-linked sialic acids (DP ~3) (28), which likely explains why ch735 and mo735 both bind to the shorter polySia structures on the glycan array. It was proposed by these authors that mo735 recognizes sialic acid trisaccharide units in a paired manner and that this lends itself to higher affinities for longer chains.
Using this chimeric human mAb, we confirmed high levels of polySia expression on several different cancer cell lines, in agreement with earlier findings that this aberrant glycan is abundantly expressed on human cancers. Importantly, polySia-positive tumor cells were observed to rapidly internalize ch735 in endosomal and lysosomal compartments. In this regard, it should be pointed out that NCAM, one of the major polySia carrier proteins, is well known to undergo internalization via the clathrin-dependent endocytic pathway in astrocytes, cortical neurons, and rat neuroblastoma cells (39,40). However, studies of Wilms tumor revealed that while NCAM was similarly present in intracellular compartments from the nuclear membrane to the plasma membrane, polySia was only detectable at the cell surface (41). Likewise, polySia was found exclusively on the surface of SW2 cells (35). A more recent investigation of polySia turnover in human rhabdomyosarcoma cells reported that small numbers of polySia-NCAM molecules were recurrently found co‐localizing with Rab5 (early endocytic marker), but only upon activation by the extracellular matrix (ECM) (42). The absence of detectable constitutive internalization of polySia in these studies leads us to believe that the rapid internalization following ch735 binding observed here is an instance of antibody-induced receptor internalization (45). Interestingly, polySia-binding Escherichia coli bacteriophages were similarly reported to induce endocytosis of polySia in human neuroblastoma cells, whereas polySia remained at the cell surface if no phage was added (46).
The ability of experimental and therapeutic antibodies to induce endocytosis of their antigens is a commonly observed phenomenon that has been leveraged as a strategy to internalize oncogenic (or survival-mediating) antigens for eliciting anti-tumor effects or to deliver cytotoxic payloads directly into cancer cells (45). In the case of the latter, polySia possesses a number of attributes that make it an ideal target for an ADC including: (1) it is abundantly and selectively expressed on cancer cells as discussed above; (2) it is not detected in extracellular supernatants (42), and the NCAM ectodomains that are shed from the cell surface are devoid of polySia (47); (3) it possesses an appropriate rate of endocytosis, comparable to that measured previously for trastuzumab (43); and (4) it is trafficked to the endolysosomal degradation pathway and retained in a maturing endosome (rather than being recycled back to the plasma membrane) (42) until finally being delivered to the lysosome, an appropriate intracellular trafficking route when using a non-cleavable linker (48). To harness these traits, we synthesized an ADC in which Tz-modified ch735 was bioorthogonally conjugated to the TCO-maleimide-DM1 drug linker. The resulting ch735-Py-DM1 conjugate exhibited potent polySia-specific cytotoxicity in vitro, rivaling the potency of a similarly synthesized T-Py-DM1 conjugate. To our knowledge, this is the first ADC that targets an N-linked glycan epitope on the surface of cancer cells and one of few to leverage the Tz/TCO bioorthogonal click chemistry described here. The relative ease of component synthesis and fast reaction kinetics of this two-step method allows for rapid generation of ADCs against new targets. Additionally, the aromatic stability of the pyridazine product formed could aid in stability (49). This is even more significant when one considers that unconjugated (‘naked’) mo735 exhibited only limited complement-dependent cytotoxicity (CDC) against cultured neurons (50) while mAb 5A5, a polySia-specific IgM, exhibited no measurable CDC against several different SCLC cell lines (37).
It is worth mentioning that the IC50 value measured for ch735-Py-DM1 compared favorably to a number of previously reported ADCs against protein antigens including HER2 and NCAM, as well as a small handful of ADCs that target cell surface O-glycans including STn, Tn, and T, the blood group-related Lewis Y antigen (Supplementary Table 2). This latter group, together with our ch735-Py-DM1 conjugate, represents a new class of glycan-directed ADCs that hold promise for anti-tumor therapy. We anticipate that the availability of antibodies such as ch735 that recognize aberrantly expressed tumor glycans should aid the development of novel glycan-directed synthetic immunotherapies for specifically focusing immune or immune-like responses on the tumor glycocalyx. While we focused here on engineering a glycan-specific ADC, it is envisioned that molecular reformatting of antibodies or antibody domains could be used to create next-generation glycan-directed immunotherapies including BsAbs or CAR-T cells.
Collectively, the results described here validate polySia as a therapeutically tractable target for ADC and pave the way for achieving selective cytotoxic effects against tumors that aberrantly express this unique oncodevelopmental antigen. The choice of polySia as a therapeutic target is supported by the fact that polySia is expressed throughout the fetus and during embryonic development, but in adults polySia expression is highly restricted (16,31). Specifically, according to previously published IHC results, the mo735 mAb reacted with only a limited number of cells and tissues including gray matter of brain, bronchial epithelia and pneumocytes, and capillary endothelial cells and ganglion neurons in the colon (31). Importantly, polySia is re-expressed in many types of cancer including SCLC (17), NSCLC (18), pancreatic cancer (19), Wilm’s tumor (20), neuroblastoma (21), and glioma (22), and its increased expression typically correlates with later stages and increased invasive and metastatic potential (25). While recent reports indicate that polySia is also expressed on certain human immune cells (51,52), this expression appears to be quite heterogenous and is progressively down-regulated in wild-type monocytes and monocyte-derived cells during migration from bone marrow (BM) through peripheral blood (PB) to pulmonary and peritoneal sites of inflammation, with levels in PB and inflammation sites reported to be extremely low or absent relative to the levels detected in BM (53). Regarding polySia’s occurrence on healthy cells, it should be pointed out that Schneerson and colleagues conducted a thorough review of published data looking for evidence that anti-polySia IgG antibodies caused immunopathology in humans. From their study, they found no evidence of increased autoimmunity and urged that the use of anti-polySia immunotherapies be considered (54). We concur that further studies on the therapeutic targeting of polySia and the safety of such an approach are highly warranted, especially in light of the results presented here.
Supplementary Material
Statement of Significance: Findings describe a glycan-specific antibody-drug conjugate that establishes polySia as a viable cell surface target within the tumor glycocalyx.
Acknowledgements.
We thank Dr. Gaurang Bhide and Dr. Karen Colley (University of Illinois at Chicago) for SW2 and A549 cell lines as well as purified endoN used in this work. We thank Carolyn Shurer (Cornell University) for technical assistance and training in confocal microscopy. We thank Cureline for performing IHC and Dr. Andrew Miller (Cornell University) for assistance with the IHC data analysis. We thank Dr. Cynthia Leifer (Cornell University) for technical assistance with endocytosis experiments. We thank Dr. Brian Green and Dr. Joshua Wilson (Glycobia, Inc.) for helpful discussions of the manuscript. This work was supported by the Defense Threat Reduction Agency (GRANT11631647 to M.P.D.), the National Science Foundation (CBET-1605242 to M.P.D.), Cornell University Startup funds (to C.A.A. and M.J.P.), the National Cancer Institute (U54 CA210184 to M.J.P.), and the Nancy and Peter Meinig Investigator Fellowship (to C.A.A.). The work was also supported by seed project funding through the National Institutes of Health-funded Cornell Center on the Physics of Cancer Metabolism (supporting grant 1U54CA210184–01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. E.C.C. was supported by a National Institutes of Health Chemical-Biology Interface (CBI) training fellowship (supporting grant T32GM008500). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. D.N.T. was supported by a National Science Foundation Graduate Research Fellowship and a Cornell Fleming Graduate Scholarship. We acknowledge instrumentation grants NYSTEM C029155 and NIH S10OD018516 for the Zeiss LSM880 microscope. We acknowledge the participation of the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics (NCFG) at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grant P41 GM103694).
Footnotes
Competing Interests. M.P.D. has a financial interest in Glycobia, Inc. and Versatope, Inc. M.P.D.’s interests are reviewed and managed by Cornell University in accordance with their conflict of interest policies. All other authors declare no other competing interests.
References
- 1.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nature reviews Cancer 2005;5:526–42 [DOI] [PubMed] [Google Scholar]
- 2.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol 2008;8:874–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol 2015;10:473–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell 2006;126:855–67 [DOI] [PubMed] [Google Scholar]
- 5.Hakomori SI, Murakami WT. Glycolipids of hamster fibroblasts and derived malignant-transformed cell lines. Proc Natl Acad Sci U S A 1968;59:254–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladenson RP, Schwartz SO, Ivy AC. Incidence of the blood groups and the secretor factor in patients with pernicious anemia and stomach carcinoma. Am J Med Sci 1949;217:194–7 [DOI] [PubMed] [Google Scholar]
- 7.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov 2005;4:477–88 [DOI] [PubMed] [Google Scholar]
- 8.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540–55 [DOI] [PubMed] [Google Scholar]
- 9.Transforming Glycoscience: A Roadmap for the Future. The National Academies Press; 2012. [PubMed] [Google Scholar]
- 10.Sterner E, Flanagan N, Gildersleeve JC. Perspectives on anti-glycan antibodies gleaned from development of a community resource database. ACS Chem Biol 2016;11:1773–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst 2009;5:1087–104 [DOI] [PubMed] [Google Scholar]
- 12.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology 2007;17:17C–23C [DOI] [PubMed] [Google Scholar]
- 13.Majzner RG, Heitzeneder S, Mackall CL. Harnessing the immunotherapy revolution for the treatment of childhood cancers. Cancer Cell 2017;31:476–85 [DOI] [PubMed] [Google Scholar]
- 14.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med 2010;363:1324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frosch M, Gorgen I, Boulnois GJ, Timmis KN, Bitter-Suermann D. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc Natl Acad Sci U S A 1985;82:1194–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colley KJ, Kitajima K, Sato C. Polysialic acid: biosynthesis, novel functions and applications. Crit Rev Biochem Mol Biol 2014;49:498–532 [DOI] [PubMed] [Google Scholar]
- 17.Kibbelaar RE, Moolenaar CE, Michalides RJ, Bitter-Suermann D, Addis BJ, Mooi WJ. Expression of the embryonal neural cell adhesion molecule N-CAM in lung carcinoma. Diagnostic usefulness of monoclonal antibody 735 for the distinction between small cell lung cancer and non-small cell lung cancer. J Pathol 1989;159:23–8 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka F, Otake Y, Nakagawa T, Kawano Y, Miyahara R, Li M, et al. Expression of polysialic acid and STX, a human polysialyltransferase, is correlated with tumor progression in non-small cell lung cancer. Cancer Res 2000;60:3072–80 [PubMed] [Google Scholar]
- 19.Kameda K, Shimada H, Ishikawa T, Takimoto A, Momiyama N, Hasegawa S, et al. Expression of highly polysialylated neural cell adhesion molecule in pancreatic cancer neural invasive lesion. Cancer Lett 1999;137:201–7 [DOI] [PubMed] [Google Scholar]
- 20.Roth J, Zuber C, Wagner P, Taatjes DJ, Weisgerber C, Heitz PU, et al. Reexpression of poly(sialic acid) units of the neural cell adhesion molecule in Wilms tumor. Proc Natl Acad Sci U S A 1988;85:2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livingston BD, Jacobs JL, Glick MC, Troy FA. Extended polysialic acid chains (n greater than 55) in glycoproteins from human neuroblastoma cells. J Biol Chem 1988;263:9443–8 [PubMed] [Google Scholar]
- 22.Suzuki M, Suzuki M, Nakayama J, Suzuki A, Angata K, Chen S, et al. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology 2005;15:887–94 [DOI] [PubMed] [Google Scholar]
- 23.Daniel L, Durbec P, Gautherot E, Rouvier E, Rougon G, Figarella-Branger D. A nude mice model of human rhabdomyosarcoma lung metastases for evaluating the role of polysialic acids in the metastatic process. Oncogene 2001;20:997–1004 [DOI] [PubMed] [Google Scholar]
- 24.Hromatka BS, Drake PM, Kapidzic M, Stolp H, Goldfien GA, Shih I-M, et al. Polysialic acid enhances the migration and invasion of human cytotrophoblasts. Glycobiology 2013;23:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falconer RA, Errington RJ, Shnyder SD, Smith PJ, Patterson LH. Polysialyltransferase: a new target in metastatic cancer. Curr Cancer Drug Targets 2012;12:925–39 [DOI] [PubMed] [Google Scholar]
- 26.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 2009;15:5323–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med 2012;367:1783–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagae M, Ikeda A, Hane M, Hanashima S, Kitajima K, Sato C, et al. Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: insight into the binding preference for polysialic acid. J Biol Chem 2013;288:33784–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dodev TS, Karagiannis P, Gilbert AE, Josephs DH, Bowen H, James LK, et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci Rep 2014;4:5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Cordon-Cardo C, Zhang HS, Reuter VE, Adluri S, Hamilton WB, et al. Selection of tumor antigens as targets for immune attack using immunohistochemistry: I. Focus on gangliosides. Int J Cancer 1997;73:42–9 [DOI] [PubMed] [Google Scholar]
- 32.Chen Y Drug-to-antibody ratio (DAR) by UV/Vis spectroscopy. Methods Mol Biol 2013;1045:267–73 [DOI] [PubMed] [Google Scholar]
- 33.Kim MT, Chen Y, Marhoul J, Jacobson F. Statistical modeling of the drug load distribution on trastuzumab emtansine (Kadcyla), a lysine-linked antibody drug conjugate. Bioconjug Chem 2014;25:1223–32 [DOI] [PubMed] [Google Scholar]
- 34.Hayrinen J, Haseley S, Talaga P, Muhlenhoff M, Finne J, Vliegenthart JFG. High affinity binding of long-chain polysialic acid to antibody, and modulation by divalent cations and polyamines. Mol Immunol 2002;39:399–411 [DOI] [PubMed] [Google Scholar]
- 35.Martersteck CM, Kedersha NL, Drapp DA, Tsui TG, Colley KJ. Unique alpha 2, 8-polysialylated glycoproteins in breast cancer and leukemia cells. Glycobiology 1996;6:289–301 [DOI] [PubMed] [Google Scholar]
- 36.Valentiner U, Muhlenhoff M, Lehmann U, Hildebrandt H, Schumacher U. Expression of the neural cell adhesion molecule and polysialic acid in human neuroblastoma cell lines. Int J Oncol 2011;39:417–24 [DOI] [PubMed] [Google Scholar]
- 37.Livingston PO, Hood C, Krug LM, Warren N, Kris MG, Brezicka T, et al. Selection of GM2, fucosyl GM1, globo H and polysialic acid as targets on small cell lung cancers for antibody mediated immunotherapy. Cancer Immunol Immunother 2005;54:1018–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol 1985;1:1–39 [DOI] [PubMed] [Google Scholar]
- 39.Diestel S, Schaefer D, Cremer H, Schmitz B. NCAM is ubiquitylated, endocytosed and recycled in neurons. J Cell Sci 2007;120:4035–49 [DOI] [PubMed] [Google Scholar]
- 40.Minana R, Duran JM, Tomas M, Renau-Piqueras J, Guerri C. Neural cell adhesion molecule is endocytosed via a clathrin-dependent pathway. Eur J Neurosci 2001;13:749–56 [DOI] [PubMed] [Google Scholar]
- 41.Zuber C, Roth J. The relationship of polysialic acid and the neural cell adhesion molecule N-CAM in Wilms tumor and their subcellular distributions. Eur J Cell Biol 1990;51:313–21 [PubMed] [Google Scholar]
- 42.Monzo HJ, Park TI, Dieriks BV, Jansson D, Faull RL, Dragunow M, et al. Insulin and IGF1 modulate turnover of polysialylated neural cell adhesion molecule (PSA-NCAM) in a process involving specific extracellular matrix components. J Neurochem 2013;126:758–70 [DOI] [PubMed] [Google Scholar]
- 43.Li JY, Perry SR, Muniz-Medina V, Wang X, Wetzel LK, Rebelatto MC, et al. A biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell 2016;29:117–29 [DOI] [PubMed] [Google Scholar]
- 44.Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res 2008;68:9280–90 [DOI] [PubMed] [Google Scholar]
- 45.Tarcic G, Yarden Y. Antibody-mediated receptor endocytosis: harnessing the cellular machinery to combat cancer In: Y. Y, G. T, editors. Vesicle Trafficking in Cancer. New York, NY: Springer; 2013. [Google Scholar]
- 46.Lehti TA, Pajunen MI, Skog MS, Finne J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat Commun 2017;8:1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinkle CL, Diestel S, Lieberman J, Maness PF. Metalloprotease-induced ectodomain shedding of neural cell adhesion molecule (NCAM). J Neurobiol 2006;66:1378–95 [DOI] [PubMed] [Google Scholar]
- 48.Ritchie M, Tchistiakova L, Scott N. Implications of receptor-mediated endocytosis and intracellular trafficking dynamics in the development of antibody drug conjugates. mAbs 2013;5:13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selvaraj R, Fox JM. trans-Cyclooctene--a stable, voracious dienophile for bioorthogonal labeling. Curr Opin Chem Biol 2013;17:753–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pon RA, Biggs NJ, Jennings HJ. Polysialic acid bioengineering of neuronal cells by N-acyl sialic acid precursor treatment. Glycobiology 2007;17:249–60 [DOI] [PubMed] [Google Scholar]
- 51.Drake PM, Nathan JK, Stock CM, Chang PV, Muench MO, Nakata D, et al. Polysialic Acid, a Glycan with Highly Restricted Expression, Is Found on Human and Murine Leukocytes and Modulates Immune Responses. J Immunol 2008;181:6850–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem 2007;282:30346–56 [DOI] [PubMed] [Google Scholar]
- 53.Stamatos NM, Zhang L, Jokilammi A, Finne J, Chen WH, El-Maarouf A, et al. Changes in polysialic acid expression on myeloid cells during differentiation and recruitment to sites of inflammation: role in phagocytosis. Glycobiology 2014;24:864–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein DM, Robbins J, Miller MA, Lin FY, Schneerson R. Are antibodies to the capsular polysaccharide of Neisseria meningitidis group B and Escherichia coli K1 associated with immunopathology? Vaccine 2006;24:221–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.