Abstract
H-scan ultrasound (US) is a new imaging technique that relies on matching a model that describes US image formation to the mathematics of a class of Gaussian-weighted Hermite polynomials (GH). In short, H-scan US (where the ‘H’ denotes Hermite or hue) is a tissue classification technique that images the relative size of acoustic scatterers. Herein, we detail development of a real-time H-scan US imaging technology that was implemented on a programmable US research scanner (Vantage 256, Verasonics Inc, Kirkland, WA). This custom US imaging system has a dual display for real-time visualization of both the H-scan and B-scan US images. This MATLAB-based (Mathworks Inc, Natick, MA) system includes a graphical user interface (GUI) for controlling the entire US scan sequence including the raw radio frequency (RF) data acquisition parameters, image processing, variable control of a parallel set of convolution filters used to derive the H-scan US signal, and data (cine loop) save. The system-level structure used for software-based image reconstruction and display is detailed. Imaging studies were conducted using a series of homogeneous and heterogeneous tissue-mimicking phantom materials embedded with monodisperse spherical US scatterers of size 15–40 μm in diameter. Relative to H-scan US image measurements from a phantom with 15 μm-sized scatterers, data from phantoms with the 30 and 40 μm-sized scatterers exhibited mean intensity increases of 5.2% and 11.6%, respectively. Overall, real-time H-scan US imaging is a promising approach for visualizing the relative size and distribution of acoustic scattering objects.
Keywords: Acoustic scatterers, H-scan, Plane waves, Spatial angular compounding, Tissue characterization, Ultrasound
1. Introduction
Many pathologies and diseases manifest as a change in normal tissue function or structure. For example, fatty liver disease (hepatic steatosis) is a condition in which fat builds up in the liver. Earlier stages of this diffuse disease are characterized by in part lipid accumulation in the hepatocytes, causing them to become progressively larger than normal liver cells [1]. On the therapeutic front, many traditional anticancer drugs disrupt cell proliferation to arrest tumor growth. A major consequence of this mitotic disruption is that cancer cells undergo apoptosis. During this programmed cell death, cancer cells shrink in size representing a morphological hallmark of this process [2]. Collectively, these examples and others suggest that any noninvasive diagnostic test or tissue characterization technique that can provide insight on cell size, including relative changes, would potentially have tremendous clinical value and worthy of investigation.
The use of noninvasive ultrasound (US) for quantitative tissue characterization has been an exciting research prospect for several decades now. Herein the challenge is to find hidden patterns in the US data to reveal more information about tissue function and pathology that cannot be seen in the more conventional US images [3]. To that end, several different US-based tissue characterization methods have been introduced [4–12]. A limitation of some of these approaches is that they necessitate a calibration step before measurement or use a relatively large kernel of US data to improve tissue characterization accuracy. The latter could negatively impact spatial resolution. Bypassing some of the limitations associated with traditional tissue characterization approaches, a new modality has emerged for the US classification of acoustic scatterer. Termed H-scan, this US technique relies on matching a model of image formation to the mathematics of a class of Gaussian-weighted Hermite polynomial (GH) functions [13]. H-scan US is a simplified approach for detecting the frequency dependence of US scattering within different tissue types. In short, the backscattered US signal can be modeled as a convolution of an incident pulse with a sequence of tissue reflections [14]. Parallel convolution filters using two GH of preset order are applied to the backscattered radio frequency US data to capture the relatively low and high frequency signal components, respectively. The resultant images are termed H-scans, where ‘H’ denotes Hermite (or hue for simplicity) to differ from the traditional B-scan US technique. H-scan US image intensity can be interpreted to describe different scattering structures or objects. An overview of the H-scan concept was given in [15–17].
To date, H-scan US images have been processed offline. To overcome that limitation, while standardizing the data acquisition and image processing, the purpose of the research project detailed herein was to develop real-time H-scan US imaging functionality using a MATLAB-based programmable US system equipped with a linear array transducer. Testing and optimization of this clinically-translatable US system was performed using tissue-mimicking phantom materials embedded with different micrometer-sized acoustic scattering objects.
2. Methodology
2.1. Programmable US system considerations
The class of programmable US systems manufactured by Verasonics (Vantage 128 or 256, Verasonics Inc, Kirkland, WA) use a system architecture that provides researchers and developers a platform with performance and flexibility for medical US innovation. The systems use proprietary hardware and software technologies to provide direct access to raw US data, while preserving the ability to perform real-time imaging with custom software, at clinically useful frame rates. The MATLAB language (Mathworks Inc, Natick, MA) is used for specifying the sequence programming of the Vantage systems and for user interface controls. Acquisition of US data is controlled by a hardware sequencer that operates independently from the sequencing software for deterministic timing of acquisition events. All software processing functions provided by Verasonics are programmed in C or C++ for speed of execution and incorporated into a MATLAB external (MEX) function that is called at the start of sequence execution. In the C environment, processing is optimized using multiple threads of execution and single instruction multiple data (SIMD) vector instructions, enabling real-time software image reconstruction, Doppler processing, and image display processing. The MATLAB programming environment is reentered for changes in system programming or execution of user provided functions.
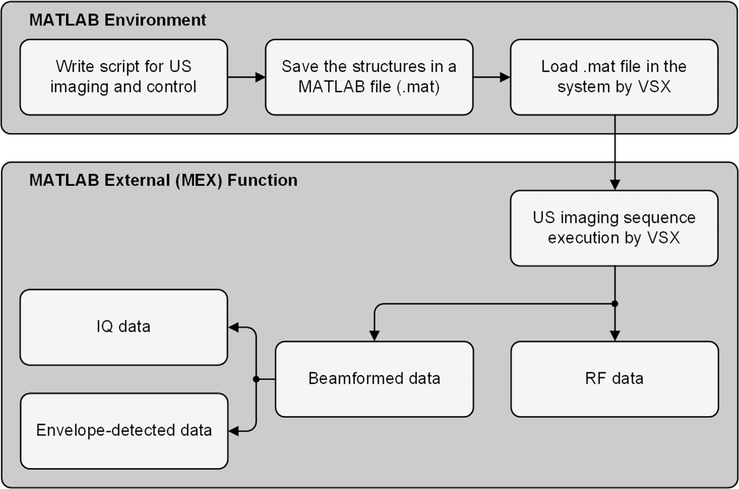
Programming a Vantage research system essentially comprises of characterizing different framework parameters and attributes, and the sequence of events to be carried out by the hardware and software. These parameters are defined using structured data in a setup file written and saved in the MATLAB environment. To execute any US imaging program, proprietary Verasonics software (Verasonics Sequence eXecution, VSX) loads the sequence programming script detailed in the MATLAB file during runtime. Raw radio frequency (RF) data is accessible throughout the imaging sequence. All beamformed inphase and quadrature (IQ) data and envelope detected US image data can be accessed as well (see Fig. 1).
Fig. 1.
Programming a Vantage ultrasound (US) research scanner essentially comprises of characterizing different framework parameters and attributes, and the sequence of events to be carried out by the Verasonics system hardware and software (MATLAB or the proprietary Verasonics Sequence eXecution, VSX) to collect requisite datatype (radiofrequency, RF, or inphase and quadrature, IQ, format) required for any US imaging mode.
2.2. H-scan US imaging system
For the context of H-scan imaging, we briefly consider three simple types of acoustic scattering objects with an assumption of small spatial variations in medium density and compressibility. A more detailed overview of the H-scan format for classification of US scatterers can be found in the referenced publications [15–17]. During US imaging, the received signals from a scattering medium can be classified by their similarity to GH, namely, GH4(t), GH5(t), or GH6(t), where the subscript denotes the increasing order of the three select polynomials. To minimize correlation between the overlapping GH spectra, one could employ more disparate functions for the convolution filtering, for example, GH2(t) and GH8. Regardless, we scale the polynomial functions to position them within the bandwidth of the RF spectrum, e.g., GH2(bt/τ) andGH8(at/τ) where τ is a constant scaling factor and a and b are parameters for center frequency shifting. To generate an H-scan US image, two parallel convolution filters are applied to the raw RF data sequences to measure the strength of the received signals relative to GH2 and GH8 after normalization by the signal energy where n denotes the GH order [15–17]. The signal envelope for each of these filtered and compounded data sequences is then calculated using a Hilbert transformation. The ensemble of filter outputs, involving all scan lines used for image formation, are then color coded whereby the lower frequency (GH2) backscatter signals are assigned to the red color channel and the higher frequency (GH8) components to the blue color channel. The envelope of the original unfiltered dataset is assigned to the green color channel to complete the RGB colormap scheme.
An important question with respect to clinical applications involves the change in H-scan US outputs with respect to small changes in scatterer size. To that end, we have examined the backscattered pressure from spherical scatterers in phantoms [18]. Assuming a convolution model of pulse-echo US imaging, the combination of transmit, scatter, and receive components can be expressed within the context of a GHn matched filter. The maximum echo amplitude for each H-scan US channel can then be computed as a function of the scatterer radius and transducer center frequency to calculate the actual physical size of the scatterer. Theoretical findings suggest that H-scan US sensitivity is sufficient to visualize differences in subwavelength scatterer size in increments of 20–30 μm using a conventional broadband transducer with a center frequency of 5 MHz. In the context of an H-scan US analysis, such scatterers would increase the discrimination between GHn channels. Second, as the transducer bandwidth increases, the adjustment of the GHn channels is possible, leading to less overlap between Hermite spectra. These factors can modify results and thus, they should not be construed as absolute upper limits on sensitivity. Higher frequency transducers would have similar results but shifted toward detection of smaller scatterers.
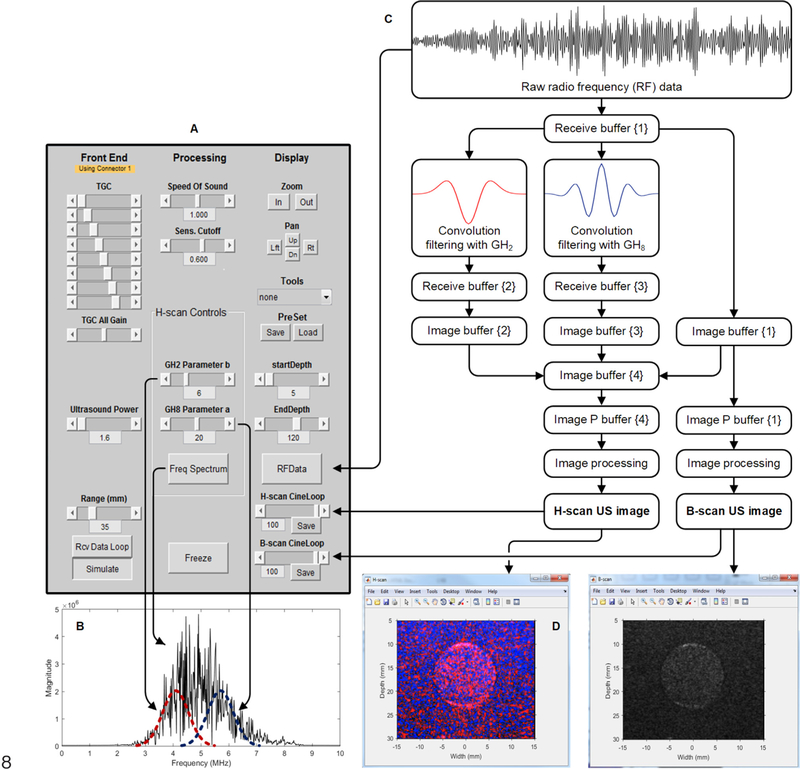
Using a Vantage 256 US scanner (Verasonics Inc) equipped with an L11–4v linear array transducer (128 elements), we implemented real-time H-scan US imaging using ultrafast plane waves with angular compounding. For spatial angular compounding, successively steered and overlapping plane wave transmissions were performed using five equally spaced angles in the ±18° range. For each image spatial location, spatial angular compounding is performed by averaging the acquisitions over all steered plane wave transmissions. As demonstrated previously, the use of more frames for compounding has been shown to only marginally impact image quality [17]. While the system permits variable control, the use of a larger angulated range can compromise compounding results due to image decorrelation artifacts. The maximum range that the plane waves can be steered is ±38° [19]. Our system-level data flow chart used for the software-based image construction and display is detailed in Fig. 2. This figure also summarizes the parallel data processing strategy and final display of the H-scan US image. Custom features of the real-time H-scan US imaging system include the convolutional spatial filtering used to derive the H-scan US signal. Multiple image displays allow visualization of both H-scan and B-scan images and R channel and B channel image data when needed. A real-time plot of the RF spectrum can be activated to allow visualization of GH kernel placement and position, which is controlled through a graphical user interface (GUI).
Fig. 2.
Schematic diagraming our real-time H-scan US imaging scanner. This clinically-translatable and programmable MATLAB-based system incorporates a (A) graphical user interface (GUI) for controlling RF data acquisition and image processing including (B) variable control of the convolution filters used to derive the H-scan US signal. (C) System-level data flow chart details US image construction and dual (D) H-scan (left) and B-scan (right) display.
The Vantage 256 programmable scanner contains a 12-core processing unit and 132 GB of memory (RAM). Plane wave imaging was performed using a center frequency of 5.2 MHz on transmission and backscattered RF data was quantized at 14 bits and sampled at a rate of 20.8 MHz (200% bandwidth Nyquist sampling). Since the −6-dB bandwidth of the L11–4v transducer used in this study was 6.8 MHz, this sampling rate was utilized to capture the full spectrum of the transducer and required for image reconstruction processing.
For H-scan US to be a robust and reliable modality for human imaging, the matched GH polynomial functions must be properly set (i.e., scaled) for the initial pulse sequence and then continuously readjusted at tissue depth as higher frequency spectral content is progressively attenuated. To compensate for frequency dependent attenuation and bandwidth reduction at tissue depth, we linearly scale the raw RF data by a tissue attenuation value α of 0.3 dB/cm/MHz. This value was derived from phantom material samples using methods recommended by the Technical Standards Committee of the American Institute of US in Medicine (AIUM) [20]. Fig. 2 shows a single plane wave H-scan US image. To generate H-scan US images with angular compounding, each angulation undergoes convolutional filtering with the scaled GHn polynomial functions before summation to form the final compounded images. The main impact of H-scan US using plane wave and spatial angular compounding include higher image contrast, speckle reduction, artifact reduction (e.g., acoustic shadowing, echo drop-out, clutter, etc.) and better visualization of lesion margins [17,21–23]. For any H-scan technique utilizing focused US data acquisitions, spatial resolution degrades away from the focal region and inherently impacts relative scatterer size estimation compared with the use of plane wave US transmission. The latter exposes the entire image field with nearly uniform acoustic intensity except for depth-dependent attenuation. While the resolution of plane wave US imaging can be inferior to that of traditional focused US approaches, the former exhibits homogeneous spatial resolution throughout the image plane that can be improved through spatial angular compounding [17,24,25]. The acoustic output (i.e., mechanical index, MI) from this system was measured and spatially mapped using a mechanized hydrophone scanning system (AIMS III, Onda Corp, Sunnyvale, CA).
2.3. Sequence objects
The sequence object is simply the collection of objects used to define all the events and attributes needed to acquire and process a sequence of US acquisitions. The events of the sequence object are read sequentially to determine the actions to be carried out by the acquisition and processing system. Each event contains other objects that describe in detail the various components of the acquisition and processing of the US information. Typically, a sequence of events is generated to specify the acquisition of one or more frames of US data. The end of a sequence typically has a jump back to the first event, so that the sequence is repeated indefinitely. The sequences of events to be executed for acquiring one or more frames of US data are defined by a collection of event objects, transmit waveform (TW) objects, transmit events structure (TX) objects, and receive event objects (Table 1).
Table 1.
Sequence objects including event objects, transmit waveform (TW) objects, transmit events structure (TX), and receive objects.
| Parameter | Description or value |
|---|---|
| TW objects | |
| Transmit pulse | [A, B, C, D] = [5.2, 0.67, 2, 1] |
| A: Frequency of transmit burst | |
| B: On time of half-cycle | |
| C: Number of half cycle periods in waveform | |
| D: Polarity | |
| TX objects | |
| Origin | (0, 0, 0) |
| Focus | 0 |
| Steer | 0 |
| Apodization | Kaiser(128, 1) |
| Delay | Zeros(1, 128) |
| Receive objects | |
| Apodization | Ones(1, 128) |
| Start depth | Data acquisition start depth |
| End depth | Data acquisition end depth |
| Acquisition number | Number of angles |
| Sample per wave | 4, NS200BW |
| Mode | 0, RF data replaces data in local memory |
2.4. Resources used by the sequence
These objects give information about the system configuration and the storage buffers. The system contains three receive buffers to receive the RF data transferred from the local memories on the Verasonics Data Acquisition System (VDAS) modules. The raw data contains 100 frames and each frame consists of 128 columns that correspond to the number of channels used to acquire the frame. Note the number of frames was arbitrarily choosen and only limited by system memory. The row dimension is dependent on the acquisition depth. For our system, they were a set of 4096 × 5 acquisitions with time increasing down the column (image depth). There are multiple transmit and receive acquisition events required to compose a frame comprised of the five-different plane wave angulations with the data samples for each event stacked vertically down the column ((numbers of rows × number of angles) × number of columns × number of frames) equal to ((4096 × 5)×128 ×100). Since image reconstruction is done by software, a depth of 4096 samples per acquisition ensures complete acquisition at more extreme depths and minimizes likelihood of data loss due to buffer overflow.
RF data is acquired and temporarily stored in three different receive buffers for processing and generation of the B-scan and H-scan US images for display. More specifically, the first receive buffer {1} contains RF data for B-scan US image formation, the second receive buffer {2} and third receive buffer {3} stores RF data from buffer {1} after convolution filtering with GH2 and GH8, respectively. These convolutions are implemented using external MATLAB processing functions. In short, the resource and destination buffers for the convolution filtering with GH2 were buffer {1} and buffer {2} respectively, and buffer {1} and buffer {3} for the convolution processing with GH8 (see Fig. 2).
2.5. Pixel-based image reconstruction
Image reconstruction is performed using the data stored in the three receive buffers to compute intensity data, which is used for 2-dimensional (2D) US imaging. The pixel data region (PData) used for reconstruction is processed by the image reconstruction software. The PData array parameters used by the US system are detailed in Table 2. The image reconstruction region is typically defined to be the same width as the transducer with a height equal to the scan depth required. The spacing between pixels (in wavelengths) is set to 1.09 in the X dimension and 0.5 in the Z dimension. US imaging transducers with a relatively broad bandwidth typically exhibit an axial resolution on the order of a wavelength and a lateral resolution of several wavelengths. Pixel spacing during image reconstruction is comparable to transducer resolution, i.e., half wavelength in the axial direction and one wavelength in the lateral direction. The number of rows and columns in the PData array will determine the overall size of the pixel data array in the image field. It is useful to specify the row size in terms of image start and end depths (in wavelengths). The former can be set a few wavelengths distal to the transducer face to avoid display of any reverberation artifacts whereas the latter determines the desired imaging depth. The spacing of PData columns is set to match the spacing of elements in the transducer.
Table 2.
List of pixel data region (PData) variables used for US image reconstruction.
| Parameter | Description |
|---|---|
| PData format | Scan format structure, rectangle |
| PDelta | Spacing between pixels in all dimensions |
| PDeltaX | x-dimension pdelta |
| PDeltaY | y-dimension pdelta |
| PDeltaZ | z-dimension pdelta |
| Size | Scan depth (rows), width (columns) equal to number of active transducer elements |
| Origin (x, y, z) | x, y, z of reconstruction volume with respect to the transducer origin |
As illustrated in Fig. 2, there are four image buffers used by the US system to receive the intensity outputs from the pixel-based image reconstructions, which can also be as target output buffers for any signal processing methods. The number of columns in the image buffer is always the same as the number of pixels on a row of the defined rectangular pixel grid. Data type in the image buffer is double-precision floating-point and intensity ranges from 0 to 2 × 106. In the processing of image buffer to display image data, a secondary buffer of the same size is created, called ImgDataP, which is used for spatial filtering and any persistence processing. The ImgDataP buffer is then used to render pixels to the display window. This allows the image buffer (ImgData) to retain the original reconstructed output data for possible reprocessing. One frame from each of the three image buffers is transferred to the image buffer {4} and then an external processing object is used for generation of the H-scan US image.
Data in the image buffer (e.g. rows × columns = 230 × 128) is transformed to uniformly spaced pixel data, which is then transferred to a display window buffer (e.g. rows × columns =329 × 372) consisting of a single frame of pixel data that is copied directly to the output window. The display window typically has more pixels than specified in the corresponding image buffer as pixel data often needs to be resized via interpolation. This step produces a higher spatial density and provides an appropriate image size more useful for high resolution display. The customized US system has two windows for real-time display of the B-scan (for anatomical guidance) and H-scan US images. The system also has an option to display two additional windows for visualization of the blue channel (high frequency) and red channel (low frequency) data using to produce the H-scan US images. All image data is intensity normalized and threshold limited to 8-bit before logarithmic compressed to help accentuate the brightness of lower level intensity values during display.
2.6. Process objects
One of these processing types used to form the final displayed image is persistence, which was set to a 20-frame moving average. Also, the process objects can be external MATLAB processing functions like the convolutional filters that we implemented for generation of H-scan US images. Note that before the final display is presented, the H-scan US images undergo a sequence of processing steps, namely, intensity scaling, threshold limiting, compression and mapping. As an optimization feature, the blue and red channel image data is also used to measure the power spectral density in each (via a fast Fourier transform, FFT) to visualize in real-time any scaling adjustments made to the Hermite functions used for convolutional filtering and H-scan US image formation. Lastly, the Verasonics GUI was modified and integrated on the H-scan US imaging system allowing the user to control the entire US scan sequence including the raw RF data acquisition parameters, image processing, variable control of the convolution filters (to maximize spectral coverage and minimize correlation between kernels), and data (cine loop) save. The frame rate for the real-time H-scan US system was 50 Hz and sufficient for future clinical imaging considerations.
2.7. In vitro testing
Testing of H-scan US imaging system functionality was conducted using a series of tissue-mimicking phantoms materials. These US test phantoms were used to test the sensitivity of H-scan US imaging to different-sized scatterer distributions [26]. Briefly, homogenous phantom materials were prepared by heating a 10% gelatin (300 Bloom, Sigma Aldrich, St. Louis, MO) in water solution to 45 °C. Silica microspheres (0.4% concentration, US Silica, Pacific, MO) were slowly introduced during constant stirring. The monodisperse silica microspheres were arbitrarily chosen to be either 15, 30, or 40 μm in diameter. All gelatin blocks were placed in a 4 °C refrigerator and allowed to cool for at least 12 h before use. Final phantom material size was about 12 cm × 12 cm × 8 cm (length × width × depth). H-scan US imaging of these homogeneous phantom materials (at room temperature or 25 °C) was then performed to detect the different-sized scattering objects and help optimize real-time H-scan US system imaging and control. A second phantom study was performed using a series of phantom materials with different-sized US scatterers. Using the above recipe, these phantoms were made using the following microsphere concentrations: 100% of 40 μm scatterers, 75% of 40 μm and 25% of 15 μm scatterers, 50% of 40 μm and 50% of 15 μm scatterers, 25% of 40 μm and 75% of 15 μm scatterers, and 100% of 15 μm scatterers.
3. Results and discussion
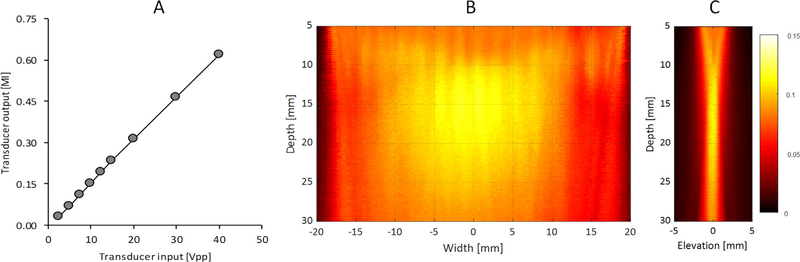
The acoustic output of the H-scan US scanner was measured using a hydrophone to ensure the system remained within limitations established by the United States’ Food and Drug Administration (FDA), specifically, that the MI does not exceed 1.9. This is an important consideration for any translational imaging studies in human. Fig. 3 details the relationship between the transducer voltage (used for transducer excitation) and pressure output (MI). As these measurements indicate, the acoustic output is linearly related to transducer voltage (R2 > 0.99, p < 0.001) and well within FDA limits for safety at the maximum allowed transducer voltage of 40 V. Because our H-scan US system uses plane wave imaging, the acoustic output is considerably lower compared to the more traditional focused US imaging approaches. However, as Fig. 3 also illustrates, spatial variance of the US pressure field is relatively low in the central image area and suited for scatterer size estimation throughout this region using H-scan US imaging [18]. While some banding is visible in the US pressure field when using a transmit angle of zero degrees (i.e., normal transmission), any resultant H-scan US image artifacts will be minimized with the use of spatial angular compounding due to field averaging.
Fig. 3.
(A) Acoustic output measurements (mechanical index, MI) from a Vantage 256 US scanner equipped with a L11–4v linear array transducer with variable excitation at a fixed frequency of 5.2 MHz. Spatial maps detail a single plane wave transmission in both the (B) transverse and (C) elevational directions. Note that image amplitude reflects the measured mechanical index (MI) at a spatial resolution of 0.3 mm.
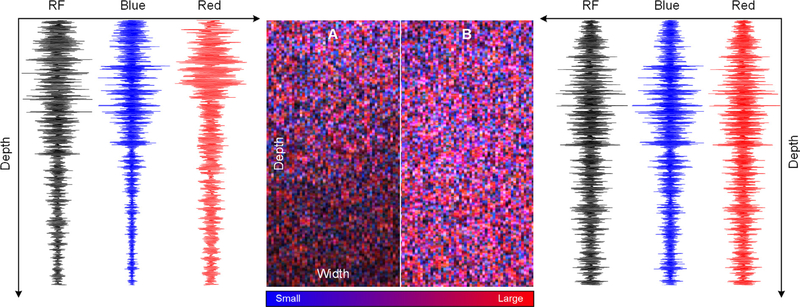
To compensate for frequency dependent attenuation and bandwidth reduction in tissue, the GH-based convolutional filters should be adjusted as a function of image depth. This is analogous to the time gain compensation (TGC) used in traditional diagnostic US imaging. Fig. 4 details a set of RF data signals and H-scan US images acquired using an attenuating homogeneous tissue-mimicking phantom before and after attenuation correction using expected values (i.e., 0.3 dB/cm/MHz). Note the higher gain compensation for the GH8-filtered data (high frequency) versus the GH2-filtered data (low frequency) and more uniform H-scan US image quality at depth. To further help improve image quality, real-time power spectral density measurements are measured and displayed. When enabled, this feature allows visualization of any scaling adjustments made to the Hermite functions used for convolutional filtering and H-scan US image formation. As illustrated in Fig. 5, if there is considerable overlap between the two filtering kernels, then H-scan US image contrast is compromised.
Fig. 4.
Example of an H-scan US image and RF data signal in a highly attenuating homogeneous tissue-mimicking phantom (A) before and (B) after attenuation correction using expected values. Image widths and depths are 20 and 40 mm, respectively. Note the improved H-scan US image quality and more uniform intensity at tissue depth.
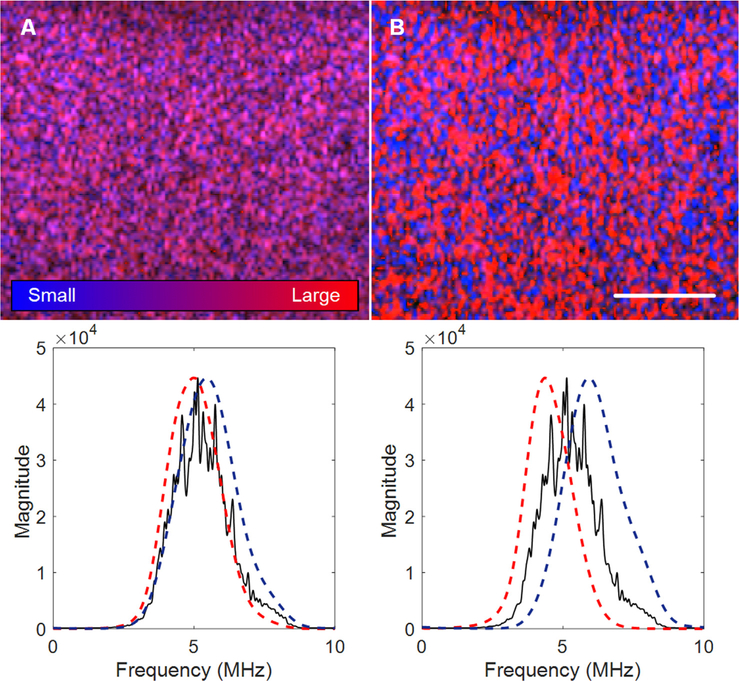
Fig. 5.
H-scan US images (top) and corresponding power spectral density measurements (bottom) for visualizing any scaling adjustments made to the Hermite functions used for convolutional filtering and H-scan US image formation. The scale bar =5 mm. (A) Considerable overlap between the two filtering kernels compromises H-scan US image contrast, which can be alleviated by (B) increased kernel spacing.
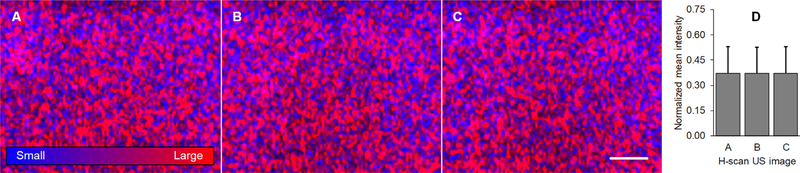
A series of soft tissue-mimicking phantom materials were used to test our new real-time H-scan US imaging system. Co-registered H-scan and B-scan US images were collected from phantoms with three distinctly different-sized (monodisperse) spherical US scatterers, namely, 15 μm, 30 μm, or 40 μm, Fig. 6. 3D histograms summarize mean image intensity for the respective R, G, and B channels used to generate the final H-scan US images. Relative to image measurements from the phantom with 15 μm-sized scatterers, data from the phantom material with the 30 and 40 μm-sized scatterers exhibited mean intensity increases of 26.8% and 29.3% for the R channel and decrease of 8.6% and 10.2% for the B channel, respectively. Overall, the H-scan US image intensity increased by 5.2% and 11.6%, respectively. Collectively, these findings indicate there is a progressive red color (hue) shift as the size of the acoustic scatterers are increased. This agrees with the H-scan US theory whereas larger scatterers dominate the red (low frequency) spectrum. In comparison, the mean B-scan US image intensity increased by 1.3% and 9.4% for the phantom materials containing the 30 and 40 μm-sized scatterers, respectively. Since the backscattered US signal is proportional to the diameter of the scattering object, an increase in B-scan US image intensity is expected. However, these results from the homogeneous phantom materials indicate that H-scan US is more sensitive to physical changes in acoustic scatterer size. Using a homogeneous phantom material containing 40 μm-sized scatterers, H-scan US images were randomly acquired at three different spatial locations to analyze intrasample reproducibility. As Fig. 7 details, real-time H-scan US is a precise imaging tool and repeat measurements (i.e., mean image intensity) are comparable.
Fig. 6.
H-scan US images from a series of tissue-mimicking phantom materials containing a homogeneous mixture of different-sized spherical microparticles, namely, (A) 15 μm scatterers, (B) 30 μm scatterers, and (C) 40 μm scatterers. The co-registered B-scan US images are provided for comparison and the white scale bar denotes 5 mm. Notably, as the acoustic scatterer size was progressively increased in the phantom materials, 3D histogram analysis of the entire US image reveals a marked increase in the red (R) channel signal used to generate the H-scan US images. These histograms summarize the color distribution for the images within a 3D RGB color space.
Fig. 7.
Repeat H-scan US images acquired in a homogeneous tissue-mimicking phantom material at three randomly selected spatial locations (A–C). The scale bar =5 mm. (D) Mean image intensity measurements illustrates reproducibility of H-scan US imaging results.
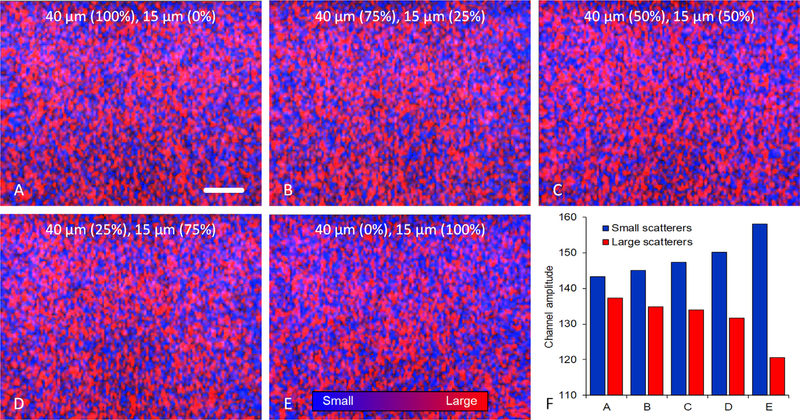
Lastly, we conducted a follow-up study using a series of homogeneous phantoms materials containing of two different-sized acoustic scatterers, namely, 100% of 40 μm scatterers, 75% of 40 μm and 25% of 15 μm scatterers, 50% of 40 μm and 50% of 15 μm scatterers, 25% of 40 μm and 75% of 15 μm scatterers, and 100% of 15 μm scatterers. Review of the corresponding H-scan US imaging results in Fig. 8 reveals progressive shifts in scatterer size as the relative size concentrations are varied. More specifically, as the relative concentration of smaller scattering objects was increased in these phantom materials, the H-scan US images exhibited both an increased blue channel shift (R2 = 0.87, p =0.02) and decreased red channel shift (R2 = 0.80, p = 0.04), corresponding to the high and low frequency signals, respectively. Note that Fig. 8F details the mean 8-bit B and R channel amplitudes to more clearly visualize the change in scattering object size and concentration. This data suggests that H-scan US imaging may be useful for qualitatively tracking relative changes in scatterer size or distribution.
Fig. 8.
H-scan US images from a series of tissue-mimicking phantom materials containing a homogeneous mixture of two different monodisperse silica microparticles, namely, (A) 100% of 40 μm scatterers, (B) 75% of 40 μm and 25% of 15 μm scatterers, (C) 50% of 40 μm and 50% of 15 μm scatterers, (D) 25% of 40 μm and 75% of 15 μm scatterers, and (E) 100% of 15 μm scatterers. The scale bar = 5 mm. (F) Mean spatial analysis of the red and blue channel signals used to create the H-scan US images (denoting the relative size of small and large US scatterers, respectively) reveals that this new tissue characterization technique can detect subtle changes in the distribution of scatterer sizes.
One limitation of the current H-scan US system is it does not compensate for frequency dependent attenuation, which lessens higher frequency data at tissue depth. To make H-scan US more robust and reliable across many transducers and organs, the matching GHnmust be set for the initial pulse and its evolution through deeper tissue compensated for as frequency dependent attenuation filters some of the higher frequency content. Development and implementation of frequency-dependent attenuation correction methods will improve H-scan US image quality and help maximize findings during clinical translational studies. A detailed preclinical study should be conducted to compare H-scan US image findings to physical measures of scatterer size (e.g., from high resolution histologic sections) and include a more detailed assessment of measurement reproducibility. Future work should also investigate the utility of H-scan US imaging in volume space, as that technology could improve sensitivity to subtle changes in tissue scatterers, such as cancer cell shrinkage associated with anticancer drug treatment and early apoptotic activity [17,26].
4. Conclusion
H-scan US reveals the local frequency dependence of different sized scatterers in soft tissue. Herein, we detailed implementation of real-time H-scan US imaging using a programmable US scanner and ultrafast plane wave techniques with angular compounding. H-scan US imaging may prove useful in the diagnosis and management of a wide range of both focal diseases (e.g., cancer) and diffuse diseases (e.g., liver steatosis) where pathology has changed the underlying tissue structures.
Acknowledgements
This work was supported in part by NIH grants K25EB017222 and R21CA212851 and Cancer Prevention and Research Institute of Texas (CPRIT) grant RP180670. The authors would like to thank Verasonics Inc for their technical support of this research project and review of this manuscript.
Footnotes
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultras.2018.12.010.
References
- [1].Brunt EM, Tiniakos DG, Histopathology of nonalcoholic fatty liver disease, World J. Gastroenterol 16 (42) (2010) 5286–5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Elmore S, Apoptosis: a review of programmed cell death, Toxicol. Pathol 35 (4) (2007) 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thijssen JM, Ultrasonic tissue characterisation and echographic imaging, Phys. Med. Biol 34 (1989) 1667–1674. [DOI] [PubMed] [Google Scholar]
- [4].Hoyt K, Forsberg F, Ophir J, Analysis of a hybrid spectral strain estimation technique in elastography, Phys. Med. Biol 51 (2006) 197–209. [DOI] [PubMed] [Google Scholar]
- [5].Hall CS, Verdonk ED, Wickline SA, Perez JE, Miller JG, Anisotropy of the apparent frequency dependence of backscatter in formalin fixed human myocardium, J. Acoust. Soc. Am 101 (1997) 563–568. [DOI] [PubMed] [Google Scholar]
- [6].Kolios MC, Czarnota GJ, Lee M, Hunt JW, Sherar MD, Ultrasonic spectral parameter characterization of apoptosis, Ultrasound Med. Biol 28 (2002) 589–597. [DOI] [PubMed] [Google Scholar]
- [7].Lizzi FL, Astor M, Feleppa EJ, Shao M, Kalisz A, Statistical framework for ultrasonic spectral parameter imaging, Ultrasound Med. Biol 23 (1997) 1371–1382. [DOI] [PubMed] [Google Scholar]
- [8].Oelze ML, O’Brien WD, Method of improved scatterer size estimation and application to parametric imaging using ultrasound, J. Acoust. Soc. Am 112 (2002) 3053–3063. [DOI] [PubMed] [Google Scholar]
- [9].Doherty JR, Trahey GE, Nightingale KR, Palmeri ML, Acoustic radiation force elasticity imaging in diagnostic ultrasound, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 60 (2013) 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gennisson JL, Deffieux T, Fink M, Tanter M, Ultrasound elastography: principles and techniques, Diagn. Interv. Imaging 94 (2013) 487–495. [DOI] [PubMed] [Google Scholar]
- [11].Hoyt K, Kneezel T, Castaneda B, Parker KJ, Quantitative sonoelastography for the in vivo assessment of skeletal muscle viscoelasticity, Phys. Med. Biol 53 (2008) 4063–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parker KJ, Doyley MM, Rubens DJ, Imaging the elastic properties of tissue: the 20 year perspective, Phys. Med. Biol 56 (2011) R1–R29. [DOI] [PubMed] [Google Scholar]
- [13].Poularikas A, Transforms and Applications Handbook, CRC Press, Boca Raton, Florida, USA, 2010. [Google Scholar]
- [14].Szabo TL, Diagnostic Ultrasound Imaging: Inside Out, Academic Press, Boston, 2014. [Google Scholar]
- [15].Parker KJ, Scattering and reflection identification in H-scan images, Phys. Med. Biol 61 (2016) L20–L28. [DOI] [PubMed] [Google Scholar]
- [16].Parker KJ, The H-scan format for classification of ultrasound scattering, OMICS J. Radiol 5 (2016) 1–7. [Google Scholar]
- [17].Khairalseed M, Xiong Kim JW, Mattrey R, Parker KJ, Hoyt K, Spatial angular compounding technique for H-Scan ultrasound imaging, Ultrasound Med. Biol 44 (2018) 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Khairalseed M, Hoyt K, Ormachea J, Terrazas A, Parker KJ, H-scan sensitivity to scattering size, J. Med. Imaging 4 (2017) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jensen J, Stuart MB, Jensen JA, Optimized plane wave imaging for fast and high-quality ultrasound imaging, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 63 (11) (2016) 1922–1934. [DOI] [PubMed] [Google Scholar]
- [20].American Institute of Ultrasound in Medicine TSC, Methods for Specifying Acoustic Properties of Tissue-Mimicking Phantoms and Objects, American Institute for Ultrasound in Medicine, Laurel, MD, 1995, 14 p. [Google Scholar]
- [21].Montaldo G, Tanter M, Bercoff J, Benech N, Fink M, Coherent plane-wave compounding for very high frame rate ultrasonography and transient elastography, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 56 (2009) 489–509. [DOI] [PubMed] [Google Scholar]
- [22].Entrekin R, Porter B, Sillesen H, Wong A, Cooperberg P, Fix C, Real-time spatial compound imaging: application to breast, vascular, and musculoskeletal ultrasound, Semin. Ultrasound CT MR 22 (2001) 50–146. [DOI] [PubMed] [Google Scholar]
- [23].Forsberg F, Ultrasonic biomedical technology; marketing versus clinical reality, Ultrasonics 42 (2004) 17–27. [DOI] [PubMed] [Google Scholar]
- [24].Couture O, Fink M, Tanter M, Ultrasound contrast plane wave imaging, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 59 (2012) 2676–2683. [DOI] [PubMed] [Google Scholar]
- [25].Tanter M, Fink M, Ultrafast imaging in biomedical ultrasound, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 61 (1989) 102–119. [DOI] [PubMed] [Google Scholar]
- [26].Xiong M. Khairalseed, Kim JW, Mattrey R, Parker KJ, Hoyt K, Detection of early tumor response to Abraxane using H-scan imaging: preliminary results in a small animal model of breast cancer, Proc. IEEE Ultrasonics Sympos. (2017) 1–4. [Google Scholar]