Abstract
Methods have been developed to allow quantitative connectivity of the whole fixed mouse brain by means of magnetic resonance imaging (MRI). We have translated what we have learned in clinical imaging to the very special domain of the mouse brain. Diffusion tensor imaging (DTI) of perfusion fixed specimens can now be performed with spatial resolution of 45 um3, i.e. voxels that are 21,000 times smaller than the human connectome protocol. Specimen preparation has been optimized through an active staining protocol using a Gd chelate. Compressed sensing has been integrated into high performance reconstruction and post processing pipelines allowing acquisition of a whole mouse brain connectome in < 12 hrs. The methods have been validated against retroviral tracer studies. False positive tracts, which are especially problematic in clinical studies, have been reduced substantially to ~ 28 %. The methods have been streamlined to provide high-fidelity, whole mouse brain connectomes as a routine study. The data package provides holistic insight into the mouse brain with anatomic definition at the meso scale, quantitative volumes of subfields, scalar DTI metrics, and quantitative tractography.
Keywords: MRI, Connectomes, Mouse Brain, MR Histology
Graphical Abstract
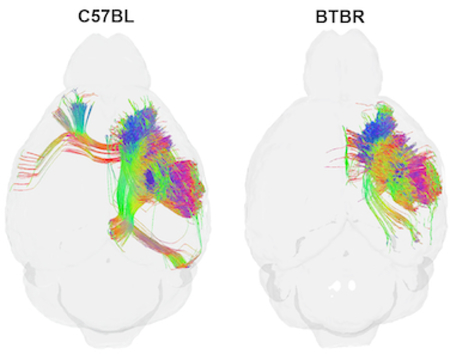
Comparison of diffusion tractography within the forelimb representation of the right primary somatosensory cortex in two murine strains: C57BL and BTBR (a murine model that bears phenotypic resemblance to human autism spectrum). Note strain-specific differences in commissural connections and longitudinal associational connections in the ipsilateral hemispheres.
INTRODUCTION
The development of diffusion MRI (dMRI), tractography and MR connectomics have spawned an enormous number of technical developments, applications, new insights into brain organization, and new questions for ongoing theoretical and empirical investigation. The field is only 25 years old; but continues to grow and evolve at a rapid rate (e.g., a literature search on “MRI”, “connectome”, and “2017” yielded 125 results).
The majority of the methods and applications have come in the clinical domain. The human connectome project(Ugurbil et al., 2013, Van Essen et al., 2013), the ENIGMA project (Thompson et al., 2013) (Kochunov et al., 2015) (Thompson et al., 2017) and the ADNI consortium (Jack et al., 2008) (Jack et al., 2015) (Kauppi et al., 2018) have all exploited dMRI to reveal new insights into brain structure, genetics, and pathology. Multiple groups are working at higher magnetic field (>7.0T). New acquisition hardware and pulse sequences abound. New post processing statistical methods are addressing some of the known deficiencies in the present state-of-the-science; however, several recent studies addressing dMRI applications in mapping connections in the human brain urge caution. Thomas et al. concluded that “a diffusion tractogaphy technique that produces anatomically accurate results remains elusive …” (2014). A recent paper by Maeir-Hein coauthored by 78 of the world’s experts in clinical tractography (2017) drew additional disconcerting conclusions:
“There has been no independent validation of human tractography”; the majority of the methods tested in this paper detected at least 90% of the true paths; but the methods tested detected far more false-positive than true positive connections.”
There have been parallel efforts, at a much lower level of activity, to generate novel insights into the structure and organization of the mouse brain. Pioneering work has been done by Zhang and Mori (2002) and Jiang (2009) (2011) in the mouse. But the mouse is not a human and DTI is not a connectome. Scale really matters. The mouse is 3000 times smaller than the human so one might expect that the spatial resolution (volume of the imaging voxel) needs to be 3000X smaller for comparable anatomic definition. The smaller imaging volume of the mouse (~ 10×10×20 mm) makes many human methods (multicoil, multiband) less applicable. Work in the clinical domain can provide a good starting point if one is careful to appreciate the challenges, the differences between the two domains, and the opportunities for translation to whole mouse brain connectomics. With careful attention to protocol development, some of the limitations that have been described in clinical connectomics can be addressed.
Our first diffusion atlas of the mouse brain was published in 2009 (Jiang and Johnson, 2011). This work was limited to six angular samples yielding diffusion scalar images based on the simple diffusion tensor model(Basser et al., 1994). But there was no connectome. Since that time the methods and our understanding of their limitations have undergone significant evolution. In 2015 we published a whole mouse brain connectome that serves as a foundation for many of our subsequent studies (Calabrese et al., 2015a).Those data were acquired with what we believe to be the highest spatial resolution yet attained (43 um isotropic resolution). The volume of each voxel is < 80 pl, which is nearly 25,000 times smaller than the (~ 2 mm3) voxels specified in the human connectome protocol (Ugurbil et al., 2013). These foundational data were acquired with 120 different angular samples using the HARDI (Tournier et al., 2004) and q ball methods (Tuch, 2004). A 3 dimensional anatomic parcellation was developed defining 296 different seeding regions of the brain. Probabilistic tracts were generated for the whole brain and connectivity maps generated between each seed region. Most importantly, the data were validated against the Allen Brain Connectivity Atlas (ABCA) of Oh et al. (2014). This connectomic atlas was based on block face light microscopy images of in vivo tract-tracing experiments using retroviral constructs. Acquisition of our foundational MR connectome atlas took ~ 10 days and the post processing required two weeks of computational time using a high-end cluster. In contrast, acquisition of the data (from 1231 brains) for the ABCA took more than a year.
More recently we have advanced the methodology for more routine throughput with the use of compressed sensing (Wang, 2018). Compressed sensing is a novel acquisition/reconstruction strategy that allows one to acquire far less data; i.e., accelerate acquisition and still recover the same spatial resolution (Lustig, 2007). The raw data (Fourier) space for MRI is sparsely sampled using a probabilistic weighting function. Data is subsequently processed using an iterative reconstruction. Simplistically, one reduces the acquisition time at the expense of reconstruction time. The increased reconstruction time has been addressed by breaking the process into many individual 2D slices that can be reconstructed in parallel. The entire process, acquisition and reconstruction, have been streamlined through an image pipeline using a cluster computer with 350 virtual cores. The streamlined protocol yields 51 3D volumes at isotropic spatial resolution of 45 microns (voxel volume of 91 pl). As acquisition of a volume is completed, it is streamed to the cluster. The entire 4-dimensional volume can be acquired and reconstructed in <12 hrs. A second imaging pipeline registers the multiple volumes to correct for eddy currents during acquisition, registers the parcellation (labels) to the 4D volumes (~ 3GB total), generates the tracts, and computes the connectome.
The infrastructure we have engineered for whole brain mouse connectomics has now made possible neurobiological investigations of brain organization and connectivity in a species that has become pre-eminent in the biomedical sciences. This same infrastructure could apply to high-efficiency studies of connectomics in other species that present unique comparative and/or translational value. This paper describes this infrastructure, our streamlined protocol, the elements that are unique to whole mouse brain connectomic histology, validation of our methods, sources of errors, and future directions.
METHODS
Animal Preparation- Active Staining
All animal studies were performed under approval form the Duke Institutional Animal Care and Use Committee (IACUC). Most whole brain imaging methods start with tissue preparation. Getting this step right is key to final success. The impact of formalin fixation on tissue relaxation was first reported in the early 1990’s(Johnson et al., 1993) (Boyko et al., 1994). Hsu et al. reported quantitative changes in T1 and T2 with fixation; i.e., that both parameters are significantly reduced by fixation and that longer fixation causes additional reduction(1994). Recognizing these effects, we developed active staining, a method of perfusion fixation for the whole brain with buffered formalin and MR contrast agent that enhances the MRI signal and stabilizes the tissue for MR histology (Johnson and Hedlund, 2000). The contrast agent reduces the T1 which enhances the signal. At the same time, the contrast agent and fixation reduce T2 and diffusivity, which are both detrimental. Active staining has been described in detail previously (Johnson et al., 2002) (Johnson et al., 2007). A catheter is inserted into the left ventricle allowing controlled infusions of saline/heparin followed by buffered formalin with Prohance, a Gd contrast agent. Hukkanen et al. (1987) reported the changes in white matter arising from autolysis. D’Arceuil et al. (2007) reported that these changes have significant impact on the diffusion properties that are crucial for generating tractography. Our active staining provides the best possible fixation, with uniform distribution of fixative and the contrast agent while minimizing autolysis. Following perfusion, the head is removed and placed in formalin for 24 hrs to complete fixation. The brain, still in the cranial vault is then transferred to phosphate-buffered normal saline (PBS) with 0.5 % Prohance (Gadoteridol). To clarify the temporal stability of fixation/staining nine specimens were actively stained. Each of the specimens was scanned six times over a period of two months. Resulting data were processed through the pipelines yielding scalar images, tractography and connectomes on nine specimens at each of six different times.
Image Acquisition
MR images are acquired on a 9.4T vertical bore magnet coupled to an Agilent console. Key to the hardware is a gradient set sufficient to apply gradients up to 2000 mT/m. Clinical scanners are usually limited to gradients less than 50 mT/m. A limited number of specialized “human connectome” scanners have gradients of 300 mT/m(McNab) (A et al., 2017, Fan et al., 2016). Stronger gradients are required to achieve higher spatial resolution and impose heavier diffusion weighting to mitigate the consequences of chemical fixation. Specialized radiofrequency coils have been developed to capture the very weak signal from the small voxels used in this microscopic imaging method(Hurlston et al., 1997).
The demands of high spatial resolution, strong diffusion weighting, signal decay from formalin fixation, and physically small specimens have driven us to imaging protocols that are far removed from those used clinically. The short T2 (<30ms) from fixation makes echo planar methods used clinically less viable. Our protocol uses a three dimensional Stesjkal Tanner spin echo sequence that is particularly immune to many artifacts that occur at higher field strength (Stejskal and Tanner, 1963). The diffusion encoding gradients are on for ~ 5 ms at 1000 mT/m. Half sine gradients are used to minimize mechanical forces on the gradients since the protocol runs continuously for 12 hrs. The bandwidth and encoding intervals have been chosen to minimize the echo time (TE=12.7 ms) thus minimizing the impact of T2 decay. Additional details of this approach have been published recently (Wang, 2018).
The fidelity of the tractography and connectome are intimately tied to the spatial resolution, the signal to noise in the composite images, the strength of the diffusion encoding gradients, and the number of different gradient angles acquired. Numerous authors have explored the topic in the context of clinical scanning where the spatial resolution is >1 mm and scan times must remain clinically realistic (<30 min) (Jones et al., 1999) (Zhan et al., 2013) (Vos et al., 2016). We have previously explored the question in the macaque brain where a fixed specimen was scanned with a fixed time (60 hrs) using protocols with spatial resolution of 130 um and 12 directions to 600 um resolution with 257 directions(Calabrese et al., 2014). The conclusions for the majority of these studies are that the spatial and angular resolution required to accurately define a specific fiber tract are very dependent on that fiber tract. We addressed the question in the specific context of the mouse brain by scanning with the highest spatial and angular resolution we could achieve (43 um, 120 angles), yielding a foundational data set for comparison to other types of data. Through a systematic down sampling of both angular and spatial resolution and validation against tract-tracing experiments in the ABCA, we arrived at our current protocol (45 um, 46 angular samples)
Reconstruction/Postprocessing
Acquisition of the first whole mouse brain connectome (43 um 120 angles) took 235 hrs. The method used traditional Fourier encoding and reconstruction via an analytical 3D Fourier transform. This approach relies on fully sampling the data (Fourier) space requiring the exceedingly long acquisition times. Lustig’s development of compressed sensing (2007)allowsus to accelerate the acquisition by 8 fold by sparsely sampling Fourier space. This was combined with a reduction in angular sampling providing a net acceleration of 20X. Reconstruction is done through the transformation to an alternative (wavelet) space followed by an iterative technique. This iterative reconstruction takes considerably longer than the simple Fourier inversion. But the tradeoff off in reduced acquisition time for increased reconstruction time can be compensated through the use of a high-end computational resources. A processing pipeline has been constructed to receive the compressed data during acquisition. An initial Fourier transform along the readout axis converts the problem to 420 individual 2D slices (256×256), which are parsed out to 4 nodes (64 processors) of the cluster. The algorithm completes reconstruction of one 3D volume in ~ 25 minutes.
The 4D data set that emerges from the reconstruction pipeline consists of 51 3D image volumes. The native space of the scanner is defined by five baseline (b0) 3D volumes with no applied gradient, dispersed throughout the 11.6 hr acquisition. The second stage of the pipeline registers the 46 diffusion weighted 3D volumes to that native space using registration tools from the ANTs software package (Avants et al., 2008) yielding a single 4D image array of ~ 3GB. The pipeline passes this volumetric image array to Diffusion Toolkit (http://trackvis.org) where the data is fit to the simple tensor model to yield scalar images characterizing the anatomy(Basser and Pierpaoli, 1996). Labels are warped to these anatomic images and the 4D package (now with labels) is passed to DSI studio (http://dsi-studio.labsolver.org/) for connectomic analysis. Tracts are generated from the 4D arrays using the HARDI/generalized q space sampling approach (Tournier et al., 2004) (Yeh, 2010). This package relies on a normalized quantitative anisotropy (NQA) analogous to the fractional anisotropy (FA). NQA provides an improved performance over FA in resolving crossing fibers when performing deterministic tractography (Yeh et al., 2013).
RESULTS
Multicontrast 3D Anatomic Images
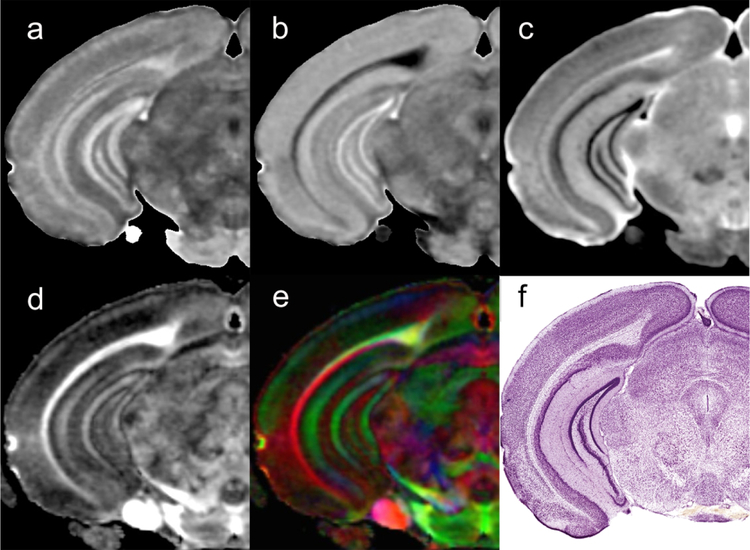
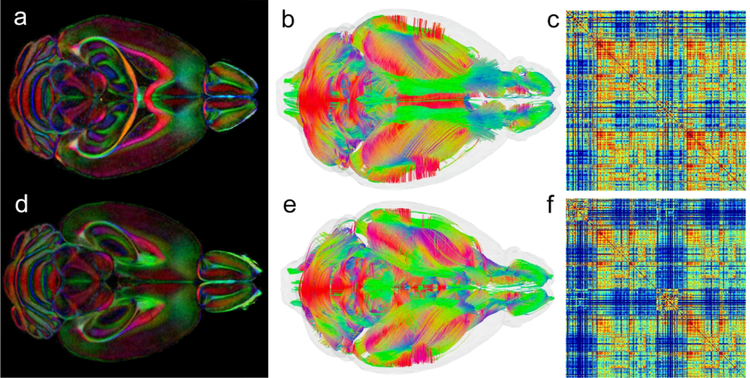
Figure 1 shows representative scalar images from a C57BL/6J male mouse scanned with the connectome protocol. The axial diffusivity (AD), radial diffusivity (RD), mean diffusivity (MD), fractional anisotropy (FA) and color fractional anisotropy (clrFA) all provide different means for visualizing neuroanatomical structures and landmarks. An early step in generating the connectome from a specimen is mapping the seed regions onto the unknown specimen. Our pipeline starts with a masking step to remove extraneous external tissue followed by ANTs-based registration using mutual information as the chosen similarity measure (Badea et al., 2012). Initial alignment via linear transforms (rigid plus affine) employ the diffusion-weighted images. Spatial normalization is further refined by a diffeomorphic transform (i.e., Symmetric Normalization (SyN)) using the FA volumes. These registration stages establish the spatial correspondence between our labeled image and any unlabeled target image thereby allowing propagation of label information from one domain to the other.
Figure 1.
Scalar images from the Trackvis pipeline are used to drive the registration of the seed labels for the connectome: a) axial diffusivity (AD); b) radial diffusivity(RD); c) mean diffusivity; d) fractional anisotropy (FA); e) color fractional anisotropy (clrFA). For comparison, a histological section stained for Nissl substance (Plate 59 reprinted from (Paxinos and Franklin, 2012) , with permission from Elsevier).
Reference Labels
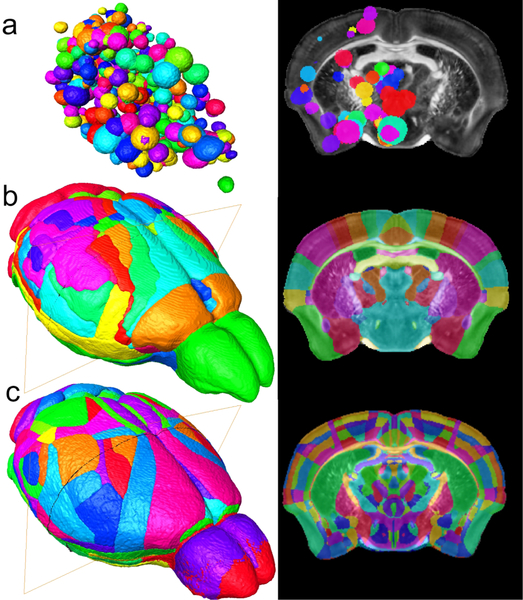
The label set that defines the seed regions for the connectome is one of the single biggest sources of variability. It defines the reference base for determining anatomic connectivity. The MNI atlas, a 3D digital atlas based on human MRI has been crucial to the development of (human) functional and structural connectomics (Evans, 1993). The mouse equivalent of the MNI atlas has undergone significant evolution in the last twenty years. Waxholm Space (WHS) was one of the first 3D digital atlases of the mouse brain based on MRI data(Johnson et al., 2010). The Allen Brain Atlas (ABA) with considerably more anatomic definition (Dong, 2008)is based on conventional 2D histology (Dong, 2008).The Allen Mouse Brain Connectivity Atlas (ABCA),with parcellation derived from the original ABA is a meso scale map of axonal projections based on 469 stereotactic injections of anterograde retroviral tracer constructs. Connections were mapped for 295 non overlapping anatomical sites defined in the Allen Brain Atlas(Dong, 2008). Our diffusion data was registered to the ABCA to allow validation of the MR connectome against the retroviral data(Calabrese et al., 2015b). The volume of each injection site was assumed to be spherical and was estimated from the quantity of tracer injected (Calabrese et al., 2015b). Figure 2a shows a volume rendered image of the ABCA seed regions registered to the FA image of one of our higher throughput FA data sets. Data can be resectioned to see the superposition of the ABCA seed regions along any plane (e.g., a coronal section on the right in Figure 2a). Our current protocol uses seed regions defined in Waxholm Space (WHS) (Johnson et al., 2010), based on anatomical landmarks. These landmarks have been defined using the multiple scalar images (Figure 1). WHS was augmented in our foundation set to include cortical labels from Ullmann et al (2013). This modified WHS includes 166 labels on both hemispheres. The volume rendered image and coronal section comparable to the ABCA labels is shown in Figure 2b. Figure 2c shows the direction for the future. The most recent version of the ABA anatomic labels (vs 3) has been recently registered to the MR images of WHS. This new label set will harmonize the connectomic data we are generating with the gene expression and connectome data of the ABA. Defining the seed regions using anatomic landmarks provides a way to compare connectomes from multiple animals. It does come with a potential downside, i.e. the reliability of the registration (Avants BB, 2011). All the work demonstrated here used the WHS labels. But moving to the new labels in ABA provides a significant opportunity to merge our approach to the gene expression data in the ABA.
Figure 2.
Seed regions are a critical determinant of the connectome. a) Seed regions (i.e., retroviral injection sites) from the ABCA have been mapped into the MR images of Waxhom Space, with spherical dimensions estimated from the volume of injected tracer. b) Seed regions from Waxholm Space have been derived from anatomic landmarks visible in the scalar images (Figure 1). c) Recent work has resulted in a merger of Waxholm Space canonical MR images with a new set of delineations from ABA (version 3). This will facilitate harmonization of connectomics data with gene expression.
Tissue Stability
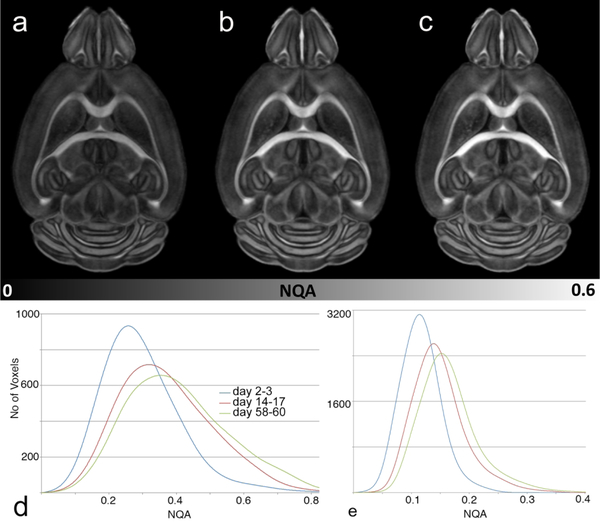
The fidelity of the registration, and to a much greater extent, the tractography that underlies the connectome are driven by the FA (for anatomic registration) and NQA (for tractography). The active staining protocol provides an increase in signal to noise ratio (SNR) of more than 8X over an unstained specimen. But it too brings a source of variability as fixation and equilibration of the stain evolve over time (days to months). Figure 3 shows a comparison of the NQA at 2–3 days, 14–17 days and 58–60 days. In a typical whole brain study a cohort (n=6–12) is perfused. The questions of concern are: When is the best time to scan? How long does it take for the fixation/staining to stabilize? How significant are the changes over time? To generate the tracts, a threshold NQA is chosen (typically ~ 0.1) to limit the generation of spurious tracts. The grayscale in both images is identical. Over the first two weeks the mean NQA histogram of all the white matter shows significant shift with the peak of the histogram moving from ~0.26 to ~ 0.33. This is initially somewhat surprising. One might expect that as fixation progresses, there might be a loss of contrast. There is instead a considerable increase at two weeks and the NQA continues to increase even out to two months where the peak has shifted to ~0.36. Figure 1e, a histogram of the NQA in the isocortex is particularly informative. At 2–3 days the peak of the histogram is ~0.11. If the threshold for tracking is set at 0.1, 32% of the voxels in the isocortex will not be included in the tractography. At two weeks, the peak of the histogram has shifted well above 0.1 leaving ~12% of the voxels below the threshold. It is particularly surprising that the NQA continues to increase even out to two months suggesting that we should wait at least two weeks to scan after perfusion but that we might have a broad window of opportunity after that. Our best practice is to allow at least three weeks for stabilization and to scan all the specimens in a group within 8–10 days of the same mean fixation time.
Figure 3.
Fixation and diffusion of the contrast agent induce change in tissue properties. a) NQA image at 2–3 days after perfusion fixation, b) 14–17 days after fixation; c) 58–60 days after fixation show increase of the NQA over time. d) Histogram of all the white matter shows a shift in NQA from day 2–3 to day 14–17 and day 58–60. d) Histogram of NQA in the isocortex demonstrates a shift even in the cortical areas that has profound impact on the resulting tractography.
Angular Sampling
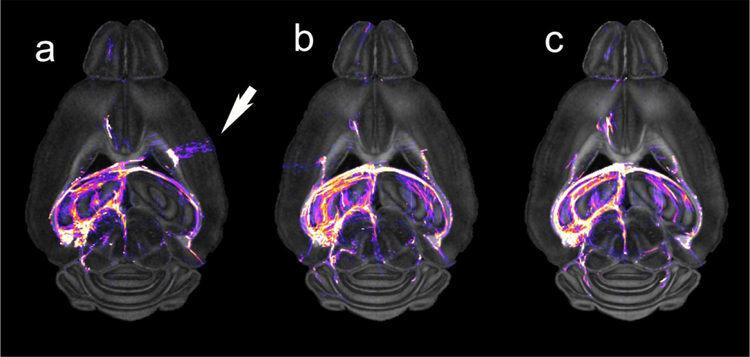
The impact of crossing and merging fibers on the validity of the connectome cannot be over stated. To generate a connectome one must acquire multiple image volumes. The 1.25 mm3 voxels in the human connectome protocol contain more than 100,000 neurons (Herculano-Houzel, 2009). Using advanced post processing methods permits resolution of crossing and merging fibers in many voxels. But even small errors can have outsized effects on the connectome. A number of creative solutions, e.g., HARDI (Tournier et al., 2004) and Q Ball imaging (Tuch, 2004), have been proposed. All require acquisition of a “sufficient” number of angular samples. Many studies have probed the question of what is “sufficient” which is in turn dependent on the algorithm, signal to noise of the data and spatial resolution(Tuch et al., 2002, Jones, 2004, Jones and Cercignani, 2010, Jones et al., 2013, Tournier et al., 2013, Vos et al., 2016). Maeir-Hein et al. explored the performance of 96 different approaches in mapping human connectomes (2017). They concluded that even the best methods had a significant number of false positive results; i.e., connections that are not really there, exceeding the true positive results. The human setting is further complicated by the fact that there is no independent standard that can be used to test MRI results. Increasing spatial and angular resolution both contribute to the total acquisition time. Figure 4 demonstrates the problem for the mouse brain by comparing tract density images from three data sets on the same specimen with 16 , 46, and 120 angular samples. The tracts from seeding the left hippocampus are overlaid on the NQA images for anatomic reference. The tract density color scale is logarithmic over nearly five orders of magnitude. The white arrow in Figure 4a (16 angles) shows a false positive streamline. Because of the limited number of samples, the algorithm has broken down and defined a streamline that has no biological validity.
Figure 4.
Dorsal NQA image from a C57BL/6J mouse acquired with a) 16 angles; b) 46 angles; c) 120 angles demonstrate the value of increased angular sampling. Tractography was generated by seeding the left hippocampus. The white arrow highlights a false positive tract in the 16-angle data that does not appear in the 46 and 120 angle data.
Connectome Validation
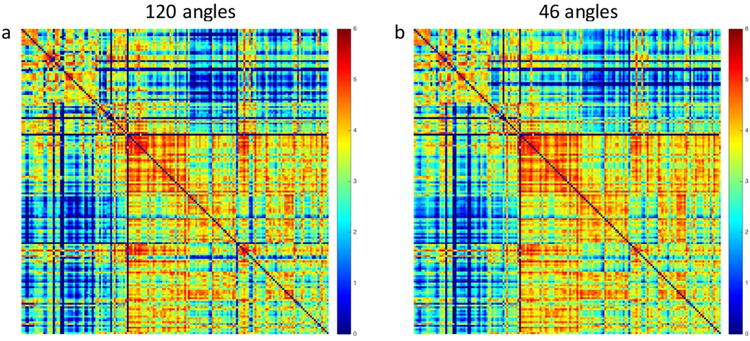
We have undertaken a systematic assessment of the “sufficient” number of angles and the spatial resolution with specific focus on the whole mouse brain (Wang, 2018) by comparison to the foundational (MR) connectome in our accelerated protocol. Our foundational connectome acquired at 43 um spatial resolution and 120 angular samples, represents the best data we have been able to acquire. Validation against the histological tract-tracing experiments presented in the ABCA yielded a false positive rate of ~ 28%, far below that of clinical scan(Calabrese et al., 2015b). As Maier-Hein have noted, it is not uncommon to have more false positive than true positive tracts in clinical data(2017). The accelerated protocol acquired with 45 um spatial resolution and 46 angular samples can be acquired in <12 hrs. Figure 5 shows a comparison of the connectomes with the two strategies (120 different angular samples over 235 hrs of acquisition vs 46 angles over 11.6 hrs). The correlation coefficient between a and bis 97%. The acceleration (by nearly 20 fold) comes with a small penalty but we feel that penalty is negligible given that this approach makes it possible to routinely acquire a connectome over the entire mouse brain with quantitative validation against the ABCA.
Figure 5.
Connectomes generated from a) our foundation set with full k space sampling and 120 angular samples (235 hour acquisition) and b) the accelerated acquisition protocol acquired with compressed sampling and 46 angular samples (11.6 hour acquisition) have a correlation coefficient of 0.97.
Population Atlases
The ability to acquire a whole mouse brain connectome in < 12 hrs opens one more door: the potential for group studies. We have once again taken advantage of the methods developed clinically (Avants BB, 2011). We and many others have adapted atlasing techniques developed within the clinical community to understand phenotypic differences in brain morphology. Similar methods can be adapted for generating connectome atlases with a subtle but significant modification. The tractography underlying differentiation of crossing fibers is dependent on accurate knowledge of the angle of the diffusion encoding gradients, relative to the local anatomy. Small differences in how the specimen is placed in the magnet result in misalignment between specimens. The gradient directions are defined by the reference system of the magnet. Traditional registration algorithms do not take into account the differences in the gradient angles between specimens. Yeh et al. have developed elegant methods to address this problem with q space diffeomorphic reconstruction (QSDR) (2016). This allows one to create atlases from study groups by mapping data into a common space. The atlases facilitate group statistical comparisons in which local connections are tracked instead of end-to-end connectivity. Thus, these “connectometry” studies track local group differences (Yeh et al., 2016). Instead of tracking end-to-end connectivity, one can compare the local connectivity between groups mapped to the common reference. This approach provides an alternative analysis to traditional connectomics that allows a certain degree of flexibility in controlling for false discovery. Figure 6 shows tractography seeding the primary somatosensory cortex for a single male C57BL/6J mouse and for a group (n=4) mapped into a common space using QSDR. A clearer picture of cortical connection is evident in the population average.
Figure 6.
Tractography from the somatosensory cortex of a) a single C57BL/6J male mouse and b) a group average (n=4) produced with q space diffeomorphic reconstruction demonstrate improved sensitivity of group analysis.
Strain Comparisons
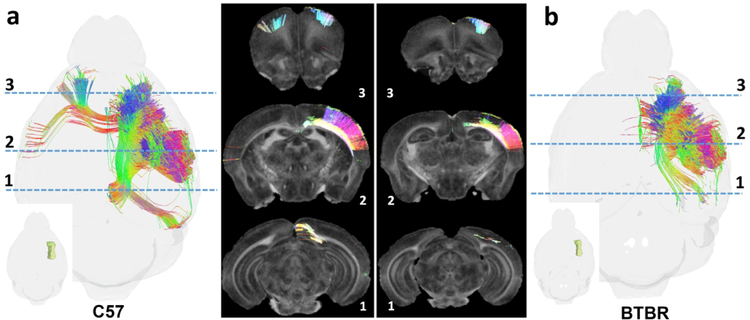
A singular advantage of diffusion-based connectomics is the ability to rapidly compare many different phenotypes from several different perspectives, including an analysis of different strains of the same species. Figure7 demonstrates one such comparison between the C57BL/6J mouse strain (top; Figure 7 a, b, c) and the BTBR strain (bottom; Figure d,e,f). These data were acquired with an earlier version of our protocol that was not as aggressive in the compression (CS=4x) resulting in 23 hour scans. Both sets have been processed through the pipelines described above. Several groups have studied the BTBR strain as a model of autism(Meyza and Blanchard, 2017, Scattoni et al., 2008). Previous MR studies of the BTBR have shown changes in white matter FA and reduced neuroanatomical volumes, including an absent corpus callosum and a reduced hippocampal commissure (Ellegood et al., 2013). Tractography and the resulting connectome have not yet been demonstrated in this model. The single dorsal slice (Figure 7a, d) with all the specimens in each group registered to the common space shows agenesis of the corpus callosum and similar absence of commissural fibers associated with the fornix. The volume tractography of the whole brain (Figure 7b,e) is viewed from the dorsal perspective. There are some striking differences in the visible cortical and subcortical connections. Figure 7b shows the whole brain tractography of the C57BL/6J group which demonstrates robust and much more broadly distributed cortico-cortical connections in the neocortex. Figure 7e shows comparable whole brain tractography of the BTBR cohort, which demonstrates more focused (less uniform) cortico-cortical connections in the neocortex, with some connections (to/from sensorimotor cortex) being particularly robust and others (to/from prefrontal cortex) much more sparse. The quantitative connectomes for each are shown in 7c,f. These matrices illustrate the relative sparseness of connectivity in BTBR group, compared to the C57BL/6J group (cf. Figure 7c and f, noting the reduction in red cells and the increased number of blue cells in the BTBR matrix).
Figure 7.
The whole brain protocol allows one to compare strains with several different perspectives. The comparison of C57BL/6J-top (a) and BTBR-bottom (d) in a dorsal color FA slice shows disruption of the corpus callosum and hippocampal commissure. Comparison of whole brain tractography (b-C57BL/6J and e-BTBR) shows significant global difference in tract organization. Connectome matrices (c-C57BL/6J and f-BTBR) shows quantitative differences in connectivity across the entire brain.
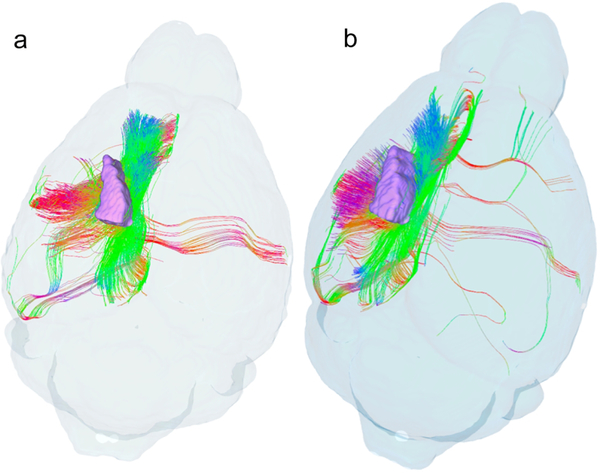
The whole brain connectome data package includes five different 3 dimensional isotropic volumes with five different contrasts for defining anatomy; tractography of the entire brain, the connectome and the tracts for each of the subfields included in the connectome calculation. Survey of the entire connectome helps highlight specific areas of interest. Figure 8 shows such an example comparing streamlines for a single specimen passing through the forelimb territory within the primary somatosensory cortex (lower insets in Figure 8a and b). Dorsal views of the connectomes associated with this cortical subdivision (far left and right images in Figure 8) reveal the absence of callosal connections and reductions in longitudinal associational connections involving more posterior parietal and occipital regions in the BTBR strain. In both species, radial tracts are prominent in the ipsilateral forelimb representation (see streamlines overlaid upon NQA images labeled 2), although the density and distribution of tracts in the primary somatosensory cortex is greater in the C57BL/J6 animal than in the BTBR animal. Both animals produced tracts that reflect connections with the ipsilateral primary motor cortex, with projections to the contralateral motor cortex largely absent in the BTBR animal (images labeled 3). Similar patterns were observed in other individuals in each strain. This representative demonstration illustrates the value of this unique approach to group-specific (strain-specific in this case) assessments of partial connectomes constrained by known subdivisions of the cerebral cortex.
Figure 8.
Comparison of tractography within the forelimb representation of the right primary somatosensory cortex in two murine strains (individual specimens): C57BL/J6 (a), a common mouse model in biomedical research, and BTBR (b) a murine model that bears phenotypic resemblance to human autism spectrum disorders (Meyza and Blanchard, 2017, Scattoni et al., 2008). Images to the far left and right in (a) and (b) show tractography as seen from the dorsal aspect of the mouse brain; Insets localize the forelimb representations. Dashed lines identify the locations of coronal NQA images from the same datasets with in-plane streamlines superimposed. Note strain-specific differences in commissural connections and longitudinal associational connections in the ipsilateral hemispheres.
DISCUSSION
We would emphasize that whole brain connectivity using dMRI is a mathematical construct based on the physical diffusion of water molecules measured using magnetic resonance imaging. Conventional neuro histological means for tracing axonal connections, such as retroviral constructs engineered for anterograde tract tracing(Oh, 2014), bear the specificity of labeling axons and their collaterals arising from a discrete population of neurons localized to the injection site. Even with the limitations of state-of-the-science tract-tracing methods (mainly, false negatives in the case of anterograde retroviral tracers), we recognize that dMRI streamlines are not labeled axons and the molecular specificity of modern neuro histological approaches (e.g., retroviral-based tracers) provides a far more accurate view of neuronal connectivity in the mammalian brain. Neuro histological tracers may be engineered to be taken up by cell bodies or synaptic terminals and thereby provide directionality information; i.e., they can map either anterograde (cell body transfection, iontophoresis or uptake) or retrograde (presynaptic endocytosis), depending on the mechanisms of uptake biased by the tracer. In contrast, the diffusion phenomenon that provide the basis of dMRI connectomics does not differentiate the polarity of neuronal (axonal) metabolism or signal propagation. Thus, the limitations of dMRI lead to major problems, including uncertainty regarding how streamlines relate to the microscopic organization of axonal projections and the ultrastructural properties of axon fascicles, lack of information regarding the polarization of neuronal connections, remaining difficulties resolving connections in gray matter and the crossing of fibers in white matter, and the persistence of relatively high rates of false positives tracts (including the present study, with its estimated false positive rate of 28%).
An enormous body of literature has addressed these problems in the settings of clinical and human subject applications, with elegant acquisition and post processing methods. But even in the most experienced hands, dMRI can lead to far more false positives than true positives(Maier-Hein et al., 2017). One might reasonably ask “is it worth the trouble?”. We believe it is. When comparing results from the clinical domain with those we can obtain in fixed specimens, size matters. Smaller voxels mean fewer crossing fibers to sort out in any given voxel. The typical 1.2–1.5 kg human brain contains around 86 billion neurons and at least that many neuroglial cells (Herculano-Houzel, 2009). Thus, a 1.25 mm3 voxel in a clinical study might contain ~ 100,000 neurons. The mouse brain contains ~ 71 million neurons (Herculano-Houzel, 2009). It follows then that the 45 um3 voxels of our protocol have ~16 neurons. Given this massive reduction in cytoarchitectural complexity compared to clinical dMRI, teasing out merging and crossing fibers in these microscopic voxels is a considerably more tractable task. In the clinical domain, an increased number of angular samples is used to help deconvolve crossing fibers. Through a novel use of compressed sensing we have been able to maintain a high angular sampling while keeping the acquisition time < 12 hrs. Differences between angular and spatial resolution and human and mouse is again about scale. To appreciate the difference, we define a resolution index (RI) which is the product of the angular and spatial resolution (1/voxel volume).
This index is artificial (dropping the unit volume) and is only meaningful as a quantitative metric when additional details of a protocol are included; e.g., signal-to-noise and acquisition time. It does however highlight the vast difference between the clinical domain and our mouse brain protocol. The RI for the human connectome is 270 angular samples with a spatial resolution of 1/1.25mm3 = 138. The RI for our protocol is 46 angular samples times a spatial resolution of 1/0.045mm3 = 504,801. Chen et al. (2014)compared DTI data in a mouse brain acquired at 62.5 um (0.0625 mm3) with six angles to the ABCA with RI=24,576. A more recent study from Aydogan et al. (2018) acquired 90 angles at 200 um resolution, yielding an RI of 11,250. Azadbakht et al. (2015) compared dMRI to previously published schemata for interareal connectivity based on neuro histological tract-tracing methods in the macaque visual cortex; in their study, RI was 1509 (120 angles and 0.43 mm spatial resolution). The disconcerting facts are that the false positive rate in human studies is greater than true positive results and the accuracy in rodent and macaque studies even at considerably higher resolution are less than one would hope when compared to retroviral methods. Our results show a much more acceptable – but still not ideal -- false positive rate of ~ 28% for our foundational dataset(Calabrese et al., 2015) and agreement between connectomes of that foundational set and our higher throughput protocol of 97% (present study).
Despite their limitations, molecular tracers of neuronal connectivity will continue to be the gold standard for the continued improvement of dMRI connectomics. We believe that whole brain connectomics using dMRI should now have an established place among the tools used to routinely interrogate the organization and neuronal connectivity of the mammalian brain. When carefully executed, with a clear understanding of the limitations, dMRI can provide a unique, comprehensive view of the mammalian brain that should be valuable for diverse exploratory or hypothesis-driven studies of neuroanatomy. Indeed, our protocol for studying the mammalian brain with dMRI yields meso-scale neuroanatomical information that is – in some respects -- considerably more accurate than that from conventional histology. For example, dMRI does not involve physical slicing, dehydrating or otherwise mechanically distorting brain tissue; in the present study, the mouse brain remained intact situated in the cranial vault with vascular and cranial nerve attachments preserved. Furthermore, three dimensional volumes of all the neuroanatomical subdivisions and resulting streamlines are readily available as are multiple means of visualizing data of interest with anatomical images projected in context (e.g., see Figure 8). The potential of quantitative connectometry and other group studies is truly appealing (e.g., genetic studies of related strains within species; comparative neurological studies; interventional studies with treatment and control cohorts). Thus, with careful consideration of technical and/or biological limitations and appropriate control conditions, quantitative comparisons are possible for diffusion parameters and streamline metrics. And finally, with the methods described here, an entire study can be completed in a day. While not exactly high throughput, the methods do allow population studies that could not be completed any other way.
The protocol described here is not static. Like most modern imaging, the technology will continue to evolve. New acquisition/reconstruction strategies will reduce the acquisition time. Improved post processing algorithms will further reduce errors. New strategies for group analysis will evolve with deep learning. But the technology is accessible now and we are eager to share our protocols and datasets with investigators ready to ask new questions concerning the organization and connectivity of mammalian brains.
ACKNOWLEDGMENTS
We thank Tatiana Johnson for help in editing the manuscript and James Cook and Lucy Upchurch for critical support in developing and maintaining the computer architecture. This work was supported by the NIH/NIBIB National Biomedical Technology Resource Center (P41 EB015897 to GA Johnson), NIH 1S10OD010683–01 (to GA Johnson), 1R01NS096720–01A1 (to JC Gee and GA Johnson).
Footnotes
CONFLICT OF INTEREST
None.
REFERENCES
- A TV, JAMISON K, GLASSER MF, SMITH SM, COALSON T, MOELLER S, AUERBACH EJ, UGURBIL K & YACOUB E 2017. Tradeoffs in pushing the spatial resolution of fMRI for the 7T Human Connectome Project. Neuroimage, 154, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVANTS BB, EPSTEIN CL, GROSSMAN M & GEE JC 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal, 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVANTS BB, T. N SONG G, COOK PA, KLEIN A, GEE JC 2011. reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage, 54, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AYDOGAN DB, JACOBS R, DULAWA S, THOMPSON SL, FRANCOIS MC, TOGA AW, DONG H, KNOWLES JA & SHI Y 2018. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct Funct, 223, 2841–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AZADBAKHT H, PARKES LM, HAROON HA, AUGATH M, LOGOTHETIS NK, DE CRESPIGNY A, D’ARCEUIL HE & PARKER GJ 2015. Validation of High-Resolution Tractography Against In Vivo Tracing in the Macaque Visual Cortex. Cereb Cortex, 25, 4299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BADEA A, GEWALT S, AVANTS BB, COOK JJ & JOHNSON GA 2012. Quantitative mouse brain phenotyping based on single and multispectral MR protocols. Neuroimage, 63, 1633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASSER PJ, MATTIELLO J & LEBIHAN D 1994. MR diffusion tensor spectroscopy and imaging. Biophys J, 66, 259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASSER PJ & PIERPAOLI C 1996. Microstructural and physiological features of tissues elucidated by quantitative diffusion tensor MRI. J. Magn. Reson. B, 111, 209–219. [DOI] [PubMed] [Google Scholar]
- BOYKO OB, ALSTON SR, FULLER GN, HULETTE CM, JOHNSON GA & BURGER PC 1994. Utility of postmortem magnetic resonance (MR) imaging in clinical neuropathology. Archives of Pathology & Laboratory Medicine, 118, 219–25. [PubMed] [Google Scholar]
- CALABRESE E, BADEA A, COE CL, LUBACH GR, SHI Y, STYNER MA & JOHNSON GA 2015a. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage, 117, 408–416. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALABRESE E, BADEA A, COE CL, LUBACH GR, STYNER MA & JOHNSON GA 2014. Investigating the tradeoffs between spatial resolution and diffusion sampling for brain mapping with diffusion tractography: time well spent? Hum Brain Mapp., 35, 5667–85. doi: 10.1002/hbm.22578. Epub 2014 July 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALABRESE E, BADEA A, COFER G, QI Y & JOHNSON GA 2015b. A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb Cortex, 25, 4628–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN H, ZHAO Y, ZHANG T, ZHANG H, KUANG H, LI M, TSIEN JZ & LIU T 2014. Construct and assess multimodal mouse brain connectomes via joint modeling of multi-scale DTI and neuron tracer data. Medical image computing and computer-assisted intervention : MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention, 17, 273–280. [DOI] [PubMed] [Google Scholar]
- D’ARCEUIL H & DE CRESPIGNY A 2007. The effects of brain tissue decomposition on diffusion tensor imaging and tractography. Neuroimage, 36, 64–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONG H-W 2008. The Allen reference atlas: A digital color brain atlas of the C57Bl/6J male mouse., John Wiley & Sons Inc. [Google Scholar]
- ELLEGOOD J, BABINEAU BA, HENKELMAN RM, LERCH JP & CRAWLEY JN 2013. Neuroanatomical analysis of the BTBR mouse model of autism using magnetic resonance imaging and diffusion tensor imaging. Neuroimage, 70, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EVANS AC, COLLINS DL, MILLS SR, BROWN ED, KELLY RL, PETERS TM 1993. 3D statistical neuroanatomical models from 305 MRI volumes. Proc. IEEE-Nuclear Science Symposium and Medical Imaging Conference, 1813–1817. [Google Scholar]
- FAN Q, WITZEL T, NUMMENMAA A, VAN DIJK KR, VAN HORN JD, DREWS MK, SOMERVILLE LH, SHERIDAN MA, SANTILLANA RM, SNYDER J, HEDDEN T, SHAW EE, HOLLINSHEAD MO, RENVALL V, ZANZONICO R, KEIL B, CAULEY S, POLIMENI JR, TISDALL D, BUCKNER RL, WEDEEN VJ, WALD LL, TOGA AW & ROSEN BR 2016. MGH-USC Human Connectome Project datasets with ultra-high b-value diffusion MRI. Neuroimage, 124, 1108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERCULANO-HOUZEL S 2009. The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci, 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HSU JC, JOHNSON GA, SMITH WM, REIMER KA & IDEKER RE 1994. Magnetic resonance imaging of chronic myocardial infarcts in formalin-fixed human autopsy hearts. Circulation, 89, 2133–40. [DOI] [PubMed] [Google Scholar]
- HUKKANEN V & ROYTTA M 1987. Autolytic changes of human white matter: an electron microscopic and electrophoretic study. Exp Mol Pathol, 46, 31–9. [DOI] [PubMed] [Google Scholar]
- HURLSTON SE, COFER GP & JOHNSON GA 1997. Optimized receiver coils for increased SNR in MR Microscopy. The International Journal of Imaging Systems and Technology, 8, 277–284. [Google Scholar]
- JACK CR JR., BARNES J, BERNSTEIN MA, BOROWSKI BJ, BREWER J, CLEGG S, DALE AM, CARMICHAEL O, CHING C, DECARLI C, DESIKAN RS, FENNEMA-NOTESTINE C, FJELL AM, FLETCHER E, FOX NC, GUNTER J, GUTMAN BA, HOLLAND D, HUA X, INSEL P, KANTARCI K, KILLIANY RJ, KRUEGER G, LEUNG KK, MACKIN S, MAILLARD P, MALONE IB, MATTSSON N, MCEVOY L, MODAT M, MUELLER S, NOSHENY R, OURSELIN S, SCHUFF N, SENJEM ML, SIMONSON A, THOMPSON PM, RETTMANN D, VEMURI P, WALHOVD K, ZHAO Y, ZUK S & WEINER M 2015. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement, 11, 740–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACK CR JR., BERNSTEIN MA, FOX NC, THOMPSON P, ALEXANDER G, HARVEY D, BOROWSKI B, BRITSON PJ, J LW, WARD C, DALE AM, FELMLEE JP, GUNTER JL, HILL DL, KILLIANY R, SCHUFF N, FOX-BOSETTI S, LIN C, STUDHOLME C, DECARLI CS, KRUEGER G, WARD HA, METZGER G,J, SCOTT KT, MALLOZZI R, BLEZEK D, LEVY J, DEBBINS JP, FLEISHER AS, ALBERT M, GREEN R, BARTZOKIS G, GLOVER G, MUGLER J & WEINER MW 2008. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging, 27, 685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG Y & JOHNSON GA 2009. Microscopic diffusion tensor imaging of the mouse brain. Neuroimage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG Y & JOHNSON GA 2011. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage, 56, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON GA, ALI-SHARIEF A, BADEA A, BRANDENBURG J, COFER G, FUBARA B, GEWALT S, HEDLUND LW & UPCHURCH L 2007. High-throughput morphologic phenotyping of the mouse brain with magnetic resonance histology. NeuroImage, 37, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON GA, BADEA A, BRANDENBURG J, COFER G, FUBARA B, LIU S & NISSANOV J 2010. Waxholm Space: An image-based reference for coordinating mouse brain research. Neuroimage, 53, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON GA, BENVENISTE H, BLACK RD, HEDLUND LW, MARONPOT RR & SMITH BR 1993. Histology by magnetic resonance microscopy. Magnetic Resonance Quarterly, 9, 1–30. [PubMed] [Google Scholar]
- JOHNSON GA, COFER GP, FUBARA B, GEWALT SL, HEDLUND LW & MARONPOT RR 2002. Magnetic resonance histology for morphologic phenotyping. Journal of Magnetic Resonance Imaging, 16, 423–429. [DOI] [PubMed] [Google Scholar]
- JOHNSON GA & HEDLUND LW 2000. Three-dimensional morphology by magnetic resonance imaging. U.S.A. patent application 09/35,424 February 8, 2000.
- JONES D, JONES D, HORSFIELD M, HORSFIELD M, SIMMONS A & SIMMONS A 1999. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magnetic Resonance in Medicine. [PubMed] [Google Scholar]
- JONES DK 2004. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: A Monte Carlo study. Magnetic Resonance in Medicine, 51, 807–815. [DOI] [PubMed] [Google Scholar]
- JONES DK & CERCIGNANI M 2010. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR in Biomedicine, 23, 803–820. [DOI] [PubMed] [Google Scholar]
- JONES DK, KNÖSCHE TR & TURNER R 2013. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. NeuroImage, 73, 239–254. [DOI] [PubMed] [Google Scholar]
- KAUPPI K, FAN CC, MCEVOY LK, HOLLAND D, TAN CH, CHEN CH, ANDREASSEN OA, DESIKAN RS, DALE AM & ALZHEIMER’S DISEASE NEUROIMAGING I 2018. Combining Polygenic Hazard Score With Volumetric MRI and Cognitive Measures Improves Prediction of Progression From Mild Cognitive Impairment to Alzheimer’s Disease. Front Neurosci, 12, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOCHUNOV P, JAHANSHAD N, MARCUS D, WINKLER A, SPROOTEN E, NICHOLS TE, WRIGHT SN, HONG LE, PATEL B, BEHRENS T, JBABDI S, ANDERSSON J, LENGLET C, YACOUB E, MOELLER S, AUERBACH E, UGURBIL K, SOTIROPOULOS SN, BROUWER RM, LANDMAN B, LEMAITRE H, DEN BRABER A, ZWIERS MP, RITCHIE S, VAN HULZEN K, ALMASY L, CURRAN J, DEZUBICARAY GI, DUGGIRALA R, FOX P, MARTIN NG, MCMAHON KL, MITCHELL B, OLVERA RL, PETERSON C, STARR J, SUSSMANN J, WARDLAW J, WRIGHT M, BOOMSMA DI, KAHN R, DE GEUS EJC, WILLIAMSON DE, HARIRI A, VAN’TENT D, BASTIN ME, MCINTOSH A, DEARY IJ, POL HEH, BLANGERO J, THOMPSON PM, GLAHN DC & VAN ESSEN DC 2015. Heritability of fractional anisotropy in human white matter: A comparison of Human Connectome Project and ENIGMA-DTI data. NeuroImage, 111, 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUSTIG M, DONOHO D, PAULY JM 2007. Sparse MRI: The Application of Compressed Sensing for Rapid MR Imaging. Magnetic Reson in Med, 58, 1182–1195. [DOI] [PubMed] [Google Scholar]
- Maier-Hein KH, NEHER PF, HOUDE J-C, CÔTÉ M-A, GARYFALLIDIS E, ZHONG J, CHAMBERLAND M, YEH F-C, LIN YC, JI Q, REDDICK WE, GLASS JO, CHEN DQ, FENG Y, GAO C, WU Y, MA J, RENJIE H, LI Q, WESTIN C-F, DESLAURIERSGAUTHIER S, GONZALEZ JOO, PAQUETTE M, ST-JEAN S, GIRARD G, RHEAULT F, SIDHU J, TAX CMW, GUO F, MESRI HY, DAVID S, FROELING M, HEEMSKERK AM, LEEMANS A, BORÉ A, PINSARD B, BEDETTI C, DESROSIERS M, BRAMBATI S, DOYON J, SARICA A, VASTA R, CERASA A, QUATTRONE A, YEATMAN J, KHAN AR, HODGES W, ALEXANDER S, ROMASCANO D, BARAKOVIC M, AURIA A, ESTEBAN O, LEMKADDEM A, THIRAN J-P, CETINGUL HE, ODRY BL, MAILHE B, NADAR MS, PIZZAGALLI F, PRASAD G, VILLALON-REINA JE, GALVIS J, THOMPSON PM, REQUEJO FDS, LAGUNA PL, LACERDA LM, BARRETT R, DELL’ACQUA F, CATANI M, PETIT L, CARUYER E, DADUCCI A, DYRBY TB, HOLLAND-LETZ T, HILGETAG CC, STIELTJES B & DESCOTEAUX M 2017. The challenge of mapping the human connectome based on diffusion tractography. Nature Communications, 8, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCNAB J, EDLOW BL, WITZEL T, et al. The Human Connectome Project and beyond: Initial applications of 300 mT/m gradients. Neuroimage, 80, 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEYZA KZ & BLANCHARD DC 2017. The BTBR mouse model of idiopathic autism - Current view on mechanisms. Neurosci Biobehav Rev, 76, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH S, HARRIS JA, NG L, et al. 2014. A mesoscale connectome of the mouse brain. Nature, 508, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAXINOS G, FRANKLIN KBJ 2012. The Mouse Brain in Stereotaxic Coordinates, New York, Academic Press. [Google Scholar]
- SCATTONI ML, GANDHY SU, RICCERI L & CRAWLEY JN 2008. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One, 3, e3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEJSKAL EO & TANNER JE 1963. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. Journal of Chemical Physics, 42. [Google Scholar]
- THOMAS C, YE FQ, IRFANOGLU MO, MODI P, SALEEM KS, LEOPOLD DA & PIERPAOLI C 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proceedings of the National Academy of Sciences of the United States of America, 111, 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMPSON PM, ANDREASSEN OA, ARIAS-VASQUEZ A, BEARDEN CE, BOEDHOE PS, BROUWER RM, BUCKNER RL, BUITELAAR JK, BULAYEVA KB, CANNON DM, COHEN RA, CONROD PJ, DALE AM, DEARY IJ, DENNIS EL, DE REUS MA, DESRIVIERES S, DIMA D, DONOHOE G, FISHER SE, FOUCHE JP, FRANCKS C, FRANGOU S, FRANKE B, GANJGAHI H, GARAVAN H, GLAHN DC, GRABE HJ, GUADALUPE T, GUTMAN BA, HASHIMOTO R, HIBAR DP, HOLLAND D, HOOGMAN M, POL HEH, HOSTEN N, JAHANSHAD N, KELLY S, KOCHUNOV P, KREMEN WS, LEE PH, MACKEY S, MARTIN NG, MAZOYER B, MCDONALD C, MEDLAND SE, MOREY RA, NICHOLS TE, PAUS T, PAUSOVA Z, SCHMAAL L, SCHUMANN G, SHEN L, SISODIYA SM, SMIT DJA, SMOLLER JW, STEIN DJ, STEIN JL, TORO R, TURNER JA, VAN DEN HEUVEL MP, VAN DEN HEUVEL OL, VAN ERP TGM, VAN ROOIJ D, VELTMAN DJ, WALTER H, WANG Y, WARDLAW JM, WHELAN CD, WRIGHT MJ, YE J & CONSORTIUM E 2017. ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide. Neuroimage, 145, 389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMPSON PM, GE T, GLAHN DC, JAHANSHAD N & NICHOLS TE 2013. Genetics of the connectome. Neuroimage, 80, 475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOURNIER JD, CALAMANTE F & CONNELLY A 2013. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed, 26, 1775–86. [DOI] [PubMed] [Google Scholar]
- TOURNIER JD, CALAMANTE F, GADIAN DG & CONNELLY A 2004. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage, 23, 1176–1185. [DOI] [PubMed] [Google Scholar]
- TUCH DS 2004. Q-ball imaging. Magnetic Resonance in Medicine, 52, 1358–1372. [DOI] [PubMed] [Google Scholar]
- TUCH DS, REESE TG, WIEGELL MR, MAKRIS N, BELLIVEAU JW & WEDEEN VJ 2002. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med, 48, 577–82. [DOI] [PubMed] [Google Scholar]
- UGURBIL K, XU J, AUERBACH EJ, MOELLER S, VU AT, DUARTE-CARVAJALINO JM, LENGLET C, WU X, SCHMITTER S, VAN DE MOORTELE PF, STRUPP J, SAPIRO G,DE MARTINO F, WANG D, HAREL N, GARWOOD M, CHEN L, FEINBERG DA, SMITH SM, MILLER KL, SOTIROPOULOS SN, JBABDI S, ANDERSSON JL, BEHRENS TE, GLASSER MF, VAN ESSEN DC, YACOUB E & CONSORTIUM WU-MH 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage, 80, 80–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ULLMANN JF, WATSON C, JANKE AL, KURNIAWAN ND & REUTENS DC 2013. A segmentation protocol and MRI atlas of the C57BL/6J mouse neocortex. Neuroimage, 78, 196–203. [DOI] [PubMed] [Google Scholar]
- VAN ESSEN DC, SMITH SM, BARCH DM, BEHRENS TE, YACOUB E, UGURBIL K & CONSORTIUM WU-MH 2013. The WU-Minn Human Connectome Project: an overview. Neuroimage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOS SB, AKSOY M, HAN Z, HOLDSWORTH SJ, MACLAREN J, VIERGEVER MA, LEEMANS A & BAMMER R 2016. Trade-off between angular and spatial resolutions in in vivo fiber tractography. Neuroimage, 129, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG N, ANDERSON RJ, BADEA A, COFER G, DIBB R, QI Y, JOHNSON GA 2018. Structural connectomics of the mouse brain using magnetic resonance histology. Brain Struct Funct, [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEH F, WEDEEN VJ, TSENG WYI 2010. Generalized q-sampling imaging. IEEE Transactions on Medical Imaging, 29, 1626–1635. [DOI] [PubMed] [Google Scholar]
- YEH F-C, VERSTYNEN TD, WANG Y, FERNÁNDEZ-MIRANDA JC & TSENG W-YI 2013. Deterministic Diffusion Fiber Tracking Improved by Quantitative Anisotropy. PLoS ONE, 8, e80713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEH FC, BADRE D & VERSTYNEN T 2016. Connectometry: A statistical approach harnessing the analytical potential of the local connectome. Neuroimage, 125, 162–171. [DOI] [PubMed] [Google Scholar]
- ZHAN L, JAHANSHAD N, ENNIS DB, JIN Y, BERNSTEIN MA, BOROWSKI BJ, JACK CR, TOGA AW, LEOW AD & THOMPSON PM 2013. Angular versus spatial resolution trade-offs for diffusion imaging under time constraints. Human brain mapping, 34, 2688–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG J, VAN ZIJL PC & MORI S 2002. Three-dimensional diffusion tensor magnetic resonance microimaging of adult mouse brain and hippocampus. Neuroimage, 15, 892–901. [DOI] [PubMed] [Google Scholar]