Abstract
While chemical analysis of contaminant mixtures remains an essential component of environmental monitoring, bioactivity-based assessments using in vitro systems increasingly are used in the detection of biological effects. Historically, in vitro assessments focused on a few biological pathways, e.g., aryl hydrocarbon receptor (AhR) or estrogen receptor (ER) activities. High-throughput screening (HTS) technologies have greatly increased the number of biological targets and processes that can be rapidly assessed. Here we screened extracts of surface waters from a nationwide survey of United States (US) streams for bioactivities associated with 69 different endpoints using two multiplexed HTS assays. Bioactivity of extracts from 38streams was evaluated and compared with concentrations of over 700 analytes to identify chemicals contributing to observed effects. Eleven primary biological endpoints were detected. Pregnane X receptor and AhR-mediated activities were the most commonly detected. Measured chemicals did not completely account for AhR and PXR responses. Surface waters with AhR and PXR effects were associated with low intensity, developed land cover. Likewise, elevated bioactivities frequently associated with wastewater discharges included endocrine-related endpoints— ER and glucocorticoid receptor. These results underscore the value of bioassay-based monitoring of environmental mixtures for detecting biological effects that could not be ascertained solely through chemical analyses.
Graphical Abstract
INTRODUCTION
Increased usage of organic chemicals in agricultural, industrial, and personal care applications has intensified concerns that aquatic organisms may be exposed to potentially hazardous mixtures of chemical contaminants. Compounds for which ecological or human health hazards are generally not well understood, which have recently been discovered in the environment, or for which public attention has greatly increased have been termed contaminants of emerging concern (CECs) and include pharmaceuticals, personal care products, natural and synthetic steroids, pesticide metabolites, persistent organics (polybrominated and perfluoro compounds), flame retardants, and plasticizers. Multiple studies since the early 2000s have demonstrated the complexity and near-ubiquitous presence of CEC mixtures in surface waters.1–3 However, even recent, dramatically expanded chemical assessments provide only partial coverage of the presumptive surface water contaminant space.4 Combined analytical chemistry methods and in vitro bioassay approaches support improved characterization of pathway-specific bioactivities and the potential hazard of complex mixtures of CECs to ecological receptors.5–7 In vitro bioassays covering a range of processes (genotoxicity,8, 9 inflammation,9 mutagenicity,8, 10 oxidative stress,9–11 photosynthetic processes10) or individual receptor-based bioactivities (reflecting androgen,9, 12, 13 aryl hydrocarbon,8, 9, 14, 15 estrogen,9, 16–18 glucocorticoid,8, 9, 19 peroxisome-proliferator,8, 9, 14 pregnane X,14, 15 progesterone,8, 9, 20 retinoic acid,9 retinoid X,9 and thyroid8, 14 signaling pathways) have been used, at least to some extent, to assess surface waters.
The Tox21 collaboration and the U.S. Environmental Protection Agency (USEPA) ToxCast programs began in 2007–2008 with the aim of advancing toxicity testing through use of in vitro high-throughput screening (HTS) assays that can rapidly measure chemical effects on hundreds of different biological pathways/processes.21 These programs generated publicly available bioactivity data for over 9,000 unique substances, including many CECs.22, 23 Recent approaches have leveraged ToxCast data to screen for potential pathway-specific effects of detected compounds in environmental samples using hazard quotients based on bioactivity.9, 24, 25 Such computational approaches are readily applied to detected environmental contaminants but are likewise constrained by extant analytical chemistry techniques and poor understanding of complex, interactive effects. Alternatively, HTS assays can be applied directly to surface water samples or extracts to screen for net effects of contaminant mixtures on targeted biological endpoints.
The present study is among a growing number of studies to apply a subset of ToxCast HTS assays to directly assess the biological effects potential of environmental samples.9, 14, 26, 27 Specifically, the U.S. Geological Survey (USGS) and USEPA conducted a collaborative investigation of both chemical occurrence and bioactivity in streams across the continental US and Puerto Rico.28 The target-chemical coverage included over 700 unique organic analytes from many functional classes (pesticides, wastewater indicators, steroid hormones, volatile organics, pharmaceuticals, and halogenated organics).29 Initial assessment of split-sample bioactivity targeted in vitro transcriptional reporter assays for the estrogen receptor (ER), androgen receptor (AR), and glucocorticoid receptor (GR).30 Here, we expand the characterization of split-sample bioactivity using two multiplexed HTS assays (Attagene cis-Factorial and trans-Factorial assays)31, 32 that encompass 69 biological endpoints. The selected HTS assays had been used in the ToxCast program to screen over 3600 compounds, providing a robust data set to compare against known chemical composition of the samples.31 Bioactivity results were compared with measured concentrations of compounds in the samples using existing HTS data for the individual chemicals to assess how well target-chemical analysis captured compounds responsible for observed bioactivities. Additional statistical analyses were then performed to identify potential relationships between bioactivity measures and chemicals detected in the test samples. Finally, ER, AR, and GR results from the HTS assays and from the previously reported in vitro assays30 were compared to assess the performance of the HTS system for routine environmental monitoring. Results of this work intended to identify the most prevalent bioactivities (among the 69 endpoints monitored), important contaminant and land-use drivers of bioactivity, and potential adverse ecological effects of CECs in streams across the US.
MATERIALS AND METHODS
Sample Collection and Processing
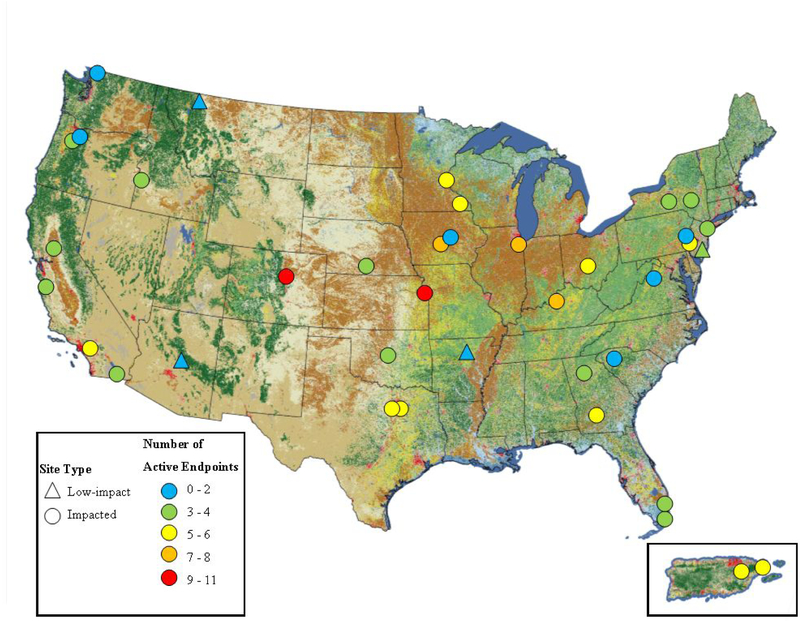
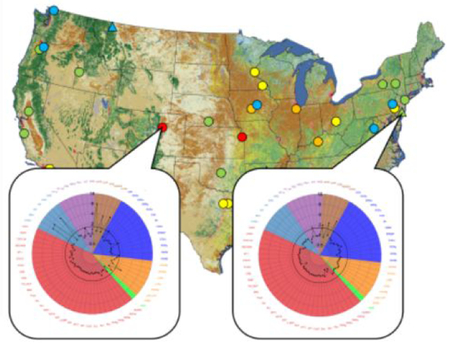
Surface water samples were collected from a total of 38 sites (4 low-impact sites – those with minimal anthropogenic inputs; 34 impacted sites – those with increased anthropogenic inputs)29 between December 2012 and June 2014 (Figure 1; Supporting Information [SI] Table S1). Sites were selected to cover a range of land use including largely undeveloped, agricultural, and heavily urban developed watersheds. Water from each site was collected into a Teflon churn prior to splitting between prepared sample bottles according to established trace-level sampling methods.29 Samples were shipped on ice to USGS and USEPA laboratories for chemical or bioassay analysis. Upon arrival at the USEPA-Duluth laboratory, samples in 1 L amber glass bottles were logged and stored at 4°C for no more than 24 h prior to filtration and extraction.
Figure 1.
Site location, classification, and number of active (AUC ≥1.5-fold above extract blank) bioassay endpoints for each site sampled from 2012–2014. Only endpoints tested across all 38 sites are included. See Table S1 for site details and Table S4 for bioassay response values.
Sample Preparation
All solvents were HPLC grade or better and purchased from Fisher Scientific (Waltham, MA, USA). Dimethyl sulfoxide (DMSO) was >99.7% purity and purchased from Sigma-Aldrich (St. Louis, MO, USA). Water samples were processed by solid-phase extraction (SPE) using Waters Oasis HLB cartridges (200 mg sorbent, 5 mL glass cartridge; Milford, MA, USA). Field blanks were not received for this bioassay analysis, but an extraction method blank (HPLC grade water) was generated with each sample batch and used to assess background activity in the method. Cartridges were preconditioned sequentially using 5 mL each of ethyl acetate, 50:50 methanol (MeOH):dichloromethane (DCM), 100% MeOH, and, finally, ultrapure water. Prior to SPE, approximately 1 L of whole water was filtered through a baked 47 mm glass-fiber filter (0.7 μm porosity). After filtration, 0.5 L of each sample was loaded onto preconditioned cartridges. Cartridges were aspirated for approximately 30 min to remove residual water then eluted sequentially with 6 mL of MeOH followed by 6 mL of 50:50 MeOH:DCM. Extracts were evaporated to dryness under a gentle stream of nitrogen at 30°C. Samples were reconstituted in 500 μL of DMSO for a final 1000-fold relative enrichment factor (REF). Samples were extracted as they were received and sent for analysis in three batches ranging 8 – 18 samples per batch. Extracts were frozen at −20°C and shipped overnight on ice for bioassay analysis.
HTS Bioassays
Sample extracts were tested by Attagene, Inc. (Morrisville, NC, USA) under contract to the USEPA, using the cis-Factorial and trans-Factorial assays.31, 32 Each assay uses HepG2 cells to assess transcription factor activity (cis-Factorial) or transfected nuclear receptor activity (trans-Factorial). Together, these multiplexed assays comprise a total of 69 target endpoints covering a range of biological pathways including xenobiotic metabolism (e.g., aryl hydrocarbon receptor [AhR], pregnane X receptor [PXR]), lipid metabolism (peroxisome proliferator-activated receptor [PPAR],) endocrine signaling (e.g., AR, ER, GR), and cellular stress (e.g., antioxidant response element [NRF2]; SI Table S2). Additionally, a subset of eight samples (the initial eight samples screened through the assays) was analyzed using a separate assay (trans-Factorial-2), which covers 24 additional nuclear receptors beyond the 69 targets from the cis-Factorial and trans-Factorial assays (SI Table S2). Sample extracts were tested at six dilutions from a maximum REF of 10 followed by five, 3-fold serial dilutions (i.e., 10, 3.3, 1.1, 0.37, 0.12, 0.04 REF). The highest REF was tested at 1% DMSO, while all remaining dilutions were at 0.33% DMSO. Positive control compounds for several of the targets (e.g., E2 for ER) were run in triplicate at a single concentration with each batch (see Table S2), and six replicates of DMSO solvent controls (both 1.0 and 0.33%) were run with each sample set. Positive controls inducing responses >2 fold passed quality assurance criteria
For the cis-Factorial assay, a set of cis-Factorial reporters as well as PXR and ER-alpha expression vectors were transiently transfected into HepG2 cells (12-well plates) using TransIT-LT1 (Mirus Bio; Madison, WI, USA) transfection reagent and according to the manufacturer’s protocol. The next day, the transfected cells were supplied with 1% charcoal-stripped HyClone FBS (GE Healthcare Life Sciences; Marlborough, MA, USA) and incubated with sample extracts for 24 h. For the trans-Factorial assay, each of the trans-Factorial reporter GAL4-NR expression vector pairs were transiently transfected into separate pools of HepG2 cells using TransIT-LT1 transfection reagent and according to the manufacturer’s protocol. The next day, the transfected cells were mixed and plated into 12-well plates for compound analysis. Twenty-four h after plating, the transfected cells were supplied with 1% charcoal-stripped HyClone FBS and incubated with evaluated compounds for 24 h. For both assays, cell viability was measured in parallel using XTT Cell Proliferation Assay Kit (ATCC; Manassas, CT, USA) with cell viability >70% required to generate acceptable data quality. A standard set of six control reporters were used to normalize response of the transfected endpoints. To assess Factorial reporter activity, total RNA was isolated using the PureLink RNA isolation kit (Invitrogen; Carlsbad, CA, USA). Total RNA from each well was reversely transcribed into cDNA, and reporter cDNA was amplified in a single PCR reaction using a pair of common primers. The PCR products were labeled using a 6-Fam-labeled primer and digested with HpaI. The resulting labeled DNA fragments were separated using capillary electrophoresis (Genetic Analyzer 3130xl; Applied Biosystems; Waltham, MA, USA). The relative activities of endpoints were calculated from the electropherogram peaks using proprietary software.29
Statistical Analysis
Bioassay responses were expressed as fold-change relative to DMSO control by dividing expression of treated cells by that of appropriate DMSO control. The use of fold-change relative to DMSO controls does influence the potential magnitude of response for each endpoint, so the relative magnitude of response across different endpoints should not be directly compared. Dose-response data were plotted and response calculated as the area under the curve (AUC) using GraphPad Prism (v5.02). Other metrics could have been used, such as a more traditional EC50. However, since many endpoints are lacking positive controls, AUC allowed for consideration of the full dose-response relative to extraction method blanks, which could be applied consistently for all endpoints regardless of a positive control. To further correct response across multiple assay batches, sample AUC was divided by the mean extraction method blank (n=1–3 per batch) AUC within each sample batch. A cutoff of 1.5-fold response (50% increase) relative to the blank baseline was used to identify endpoints clearly responding to environmental extracts. Pearson correlation was used to assess relations between blank-corrected AUC responses, exposure-activity ratios (EARs),25 or land use metrics. A nonparametric analysis of ranked, normalized AUC responses and bioanalytical equivalent concentrations (i.e. 17β-estradiol equivalents [E2Eqs, ng L−1]), previously reported by Conley et al.,30 was performed using Spearman Rank correlation.
Exposure-activity ratio (EAR) analysis25 was used to estimate whether bioactivity would be expected based on the measured concentrations of detected compounds29 and their corresponding activity concentration at cutoff (ACC) values for Attagene assays, where available (i.e., for compounds previously tested in ToxCast).22 The ACC was chosen as a metric of chemical potency as this value compares chemical response with a defined baseline and is independent of chemical efficacy, though other metrics of potency, such as the half-maximal activity concentration (AC50), could be used. An EAR value was calculated for each site and assay by summing EAR values of individual compounds (Eq. 1) detected in a given mixture, thus assuming non-interactive concentration addition. Theoretically, EAR values ≥1 indicate measured compounds are at a concentration expected to produce bioactivity for a given endpoint. Comparison of this predicted bioactivity with that observed in the Attagene bioassay can be used to infer whether the known composition of the sample likely accounts for the activity observed, or whether unidentified compounds may be contributing. Likewise, if observed bioactivity is less than predicted, the results may suggest non-additive interactions are occurring between compounds present in the mixture.
| (Eq. 1) |
To identify other potential associations between chemical occurrence and bioassay responses, a sparse partial least squares (SPLS) analysis33 was performed to identify correlations between measured chemicals and bioassay response in order to identify potentially bioactive compounds. The SPLS analysis is a partial least squares analysis with the addition of elastic net regulation, which allows for the removal of variables.34 Analysis was performed using the SPLS package35 in R version 3.3.3.36 After SPLS analysis was performed, bootstrapped confidence intervals (also using the SPLS package35) were estimated for each chemical-assay combination, and those with intervals that contained zero were removed.
Prior to SPLS analysis, the full chemical dataset37 was filtered to remove chemicals with no detections above the reporting limit, chemicals with zero variance in concentration, or chemicals not reported at three or more sites. Both chemical concentration and assay AUC were then centered and scaled across sites. For analytes with missing values at two or fewer sites, Multiple Imputation by Chained Equations (MICE)38 was used to generate missing values for SPLS analysis using the MICE R package.39 Finally, based on prior knowledge of analyte loss during the evaporation step of extract preparation (see SI for more information; Figure S1, Table S3), chemicals with vapor pressure above 0.01 mmHg (as calculated using EpiSuite v4.1140) were removed from the SPLS dataset. These more volatile compounds are lost during evaporation and solvent exchange and are not expected to be present in the final bioassay extract. A total of 386 analytes and 69 assay endpoints were used for SPLS analysis.
RESULTS AND DISCUSSION
Bioactivity in surface water extracts
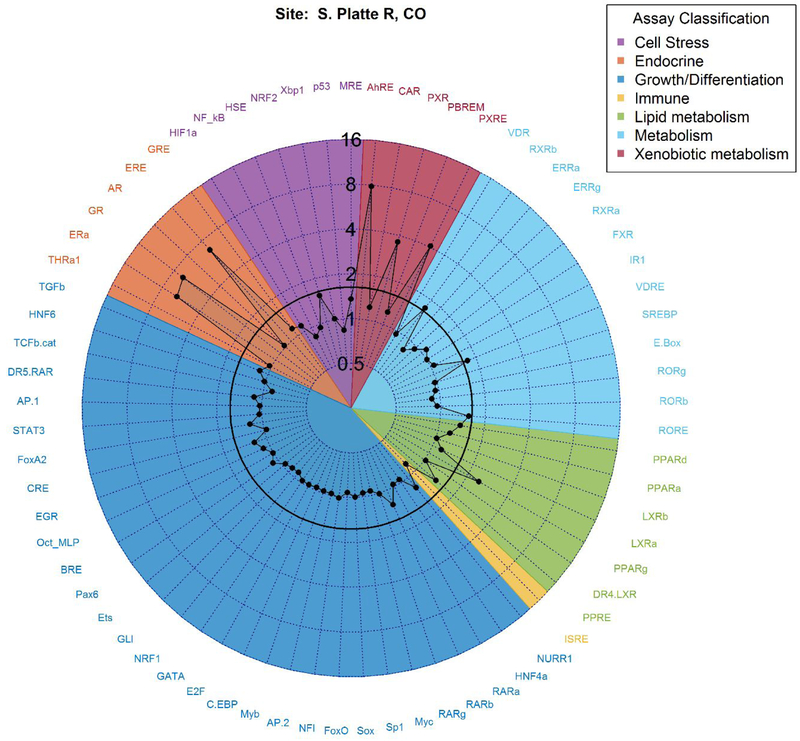
Activity ≥1.5-fold above baseline (hereafter referred to as ‘active’) was detected in the cis-Factorial and trans-Factorial assays at two or more sites for 11 endpoints (Table S4). The site with the greatest number of active endpoints of any sampled site (South Platte River, CO), is shown in Figure 2 while all remaining sites are provided in the SI (Figures S2–S38). The 11 active endpoints reflected nine different receptor-mediated pathways. In the subset of eight samples that were analyzed using the trans-Factorial-2 assay, three more endpoints (ERβ_TRANS2, PR_TRANS2, MR_TRANS2) were active (Figure S39, Table S4). Most active endpoints are associated with regulation of metabolism of xenobiotic (e.g., AhR_CIS, PXR_TRANS, PXRE_CIS) or endogenous compounds (PPARγ_TRANS, RORE_CIS, RXRβ_TRANS, VDRE_CIS) and one endpoint (NRF2_CIS) is indicative of oxidative stress. The remaining endpoints (ERα_TRANS, ERE_CIS, and GR_TRANS) and the endpoints active from the subset of samples analyzed through the trans-Factorial-2 assay (ERβ_TRANS2, MR_TRANS2, PR_TRANS2) are associated with different endocrine pathways.
Figure 2.
An example radar plot of blank-normalized area under the curve (AUC) values of both cis_Factorial and trans_Factorial endpoints for the South Platte River, CO site. Assay endpoint names correspond to those in Table S2 and are grouped in colors by general function. The radar axis represents AUC values for each measured endpoint. The activity cutoff value of 1.5 is shown in bold, and activities at or above this value were identified as active.
Several of these receptor-based pathways have well-defined ligands known to induce activity. For example, AhR ligands include polycyclic and halogenated hydrocarbons including many current use pesticides;41 AhR activity has previously been observed in different studies with environmental samples (often through induction of CYP1A1).9, 10, 42 PXR is a relatively promiscuous receptor with a large, hydrophobic binding pocket capable of interacting with a broad range of organic compounds.43 PPARγ ligands include endogenous fatty acids, prostaglandins, and thiazolidinedione pharmaceuticals (e.g., rosiglitazone, pioglitazone). Comparatively few ligands have been identified for the retinoid X receptor (RXR), the RAR-related orphan receptor (ROR), or vitamin-D receptor (VDR) aside from the endogenous molecules such as retinoic acids or vitamin-D3. NRF2 activity is associated with oxidative stress and can be induced by a wide range of compounds, such as reactive oxygen species and electrophilic chemicals. Activation of NRF2 has been previously observed in environmental samples, but specific compounds responsible for activity were not identified.11 Progesterone receptor (PR), GR and ER activation have previously been documented in environmental samples26 and several known natural (e.g., estradiol, cortisol, progesterone) and synthetic (e.g., 17α-ethinylestradiol, dexamethasone, levonorgestrel) ligands detected in surface waters. Mineralocorticoid receptor (MR) activity, which was observed at one of eight tested sites, has not been commonly reported in surface waters. The MR is known to bind two endogenous classes of corticosteroid ligands; the mineralocorticoid aldosterone; and the glucocorticoids cortisol or corticosterone, in the human or rat, respectively.44 The trans-Factorial-2 assay, which contains the MR endpoint, is not available in the ToxCast database;22 however, another assay NVS_NR_rMR, which uses the rat MR, has been used to screen over 2700 chemicals. Twenty-one compounds were active, including 17 steroidal compounds. Those with the highest binding affinity include three glucocorticoids (corticosterone, dexamethasone, and 17α-hydroxyprogesterone), which are also active in GR-related assays. The single site with elevated MR activity induced the highest GR activity observed at any site, suggesting that the observed MR activity reflects glucocorticoids present at this site.
Distribution of Activity Across US Streams
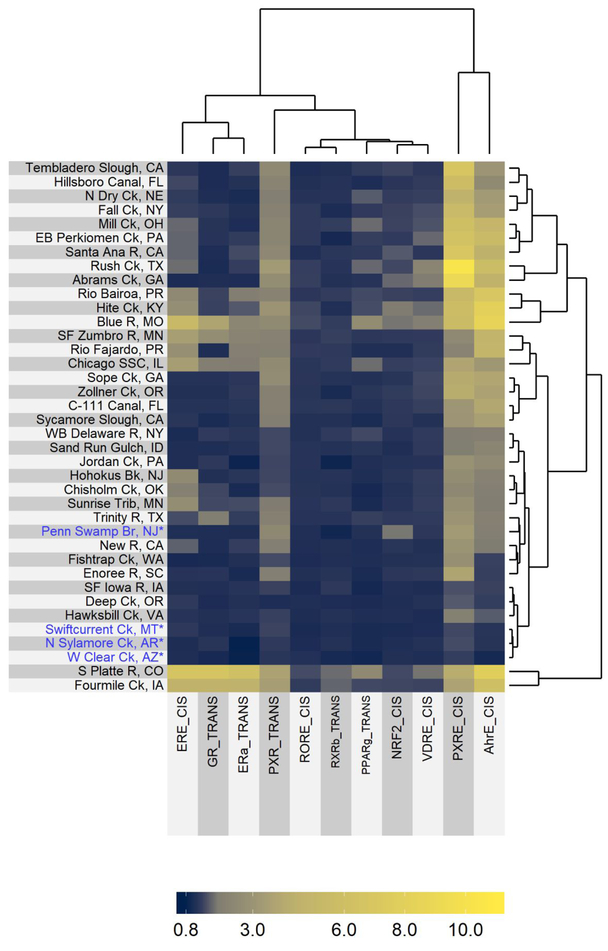
Biological activity varied widely across sites with the number of active endpoints ranging from zero to 11 (Figure 1) and the magnitude of response within individual active endpoints ranging from an AUC of 1.55 to 10.3 (Figure 3). Activation of endpoints associated with xenobiotic metabolism were the most frequently induced. Elevated PXRE_CIS, AhRE_CIS, and PXR_TRANS occurred at 36, 31, and 30 sites, respectively, demonstrating near-ubiquitous activity across surface waters. The remaining endpoints responded at fewer sites. For example, ERE_CIS was active at 17 sites, ERα_TRANS and PPARγ_TRANS at 10 sites, VDRE_CIS at eight sites, GR_TRANS at seven sites, NRF2_CIS at six sites, and RORE_CIS and RXRβ_TRANS at two sites. Among the eight samples tested using the trans-Factorial-2 assay, ERβ_TRANS2 activity was detected at two sites, while MR_TRANS2 and PR_TRANS2 activity were detected at one (Figure S39).
Figure 3.
Hierarchical clustering of blank normalized area under the curve (AUC) of site extracts for individual assay endpoints. Only endpoints with AUC response ≥1.5 at one or more sites are included. Sites noted with an asterisk (*) and in blue are low-impact sites, with limited anthropogenic inputs compared to other sites. See Table S4 for bioassay response values.
Samples from two low-impact sites (West Clear Creek, AZ; Swiftcurrent Creek, MT) did not elicit any responses above baseline. Some responses associated with xenobiotic metabolism or cell stress were detected at the two other low-impact locations (PXRE_CIS at North Sylamore Creek, AR; PXRE_CIS, AhRE_CIS, PXR_TRANS, and NRF2_CIS at Penn Swamp Branch, NJ), but neither induced any endocrine-related bioactivities, such as ER or GR (Figure 3). Penn Swamp Branch, NJ is noted as unique compared to other low-impact sites in that it drains a wetland area, is acidic (pH 4.18), and contains high dissolved organic carbon (15.9 mg L−1; Table S1).29 Natural humic acids, which would be expected to be present at high concentrations in an acidic, wetland-drained watershed, have been previously identified as activators of AhR, though at concentrations several orders of magnitude higher than TCDD, the prototypical ligand for AhR.45 Many natural product ligands have been identified for PXR, a very promiscuous xenobiotic-sensing nuclear receptor.46 Thus, the presence of natural dissolved organic compounds at the low-impact Penn Swamp Branch, NJ site (and possibly at other “impacted” sites) may contribute to increased activity observed in AhR and PXR related endpoints. If ubiquitous organic compounds are indeed a primary driving force for these endpoints, the utility of these endpoints for predicting potential adverse effects will be reduced.
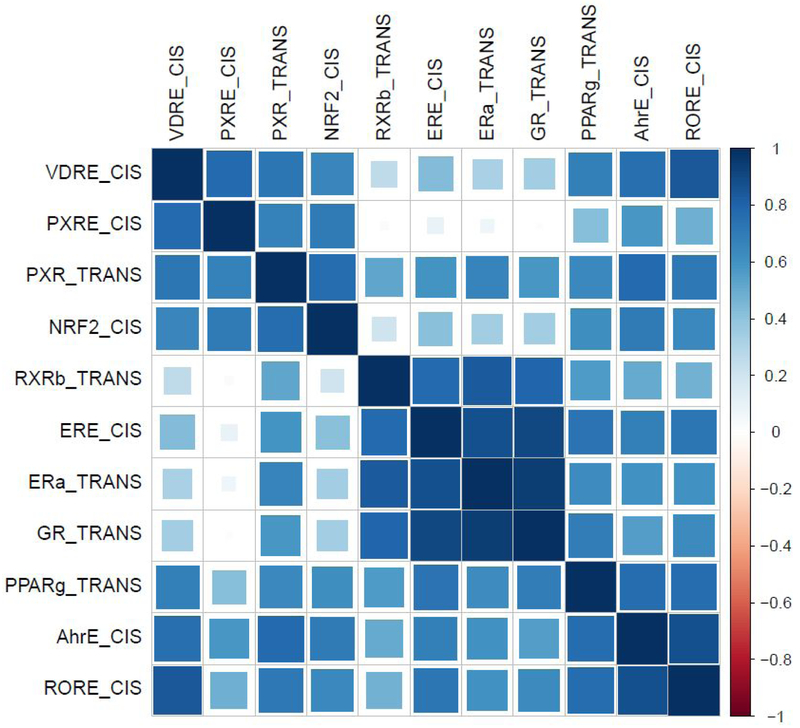
Overall, most of the impacted sites cluster closely with low-impact sites and were characterized by low to moderate PXR and AhR bioactivity and little to no ER or GR bioactivity (Figure 3). Remaining sites show moderate to high ER and GR activity and generally elevated PXR and AhR activity. The wide occurrence of elevated PXR and AhR activity suggests somewhat non-specific effects associated with a wide variety of natural and manmade ligands, which is consistent with a biological role of xenobiotic sensing and induction of metabolizing enzymes.47 Both AhR and PXR can interact with a wide range of structurally diverse compounds, thus the wide occurrence of both bioactivities is not surprising and aligns with previous studies of environmental extracts.15, 48 The promiscuity of both receptors is also reflected in the ToxCast database as AhRE_CIS and PXRE_CIS are active in 13.8% and 46.7%, respectively, of 3860 compounds tested in the cis-Factorial assay. Elevated ER and GR activity can be due to a variety of natural or synthetic steroidal compounds, and elevated activity has been previously observed in water samples associated with wastewater treatment plant (WWTP) discharges.9, 16, 49 Activation of GR_TRANS was highly correlated with ERE_CIS (p<0.0001, r=0.90; 95% CI [0.82, 0.95]) and ERα_TRANS endpoints (p<0.0001, r=0.94; 95% CI [0.90, 0.97]), and ERE_CIS was significantly correlated with PPARγ_TRANS (p<0.0001, r=0.74; 95% CI [0.54, 0.85]), consistent with a common point source of chemicals contributing to ER, GR, and PPARγ bioactivities across sampled streams (Figure 4). Because no individual detected compound was identified in the ToxCast database22 as a strong agonist of more than one of these receptors (data not shown), the observed bioactivities may be due to a mixture of compounds (as would be expected from WWTP effluents). Anecdotally, the three sites showing the greatest ER, GR, and PPARγ activity (South Platte River, CO; Blue River, MO; Fourmile Creek, IA) are sites with documented WWTP effluent sources.
Figure 4.
Correlation matrix of eleven endpoints with AUC response ≥1.5. Cell color and area of shading represent the correlation coefficient between endpoints.
Lastly, the inclusion of endpoints for the same pathway across both Attagene assays allows for a comparison across assays. Both ER- (ERE_CIS, ERα_TRANS) and PXR-related (PXRE_CIS, PXR_TRANS) endpoints from the two separate assays were well correlated, demonstrating a similar response from each individual assay (Figure 4). These two sets of endpoints are the only paired endpoints that showed consistent activation across both assays. On the contrary, the PPAR- and GR-related endpoints (PPRE_CIS, GRE_CIS) were not active at any site, while their counterparts (PPARγ_TRANS, GR_TRANS) were active at 10 and 7 sites, respectively. This demonstrates that each endpoint may have differential response across both assays.
Exposure-activity ratio analysis
An EAR analysis was performed to identify how well measured compounds explained the observed bioactivity of surface waters. Of the 406 unique compounds analytically detected at one or more sites,29 297 (73%) were tested in ToxCast, and 201 (49.5%) were tested in the Attagene HTS assays used in the current study. Where there were co-occurring chemical concentration and ToxCast bioactivity data, EAR values were calculated for each assay endpoint at each site (Table S5). An EAR value could be calculated for a total of 53 endpoints (one or more chemicals active for endpoint). Two endpoints, ERE_CIS and ERα_TRANS, had EAR values above 0.1 and ERE_CIS showed EAR values above 1.0. Four other endpoints (AhR_CIS, NRF2_CIS, PXRE_CIS, Sox_CIS) had a maximum EAR >0.01, while the remaining 47 endpoints had maximum EARs <0.006. For both ERα_TRANS and ERE_CIS, over 50 compounds contributed to some degree to the calculated EARs, but estrone, 17β-estradiol, bisphenol A, and 4-nonylphenol account for much of the EAR. At sites where it was detected above the reporting limit, estrone alone explained 55–100% and 80–100% of the total EAR for the ERα_TRANS and ERE_CIS assays, respectively. Observed bioactivity and EARs for both ERα_TRANS (p<0.0001, r=0.83; 95% CI [0.70, 0.91]) and ERE_CIS (p<0.0001, r=0.68; 95% CI [0.46, 0.82]) were also significantly correlated, indicating the measured chemicals largely account for observed ER activity. The remaining four endpoints with EARs >0.01 were not significantly correlated with observed bioactivity, indicating that measured chemicals likely do not adequately explain any other observed bioactivities.
Sparse Partial Least Squares Analysis
Sparse partial least squares (SPLS) analysis was performed to identify possible relationships between detected chemicals and bioassay responses. Through this technique, it was possible to take more chemicals into account (386 chemicals) compared to EAR calculations (201 chemicals); however, results are inferred from statistical (correlative) relationships, not empirical assay data. Among the 386 chemicals in the dataset, 48 unique compounds were identified by SPLS as predictors of bioassay responses (Table S6). Out of 70 total assay endpoints, 27 were related to observed chemical occurrence patterns. These included 10 of the 11 endpoints activated by the tested water samples (the exception was PXRE_CIS). Other than estrone, 17β-estradiol, and androstenedione, most of the compounds identified through SPLS analysis are not known to directly interact with receptors responsible for the commonly observed bioactivities (AhR, ER, GR, PXR, etc.). Many identified compounds are classified as pharmaceuticals or wastewater indicators, both of which can be broad indicators of anthropogenic waste. Two specific compounds, HHCB and triclosan, were previously shown to have a positive relationship between their concentrations and the total number of compounds detected across these same 38 sites.29 Therefore, we hypothesize, that most compounds identified by SPLS are likely not causing observed bioactivities directly but rather covary with other bioactive compounds present in these systems. Regardless, the identification of multiple pharmaceuticals and other wastewater indicators as covariates of bioeffects does provide a reduced set of targeted analytical methods to monitor as a potential surrogate of effects.
Comparison of HTS and Targeted Reporter Assays
An important attribute of the current study was the use of split samples for all chemical and biological analyses, allowing for direct comparison of data produced at different laboratories. Conley et al. measured bioactivity in these same samples using targeted transcriptional reporter assays with ER, GR and AR,30 providing an opportunity to compare the performance of the HTS assays. Three samples were lost during shipment and were not analyzed by Conley et al.30 but comparisons could be made for 35 of 38 sites presented in the current work.
Five samples analyzed by Conley et al. elicited AR-dependent responses in the MDA-kb2 assay,30 but no samples induced AR activity above baseline in the HTS assay. The MDA-kb2 assay can detect both AR or GR agonists, although the assay is ~100-fold more sensitive to AR agonists.30 One site (Blue River, MO), had detectable androgenic compounds that potentially account for AR activity in the MDA-kb2 assay; however, four of five sites inducing MDA-kb2 activity also showed elevated GR activity in both the GR transcriptional assay employed by Conley et al. and the HTS assays described herein. Overall, while it appears that the HTS Attagene assay may be less sensitive to AR agonists than the MDA-kb2 system, it remains unclear whether MDA-kb2 activity observed at most sites was actually AR- or GR-dependent.
Elevated GR activity was identified at nine sites using CV-1 cells transduced with human GR26. Using the HTS assays, seven samples were identified as active, five of which were in common with the CV-1 assay results. Conley et al. reported relative GR-mediated potency in terms of an equivalent concentration of dexamethasone (dexamethasone equivalents; DexEq), a potent GR agonist, that produces the same magnitude of bioassay response. Three sites with reported DexEq greater than 30 ng L−1 were active in the HTS assays. In contrast, for samples estimated to contain 6.0 – 30 ng DexEq L−1, only three of six samples were active. Overall, the HTS Attagene assays appear somewhat less sensitive to GR agonists compared to the CV-1 reporter assay, but the sites inducing the greatest activity were identified using both systems.
Measures of ER activity provide the most complete comparison between the Attagene HTS system and a targeted in vitro reporter assay. Using the T47D-KBluc assay,26 Conley et al. detected elevated ER activity in 34 samples, while the HTS endpoints ERE_CIS and ERα_TRANS identified 17 and 10 active samples, respectively. This again indicates a lower sensitivity to ER agonists in the HTS assays compared to the T47D-KBluc reporter gene assay. Dose-dependent responses to a 17β-estradiol positive control in the T47D-KBluc assays allowed the determination of equivalent concentration (E2Eq) values for the test samples, but positive controls for the HTS assays were not run in concentration-response, so E2Eq could not be calculated for the HTS results. However, the rank order of E2Eq concentration in the T47D-KBluc assay and normalized AUC values for both ERE_CIS and ERα_TRANS were significantly correlated (r=0.73; r=0.69, respectively; Figure S40). Thirteen of 16 sites with >0.8 ng E2Eq L−1 were active for one or both ER endpoints, demonstrating the ability of the HTS assay to identify sites with low ng L−1 E2Eq activity.
As with any in vitro system, there are strengths and weaknesses of the presented approach using the Attagene assay systems. The HTS assays utilized in the current study appear to be less sensitive overall compared to the three individual targeted in vitro assays. The NRF2 pathway, which provides a measure of oxidative stress, has been commonly observed in rivers in Europe, but was observed less frequently using the multiplexed assays in the current study. This again may be indicative of lower sensitivity in the multiplexed assay relative to targeted in vitro assays. Nonetheless, sites with the greatest GR and ER bioactivities were identified by both targeted and multiplexed assay approaches. The multiplexed nature of the Attagene systems may partially explain the differential sensitivity between assay systems, but the multiplexed nature of the assays (one of the greatest strengths of this approach) does additionally provide for an integrated system response accounting for potential interactions between transcriptional pathways. Other disadvantages of the current approach include a lack of positive control compounds for all endpoints and the lack of dose-response data for positive controls. This ultimately limits the ability to quantitatively compare extract response to that of a reference compound. Finally, for this specific study, screening was limited to a single replicate due to cost constraints. While issues of positive controls and replication could be overcome, it would come at additional project costs. Taken together, we propose the use of these multiplexed assays as a screening and prioritization tool in environmental monitoring. Where warranted, further site-specific follow-up employing the more sensitive targeted assays could be performed at high priority sites to more quantitatively assess relevant bioactivities.
Associating bioactivity with land use land cover
Another objective of the overall mixtures study was to identify potential associations between chemical occurrence or bioassay response and land use and land cover (LULC) attributes.29 The National Land Cover Database (NLCD) classification of site watershed-scale LULC attributes (previously reported29) were compared with AUC values for the 11 active endpoints to better evaluate whether specific point sources or land use patterns were related to bioactivity. Of 38 watershed-scale LULC parameters evaluated, two showed weak but significant correlation with bioactivity. Percent of watershed area classified as developed, low intensity (NLCD category 22) was significantly correlated with AhRE_CIS (p=0.016, r=0.39, 95% CI [0.080, 0.63]), ERE_CIS (p=0.037, r=0.34, 95% CI [0.021, 0.59]), PPARγ_TRANS (p=0.027, r=0.36, 95% CI [0.044, 0.61]), PXR_TRANS (p=0.016, r=0.39, 95% CI [0.077, 0.63]), PXRE_CIS (p=0.026, r=0.36, 95% CI [0.047, 0.61]), RORE_CIS (p=0.009, r=0.42, 95% CI [0.12, 0.65]), and VDRE_CIS (p=0.001, r=0.51, 95% CI [0.23, 0.71]). Additionally, the number of registered National Pollution Discharge Elimination System (NPDES) facilities was correlated with ERE_CIS (p=0.020, r=0.38, 95% CI [0.066, 0.63]), ERα_TRANS (p=0.016, r=0.39, 95% CI [0.078, 0.64]), GR_TRANS (p=0.012, r=0.41, 95% CI [0.096, 0.65]), PPARγ_TRANS (p=0.020, r=0.38, 95% CI [0.064, 0.63]), and RXRβ_TRANS (p=0.048, r=0.33, 95% CI [0.0045, 0.59]). Correlation of mainly xenobiotic metabolism related assays with low intensity development suggests non-point sources of bioactivity. Increased chemical burdens, either of natural organics (as seen at Penn Swamp Branch, NJ) or anthropogenic-related contaminants, may be a primary contributor to activity of xenobiotic-metabolism related endpoints such as AhR and PXR. AhR activity in surface water was previously identified at the highest levels in urban streams in Southern California,27 suggesting anthropogenic related contaminants are likely the primary contributors of elevated AhR activity. Correlation of more specific, endocrine-related activity (ER and GR) with NPDES inputs suggests wastewater as a source of bioactive contaminants.
While significant, no single LULC showed a strong predictive relationship with bioactivity (R2 ranging 0.108 to 0.262). Both significantly correlated features relate to urban use and wastewater inputs, which does strengthen the case that urban, anthropogenic inputs are the primary contributing factors of observed bioactivity. To supplement the watershed-scale analysis, catchment-scale LULC attributes were generated for each site to determine if increased resolution would provide better indicators of bioactivity, but similar relationships between catchment-scale LULC and bioactivity were observed. Catchment-scale medium and high intensity development were additionally correlated with metabolism and endocrine-related endpoints, further suggesting urban, anthropogenic inputs are the primary drivers of observed effects.
Linking Observed Bioactivities to Adverse Effects
The ultimate purpose of identifying pathway-based bioactivity in samples is to gain insights into potential hazards of exposure to contaminants present in surface waters. The adverse outcome pathway (AOP) framework50 provides the means to link pathway-based activity, such as data from in vitro bioassays, with higher organismal responses and relevant adverse outcomes. Some molecular targets, such as AhR and ER, have been studied extensively and AOPs associated with their activation are well-defined for aquatic vertebrates.51 However, clear and well supported associations with apical hazards are still lacking for a number of the active HTS endpoints identified in the present work. Efforts are being made to link all ToxCast assay targets, which includes those used in the current study, with existing AOP descriptions publicly available through the AOP-Wiki,51 a repository for developed or in-progress AOPs. Relevant AOPs in terms of species (e.g., fish) and endpoints (growth, development, reproduction) are available only for pathways associated with the AhR and ER (Table S7).51 Potential apical hazards associated with activation of those pathways include early life stage mortality and reproductive dysfunction. A search of the AOPWiki also identified existing AOPs for GR, NRF2, PPARγ, and PXR, although these have been primarily demonstrated in mammalian species and may not be applicable to aquatic species of concern (Table S7). As such, these four pathways represent targets for which AOP development should be prioritized. However, as with all in vitro based monitoring, in vitro bioactivity alone does not necessarily indicate adverse effects will occur in the environment. Much research has focused on establishing trigger values, or a value of bioactivity at which there is reasonable concern of adverse effects in the environment, for assays commonly used in environmental monitoring (e.g. ER reporter assays).11, 48 Additional characterization of the Factorial assays is needed before similar trigger values could be established.
No AOPs linking RXR, ROR, or VDR activation to apical outcomes have been described to date on the AOPWiki.51 These three activities were not as frequently observed in the environment relative to other targets, but they could be prioritized for AOP development, particularly if they are detected for a growing number of samples. It is notable that VDR response element, which is monitored by the VDRE_CIS endpoint, has been previously identified as a binding target of PXR.52 VDRE_CIS is significantly correlated with PXRE_CIS (p< 0.001, r=0.77; 95% CI [0.60, 0.88]) and PXR_TRANS activity (p< 0.001, r=0.73; 95% CI [0.53, 0.85]). The TRANS assay counterpart to VDRE_CIS, VDR_TRANS, which measures direct VDR-mediated activity, is not active at any site. This supports observed VDRE_CIS activity as an indicator of PXR-mediated activity and not indicative of specific VDR-mediated responses. As such, even if available, VDR-related AOPs are likely not relevant.
With regard to AOPs, for complex mixtures multiple relevant AOPs may be triggered simultaneously. Consequently, it has been proposed that consideration of AOP networks may be important for hazard prediction in these scenarios.53, 54 Knapen et al.53 provided an example of how Attagene bioactivity screening data for a complex environmental sample could be linked to relevant AOPs, assembled into a relevant AOP network, refined based on problem formulation, and used to both identify potential hazards of concern and endpoints that could be employed in effects-based monitoring. Based upon activation of eight endpoints in the Attagene assays, all of which were active in the current study, Knapen et al. identified potential adverse outcomes of impaired reproduction, early life stage mortality, and cardiotoxicity.53 The utility of this integrated approach will increase with continued linkage of Attagene endpoints and improved tools for assembling, filtering, and analyzing AOP networks.53
Finally, when considering in vitro assays in the context of AOPs for ecologically relevant species, the conservation of molecular targets must be considered. The Attagene assays used in the current work use only human-based receptors, which may or may not be representative of ecological species of concern. The AhR, for example, is broadly conserved in vertebrates, but AhR-mediated embryo toxicity can vary across teleost fish species by multiple orders of magnitude (see review by Doering et al.).55 Techniques now exist to help assess the similarity of molecular targets across various taxa. The SeqaPASS tool was recently used to compare all targets present in the ToxCast database with currently known sequences for non-human taxa.56 This data source could be used to compare the targets identified through the current study with those of ecologically relevant species of concern and predict if human-based assay results are representative of the target species. Ultimately, as extrapolations move from the molecular level, as measured through in vitro cell-based techniques, to apical-level effects identifications, the information required to make confident assessments of the potential effects will continue to grow.
The use of HTS assays to characterize bioactivity in US streams, along with parallel chemical characterization, identified multiple bioactivities of concern, potential chemicals of concern, and sites with the greatest potential for adverse effects to ecological receptors. Further, comparison with more traditional, cell-based reporter assays aligned well with the HTS assay results, providing evidence supporting the use of these HTS assays for environmental surveillance efforts. Though coverage of the chemical space (>700 compounds monitored) and biological space (69 endpoints) was more extensive than many studies, the majority of bioactivity could not be explained by measured chemicals, and only estrogenic activity could be reasonably predicted. The expanded endpoint coverage of the HTS assays employed here revealed additional biological pathways that may be commonly impacted in the environment. Because these assays cover vertebrate-centric (especially mammalian) endpoints, other relevant targets (i.e. those pertaining to invertebrates and plants) should be examined to more realistically evaluate potential ecological effects of complex, environmental mixtures.
Supplementary Material
Acknowledgments
We thank Dr. Anthony Schroeder for early discussions of data processing and interpretation, and Dr. William Foreman for supplying analyte loss data for inclusion in data analysis. This research was performed as part of the Chemical Safety for Sustainability (CSS) research program of the US Environmental Protection Agency (EPA) Office of Research and Development (ORD). This manuscript has been reviewed in accordance with the requirements of EPA ORD. The findings and conclusions in this article do not necessarily represent the views or policies of the EPA. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the US Government.
Footnotes
Supporting Information
Additional details on analyte recovery following evaporation, seven tables, and forty figures (PDF, XLSX) are provided. Complete USGS water quality data are downloadable from USGS ScienceBase (https://doi.org/10.5066/F7GF0RPH)37 and USGS National Water Information System (NWIS) (http://dx.doi.org/10.5066/F7P55KJN).57
References
- 1.Bradley PM; Battaglin WA; Clark JM; Henning FP; Hladik ML; Iwanowicz LR; Journey CA; Riley JW; Romanok KM, Widespread occurrence and potential for biodegradation of bioactive contaminants in Congaree National Park, USA. Environ Toxicol Chem 2017, 36, (11), 3045–3056. [DOI] [PubMed] [Google Scholar]
- 2.Kolpin DW; Furlong ET; Meyer MT; Thurman EM; Zaugg SD; Barber LB; Buxton HT, Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol 2002, 36, (6), 1202–11. [DOI] [PubMed] [Google Scholar]
- 3.Lee KE; Langer SK; Menheer MA; Hansen DS; Foreman WT; Furlong ET; Jorgenson ZG; Choy SJ; Moore JN; Banda J; Gefell DJ Chemicals of emerging concern in water and bottom sediment in the Great Lakes Basin, 2012: collection methods, analytical methods, quality assurance, and study data; 910; Reston, VA, 2015; p 14. [Google Scholar]
- 4.Tang JY; McCarty S; Glenn E; Neale PA; Warne MS; Escher BI, Mixture effects of organic micropollutants present in water: towards the development of effect-based water quality trigger values for baseline toxicity. Water research 2013, 47, (10), 3300–14. [DOI] [PubMed] [Google Scholar]
- 5.Altenburger R; Ait-Aissa S; Antczak P; Backhaus T; Barcelo D; Seiler TB; Brion F; Busch W; Chipman K; Lopez de Alda M; Umbuzeiro Gde A; Escher BI; Falciani F; Faust M; Focks A; Hilscherova K; Hollender J; Hollert H; Jager F; Jahnke A; Kortenkamp A; Krauss M; Lemkine GF; Munthe J; Neumann S; Schymanski EL; Scrimshaw M; Segner H; Slobodnik J; Smedes F; Kughathas S; Teodorovic I; Tindall AJ; Tollefsen KE; Walz KH; Williams TD; Van den Brink PJ; van Gils J; Vrana B; Zhang X; Brack W, Future water quality monitoring--adapting tools to deal with mixtures of pollutants in water resource management. Sci Total Environ 2015, 512–513, [DOI] [PubMed] [Google Scholar]
- 6.Brack W; Altenburger R; Schuurmann G; Krauss M; Lopez Herraez D; van Gils J; Slobodnik J; Munthe J; Gawlik BM; van Wezel A; Schriks M; Hollender J; Tollefsen KE; Mekenyan O; Dimitrov S; Bunke D; Cousins I; Posthuma L; van den Brink PJ; Lopez de Alda M; Barcelo D; Faust M; Kortenkamp A; Scrimshaw M; Ignatova S; Engelen G; Massmann G; Lemkine G; Teodorovic I; Walz KH; Dulio V; Jonker MT; Jager F; Chipman K; Falciani F; Liska I; Rooke D; Zhang X; Hollert H; Vrana B; Hilscherova K; Kramer K; Neumann S; Hammerbacher R; Backhaus T; Mack J; Segner H; Escher B; de Aragao Umbuzeiro G, The SOLUTIONS project: challenges and responses for present and future emerging pollutants in land and water resources management. Sci Total Environ 2015, 503–504, [DOI] [PubMed] [Google Scholar]
- 7.Di Paolo C; Ottermanns R; Keiter S; Ait-Aissa S; Bluhm K; Brack W; Breitholtz M; Buchinger S; Carere M; Chalon C; Cousin X; Dulio V; Escher BI; Hamers T; Hilscherova K; Jarque S; Jonas A; Maillot-Marechal E; Marneffe Y; Nguyen MT; Pandard P; Schifferli A; Schulze T; Seidensticker S; Seiler TB; Tang J; van der Oost R; Vermeirssen E; Zounkova R; Zwart N; Hollert H, Bioassay battery interlaboratory investigation of emerging contaminants in spiked water extracts - Towards the implementation of bioanalytical monitoring tools in water quality assessment and monitoring. Water research 2016, 104, 473–484. [DOI] [PubMed] [Google Scholar]
- 8.Jia A; Escher BI; Leusch FD; Tang JY; Prochazka E; Dong B; Snyder EM; Snyder SA, In vitro bioassays to evaluate complex chemical mixtures in recycled water. Water research 2015, 80, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konig M; Escher BI; Neale PA; Krauss M; Hilscherova K; Novak J; Teodorovic I; Schulze T; Seidensticker S; Kamal Hashmi MA; Ahlheim J; Brack W, Impact of untreated wastewater on a major European river evaluated with a combination of in vitro bioassays and chemical analysis. Environ Pollut 2017, 220, (Pt B), 1220–1230. [DOI] [PubMed] [Google Scholar]
- 10.Neale PA; Altenburger R; Ait-Aissa S; Brion F; Busch W; de Aragao Umbuzeiro G; Denison MS; Du Pasquier D; Hilscherova K; Hollert H; Morales DA; Novak J; Schlichting R; Seiler TB; Serra H; Shao Y; Tindall AJ; Tollefsen KE; Williams TD; Escher BI, Development of a bioanalytical test battery for water quality monitoring: Fingerprinting identified micropollutants and their contribution to effects in surface water. Water research 2017, 123, 734–750. [DOI] [PubMed] [Google Scholar]
- 11.Escher BI; van Daele C; Dutt M; Tang JY; Altenburger R, Most oxidative stress response in water samples comes from unknown chemicals: the need for effect-based water quality trigger values. Environ. Sci. Technol 2013, 47, (13), 7002–11. [DOI] [PubMed] [Google Scholar]
- 12.Blake LS; Martinovic D; Gray LE; Wilson VS; Regal RR; Villeneuve DL; Ankley GT, Characterization of the androgen-sensitive MDA-KB2 cell line for assessing complex environmental mixtures. Environ Toxicol Chem 2010, 29, (6), 1367–1376. [DOI] [PubMed] [Google Scholar]
- 13.Cavallin JE; Durhan EJ; Evans N; Jensen KM; Kahl MD; Kolpin DW; Kolodziej EP; Foreman WT; LaLone CA; Makynen EA; Seidl SM; Thomas LM; Villeneuve DL; Weberg MA; Wilson VS; Ankley GT, Integrated assessment of runoff from livestock farming operations: Analytical chemistry, in vitro bioassays, and in vivo fish exposures. Environ Toxicol Chem 2014, 33, (8), 1849–57. [DOI] [PubMed] [Google Scholar]
- 14.Escher BI; Allinson M; Altenburger R; Bain PA; Balaguer P; Busch W; Crago J; Denslow ND; Dopp E; Hilscherova K; Humpage AR; Kumar A; Grimaldi M; Jayasinghe BS; Jarosova B; Jia A; Makarov S; Maruya KA; Medvedev A; Mehinto AC; Mendez JE; Poulsen A; Prochazka E; Richard J; Schifferli A; Schlenk D; Scholz S; Shiraishi F; Snyder S; Su G; Tang JY; van der Burg B; van der Linden SC; Werner I; Westerheide SD; Wong CK; Yang M; Yeung BH; Zhang X; Leusch FD, Benchmarking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ. Sci. Technol 2014, 48, (3), 1940–56. [DOI] [PubMed] [Google Scholar]
- 15.Neale PA; Ait-Aissa S; Brack W; Creusot N; Denison MS; Deutschmann B; Hilscherová K; Hollert H; Krauss M; Novák J; Schulze T; Seiler T-B; Serra H; Shao Y; Escher BI, Linking in Vitro Effects and Detected Organic Micropollutants in Surface Water Using Mixture-Toxicity Modeling. Environmental Science & Technology 2015, 49, (24), 14614–14624. [DOI] [PubMed] [Google Scholar]
- 16.Cavallin JE; Jensen KM; Kahl MD; Villeneuve DL; Lee KE; Schroeder AL; Mayasich J; Eid EP; Nelson KR; Milsk RY; Blackwell BR; Berninger JP; LaLone CA; Blanksma C; Jicha T; Elonen C; Johnson R; Ankley GT, Pathway-based approaches for assessment of real-time exposure to an estrogenic wastewater treatment plant effluent on fathead minnow reproduction. Environ Toxicol Chem 2016, 35, (3), 702–16. [DOI] [PubMed] [Google Scholar]
- 17.Conley JM; Evans N; Mash H; Rosenblum L; Schenck K; Glassmeyer S; Furlong ET; Kolpin DW; Wilson VS, Comparison of in vitro estrogenic activity and estrogen concentrations in source and treated waters from 25 U.S. drinking water treatment plants. Sci Total Environ 2017, 579, 1610–1617. [DOI] [PubMed] [Google Scholar]
- 18.Wehmas LC; Cavallin JE; Durhan EJ; Kahl MD; Martinovic D; Mayasich J; Tuominen T; Villeneuve DL; Ankley GT, Screening complex effluents for estrogenic activity with the T47D-KBluc cell bioassay: assay optimization and comparison with in vivo responses in fish. Environ Toxicol Chem 2011, 30, (2), 439–45. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q; Jia A; Snyder SA; Gong Z; Lam SH, Glucocorticoid activity detected by in vivo zebrafish assay and in vitro glucocorticoid receptor bioassay at environmental relevant concentrations. Chemosphere 2016, 144, 1162–9. [DOI] [PubMed] [Google Scholar]
- 20.Scott PD; Bartkow M; Blockwell SJ; Coleman HM; Khan SJ; Lim R; McDonald JA; Nice H; Nugegoda D; Pettigrove V; Tremblay LA; Warne MS; Leusch FD, An assessment of endocrine activity in Australian rivers using chemical and in vitro analyses. Environ Sci Pollut Res Int 2014, 21, (22), 12951–67. [DOI] [PubMed] [Google Scholar]
- 21.Dix DJ; Houck KA; Martin MT; Richard AM; Setzer RW; Kavlock RJ, The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 2007, 95, (1), 5–12. [DOI] [PubMed] [Google Scholar]
- 22.USEPA, ToxCast & Tox21 Summary Files from invitrodb_v2. In October 2015. ed.; https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data (
- 23.Richard AM; Judson RS; Houck KA; Grulke CM; Volarath P; Thillainadarajah I; Yang C; Rathman J; Martin MT; Wambaugh JF; Knudsen TB; Kancherla J; Mansouri K; Patlewicz G; Williams AJ; Little SB; Crofton KM; Thomas RS, ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chemical research in toxicology 2016, 29, (8), 1225–51. [DOI] [PubMed] [Google Scholar]
- 24.Becker RA; Friedman KP; Simon TW; Marty MS; Patlewicz G; Rowlands JC, An exposure:activity profiling method for interpreting high-throughput screening data for estrogenic activity--proof of concept. Regulatory toxicology and pharmacology : RTP 2015, 71, (3), 398–408. [DOI] [PubMed] [Google Scholar]
- 25.Blackwell BR; Ankley GT; Corsi SR; DeCicco LA; Houck KA; Judson RS; Li S; Martin MT; Murphy E; Schroeder AL; Smith ER; Swintek J; Villeneuve DL, An “EAR” on Environmental Surveillance and Monitoring: A Case Study on the Use of Exposure-Activity Ratios (EARs) to Prioritize Sites, Chemicals, and Bioactivities of Concern in Great Lakes Waters. Environ. Sci. Technol 2017, 51, (15), 8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehinto AC; Jia A; Snyder SA; Jayasinghe BS; Denslow ND; Crago J; Schlenk D; Menzie C; Westerheide SD; Leusch FD; Maruya KA, Interlaboratory comparison of in vitro bioassays for screening of endocrine active chemicals in recycled water. Water research 2015, 83, 303–9. [DOI] [PubMed] [Google Scholar]
- 27.Mehinto AC; VanDervort DR; Lao W; He G; Denison MS; Vliet SM; Volz DC; Mazor RD; Maruya KA, High throughput in vitro and in vivo screening of inland waters of Southern California. Environmental Science: Processes & Impacts 2017, 19, (9), 1142–1149. [DOI] [PubMed] [Google Scholar]
- 28.Buxton HT, Reilly TJ, Kuivila KM, Kolpin DW, Bradley PM, Villeneuve DL, and Mills MA, Chemical mixtures and environmental effects—A pilot study to assess ecological exposure and effects in streams. U.S. Geological Survey Open-File Report 2015, 2015–1113. [Google Scholar]
- 29.Bradley PM; Journey CA; Romanok KM; Barber LB; Buxton HT; Foreman WT; Furlong ET; Glassmeyer ST; Hladik ML; Iwanowicz LR; Jones DK; Kolpin DW; Kuivila KM; Loftin KA; Mills MA; Meyer MT; Orlando JL; Reilly TJ; Smalling KL; Villeneuve DL, Expanded Target-Chemical Analysis Reveals Extensive Mixed-Organic-Contaminant Exposure in U.S. Streams. Environ. Sci. Technol 2017, 51, (9), 4792–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conley JM; Evans N; Cardon MC; Rosenblum L; Iwanowicz LR; Hartig PC; Schenck KM; Bradley PM; Wilson VS, Occurrence and In Vitro Bioactivity of Estrogen, Androgen, and Glucocorticoid Compounds in a Nationwide Screen of United States Stream Waters. Environ. Sci. Technol 2017, 51, (9), 4781–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin MT; Dix DJ; Judson RS; Kavlock RJ; Reif DM; Richard AM; Rotroff DM; Romanov S; Medvedev A; Poltoratskaya N; Gambarian M; Moeser M; Makarov SS; Houck KA, Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA’s ToxCast program. Chemical research in toxicology 2010, 23, (3), 578–90. [DOI] [PubMed] [Google Scholar]
- 32.Romanov S; Medvedev A; Gambarian M; Poltoratskaya N; Moeser M; Medvedeva L; Gambarian M; Diatchenko L; Makarov S, Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nature methods 2008, 5, (3), 253–60. [DOI] [PubMed] [Google Scholar]
- 33.Chun H; Keles S, Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Stat Soc Series B Stat Methodol 2010, 72, (1), 3–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou H; Hastie T, Regularization and Variable Selection via the Elastic Net. Journal of the Royal Statistical Society. Series B (Statistical Methodology) 2005, 67, (2), 301–320. [Google Scholar]
- 35.Chung D; Chun H; Keles S spls: Sparse Partial Least Squares (SPLS) Regression and Classification, R package version 2.2–1; https://CRAN.R-project.org/package=spls, 2013.
- 36.Team RDC, R: A language and environment for statistical computing. In Vienna, Austria, 2008. [Google Scholar]
- 37.Romanok KM; Bradley PM; Journey CA, Inorganic and organic concentration data collected from 38 streams in the United States, 2012–2014, with supporting data, as part of the Chemical Mixtures and Environmental Effects Pilot Study. U.S. Geological Survey data release 2017. 10.5066/F7GF0RPH [DOI]
- 38.Azur MJ; Stuart EA; Frangakis C; Leaf PJ, Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res 2011, 20, (1), 40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Buuren S; Groothuis-Oudshoorn K, mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software; Vol 1, Issue 3 (2011) 2011. [Google Scholar]
- 40.EPA, U., Estimations Program Interface Suite for Microsoft Windows, v4.11. United States Environmental Protection Agency, Washington, DC, USA: 2012. [Google Scholar]
- 41.Long M; Laier P; Vinggaard AM; Andersen HR; Lynggaard J; Bonefeld-Jørgensen EC, Effects of currently used pesticides in the AhR-CALUX assay: comparison between the human TV101L and the rat H4IIE cell line. Toxicology 2003, 194, (1), 77–93. [DOI] [PubMed] [Google Scholar]
- 42.Behnisch PA; Hosoe K; Sakai S, Bioanalytical screening methods for dioxins and dioxin-like compounds a review of bioassay/biomarker technology. Environ Int 2001, 27, (5), 413–39. [DOI] [PubMed] [Google Scholar]
- 43.Ekins S; Chang C; Mani S; Krasowski MD; Reschly EJ; Iyer M; Kholodovych V; Ai N; Welsh WJ; Sinz M; Swaan PW; Patel R; Bachmann K, Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol Pharmacol 2007, 72, (3), 592–603. [DOI] [PubMed] [Google Scholar]
- 44.Fuller PJ; Yao Y; Yang J; Young MJ, Mechanisms of ligand specificity of the mineralocorticoid receptor. Journal of Endocrinology 2012, 213, (1), 15–24. [DOI] [PubMed] [Google Scholar]
- 45.Bittner M; Janošek J; Hilscherová K; Giesy J; Holoubek I; Bláha L, Activation of Ah receptor by pure humic acids. Environmental Toxicology 2006, 21, (4), 338–342. [DOI] [PubMed] [Google Scholar]
- 46.Staudinger JL; Ding X; Lichti K, Pregnane X receptor and natural products: beyond drug-drug interactions. Expert Opin Drug Metab Toxicol 2006, 2, (6), 847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omiecinski CJ; Vanden Heuvel JP; Perdew GH; Peters JM, Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol Sci 2011, 120 Suppl 1, S49–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escher BI; Neale PA; Leusch FD, Effect-based trigger values for in vitro bioassays: Reading across from existing water quality guideline values. Water research 2015, 81, 137–48. [DOI] [PubMed] [Google Scholar]
- 49.Roberts J; Bain PA; Kumar A; Hepplewhite C; Ellis DJ; Christy AG; Beavis SG, Tracking multiple modes of endocrine activity in Australia’s largest inland sewage treatment plant and effluent-receiving environment using a panel of in vitro bioassays. Environ Toxicol Chem 2015, 34, (10), 2271–81. [DOI] [PubMed] [Google Scholar]
- 50.Ankley GT; Bennett RS; Erickson RJ; Hoff DJ; Hornung MW; Johnson RD; Mount DR; Nichols JW; Russom CL; Schmieder PK; Serrrano JA; Tietge JE; Villeneuve DL, Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 2010, 29, (3), 730–41. [DOI] [PubMed] [Google Scholar]
- 51.Adverse Outcome Pathway Wiki (AOP-Wiki). https://aopwiki.org/ (April/26/2018),
- 52.Echchgadda I; Song CS; Roy AK; Chatterjee B, Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol 2004, 65, (3), 720–9. [DOI] [PubMed] [Google Scholar]
- 53.Knapen D; Angrish MM; Fortin MC; Katsiadaki I; Leonard M; Margiotta-Casaluci L; Munn S; O’Brien JM; Pollesch N; Smith LC; Zhang X; Villeneuve DL, Adverse outcome pathway networks I: Development and applications. Environ Toxicol Chem 2018, 37, (6), 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villeneuve DL; Crump D; Garcia-Reyero N; Hecker M; Hutchinson TH; LaLone CA; Landesmann B; Lettieri T; Munn S; Nepelska M; Ottinger MA; Vergauwen L; Whelan M, Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci 2014, 142, (2), 312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doering JA; Giesy JP; Wiseman S; Hecker M, Predicting the sensitivity of fishes to dioxin-like compounds: possible role of the aryl hydrocarbon receptor (AhR) ligand binding domain. Environmental Science and Pollution Research 2013, 20, (3), 1219–1224. [DOI] [PubMed] [Google Scholar]
- 56.LaLone CA; Villeneuve DL; Doering JA; Blackwell BR; Transue TR; Simmons CW; Swintek J; Degitz SJ; Williams AJ; Ankley GT, Evidence for Cross Species Extrapolation of Mammalian-Based High-Throughput Screening Assay Results. Environmental Science & Technology 2018, 52, (23), 13960–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.U.S. Geological Survey, National Water Information System—Web interface. 10.5066/F7P55KJN (accessed Feb 27, 2018). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.