Abstract
For most cancers, metastasis is the point at which clinical treatment shifts from curative intent to extending survival. Biomaterial implants acting as a synthetic pre-metastatic niche recruit metastatic cancer cells and provide a survival advantage, and their use as a diagnostic platform requires assessing their relevance to disease progression. Here we showed that scaffold-captured tumor cells (SCAF) were 30 times more metastatic to the lung than primary tumor cells (PT), similar to cells derived from lung micrometastases (LUNG). SCAF cells were more aggressive in vitro, demonstrated higher levels of migration, invasion, and mammosphere formation, and had a greater proportion of cancer stem cells than PT. SCAF were highly enriched for gene expression signatures associated with metastasis and had associated genomic structural changes including globally enhanced entropy. Collectively, our findings demonstrate that SCAF cells are distinct from PT and more closely resemble LUNG, indicating that tumor cells retrieved from scaffolds are reflective of cells at metastatic sites.
Keywords: cancer metastasis, biomaterial implant, breast cancer, Hi-C, RNAseq
Precis:
Biomaterial scaffold-recruited cancer cells are an aggressive population resembling metastatic cells found in the lung, and thus, biopsy of the scaffold may serve as a surrogate for the physiologic metastatic site, representing a step towards the development of a molecular staging of metastatic disease.
Introduction
In most types of cancer, the formation of distant metastases is the point at which the disease is no longer considered curable. Tumor cell colonization of distal sites occurs through a sequence of events, initiated by generation of pre-metastatic niches by cells originating in the bone marrow (1–3). A small subset of tumor cells that have successfully invaded the tumor vasculature then home to these pre-conditioned locations (4). Tumor cells recruited to metastatic sites are distinct from both primary tumor and the major population of circulating tumor cells (CTCs), and are often more aggressive (5), resistant to therapy (6), and cancer “stem cell” like (7). This suggests that neither the primary tumor nor CTCs are representative of cells with metastatic potential. At present, methods to detect metastasis involve radiologic imaging, which are capable of detecting metastatic foci 7-10 mm in size (8). Imaging is typically performed for evaluation of clinical symptoms indicating compromised tissue function, and thus foci of this size are usually associated with late stage disease.
Most early detection strategies currently developed focus on the use of blood as a “liquid biopsy”. The umbrella term liquid biopsies includes circulating tumor cell (CTC), circulating tumor DNA (ctDNA), and exosome detection in the blood: all of which are emerging as platforms to stage patients beyond the presence, size, and molecular characteristics of the primary tumor (9). However, while each of these is associated with metastatic risk the interplay between the presence and relative number of each of these markers and presence of metastatic disease is tenuous. Biomaterial scaffolds that capture metastatic tumor cells (10) extend beyond the power of the liquid biopsy to capture immune cells associated with the metastatic niche and metastatic tumor cells themselves (11–15). These technologies have been successful in mouse models of breast (11–15), ovarian (16), prostate (17), melanoma (18) and hematologic cancers (19) and have captured tumor cells, reduced metastatic burden (14, 15), and improved survival (15).
A crucial piece of scaffolds as a detection platform and alternative to liquid biopsy is understanding the phenotype of tumor cells recruited to biomaterial scaffolds in vivo testing the hypothesis that these cells are truly metastatic and similar to tumor cells that have colonized an organ. As poly(ε-caprolactone) (PCL) scaffold implantation in combination with surgical resection results in a survival advantage relative to mock surgery (15), we expect that the scaffold may be capturing an aggressive population of metastatic tumor cells. Identifying the phenotype of scaffold-captured tumor cells within the continuum of tumor cell phenotypes will inform the use of scaffolds as surrogates for metastatic sites, facilitating the development of therapeutic strategies targeting metastatic disease.
In this report, we derived cell lines from the primary tumor MDA-MB-231BR breast cancer xenografts, as well as matched lung micro-metastasis, and biomaterial scaffold captured-tumor cells. In vitro and in vivo assays were utilized to characterize phenotypic differences between these cell lines. Finally, we performed RNAseq and Hi-C to elucidate transcriptional and chromatin configuration differences that generate these phenotypic characteristics. These studies support the utility of scaffold-captured cells as a metastasis surrogate to reveal molecular mechanisms and identify potential therapeutic targets for metastatic cancer.
Materials and Methods
Scaffold fabrication and implantation
Microsphere preparation.
PCL microspheres were prepared as previously described (15). In brief microspheres were made by emulsification of a 6% (w/w) solution of PCL (Lactel Absorbable Polymers; Inherent viscosity 0.65-0.85 dL/g) in dichloromethane in a 10% (w/v) poly(vinyl alcohol) solution followed by homogenization at 10,000 rpm for 1 minute. The solution was then stirred for 3 hours to evaporate dichloromethane solvent. Microspheres were then collected by centrifugation at 2000 × g for 10 minutes and washed at least five times in deionized water. Finally, microspheres were lyophilized for 48 hours.
Scaffold fabrication.
Microporous PCL scaffolds were prepared by mixing PCL microspheres and sodium chloride crystals (250-425 μm in diameter) at a 1:30 (w/w) ratio. This salt and polymer microsphere mixture was then pressed in a steel die for 45 seconds at 1500 PSI. Polymer/salt disks were then heated at 60°C for 5 minutes per side to melt polymer microparticles around salt crystals to form a continuous structure. Salt crystals were subsequently removed by immersion in water for 1.5 hours. Scaffolds were then disinfected for animal studies using 70% ethanol, rinsed with sterile water, and dried on a sterile surface.
Scaffold implantation.
All animal studies were performed in accordance with institutional guidelines and protocols (PRO00007801) approved by the University of Michigan Institutional Animal Care and Use Committee. Scaffolds were implanted into the dorsal subcutaneous space of 8-week-old female NOD/SCID-IL2Rγ−/− (NSG) mice (Jackson Laboratory) immediately above the shoulder blades and as physically distant from the fourth-right mammary fat pad as possible. For the implantation procedure, animals were anesthetized via isoflurane (2%, inhaled), prepared with Carprofen analgesia (5 mg/kg, subcutaneous injection), the upper back was shaved and prepped using a Betadine swab followed by an ethanol swab and this procedure was repeated 3 times. A fenestrated sterile field was draped over the surgical area and a 1 cm incision was made in the upper back. Following incision, subcutaneous pockets were created perpendicular to the incision, into which sanitized scaffolds were inserted (2 scaffolds/mouse). The skin was then closed using sterile wound clips (Reflex 7 mm, Roboz Surgical Instrument Co).
Tumor inoculation
Tumor inoculations were performed by injection of 2 × 106 MDA-MB-231BR-tdTomato-luc2 (parental line, Northwestern University Developmental Therapeutics Core) or 231BR-SCAF,-PT, or -LUNG derived cell lines in 50 μL PBS (Life Technologies) into the fourth right mammary fat pads of 10-week-old female NSG mice (Jackson Laboratory). Cell lines were confirmed to be pathogen- and mycoplasma-free and authenticated by short tandem repeat DNA analysis and compared to the ATCC STR profile database (DDC Medical).
Magnetic Mouse Cell Depletion and Culture
Mice were euthanized four weeks following tumor inoculation and scaffolds and organs were retrieved and washed in Hank’s Balanced Salt Solution (HBSS, Life Technologies). Samples were minced using microscissors in Liberase TL or TM (0.38 mg/mL, Roche) and incubated at 37°C for 20 minutes. Following incubation with liberase, the enzyme was neutralized via addition of 0.5M EDTA (Life Technologies) and digested tissues were strained through a 70 μm filter in FACS buffer [PBS (Life Technologies) with 0.5% Bovine Serum Albumin (Sigma Aldrich) and 2 mM EDTA (Life Technologies)]. Cells were counted using a Countess Automated Cell Counter (Invitrogen) and magnetically sorted using a Mouse Cell Depletion Kit (Miltenyi) according to manufacturer instructions. Human cell fractions were cultured in DMEM D6429 (Sigma Aldrich) with 10% FBS and 1% penicillin/streptomycin until growth of tumor cell colonies was evident. Following the first passage of cells, culture was continued without antibiotics. Care was taken to minimize passage number of tumor cells, for all experiments the passage number was less than ten.
Flow cytometry to identify tdTomato+ tumor cells
For quantification of in vivo metastatic ability of 231BR-SCAF, -PT, and –LUNG derived cell lines, mice were euthanized three weeks after tumor inoculation and retrieved scaffolds and lungs were washed in HBSS (Life Technologies) and processed with liberase as described for Magnetic Mouse Cell Depletion above. Following isolation of a single cell suspension, tdTomato+ tumor cells were quantified using a MoFlo Astrios Flow Cytometer (Beckman Coulter). The detection sensitivity for cancer cells via flow cytometry was 0.002% (i.e. 5 cancer cells in 250,000 total cells) (14). Gating strategy is provided in Figure S1A. Metastasis assays were repeated twice.
Scratch assays
Scratch assays were performed as described by Justus et al (20). Briefly, cells were seeded in 24 well plates and allowed to grow to form a confluent monolayer. A sterile 200 μL pipette tip was used to create a wound in the monolayer. The surface was washed with PBS (Life Technologies) to remove any detached cells and replaced with media. Images were taken immediately after wounding, 5h and 10-12h after wounding. Results were quantified by identification of the proportion of scratches that had no closure at endpoint (no cells touching from either side of wound), partial closure (at least 2 cells from either side of wound in direct contact), or complete closure (no evidence of wound remains). Assays were repeated 3 times.
Transwell invasion assays
Transwell assays were performed as described by Justus et al (20). Briefly, Matrigel invasion chambers (BD Biosciences) were rehydrated for 30 min at 37°C using 500 μL serum-free media. 750 μL of chemoattractant media (serum-containing) was placed in the receiver well plate. Cells were harvested using TryplE (Life Technologies), counted and suspended at 50,000 cells/mL in serum-free media. Rehydrated invasion chambers were transferred to receiver well plate containing chemoattractant and 500 μL of cell suspension was added to each insert. Care was taken to prevent trapped air between the insert and chemoattractant media. Assays were incubated for 24h at 37°C and subsequently stained and fixed using crystal violet in ethanol. Invaded cells were imaged directly on the membrane using a Nikon Eclipse inverted microscope and imaged at 10×. Four images per well were captured and cell numbers were quantified in ImageJ using automatic particle counting with constant image thresholds. Assays were repeated three times.
Cancer stem cell marker flow cytometry and mammosphere assay
Proportion of cancer stem cell like tumor cells in each cell line was determined by quantification of EpCAM+CD44+CD24- cell population by flow cytometry. Tumor cells were harvested using TryplE (Life Technologies), counted and suspended at 1×107 cells/mL. Cells were evenly split between control and test conditions and stained with anti-human BV421 EpCAM (clone EBA-1, BD Biosciences), PerCP-Cy5.5 CD44 (clone G44-26, BD Biosciences), and PE/Cy7 CD24 (clone ML5, Biolegend). Samples were run on MoFlo Astrios Flow Cytometer (Beckman Coulter) and data processed using FlowJo (TreeStar Inc.). Gating strategy is provided in Figure S1B. Cancer stem cell marker flow cytometry was repeated three times. Mammosphere assays were performed as described by Tarasewicz et al. (21)
RNAseq sample preparation, library construction, and sequencing
Cells were harvested using TryplE (Life Technologies), counted and suspended at 5×106 cells/mL. RNA was isolated using High Pure RNA Isolation Kit (Roche) according to manufacturer instructions. Isolated RNA concentrations were quantified using NanoDrop 2000 (Thermo Fisher) and resuspended at 80 ng/μL. RNA samples were kept at −80’C until further use. RNA quality was assessed using an Agilent 2100 Bioanalyzer. The DNA Sequencing Core at University of Michigan performed library construction and sequencing using the Hiseq-4000 platform using single-end 50-base sequence reads with a multiplex of 6 per sequencing lane.
Transcriptome analysis
Transcriptome analysis was performed as previously described (22). For RNAseq data processing the raw reads were checked with FastQC to identify potential quality issues in the raw data. Next, Tophat and Bowtie were used to align the reads to a human reference transcriptome (HG19). Alignment settings were kept on default except for “-b2-very-sensitive”, “-no-coverage-search” and “-no-novel-juncs” in order to limit the search to known transcripts only. FastQC was then performed again on the mapped reads. Cufflinks/Cuffdiff were then utilized to quantify expression and perform differential expression analysis using parameter settings “—multi-read-correct” and “upper-quartile-norm”. CummeRbund was then used to format the cufflinks output. Benjamini-Hochberg procedure was used in CummeRbund for multiple testing method correction. Gene level analysis was performed using fragments per kilobase per million reads (FPKM) values outputted by Cufflinks and log2 fold change with pseudocounts. Bin level gene expression vectors were calculated using raw counts outputted by cufflinks and adding up the counts for all the genes in each bin then normalizing by million reads to convert them to FPM. Gene Set Enrichment analysis (23, 24) was performed with GSEA v3.0 software.
Hi-C sample preparation, library construction, and sequencing
Hi-C library construction was performed as described by Chen et al (25). Briefly 2×106 cells for each condition were crosslinked with 1% formaldehyde (Fisher Scientific) in serum-free media for 10 min at room temperature and subseueqntly quenched with glycine (Sigma Aldrich) to a final concentration of 0.125 M. Crosslinked cells were flash frozen in liquid nitrogen and stored at −80°C until the construction of libararies. Crosslinked cells were thawed on ice and resuspended in 1 mL lysis buffer (10 mM Tris-HCl, 10 mM NaCL, 0.2% Igpel (Signma Aldrich) and incubated for 15 min. Cells were next homogenized on ice and the lysate was transferred to a 1.7 mL tube. Cells were collected by centrifugation (5 min at 2000×g) and washed twice in NEB buffer. HindIII digestion was then performed in NEB buffer at 37°C overnight. Following digestion, restriction overhang ends were filled and labeled with biotin. Samples were then ligated at 16°C for 4 h in a reaction containing 1× ligation buffer, 1% Triton X-100 (Sigma Aldrich), 1 mg/mL bovine serum albumin (Fisher Scientific), 10 mM ATP (Sigma Aldrich) and 50 u T4 DNA ligase (Life Technologies). Proteinase K (0.5 mg, Life Technologies) was then added and incubated for 4 h at 65°C. After this incubation an additional 0.5 mg proteinase K was added to each tube and the incubation was continued overnight at 65°C. DNA was then extracted using phenol:chloroform (1:1) and desalted using AMICON ultra centrifugal filter unit (Millipore) with 1× TE buffer. Biotin was then removed from un-ligated ends, DNA purified with signle phenol extraction and precipitated by addition of ethanol. DNA was resuspended in water and fragmentation was performed using a sonicator (Covaris). Fragments of 200-400 bp were recovered with Agencourt AMPure XP (Beckman Coulter) utilizing manufacturer protocols. DNA fragment ends were repaired and purified with a MinElute column, and A-tailing was performed. Streptavidin pull-down of biotinylated Hi-C DNA was performed. Illumina adapter ligation was then performed and then DNA was amplified by 15 PCR cycles for Illumina HiSeq Sequencing. A standard quality control protocol was performed prior to sequencing and all libraries passed. Samples were pooled and sequence in a single lane of a flow cell on HiSeq 4000 to generate paired-end sequence reads at 100 bases per end read.
Nucleome analysis
Generation of Hi-CMatrices:
A standard pipeline was used to process Hi-C sequence data using the University of Michigan Bioinformatics Core facilities. Raw sequence reads were processed with FastQC for data quality control and paired-end reads were mapped to the HG19 reference human genome using Bowtie2. HOMER was used to develop the contact matrix at 100 kb and 1 Mb resolution.
Analysis of Hi-CMatrices:
Hi-C matrices were normalized, plotted, and analyzed in conjunction with RNAseq data for cell lines using the 4D Nucleome Analysis Toolbox (4D-NAT) described by Seaman et al (26).
Whole Exome Sequencing
Sequencing:
SCAF, PT, and LUNG cell lines were cultured and passaged as described above. Roughly 2×106 cells were isolated and genomic DNA was isolated with Qiagen DNEasy kit according to manufacturer instructions. Samples were submitted to the UMich DNA sequencing core and the core performed quality control, exome capture using the Nimblegen Human Exome Capture kit, and paired-end sequencing at 150 bases per end read using the Illumina HiSeq-4000 platform with ~100× coverage for each cell line.
Analysis:
Genestack platform was used for analysis of whole exome sequencing data. Quality control was performed using FastQC algorithm Adaptors and contaminants were trimmed according to methods published by Aronesty et al (27) and then sequences were mapped onto the human reference genome HG19. Quality control of the mapped reads was again performed using the FastQC algorithm. Variant calling and effect prediction was then performed using SAMtools and SnpEff algorithms respectively and variants were selected to only include those resulting in missense or nonsense mutations with a quality >60. Gene ontology analysis of mutant genes was then performed using the Gene Ontology Consortium software (28).
Data Availability Statement
Figures 2–7 have associated raw data. Data is provided as a private link to a OneDrive folder https://1drv.ms/f/s!AmVnzTmi4yrLg8FbakFHWvhuImHOxA with password: cancerresearch until publication, at which point it will be made publicly available.
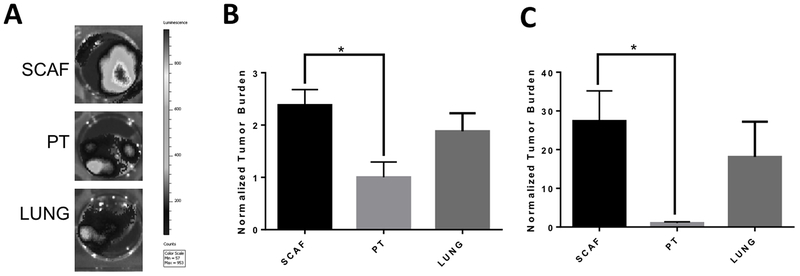
Figure 2: SCAF cell line is highly metastatic in vivo compared to the PT cell line and similar to LUNG cell line.
(A) Representative bioluminescence images for lungs of mice inoculated with SCAF (n=11 mice), PT (n=5 mice), or LUNG (n=6 mice) cells. Tumor burden as measured by flow cytometry for tdTomato+ cells normalized to average tumor burden for PT cell line for scaffolds(B) and lungs(C). Error bars, SEM. *p<0.05 by two tailed t-test.
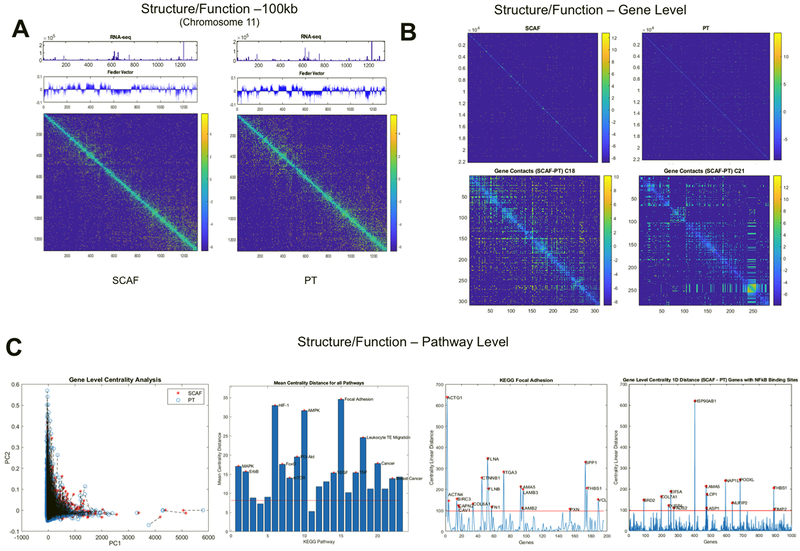
Figure 7: SCAF cells demonstrate differential structure and function relationships from PT cells.
Structure (Hi-C matrix and corresponding Fielder Vector) and function (RNAseq) investigated at the 100kB level for Chromosome 11(A). Structure relationships for the whole genome (top) for SCAF and PT and differences in gene level contacts for chromosome 18 and 21 visualized as the SCAF matrix – PT matrix (bottom)(B). Gene level centrality analysis for the whole genome comparing structure/function for SCAF and PT pairs for each gene connected by dashed black line(C). KEGG pathways investigated for difference in centrality distance from SCAF to PT for genes in a given pathway, red stars indicate pathways that are significantly different from the mean genome-wide centrality distance by two tailed t-test. Centrality distance for SCAF – PT calculated for KEGG Focal Adhesion Pathway and for genes with NFkB binding sites. Red stars indicate genes that are more than four standard deviations above the genome-wide mean centrality distance.
Results
Tumor cell lines were derived from scaffold, primary tumor, and metastatic sites in vitro
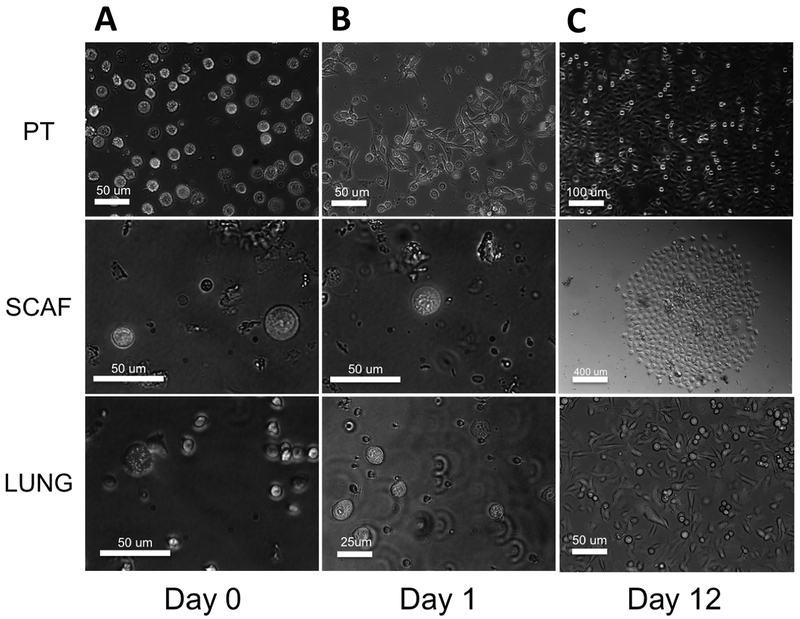
We generated cell lines from various tumor cell locations in vivo including cells derived from subcutaneously implanted scaffolds and compared them to cell lines derived from primary tumors or lung metastases derived from the same set of mice. Based on previous reports, the earliest metastatic cells should be captured in the scaffold(14, 15), with metastases arriving in the lung at subsequent times. This difference in time of arrival may underlie some variations in these metastatic cell populations. Using a mouse cell depletion kit and magnetic activated cell sorting, we isolated human MDA-MB-231BR human breast cancer cells from each location. Approximately 10,000 cells were plated for PT-derived cells, whereas ~100 and ~500 cells were isolated for scaffold and lung derived cells respectively. Representative images of each are shown (Figure 1). While PT cells attached and spread day 1 (Figure 1A), scaffold and lung-derived cells took much longer to attach and begin growing, likely due to plating density (Figure 1B). By day 12 of culture, scaffold-captured cells had attached and formed ~15 individual colonies and lung-derived cells had formed hundreds of colonies, whereas primary tumor formed confluent cultures without visible colonies (Figure 1C). Cells were passaged and cultured to develop stable cell lines, hereafter referred to as SCAF (scaffold-captured), LUNG (lung micro-metastasis derived) and PT (primary tumor-derived) cell lines.
Figure 1: Micro-porous scaffolds implanted in NSG mice recruit 231BR cells in vivo.
Tumor cells can be isolated by MACS mouse cell depletion and tdTomato+ primary tumor (PT), scaffold (SCAF), and lung (LUNG) derived tumor cells are evident initially at day 0(A), PT cells begin to attach at day 1 but SCAF and LUNG cells remain in suspension(B), and by day 12 SCAF cells attached and grew out to form ~15 colonies of cells, LUNG cells grew out from ~500 cells and PT from ~105 cells(C). Scale bars represent 25, 50, 100 or 400 um as labeled.
Scaffold-captured cells are highly metastatic in vivo
We investigated the metastatic capacity of these cell lines in vivo. Cell lines were orthotopically inoculated into NSG mice and allowed to grow for 21 days. Tumor and spleen mass were similar for all lines. Representative bioluminescent images of the lungs indicate metastasis, with differential metastatic capacity for each (Figure 2A). Metastasis was quantified by the presence of tdTomato+ cells in tissue homogenates by flow cytometry. Metastatic tumor burden was significantly higher in implanted scaffolds (Figure 2B) (2.4 ± 1.4 times higher) and lungs (Figure 2C) (27.3 ± 22.5 times higher) for SCAF tumor-bearing mice relative to PT tumor-bearing mice (p<0.05 via t-test). Metastatic tumor burden was not significantly different between LUNG and PT or LUNG and SCAF in implanted scaffolds (Figure 2B) (1.9 ± 1.2 times higher than PT) or lungs (Figure 2C) (18.1 ± 22.3 times higher than PT). As a control, we also developed a tumor cell line from the lung of a mouse that did not have scaffold implants, termed LUNG2. We did not observe a difference in metastatic ability from LUNG to LUNG2 (Figure S2). Collectively, these studies demonstrate that SCAF are more metastatic than PT and more comparable to LUNG.
Scaffold-captured cells display distinct phenotypic behavior in vitro
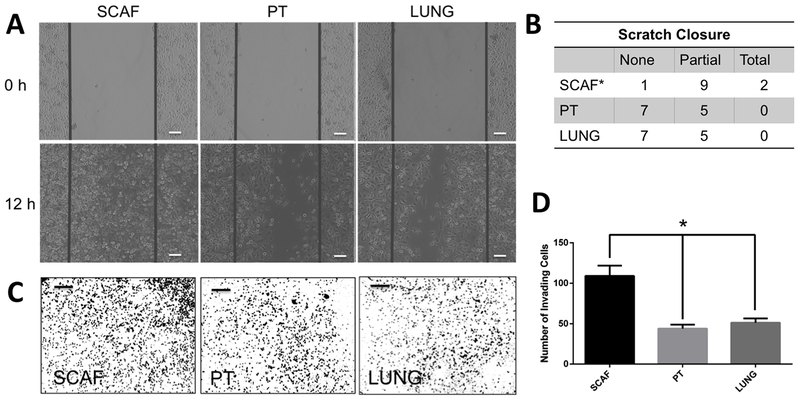
As scaffold-captured cells were highly metastatic in vivo, we next investigated the lines in vitro to identify mechanisms of enhanced metastasis. Assays were performed to analyze migration and invasion as well as cancer stem cell characteristics. Using a standard scratch assay, migration into a defect was quantified. Representative images demonstrate migrating tumor cells at 0 and 12 hours following the scratch (Figure 3A). PT and LUNG samples demonstrate failure to generate closure (7/12) or partial closure (5/12), and show no samples with full closure (Figure 3B). For the SCAF cell line, a statistically significant increase was observed in both partial (9/12) and total (2/12) closure and a significant decrease in no closure (1/12) was observed relative to PT and LUNG (p<0.05 via Fisher’s Exact Test).
Figure 3: SCAF cell line is more migratory and invasive than PT cell line and similar to metastatic cell lines.
SCAF and PT cells were analyzed in parallel for migration and invasion ability to close a scratch in 12h. SCAF cells close a scratch faster than PT and LUNG cells with representative images (scale bar 100 µm)(A) and as quantified by the number of scratched with no closure, partial closure, or complete closure(B) (*p<0.05 relative to PT and LUNG via two tailed t-test, n=12 distinct samples per condition). SCAF cells were found to be more invasive as determined by transwell invasion assay than PT and LUNG cells with representative images (scale bar 400 µm)(C) and quantified as the number of invading cells in each 5× field of view (n=6 distinct samples per condition)(D).
The invasion characteristics were measured using a transwell invasion assay. Representative images are provided (Figure 3C). PT and LUNG invaded at a rate of 44 ± 17 and 51.3 ± 18.1 cells/image respectively (Figure 3D). Consistent with the scratch assay, a significant increase in the number of SCAF cells were observed, with 109 ± 44 cells/image respectively. The results demonstrate that SCAF cells have a higher capacity for invasion and migration than either PT or LUNG.
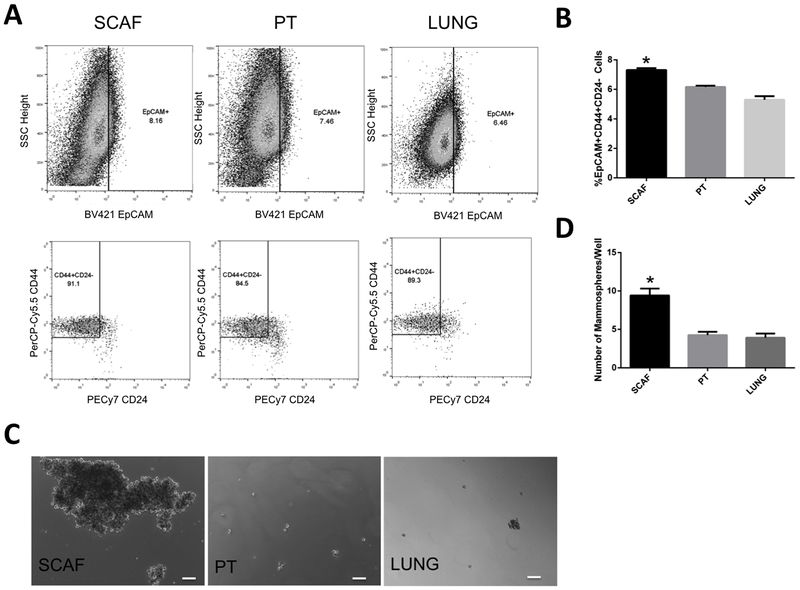
We next investigated expression of cancer stem cell markers in each cell population. Using flow cytometry, we evaluated the expression of EpCAM, CD44 and CD24. Representative plots are provided, showing relative levels of EpCAM+ and CD44+CD24- cells in cell lines (Figure 4A). PT cells had 6.2 ± 0.2% EpCAM+CD44+CD24- (CSC+) cells. A significantly increased proportion of CSC+ cells (7.3 ± 0.2%) was observed for SCAF (Figure 4B). LUNG had a significantly decreased percentage of CSC+ cells relative to both PT and SCAF (5.3 ± 0.4%) (Figure 4B).
Figure 4: Scaffold-captured tumor cell line shows higher proportion of cancer stem cell like cells.
(A) Representative flow plots for SCAF, PT, and LUNG cell lines.(B) Percentage of EpCAM+CD44+CD24- cancer stem cells (n=3 distinct samples per condition).(C) Representative images of mammospheres from SCAF, PT, and LUNG cell lines.(D) Number of mammospheres per well for each cell line. (n=12 distinct samples per condition). Error bars represent s.e.m. *p<0.01 relative to PT, and LUNG lines via two tailed t-test.
The stem cell characteristics were also investigated via mammosphere assay, which measures self-renewal in suspension. Representative images are provided (Figure 4C), demonstrating mammosphere formation for SCAF, PT, and LUNG cell lines. PT generated 4.3 ± 1.5 spheres/well of a 24 well culture dish, which was comparable to the LUNG, which was 3.9 ± 1.9 spheres/well (Figure 4D). SCAF had significantly increased mammosphere formation capability with 9.4 ± 3.1 spheres/well (p<0.05 relative to PT, and LUNG). Additionally, mammospheres in the SCAF line were much larger than PT or LUNG, however we cannot rule out that this was a result of aggregation of individual spheres thus only sphere number was quantified. Collectively, these results demonstrate that the scaffold sequesters a population of cells that have increased capacity for migration and invasion, and increased proportion of cancer stem cells, both of which could contribute to the survival advantage provided by the scaffold.
Scaffold-captured cells do not exhibit additional driver mutations from lung or primary tumor derived cells
We next investigated the hypothesis that phenotypic differences between SCAF, PT, and LUNG result from the differential accumulation of mutations in these populations. To this end we performed whole exome sequencing on SCAF, PT, and LUNG. Variant calling was performed on filtered reads mapped to the human reference genome HG19 (GRCh37) and these variants were further filtered to include only mutations resulting in a missense or nonsense mutation and compared for variants between each line (Figure S3, Table S1). The vast majority of variants relative to HG19 were shared across all lines (3191 or 95.5% of total variants) with each showing 10-13 unique variants (0.3-0.4% of total variants). Gene ontology was then used to investigate the functional impact of these differential mutations. Mutations for all lines were implicated in the same ontological terms indicating selection based on additional accumulated mutations after leaving the primary tumor is unlikely in this setting (Table S2).
Scaffold-captured cell transcriptome is more similar to Lung derived than PT derived cells
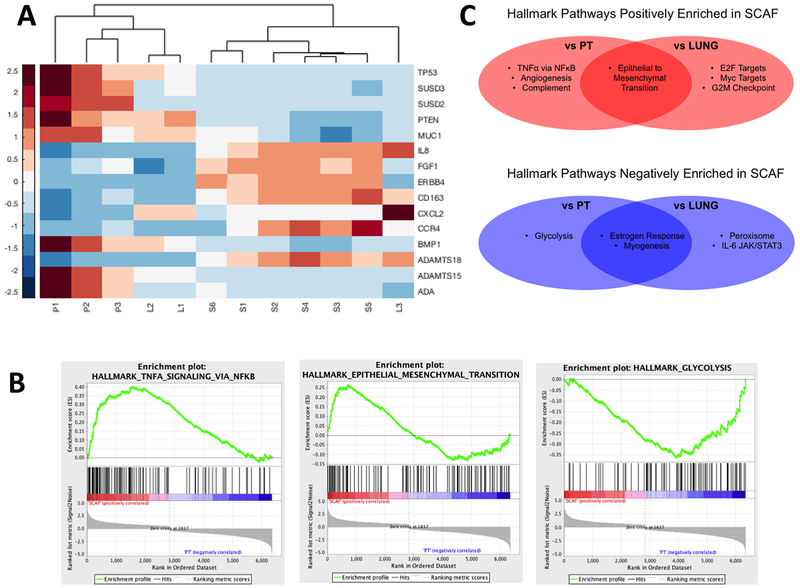
We next utilized RNAseq to characterize transcriptomic differences between the cell lines that might account for their phenotypic properties. We identified 14,232 genes with measured expression, 2901 of which were differentially expressed (Benjamini-Hochberg corrected p<0.05 and log fold change >0.6) between SCAF and PT cells and 14,572 genes with measured expression, 2,398 genes of which were differentially expressed between SCAF and LUNG cells. A small subset of the most differentially expressed genes (Figure S4A) was confirmed using qRT-PCR (Figure S4B). Of this set of genes, the vast majority (19/20) were found to be more similarly expressed between SCAF and LUNG compared to SCAF and PT. The PAM50 gene set (29) was interrogated, and the majority of genes (35/50) were more similarly expressed in SCAF and LUNG when compared to SCAF and PT. These genes included Myc, MDM2, ESR1, ERBB2, EGFR, CCNB1, CXCR1, IL8, IL6R, IL6ST. Using unsupervised hierarchical clustering with the top 15 differentially expressed genes, SCAF cells clustered separately from all PT samples and all but one LUNG sample (Figure 5A). The same result was obtained by hierarchical clustering with all genes with differential expression (Figure S4A). Using gene set enrichment analysis (GSEA), TNFα via NFκB signaling and epithelial-to-mesenchymal transition (EMT) were positively enriched and glycolysis was depleted in SCAF (false discovery rate q-value <0.25 and p<0.05) relative to PT (Figure 5B). EMT was also enriched in SCAF relative to LUNG (Figure 5C) along with E2F Targets, Myc Targets, and G2M Checkpoint. Similarly, Estrogen Response and Myogenesis were depleted in SCAF relative to both PT and LUNG and Peroxisome and IL-6, JAK/STAT3 signaling were depleted in SCAF relative to LUNG (Figure 5C). No pathways were enriched in PT relative to LUNG by GSEA, however TNFα via NFκB signaling was enriched in LUNG relative to PT (NES 1.969 with FDR q-value < 0.001). Collectively, this analysis of gene expression indicates that the SCAF phenotype aligns more closely to LUNG than to PT.
Figure 5: Scaffold-captured tumor cell line shows distinct transcriptome from PT and similar to LUNG by RNAseq.
(A) Heat map with unsupervised hierarchical clustering of top differentially expressed genes for SCAF (S1-S6), PT (P1-P3), and LUNG (L1-L3) demonstrating SCAF cells have a distinct transcriptome relative to LUNG and PT but cluster more closely with LUNG than PT.(B) Gene set enrichment analysis (GSEA) enrichment plots for pathways altered in SCAF transcriptome relative to PT (C) Gene set enrichment analysis for Hallmark pathways positively enriched in SCAF vs PT and SCAF vs LUNG (red) and negatively enriched in SCAF vs PT and SCAF vs LUNG (blue) (false discovery rate q-value <0.25 and p-value <0.05).
Scaffold-captured cells have a distinct chromatin structure resulting in functional changes consistent with enhanced aggressiveness and metastatic ability
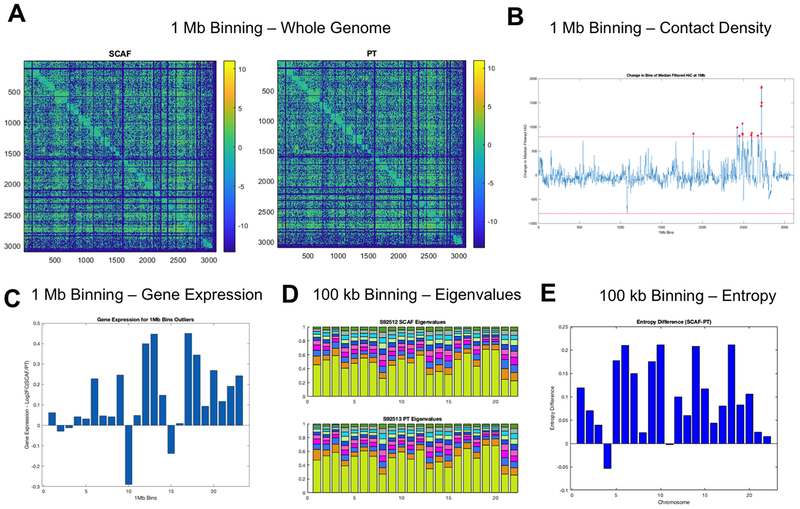
We subsequently compared SCAF and PT cell lines by Hi-C (30) to investigate chromatin structure and genome organization differences that may give rise to differential gene expression and resultant functional behavior including metastatic phenotype. Hi-C data was collected from SCAF and PT lines and analyzed in conjunction with RNAseq results at three scales: 1 Mb, 100 kb, and gene/pathway level binning (Figure S5). Overall, the Hi-C matrices appear similar with the same translocations and copy number changes visible within the Toeplitz normalized matrices at 1 Mb binning (Figure 6A). With filtering to the 95th percentile of each matrix, translocations are visible as blocks of strong trans interchromosomal interactions and unbalanced and balanced translocations have an ‘L’ or ‘X’ appearance respectively and were found to be matched between SCAF and PT (Figure S6A). Copy number changes can also be visualized by looking at the sum of Hi-C contacts at 100 Mb binning for each chromosome and these were also found to be concordant between SCAF and PT (Figure S6B). Additionally, the copy number normalization method published by Seaman et al.(22) was utilized to investigate differential breakpoints. No differential breakpoints were found between SCAF and PT samples. Normalized 1 Mb Hi-C contact matrices were first investigated for large-scale structural differences by summing the counts for each 1 Mb bin in one dimension. Bins with a value more than 4 standard deviations above the mean were identified as altered in structure and starred in red (Figure 6B) with all having higher contact density in SCAF compared to PT. These altered bins were then investigated for changes in gene expression (Figure 6C), which showed that the increased genomic interaction measured by Hi-C was accompanied by an increase in expression in SCAF relative to PT for the majority of bins. For bins with altered gene expression the average number of genes per bin that were differentially expressed with a log2 fold change >0.6 was significantly increased relative to randomly selected bins of the same number of genes (p<0.05). Next, eigenvalue decomposition (Figure 6D) and calculation of entropy difference between samples was performed on 100 kb binned matrices for each chromosome (Figure 6E) demonstrating enhanced entropy in SCAF relative to PT for the majority of chromosomes. Additionally, the Fiedler vector (the eigenvector associated with the second smallest eigenvalue of the Laplacian matrix (31)) was calculated for each 100 kb binned matrix for each chromosome and plotted with the Hi-C contact matrix and RNAseq at the same binning. Structural differences from PT to SCAF were found in the Fiedler vector for chromosomes 11 (Figure 7A), 18 and 21.
Figure 6: SCAF tumor cells show distinct genome structure identified by Hi-C.
(A) Hi-C contact matrices for SCAF and PT cells at 1 Mb binning.(B) Contact density difference (SCAF-PT) for each 1 Mb bin calculated by summing the counts for each bin in one dimension. Bins with a value more than 4 standard deviations above the mean were identified as altered in structure and starred in red.(C) Log2 fold change in gene expression for SCAF/PT plotted for each of the altered bins identified in B. For identified bins with higher counts for SCAF, most have a corresponding higher gene expression.(D) Eigenvalue decomposition of 100 kb Hi-C matrices for each chromosome showing subtle differences in the eigenvalues for both samples.(E) Entropy difference for SCAF - PT calculated for each chromosome at 100 kb resolution.
Next, Hi-C contact matrices were generated at the gene level for the whole genome (Figure 7B, top) and individual chromosomes 18 and 21 identified as altered in structure at 1 Mb and 100 kb (Figure 7B). Finally, changes in centrality were calculated for each gene (Hi-C) and combined with gene expression (RNAseq) in principal component analysis (PCA) to reduce the dimension of the data while maintaining meaningful variability. Projections of each bin onto the first two PCs are shown (Figure 7C). To quantify which genes have the greatest change, the distance in the PC space from PT to SCAF samples was calculated. Genes whose distance from PT to SCAF changed more than 4 standard deviations from the mean whole genome distance were identified as altered in structure/function. KEGG pathways were then selected based on RNAseq (Figure 5C) to investigate enrichment of structure/function changes within subsets of genes (Table S3). By centrality analysis the KEGG MAPK, ErbB, HIF-1α, FoxO, mTOR, PI3-Akt, AMPK, VEGF, Focal Adhesion, TNFα, Leukocyte Transendothelial Migration, Cancer, and Breast Cancer pathways were significantly altered from PT to SCAF (p<0.05) (Figure 7C). Similarly, distances greater than 4 standard deviations above the mean could be observed for specific genes within pathways including COL6A1, FN1, and CTNNB1 for the Focal Adhesion pathway (Figure 7C). Finally, based on GSEA implicating NFκB as a key regulatory pathway, genes with NFκB binding sites were investigated for changes in structure/function. By centrality analysis, genes with NFκB binding sites were significantly altered in structure and function. Altered genes more than 4 standard deviations above the mean were identified including HSP90AB1, COL7A1, EIF5A, and BRD2 among others (Figure 7C). Taken together, these results indicate we can identify genome structure organizational differences that are associated with development of a metastatic phenotype.
Discussion
In this study metastatic tumor cells derived from those captured by microporous PCL scaffolds in vivo were investigated for their phenotype relative to tumor cells from the primary tumor, and metastatic lung lesions. This work was motivated by the idea that scaffold-captured cells may be more easily sampled, while accessing organ-derived metastatic cells would necessitate an invasive biopsy with potential for high morbidity and false negative results dependent upon the extent of disease progression. Our work and that of others have demonstrated that scaffolds recapitulate multiple elements of the pre-metastatic niche in vivo along with the capacity to capture metastatic cells within a biomaterial implant (10). Analyzing these cells along the continuum of cancer cell phenotypes provides a foundation for using a scaffold biopsy to impact patient care, such as for the development of personalized therapies (32). We developed stable cell lines from metastatic locations in order to have sufficient numbers of cells for in depth in vitro behavioral, functional, and molecular assays and in vivo metastasis assays. Notably, the scaffold-derived tumor cells were highly metastatic in vivo, demonstrated substantial invasiveness in vitro, and molecular analysis indicated these cells were similar to cells derived from metastatic lung lesions.
SCAF and LUNG were more highly metastatic than PT in vivo, supporting the notion that cells captured by the scaffold represent a population with the ability to successfully metastasize and colonize a tissue. Interestingly, the SCAF line showed comparable ability to metastasize to both scaffold and lung tissues, which differs from literature that has reported cell proclivity to metastasize to the location of origin (33–35). The concordant metastatic tropism to lung for both SCAF and LUNG suggests that the metastatic traits of SCAF are not site specific yet are indicative of overarching traits required for metastasis. Consistent with increased aggressiveness in vivo, scaffold-captured cells were more migratory, invasive, and stem-like in vitro than cells from the primary tumor. These results provide the first evidence that tumor cells derived from biomaterial scaffolds are a functionally aggressive and metastatic population of cells, with the capacity to spread to distal sites and compromise organ function.
Unsupervised hierarchical clustering of RNAseq data identified that SCAF clustered more closely with LUNG than with PT. Additionally, upon investigation of the top differentially expressed genes between SCAF and PT, 19/20 of these genes showed greater homology between SCAF and LUNG when compared with SCAF and PT. Similarly, for the PAM50 gene set 35/50 genes had more similar expression between SCAF and LUNG than for SCAF and PT. Collectively, these results indicate that scaffold and lung-derived cells, both originating from metastatic sites, show greater homology compared to scaffold-captured cells and those derived from the primary tumor.
Further investigation of the tumor cell phenotypes by GSEA identified pathways significantly enriched/depleted in SCAF cells relative to PT and LUNG. Hallmark pathways (36) enriched in SCAF relative to PT included TNFα signaling via NFκB, Epithelial to Mesenchymal Transition (EMT), Angiogenesis, and Complement. TNFα signaling through NFκB has been shown to enhance stemness of breast cancer cells (37), angiogenesis and complement are indicators of metastasis and survival in patients (38, 39), and EMT is closely linked to metastasis and stemness (40, 41). The SCAF line is more migratory and invasive than the LUNG line in vitro, and yet SCAF and LUNG demonstrate similar metastatic ability in vivo. This apparent contracdiction may be explained by the following observations. First, EMT is enriched in SCAF relative to both PT and LUNG, indicating the SCAF line is more mesenchymal and thus more migratory and invasive in vitro. Second, in previous work it has been shown that the earliest metastatic tumor cells colonize the scaffold prior to the lung (14). Third, the SCAF cells are more homogeneous than LUNG, as demonstrated by hierarchical clustering. Fourth, IL-6 JAK/Stat3 signaling was enriched in LUNG relative to SCAF which may enhance metastasis in vivo through paracrine communication with myeloid derived suppressor cells enhancing stemness but may not influence phenotype in vitro (42). Finally, SCAF cells are more stem-like and demonstrate higher entropy/plasticity. Hallmark pathways enriched in SCAF relative to LUNG also included E2F targets (43), Myc targets (44), and G2M checkpoint (44): all important axes in breast cancer. Peroxisome and IL-6 JAK/Stat3 signaling were enriched in LUNG relative to SCAF and have been implicated in therapeutic resistance (45) and stemness (42). Importantly, TNFα signaling through NFκB was enriched in both SCAF and LUNG relative to PT, indicating this may be the crucial pathway for gaining enhanced metastatic ability for these cells. These results suggest that LUNG and SCAF may evolve toward similar phenotypes through the same dominant transcriptional program (TNFα via NFκB) but with different accessory programs facilitating site-specific adaptations (i.e. SCAF with EMT and LUNG with IL-6 JAK/Stat3 signaling) (42), which is consistent with other observations of metastatic lines (46). This supports the use of scaffold as a sampling location and also suggests that both metastatic locations are derived from a similar pool of cells yet may experience differential evolutionary pressure and adaptation at different distal sites. The scaffold has the capacity to provide a sampling site that would be clinically easy to access relative to an organ biopsy and contains a source of cells with clear phenotypic and molecular similarities to the pathologic metastatic site.
The underlying genome/chromatin organization role in the development of metastatic phenotype from PT to SCAF was identified with Hi-C. Using a multi-scale approach, we found structural changes linked to functional outcomes at all points. Critically, for all except chromosome 4, entropy for SCAF was higher than PT, indicating a more dysregulated network consistent with the concept of increased aggressiveness and phenotypic flexibility that are hallmarks of stemness and metastasis (47–49). We chose to analyze SCAF as a surrogate for other metastatic sites due to the in vivo and transcriptomic similarity of these cells to the LUNG. Importantly, no copy number changes or additional translocations were observed, indicating the karyotype from PT to SCAF was conserved. Even at 1 Mb binning analysis of whole genome contact density we found structural differences between SCAF and PT indicating areas of chromosomes 11, 18, and 21 with a higher density of intrachromosomal contacts for SCAF. Interestingly, there were no locations identified where PT had higher contact density. Investigating these altered bins further, we found the vast majority had higher expression in SCAF indicating increased connectivity resulted in enhanced gene expression. Similarly, we observed subtle changes in both eigenvalue decomposition and Von Neumann entropy for each chromosome. At 100 kb we found chromosomes 11, 18 and 21 identified by contact density at 1 Mb to have differences in Fiedler vector consistent with a change in structure from SCAF to PT. We also investigated the centrality of each chromosome at the gene level (structure) and gene expression (function) for the whole genome and identified significantly altered pathways in structure/function. Implicated genes in these pathways include many genes associated with metastasis and stemness including COL6A1, FN1, ITGA3, IL11, IL6ST, IL8, and TGFβR2. In particular, IL8 has been implicated in enhanced stemness, aggressiveness (50, 51), and resistance to therapy (52). Finally, based on GSEA of RNAseq data implicating TNFα via NFκB signaling as the common transcriptional program associated with metastatic phenotype in both SCAF and LUNG cells, we investigated structure/ function relationships for genes with NFκB binding sites. We found this gene set to be significantly altered from PT to SCAF in structure/function. This further supports NFκB as one of the major drivers of phenotypic change from non-metastatic to metastatic phenotype (53–55). Our data are among the first to connect genome/chromatin structure alterations to changes in functional gene expression driving metastatic phenotype.
Our results provide primary evidence that biomaterial scaffold-captured tumor cells are a functionally aggressive and metastatic population capable of distal organ colonization. Taken together, this work demonstrates scaffold recruited tumor cells are similar to other metastatic sites, but also represent a population of highly aggressive, stem-like cells whose capture may hold promise for reducing breast cancer mortality. Additionally, these results support the possibility that metastatic phenotype can be gained through changes in genome structure impacting functional gene expression rather than mutational alterations. Transcriptional alterations in tumor cell phenotype and plasticity that are maintained in culture may be equally as important as mutational changes for development of metastatic ability. The scaffold platform technology provides a unique opportunity to analyze the earliest metastatic cells, enabling the analysis of tumor cells that have left the primary tumor and colonized a distal site. This manuscript identifies for the first time, the many similarities and differences that are present between the metastatic cells recruited to a biomaterial scaffold and the lung. Future work should focus on the impact of heterogeneity, timing at which tumor cells arrive at the niche, and the impact of varied microenvironments on tumor cell phenotype. This platform provides an unprecedented opportunity for analysis and development of tools for personalized medicine.
Supplementary Material
Significance.
Findings suggest that metastatic tumor cells captured by a biomaterial scaffold may serve as a diagnostic for molecular staging of metastasis.
Acknowledgements and Contributions
Thanks to the NIH for their support through 5R01CA173745-06 and R01 CA214384. G.G.B. is a recipient of the NSF Graduate Research Fellowship and NRSA F31 CA224982-01. Thanks to the University of Michigan DNA Sequencing Core for assistance in sequencing and analysis. Thanks to the Developmental Therapeutics Core at the Northwestern University for donation of MDA-MB-231-BR-tdTomato+Luc2+ parental line. G.G.B., M.S.W., I.R., J.S.J., and L.D.S. conceived the presented hypotheses and experimental designs. G.G.B carried out experiments with support from H.C., T.P.H, R.S.O, Y.Z., and R.D.H. and completed computational analysis with support from S.R. and H.C. G.G.B. and L.D.S. wrote the manuscript with support from J.S.J., M.S.W., and I.R. L.D.S. supervised the project with support from J.S.J. All authors discussed the results and reviewed the final manuscript.
Grant Support: The authors acknowledge support from the National Institutes of Health NIH-Director’s Transformative Research Award-R01CA173745 (to L.D. Shea and J.S. Jeruss). G.G. Bushnell is a recipient of the NSF Graduate Research Fellowship and NRSA F31 CA224982-01.
Footnotes
Competing Interests: The scaffold as a platform for metastasis detection is described in a current patent application (US20170281798A1) by assignee Northwestern University with inventors Lonnie D. Shea, Samira M. Azarin, Robert M. Gower, Jacqueline S. Jeruss.
References
- 1.Kaplan RN, Psaila B, Lyden D. Bone marrow cells in the ‘pre-metastatic niche’: within bone and beyond. Cancer Metastasis Rev 2006;25:521–9. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005;438:820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer 2017;17:302–17. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ. Metastasis: quantitative analysis of distribution and fate of tumor emboli labeled with 125I-5-iodo-2′-deoxyuridine. Journal of the National Cancer Institute 1970;45:773–82. [PubMed] [Google Scholar]
- 5.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127:679–95. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer, Breast Cancer Chemosensitivity. Springer; 2007. p. 1–22. [DOI] [PubMed] [Google Scholar]
- 7.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 2018;173:338–54. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdi YE. Limits of tumor detectability in nuclear medicine and PET. Molecular imaging and radionuclide therapy 2012;21:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer discovery 2016. [DOI] [PubMed] [Google Scholar]
- 10.Aguado BA, Bushnell GG, Rao SS, Jeruss JS, Shea LD. Engineering the pre-metastatic niche. Nature biomedical engineering 2017;1:0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguado BA, Caffe JR, Nanavati D, Rao SS, Bushnell GG, Azarin SM, et al. Extracellular matrix mediators of metastatic cell colonization characterized using scaffold mimics of the pre-metastatic niche. Acta biomaterialia 2016;33:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguado BA, Hartfield RM, Bushnell GG, Decker JT, Azarin SM, Nanavati D, et al. A synthetic pre-metastatic niche mimic alters the primary tumor and tumor microenvironment. Advanced Healthcare Materials 2018:1700903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguado BA, Wu JJ, Azarin SM, Nanavati D, Rao SS, Bushnell GG, et al. Secretome identification of immune cell factors mediating metastatic cell homing. Scientific reports 2015;5:17566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azarin SM, Yi J, Gower RM, Aguado BA, Sullivan ME, Goodman AG, et al. In vivo capture and label-free detection of early metastatic cells. Nature communications 2015;6:8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao SS, Bushnell GG, Azarin SM, Spicer G, Aguado BA, Stoehr JR, et al. Enhanced survival with implantable scaffolds that capture metastatic breast cancer cells in vivo. Cancer research 2016;76:5209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Fuente A, Alonso-Alconada L, Costa C, Cueva J, Garcia-Caballero T, Lopez-Lopez R, et al. M-trap: exosome-based capture of tumor cells as a new technology in peritoneal metastasis. JNCI: Journal of the National Cancer Institute 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bersani F, Lee J, Yu M, Morris R, Desai R, Ramaswamy S, et al. Bioengineered implantable scaffolds as a tool to study stromal-derived factors in metastatic cancer models. Cancer research 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko C-Y, Wu L, Nair AM, Tsai Y-T, Lin VK, Tang L. The use of chemokine-releasing tissue engineering scaffolds in a model of inflammatory response-mediated melanoma cancer metastasis. Biomaterials 2012;33:876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Li M, Milwid J, Dunham J, Vinegoni C, Gorbatov R, et al. Implantable microenvironments to attract hematopoietic stem/cancer cells. Proceedings of the National Academy of Sciences 2012;109:19638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. Journal of visualized experiments: JoVE 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarasewicz E, Oakes RS, Aviles MO, Straehla J, Chilton KM, Decker JT, et al. Embryonic stem cell secreted factors decrease invasiveness of triple-negative breast cancer cells through regulome modulation. Cancer biology & therapy 2018;19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaman L, Chen H, Brown M, Wangsa D, Patterson G, Camps J, et al. Nucleome Analysis Reveals Structure–Function Relationships for Colon Cancer. Molecular Cancer Research 2017;15:821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics 2003;34:267. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Chen J, Muir LA, Ronquist S, Meixner W, Ljungman M, et al. Functional organization of the human 4D Nucleome. Proceedings of the National Academy of Sciences 2015;112:8002–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seaman L, Rajapakse I. 4D nucleome Analysis Toolbox: analysis of Hi-C data with abnormal karyotype and time series capabilities. Bioinformatics 2017;34:104–06. [DOI] [PubMed] [Google Scholar]
- 27.Aronesty E Comparison of sequencing utility programs. The Open Bioinformatics Journal 2013;7. [Google Scholar]
- 28.Consortium GO. The Gene Ontology (GO) database and informatics resource. Nucleic acids research 2004;32:D258–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastien RR, Rodríguez-Lescure Á, Ebbert MT, Prat A, Munárriz B, Rowe L, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC medical genomics 2012;5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ried T, Rajapakse I, The 4D Nucleome. 2017. [DOI] [PubMed] [Google Scholar]
- 31.Chung FR. Spectral graph theory. 1997: American Mathematical Soc. [Google Scholar]
- 32.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nature Reviews Cancer 2015;15:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459:1005–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003;3:537–49. [DOI] [PubMed] [Google Scholar]
- 35.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005;436:518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov J, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1 (6): 417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C-W, Xia W, Huo L, Lim S-O, Wu Y, Hsu JL, et al. Epithelial–mesenchymal transition induced by TNF-α requires NF-κB–mediated transcriptional upregulation of Twist1. Cancer research 2012;72:1290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linderholm B, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtiö J, et al. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Annals of oncology 2009;20:1639–46. [DOI] [PubMed] [Google Scholar]
- 39.Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. Journal of Clinical Oncology 2009;27:3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Research 2011;13:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo M, Brooks M, S Wicha M. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Current pharmaceutical design 2015;21:1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng D, Tanikawa T, Li W, Zhao L, Vatan L, Szeliga W, et al. Myeloid-derived suppressor cells endow stem-like qualities to breast cancer cells through IL6/STAT3 and NO/NOTCH cross-talk signaling. Cancer research 2016;76:3156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JL, Pillai S, Pernazza D, Sebti SM, Lawrence NJ, Chellappan SP. Regulation of matrix metalloproteinase genes by E2F transcription factors: Rb–Raf-1 interaction as a novel target for metastatic disease. Cancer research 2012;72:516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature cell biology 2010;12:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Bagheri-Yarmand R, Balasenthil S, Hortobagyi G, Sahin AA, Barnes CJ, et al. HER2 regulation of peroxisome proliferator-activated receptor γ (PPARγ) expression and sensitivity of breast cancer cells to PPARγ ligand therapy. Clinical Cancer Research 2003;9:3198–203. [PubMed] [Google Scholar]
- 46.Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. Journal of bone and mineral research 2001;16:1486–95. [DOI] [PubMed] [Google Scholar]
- 47.Tarabichi M, Antoniou A, Saiselet M, Pita JM, Andry G, Dumont JE, et al. Systems biology of cancer: entropy, disorder, and selection-driven evolution to independence, invasion and “swarm intelligence”. Cancer and Metastasis Reviews 2013;32:403–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berretta R, Moscato P. Cancer biomarker discovery: the entropic hallmark. PLoS One 2010;5:e12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerji CR, Severini S, Caldas C, Teschendorff AE. Intra-tumour signalling entropy determines clinical outcome in breast and lung cancer. PLoS computational biology 2015;11:e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin AR, Wang SE. Cytokines driving breast cancer stemness. Molecular and cellular endocrinology 2014;382:598–602. [DOI] [PubMed] [Google Scholar]
- 51.Singh JK, Simões BM, Howell SJ, Farnie G, Clarke RB. Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells. Breast Cancer Research 2013;15:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpé S, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clinical Cancer Research 2004;10:7157–62. [DOI] [PubMed] [Google Scholar]
- 53.Ali S, Lazennec G. Chemokines: novel targets for breast cancer metastasis. Cancer and Metastasis Reviews 2007;26:401–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santini D, Schiavon G, Vincenzi B, Gaeta L, Pantano F, Russo A, et al. Receptor activator of NF-kB (RANK) expression in primary tumors associates with bone metastasis occurrence in breast cancer patients. PloS one 2011;6:e19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, et al. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. The Journal of clinical investigation 2004;114:569–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Figures 2–7 have associated raw data. Data is provided as a private link to a OneDrive folder https://1drv.ms/f/s!AmVnzTmi4yrLg8FbakFHWvhuImHOxA with password: cancerresearch until publication, at which point it will be made publicly available.
Figure 2: SCAF cell line is highly metastatic in vivo compared to the PT cell line and similar to LUNG cell line.
(A) Representative bioluminescence images for lungs of mice inoculated with SCAF (n=11 mice), PT (n=5 mice), or LUNG (n=6 mice) cells. Tumor burden as measured by flow cytometry for tdTomato+ cells normalized to average tumor burden for PT cell line for scaffolds(B) and lungs(C). Error bars, SEM. *p<0.05 by two tailed t-test.
Figure 7: SCAF cells demonstrate differential structure and function relationships from PT cells.
Structure (Hi-C matrix and corresponding Fielder Vector) and function (RNAseq) investigated at the 100kB level for Chromosome 11(A). Structure relationships for the whole genome (top) for SCAF and PT and differences in gene level contacts for chromosome 18 and 21 visualized as the SCAF matrix – PT matrix (bottom)(B). Gene level centrality analysis for the whole genome comparing structure/function for SCAF and PT pairs for each gene connected by dashed black line(C). KEGG pathways investigated for difference in centrality distance from SCAF to PT for genes in a given pathway, red stars indicate pathways that are significantly different from the mean genome-wide centrality distance by two tailed t-test. Centrality distance for SCAF – PT calculated for KEGG Focal Adhesion Pathway and for genes with NFkB binding sites. Red stars indicate genes that are more than four standard deviations above the genome-wide mean centrality distance.