Abstract
Delay in the assessment of tumor response to radiation therapy continues to pose a major challenge to quality of life for patients with non-responsive tumors. Here we exploited label-free Raman spectroscopic mapping to elucidate radiation-induced biomolecular changes in tumors and uncovered latent microenvironmental differences between treatment-resistant and -sensitive tumors. We used isogenic radiation-resistant and -sensitive A549 human lung cancer cells human head and neck squamous cell carcinoma (HNSCC) cell lines (UM-SCC-47 and UM-SCC-22B, respectively) to grow tumor xenografts in athymic nude mice and demonstrated the molecular specificity and quantitative nature of Raman spectroscopic tissue assessments. Raman spectra obtained from untreated and treated tumors were subjected to chemometric analysis using multivariate curve resolution-alternating least squares (MCR-ALS) and support vector machine (SVM) to quantify biomolecular differences in the tumor microenvironment. The Raman measurements revealed significant and reliable differences in lipid and collagen content post-radiation in the tumor microenvironment, with consistently greater changes observed in the radiation-sensitive tumors. In addition to accurately evaluating tumor response to therapy, the combination of Raman spectral markers potentially offers a route to predicting response in untreated tumors prior to commencing treatment. Combined with its non-invasive nature, our findings provide a rationale for in vivo studies using Raman spectroscopy, with the ultimate goal of clinical translation for patient stratification and guiding adaptation of radiotherapy during the course of treatment.
Introduction
Radiation in conjunction with chemotherapy or other targeted therapies is used to treat the majority of lung and head and neck cancer patients. The overall radiation dose is fractionated and delivered over a period of 5-7 weeks (2 Gy/day, 5 days/week) because dose fractionation is believed to improve tumor oxygenation and, hence, overall cell kill [1, 2]. An outstanding challenge in optimizing the efficacy of such treatment resides in determining the degree of radiosensitivity associated with a specific patient’s disease and the extent of tumor response to radiation. There are no accepted methods to determine treatment response either before or during the early stages of therapy. Although Human Papilloma Virus (HPV)-negative head and neck squamous cell carcinomas (HNSCCs) are associated with significantly worse outcomes compared with HPV-positive tumors [3, 4], HPV status is not used to guide treatment of HNSCC. Currently, X-ray Computed Tomography (CT) or Magnetic Resonance Imaging (MRI) is used to determine tumor shrinkage about 2-3 weeks after completion of therapy. Positron Emission Tomography (PET) of fluorodeoxyglucose (FDG) uptake to measure functional tumor response is recommended about 8-12 weeks after completion of therapy to avoid false positives. Hence, patients who undergo the full treatment regimen and are later identified as non-responders are exposed to the toxic side effects of ineffective therapy for the full duration of the treatment regimen. Identifying patients with radiation-resistant tumors, prior to commencing treatment or immediately after, would significantly improve treatment response rates and help non-responding patients avoid the toxic side effects of ineffective radiation therapy.
Seeking to address this unmet need, molecular alterations in the tumor microenvironment in response to radiation therapy have been studied from multiple points of view including tumor hypoxia [5–7], cell repopulation [8–10], and genetic mutations involved in DNA repair pathways [11]. However, elucidation of serum and/or imaging biomarkers for accurate patient stratification and continuous assessment of therapy response, and their translation to the clinic has proven to be challenging. In an effort to develop better phenotypic strategies that could aid the clinical practice of radiation oncology, we propose an entirely complementary optical tool to the existing imaging arsenal featuring Raman scattering to non-invasively quantify the putative differences in the molecular milieu of radiosensitive and radioresistant tumors.
Raman spectroscopy offers a non-ionizing, label-free and highly specific technique for molecular characterization of the tumor and its microenvironment [12, 13]. It relies on the inelastic scattering of light, arising from its interactions with the biological specimen, to quantify the unique vibrational modes of molecules within its native context [14]. Raman spectroscopy offers the ability to probe biomolecular changes both in vivo and ex vivo, and interrogate complex molecular heterogeneity directly from cells and tissues [15]. Recent studies by us and others have harnessed vibrational profiles for objective recognition of epithelial and stromal changes in cancers [16–22]. Emerging data suggests the presence of post-radiation alterations in Raman spectral features and biomolecular differences between cell lines of varying radiosensitivity [23, 24]. Krishna and colleagues showed that radiation-induced changes in Raman spectra could be used to differentiate treatment responders and non-responders in excised cervical cancers; however, pre-treatment Raman spectra were incapable of identifying radiation response [25]. Furthermore, a recent Raman spectroscopic study on ex vivo tumor xenografts by Jirasek and co-workers identified elevated levels of glycogen in tumors exposed to a single, high radiation dose of 15 Gy [26]. While these reports underscore the promise of Raman spectroscopy in detecting radiation-induced changes, these measurements were performed on cells or tumor xenografts following a single radiation dose. More systematic studies that examine the sensitivity of Raman spectroscopy to changes in the tumor microenvironment when subjected to fractionated, clinically-relevant radiation doses have been lacking. Such measurements would provide a better understanding of molecular modifications resulting from fractionated dosing and, ultimately, facilitate a personalized treatment approach. In addition, spectral markers of intrinsic radiation resistance that can be identified in tumors even before commencing therapy could provide a paradigm shift in determining treatment regimen.
The goal of our study was to leverage Raman spectroscopy to investigate biomolecular changes within tumor xenografts in response to fractionated radiation therapy, and to determine the feasibility of differentiating treatment response from failure. Additionally, we sought to determine whether classifier models based on Raman spectral markers could be used to distinguish between untreated radiation-resistant and sensitive tumors. To accomplish our goals, we used two sets of radiation-sensitive and radiation-resistant cell lines. First, we employed a recently developed matched model of radiation resistance [27], wherein a radiation-resistant clonal population of cells (rA549) was generated from parental A549 lung cancer cells. Second, we used HNSCC cell lines – UM-SCC-22B (UM22) and UM-SCC-47 (UM47) – for which radiation resistance and sensitivity have been established in previous studies [28]. Raman spectroscopic mapping of excised tumor xenografts (control and radiated tumors) grown from all four cell lines revealed consistent compositional alterations based on tumor type and in response to a radiation dose of only 2 Gy. Using multivariate curve resolution-alternating least squares (MCR-ALS), we translated the spectral information to uncover changes in lipid, collagen, and glycogen content. Data from both lung and head and neck (henceforth referred to as HN) tumors show consistently higher changes in lipid and collagen content in radiation-sensitive tumors that were treated with radiation compared with their radiation-resistant counterparts. Definition of the tumor phenotypes in terms of quantitative spectral features corresponding to key biomolecules also enabled the development of classifier models that exhibit high accuracy in discriminating between radiation-resistant and sensitive tumors. Furthermore, our use of an isogenic radiation-resistant clone allowed, to the best of our knowledge, the first determination of discriminative Raman features in untreated tumors thereby offering fresh insights into specific molecular roles underlying intrinsic radiation resistance. Taken together, our findings highlight the potential of Raman spectroscopic imaging as a label-free, non-ionizing tool whose in vivo translation would permit monitoring of therapeutic effects with finer temporal resolution than is possible at the present time and potentially enable stratification of radiation-resistant patient cohorts.
Materials and Methods
Cell culture.
Human lung carcinoma A549 cells were purchased from American Type Culture Collection (ATCC; CCL185) and were authenticated using short tandem repeat (STR) profiling. A549 cells were grown in Ham’s F-12K (Kaighn’s) Medium mixed with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. These cells were irradiated at an average dose of 2.2 Gy every three days using an orthovoltage X-ray irradiator (CP-160, Faxitron X-Ray Corp. Wheeling, IL) for a cumulative dose of 55 Gy (25 fractions) to create the radiation-resistant cell clones (rA549) [27]. UM-SCC-22B and UM-SCC-47 were purchased from EMD Millipore and cultured in a mixture of Dulbecco’s Modified Eagle Medium (DMEM), 10% Fetal Bovine Serum (FBS), 1% Penicillin-Streptomycin, 1% non-essential amino acids (NEAA), and 1% L-Glutamine. All cell lines used in this study tested negative for mycoplasma and were authenticated using short tandem repeat (STR) profiling.
Fractionated therapy of tumor xenografts.
A schematic representation of this study design is presented in Fig. 1A. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Arkansas (Protocols 16022 and 18061). Athymic nu/nu mice were injected with a subcutaneous bolus of cells suspended in 100 μl of serum and media-free saline (10 million for A549 and rA549 cells, 2 million for UM-SCC-22B and UM-SCC-47) to grow tumor xenografts. Once tumor volume reached 200 mm3, mice were randomized to either radiation (XT) or control (NT) groups, as presented in Table 1. Fractionated radiation therapy was administered using an X-RAD 320 biological irradiator (Precision X-Ray, North Branford, CT) as four 2 Gy fractions delivered over two consecutive weeks (total dose of 8 Gy), as described by others [28]. All animals completed the treatment. Tumor volumes were monitored using Vernier calipers, and tumors were excised when the majority of untreated control tumors had reached 1500 mm3 (approx. 35-50 days after treatment commenced). Tumor volume was calculated according to the equation V = (π/6) × (length) × (width) × (height). A comparison of tumor xenograft volumes is presented in Fig. SF1. After excision, tumors were embedded in OCT mounting medium, snap-frozen, and stored at −80 °C.
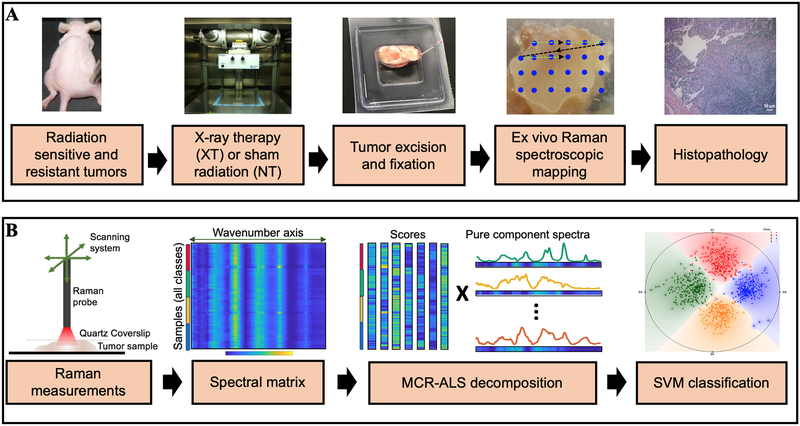
Figure 1. Raman spectroscopic study of radiation response and resistance.
Overview of the (A) experimental and (B) data analysis workflow for Raman spectroscopic mapping in lung and head and neck tumor xenografts, of differential intrinsic radioresistance, subjected to radiation therapy. Details of the individual steps are provided in the methods section.
Table 1.
Cell lines used to generate tumor xenografts in the study
| Lung Tumors | Head and neck tumors | ||||
|---|---|---|---|---|---|
| Tumor group | Class label | Number of tumors | Tumor group | Class label | Number of tumors |
| A549-NT | A549-NT | 5 | UM-SCC-22B-NT | UM22-NT | 6 |
| A549-XT | A549-XT | 4 | UM-SCC-22B-XT | UM22-XT | 7 |
| rA549-NT | rA549-NT | 5 | UM-SCC-47-NT | UM47-NT | 9 |
| rA549-XT | rA549-XT | 5 | UM-SCC-47-XT | UM47-XT | 10 |
| Total number of tumors | 19 | Total number of tumors | 32 | ||
Raman spectroscopy.
The frozen tumors were thawed and fixed in 10% neutral buffered formalin prior to making Raman measurements. The fixed tumors were rinsed in PBS and sandwiched between a quartz cover slip and aluminum block to maintain a constant distance between the tissue and probe. The flattened tumors were scanned using a fiber-optic probe-based portable Raman spectroscopy system [16, 29]. Briefly, the custom-built system consists of an 830 nm diode laser (500 mW maximum power, Process Instruments) as the excitation source, and a combination of a spectrograph (Holospec f/1.8i, Kaiser Optical Systems) and a thermoelectrically (TE)-cooled CCD camera (PIXIS 400BR, Princeton Instruments) for recording spectra. Laser delivery and collection of back-scattered light was achieved via a lensed fiber-optic bundled Raman probe (probe diameter: 2 mm; Emvision LLC) with an estimated tissue sampling volume of 1 mm3. The laser power at the tumor samples was maintained at ~ 20 mW in this study. Acquisition time for each spectrum was 10s (10 accumulations of 1 second each to prevent saturation of CCD). Tissue dehydration due to laser exposure was prevented by intermittent addition of PBS. Raster scanning of the probe using motorized translation stages (T-LS13M, Zaber Technologies Inc., travel range: 13 mm) and spectral acquisition were performed on each side of the flattened tumors (~ 100 mm2) using a LabVIEW interface. About 4100 and 7000 spectra were acquired from the nineteen lung and thirty-two HN tumor xenografts, respectively.
Data analysis.
Figure 1B illustrates the data analysis workflow in this study. All the data analysis in the present study was carried out using scripts written in MATLAB 2017a (Mathworks, Natick, MA, USA) environment unless otherwise stated. The wavenumber axis of the Raman system was calibrated using 4-acetamidophenol. The fingerprint wavenumber region (600 - 1800 cm−1) was chosen for further analysis. The Raman spectra recorded from the tumors were subjected to a fifth order best-fit polynomial-based fluorescence removal and cosmic ray removal using median filtering. The spectra were then vector normalized (such that their Euclidean norm is set to unity) to minimize the effects of potential variations in laser power at the sample. The normalized spectra were used without any spatial averaging in the analysis. Multivariate curve resolution - alternating least squares (MCR-ALS) was employed to recover the pure spectral profiles of the chemical constituents of the tissue specimen without a priori information of the composition of the specimen [30]. The decomposition is achieved through an iterative optimization routine under non-negativity constraint on pure spectral (loadings) and concentration (scores) matrices. The non-negativity constraints enable us to interpret the unresolved specimen spectra in the form of loadings that represent spectra of pure (or enriched) biochemical components and the corresponding scores that provide a measure of abundance of the particular component. Additionally, spectral equal length constraint is imposed on the pure spectra to facilitate comparison of corresponding scores across the classes (treatment groups). The normalized scores corresponding to each key biological constituent were compared across different classes through box and whisker plots with conventional thresholds. The spatial heterogeneity in the score maps of major biological constituents for each tumor was quantified in terms of distributional homogeneity index (DHI), as defined elsewhere in the literature [31]. The significance of differences in medians of constituent scores across studied classes were assessed based on two-sided Wilcoxon rank sum test statistics. A conventional criterion of p-value less than 0.05 was used to consider the medians different. The differences between the groups were quantified in terms of effect size calculated using the Wendt formula for rank biserial correlation [32].
Support vector machine (SVM) was used to develop a decision algorithm to identify radiation treatment response and to predict resistant phenotype prior to treatment. SVM is a nonlinear classification method wherein classification is achieved by solving a constrained quadratic optimization problem to obtain separating boundaries between the classes in higher dimensional spaces [33]. In this study, the LIBSVM library [34] was used to develop a C-SVM classifier. The background-corrected spectra were used along with tumor group labels for each group studied, without any spatial averaging. A radial basis function kernel with a Gaussian envelope was employed to enable nonlinear mapping of the input feature space, and the optimal C-SVM parameters (i.e. cost and kernel parameter gamma) were selected using a k-fold cross validation-based grid search algorithm. A leave-one-mouse-out analysis was conducted in which spatially distinct spectra belonging to each mouse were eliminated from the training dataset and the resulting binary SVM classifiers for pairs of classes of interest were tested using the spectra belonging to the left-out mouse. Each mouse specimen was assigned an overall predicted class label if more than 90% of its spectra were predicted as belonging to that class; otherwise, the specimen was labeled as unclassified if the desired confidence level was not achieved. Randomized class equalization was performed iteratively prior to implementing SVM classification in order to avoid skewing the model due to varying class sizes.
Histopathology.
The tumors were stored in 70% ethanol after acquisition of Raman spectra and submitted to the Phenotyping and Pathology Core at Johns Hopkins Medical Institutions. The formalin-fixed tumors were embedded in paraffin and sectioned serially onto glass slides for histology. Hematoxylin and eosin (H&E) staining, Masson’s trichrome staining for collagen and Periodic acid Schiff (PAS) staining (without Hematoxylin counterstain) for glycogen were performed by the Core according to standard protocols. The stained slides were imaged using a Leica DMi8 inverted optical microscope. In addition, Oil Red O staining for lipids was performed on frozen tumor sections according to standard IHC protocols and imaged using a Nikon fluorescence microscope.
Results
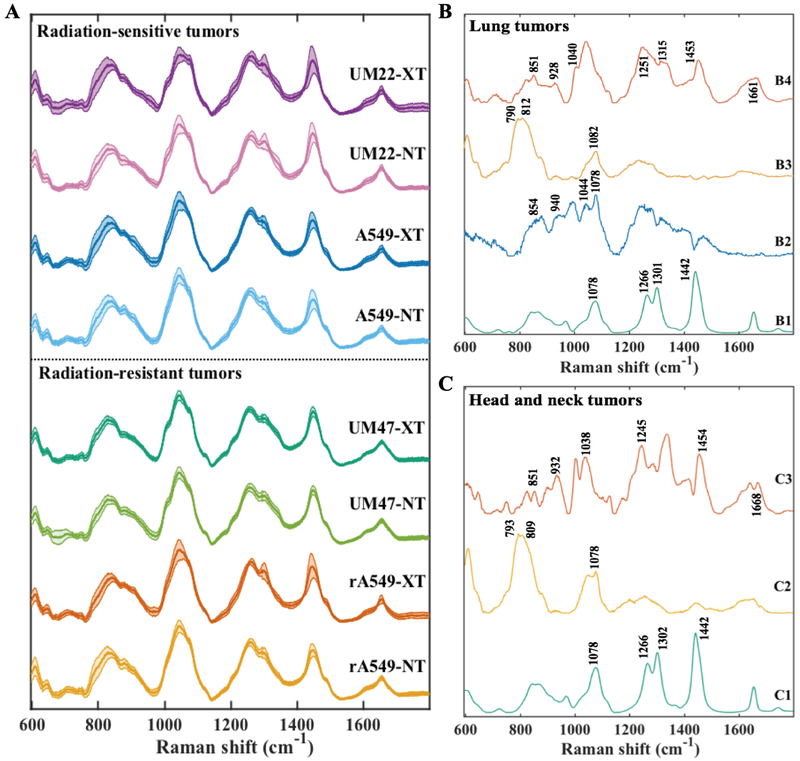
To capture the tumor heterogeneity and variance arising from differential response to treatment, lung and HN tumors in each group – radiation treated (XT) and controls (NT), were mapped to obtain spatially distinct Raman spectra from each specimen (Fig. 2A). Each map had an average of 218 spectra (ranging between 50 and 334 spectra depending on the size of the tumor). Except for this visualization (Fig. 2A), the spectra collected from each tumor specimen were treated individually without any spatial averaging in all analyses. The spectra across all the classes show prominent peaks at 1045 cm−1 (glycogen), 1256 cm−1 (glycogen), 1301 cm−1 (CH vibration in lipids), 1448 cm−1 (CH2 bending modes in lipids and collagen), and 1656 cm−1 (amide-I feature of proteins with potential contribution from C=C stretching in lipids). To discern possible molecular differences, the spectral datasets were decomposed into key compositional biomolecular signatures that were compared across the different groups.
Figure 2. Raman spectra of radiation-resistant and sensitive tumors.
(A) Mean Raman spectra (with the shadow representing 1 standard deviation) acquired from lung tumors derived from parental A549 and radiation-resistant (rA549) cells and head and neck tumors derived from radiation-sensitive UM-SCC-22B and radiation-resistant UM-SCC-47 cells. Each of the types were either exposed to fractionated radiation (XT) or sham radiation (NT). (B) A subset of relevant MCR loading vectors derived from the spectra of lung tumors belonging to all the four study classes. The spectra B1 through B4 represent lipid-rich, glycogen-rich, nucleic acid-rich and collagen-rich loadings, respectively. (C) A subset of relevant MCR loading vectors derived from the spectra of tumors belonging to head and neck tumor dataset. The spectra C1 through C3 represent lipid-rich, nucleic acid-rich and collagen-rich loadings, respectively.
To achieve this decomposition, we performed MCR-ALS with 7 components to obtain a loadings matrix containing the ‘pure component’ basis spectra and a scores matrix containing the weights of each of the seven components for all the spectra in the dataset. Figures 2B and 2C illustrate the relevant MCR loadings that present Raman features corresponding to key tissue constituents in the lung and HN tumor cohorts, respectively. The remaining loadings, that do not show direct correspondence to the vibrational signature of a prominent molecular constituent or stem from the presence of formalin (spectral contaminant in the tissue specimen), are provided in the Supporting Information Fig. SF2. As seen in Fig. 2B, spectrum B1 shows prominent peaks at 1078 cm−1, 1266 cm−1, 1301 cm−1, 1442 cm−1 and 1654 cm−1 that are characteristic of lipids, specifically triglycerides. Spectral pattern B2 resembles the Raman spectral profile of glycogen with peaks at 708 cm−1, 940 cm−1, 1044 cm−1, 1078 cm−1 and 1256 cm−1. Furthermore, B3 has spectral features corresponding to nucleic acids at 790 cm−1, 812 cm−1 and 1082 cm−1, while loading B4 has peaks at 851 cm−1, 928 cm−1, 1040 cm−1, 1251 cm−1, 1315 cm−1, 1453 cm−1 and 1661 cm−1, which are characteristic of collagen. The loadings derived from the HN tumor dataset C1, C2 and C3 presented features similar to the loadings B1, B3 and B4, respectively. The detailed peak allocations of all the features of the 7 loadings derived from lung and HN tumor datasets have been tabulated in Tables TS1 and TS2 (Supporting Information), respectively. It is worth noting that the MCR decomposition of HN tumors did not reveal a glycogen-rich loading. The minor spectral features that stem from the use of formalin as a fixative – at 1490 cm−1 and 1040 cm−1 – present themselves in a single MCR loading, thus indicating that the effects of formalin fixation may be digitally removed [35].
Representative MCR score maps (abundance maps) of the observed relevant loadings across the treatment groups have been provided in Supporting Information Fig. SF3. To characterize the spatial heterogeneity in biochemical composition of the studied tumors, we measured the distributional homogeneity index (DHI) for each MCR score map. DHI is a measure of deviation of the spatial distribution in compositional maps from their randomized counterparts [31]. Thus, DHI is positively correlated with heterogeneity in abundance maps, with a value of one representing homogeneous (or randomized) distribution and higher values indicative of localization of constituents. Therefore, from the observed large values of DHI (Fig. SF3), it is evident that the abundance maps for relevant loadings obtained for lung and HN tumors exhibit substantial spatial heterogeneity in the tumor samples.
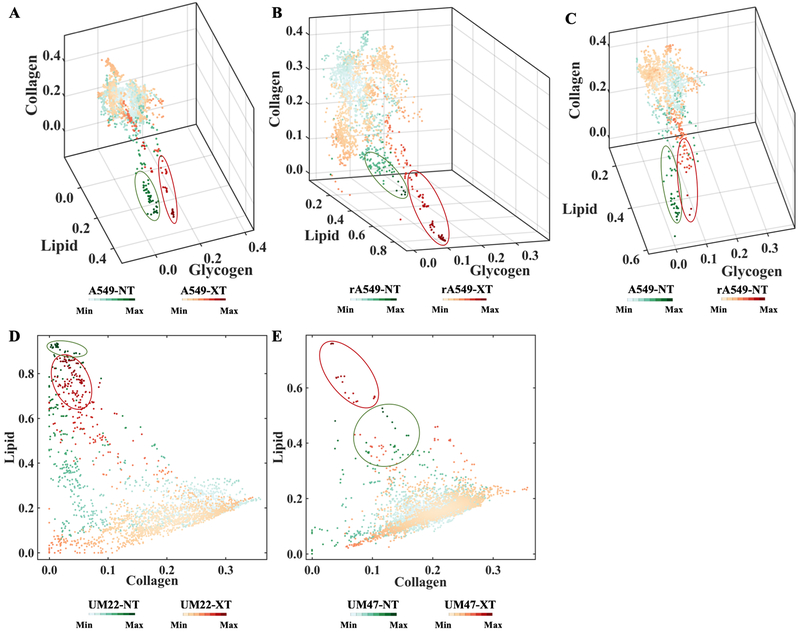
The primary objectives of our study were to investigate differences in radiation-induced microenvironmental changes between resistant and sensitive tumors, and whether such molecular alterations were consistent in tumors derived from two different sites – lung and HN. Accordingly, NT vs. XT comparisons for each of the 4 cell lines were performed to evaluate the differential response of sensitive (A549, UM-SCC-22B) and resistant (rA549, UM-SCC-47) tumors. A secondary objective was to determine if these molecular features could distinguish between untreated resistant and sensitive tumors. Since the HN tumors stemmed from different cell lines, this specific comparison was only performed in the lung cohort (A549-NT vs rA549-NT) to identify intrinsic differences underlying radiation resistance. Here, we used density scatter plots as they allow better visualization of large datasets by avoiding overlap in regions of high density. Figures 3A-C present three-dimensional (3D) density plots using the normalized MCR-ALS scores corresponding to the lipid-rich, glycogen-rich and collagen-rich loadings of A549 and rA549 tumors. Similarly, two-dimensional (2D) density plots with only lipid-rich and collagen-rich loadings are shown in Figs. 3D-E for the HN tumors. Both sets of density plots underscore expected tumor heterogeneity and critically, the presence of group-specific local spectral clustering, which is reflected in the higher density of co-located points (circled in the figure).
Figure 3. Qualitative visualization of MCR-ALS scores of Raman spectra.
(A-C) Three-dimensional density plots showing the distribution of normalized scores of lipid-rich, collagen-rich and glycogen-rich MCR-ALS loadings showing radiation induced differences in sensitive lung tumors (A549-NT vs A549-XT), radiation induced differences in resistant lung tumors (rA549-NT vs rA549-XT) and pre-radiation differences between sensitive and resistant lung tumors (A549-NT vs rA549-NT), respectively. (D-E) Two-dimensional density plots showing the distribution of normalized scores of lipid-rich and collagen-rich MCR-ALS loadings showing radiation induced differences in sensitive head and neck tumors (UM22-NT vs UM22-XT) and radiation induced differences in resistant head and neck tumors (UM47-NT vs UM47-XT), respectively. The class specific clustering in high density regions are circled.
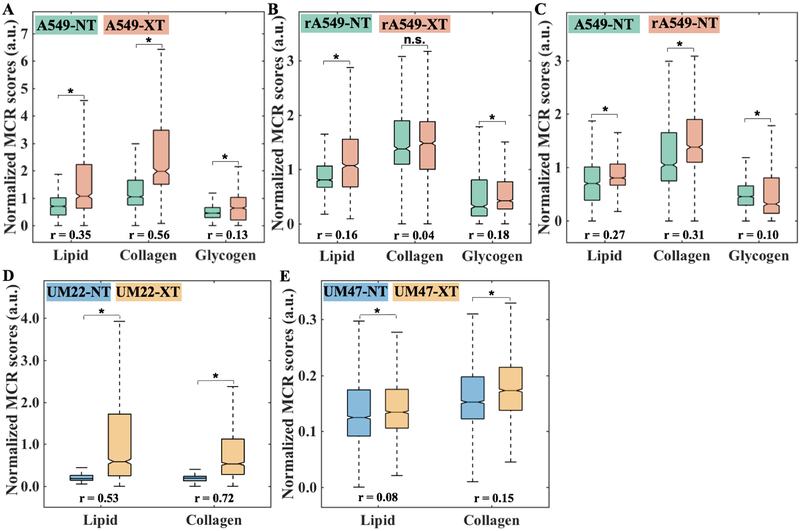
To quantitatively study the differences across the treatment groups, we compared the normalized MCR-ALS scores of lipid-rich, collagen-rich and glycogen-rich loadings. We observed an increase in lipid, collagen, and glycogen levels for both sensitive (Fig. 4A) and resistant (Fig. 4B) lung tumors that were treated with radiation, with a much larger increase in the lipid-rich and collagen-rich signatures in the radiation-sensitive tumors. To examine intrinsic radioresistance, we also compared the scores of these biomolecular components between untreated A549 and rA549 tumors (Fig. 4C). The MCR-ALS scores point to higher lipid and collagen content but lower glycogen content in the resistant tumors compared with the sensitive tumors. Furthermore, we observed very similar effects in the HN tumors with a greater increase in lipid and collagen levels in the radiation-sensitive UM-SCC-22B (Fig. 4D) compared to the radiation-resistant UM-SCC-47 tumors (Fig. 4E). Glycogen, as mentioned previously, was not observed as a significant feature in the HN tumors. Notably, the greater changes in sensitive tumors post-radiation are also borne out by the effect sizes for lipid and collagen content, which are consistently higher for the sensitive tumor cohorts (≥ 0.35 in A549 and UM-SCC-22B; ≤ 0.16 in rA549 and UM-SCC-47).
Figure 4. Quantitative MCR-ALS analysis of Raman spectra.
(A-C) Boxplots of normalized scores of lipid-rich, collagen-rich and glycogen-rich MCR-ALS loadings showing radiation induced differences in sensitive lung tumors (A549-NT vs A549-XT), radiation induced differences in resistant lung tumors (rA549-NT vs rA549-XT) and pre-radiation differences between sensitive and resistant lung tumors (A549-NT vs rA549-NT), respectively. The differences in the scores of lipid and glycogen loadings are statistically significant (indicated by * and n.s. otherwise) at p < 0.001 level (Wilcoxon rank sum test) for each of the three comparisons (A-C), whereas the differences in the scores of collagen loadings are statistically significant only for the comparisons in (A) and (C). (D-E) Boxplots of normalized scores of lipid-rich and collagen-rich MCR-ALS loadings showing radiation induced differences in sensitive head and neck tumors (UM22-NT vs UM22-XT) and radiation induced differences in resistant head and neck tumors (UM47-NT vs UM47-XT), respectively. The differences in the scores of lipid and collagen loadings are statistically significant (indicated by *) at p < 0.001 level (Wilcoxon rank sum test) for both the comparisons. The effect size (r), characterizing magnitude of differences between groups, is provided for each comparison.
While the comparison of MCR scores provides a starting point for delineating the molecular mediators of treatment response/resistance and assessing the predictive power of the spectroscopic data, comparison of the individual component scores alone may not provide a robust diagnostic framework, especially to classify prospective samples. Therefore, we developed decision models based on support vector machines (SVM), a supervised classification method that can deal with ill-posed problems and lead to unique global models [33]. We conducted a leave-one-mouse-out analysis which involved training three separate binary SVM classifiers for each tumor type (lung and HN tumors), corresponding to the three sets of comparisons – RS-NT vs RS-XT, RR-NT vs RR-XT and RS-NT vs RR-NT, respectively. RS and RR indicate radiation-sensitive and radiation-resistant tumor xenografts, respectively. Table 2 shows the aggregated number of mice accurately classified, unclassified, and misclassified (as determined by the criteria detailed in the Methods section) for each of the three comparisons. (The tumor model-specific decomposition is provided in Supporting Information Tables TS3 and TS4.) The leave-one-mouse-out protocol provides satisfactory predictions in all cases with an overall misclassification rate of only ca. 3%. While slightly higher unclassification rates were noted for the lung tumor dataset, incorporation of a larger cohort of animals in the HN tumor data allowed significant reduction in the same. In the latter set of HN tumors, slightly increased unclassification rate (albeit with zero misclassification) was observed for the comparison between treated (UM47-XT) and untreated (UM47-NT) radiation-resistant HN tumors. We attribute this increased unclassification to smaller effect size observed in MCR-ALS-based univariate comparisons and the high classification threshold used in our leave-one-mouse-out analysis. For example, relaxing the threshold to 80% level reduced the unclassification rates of comparison between UM47-NT and UM47-XT significantly (2/19 mice compared to 7/19 mice at 90% level).
Table 2.
Results of binary leave-one-mouse-out SVM analyses
| Number of mice classified accurately, unclassified and misclassified | |||||
|---|---|---|---|---|---|
| RS-NT | RS-XT | RR-NT | RR-XT | ||
| Binary SVM comparisons | RS-NT vs RS-XT | (11+0+0)/11 | (8+2+1)/11 | - | - |
| RR-NT vs RR-XT | - | - | (9+5+0)/14 | (12+2+1)/15 | |
| RS-NT vs RR-NT | (10+1+0)/11 | - | (12+2+0)/14 | - | |
| RS-XT vs RR-XT | - | (10+0+1)/11 | - | (14+1+0)/15 | |
Finally, to verify the lack of spurious correlations in the dataset [36], we repeated our leave-one-mouse-out analysis using the same spectral dataset, but with randomly assigned class labels instead of their original labels for each comparison. Average correct classification rate of ca. 57% for both lung tumor and HN tumor datasets, were obtained (comparable to the random likelihood of selection of the true class label – 50%). Taken together, the results of the SVM-derived classifier model studies demonstrate the utility of the Raman spectroscopic data in capturing distinct radiobiological responses in radiosensitive and radioresistant lung and HN tumor xenografts.
Discussion
A fundamental principle of personalized medicine is to design treatment strategies that tackle the biological heterogeneity characteristic of cancer in order to achieve maximal tumor control while minimizing toxicity. The lack of suitable imaging tools, which can identify patients unlikely to benefit from radiation and perform frequent response monitoring to better inform treatment doses and fractionation schemes, remains a major impediment in customizing radiotherapy. In contrast to existing clinical technologies, optical spectroscopy offers a non-invasive or minimally invasive route to providing real-time evaluation of treatment response based on functional and biomolecular changes in the tumor microenvironment. In this study, we demonstrate the utility of using label-free Raman spectroscopy in conjunction with chemometric analysis to reveal distinct biomolecular changes in radiation-resistant and sensitive tumors when subjected to fractionated, clinically-relevant radiation doses. Specifically, MCR-ALS analysis reveals consistent differences in lipid and collagen content post-radiation in the microenvironment of lung and head and neck tumor xenografts with especially pronounced changes in the radiation-sensitive cases. By leveraging SVM-derived classifiers, we are also able to differentiate between vibrational signatures recorded from untreated radiation-sensitive and radiation-resistant tumors indicating the potential for future Raman spectroscopic application to not only monitor but also predict radiation response in individuals.
The specific biomolecular features derived using MCR-ALS analysis have been previously studied in the context of cellular response to radiation. Hypoxia and its transcription factor, hypoxia-inducible factor (HIF-1), have been shown to promote extracellular matrix (ECM) remodeling and play an important role in promoting fibrosis [37, 38] and collagen biogenesis [39, 40]. Indeed, our recent investigation of cellular metabolism showed a significant increase in HIF-1α expression after radiation in both the A549 and rA549 cells [41]. We reason that the radiation-induced increase in HIF-1 content is, in part, responsible for promoting collagen deposition in the A549 and rA549 tumors. The increased collagen content may also be explained by the actions of growth factors, such as TGF-β, which are recruited in response to HIF-1α-stimulated macrophage accumulation [42]. Overexpression of TGF-β serves as a chemoattractant for the recruitment of fibroblasts, and may drive the increase in collagen-rich MCR scores upon irradiation for both sensitive and resistant tumors [43].
Previous work has established that de novo lipogenesis protects cancer cells from external insults, such as oxidative stress, and that inhibition of lipogenesis increases oxidative stress-induced cell death [44]. The increased lipid content observed in both groups of radiated tumors in our study could be attributed to such a cellular defense mechanism in response to radiation-induced oxidative stress. Although the exact mechanism for increased lipid content needs further investigation, studies have found elevated levels of fatty acid synthase (FASN) in radiation-resistant HN cancer cells [45]. Furthermore, inhibition of FASN decreased cellular survival of these radiation-resistant cancer cells. FASN is a key player in lipogenesis and has also been shown to be a prognostic indicator of radiation resistance in clinical nasopharyngeal carcinoma [46]. Taken together, these studies highlight the potential of lipids to serve as a powerful biomarker of radiation resistance.
Our findings of increased glycogen content in response to radiation in the radiation-resistant rA549 tumors are consistent with a recent in vitro study by Matthews et al. that reported an increase in radiation-induced glycogen in the relatively radiation-resistant MCF7 breast cancer and H460 lung cancer cell lines [23]. Jirasek and colleagues have reported substantially increased glycogen content in radiated non-small cell lung tumor xenografts compared with non-radiated xenografts [26]. However, while our study also found significantly higher glycogen content in the radiation-sensitive A549 tumors, Mathews and colleagues found no changes in glycogen content in the radiation sensitive LNCaP prostate cancer cells. These differences could be principally attributed to the different nature of the measurement specimen (cells vs. tissue). The increase in intracellular glycogen in their study was attributed to the phosphorylation of glycogen synthase kinase (GSK-3β), a negative regulator of glycogen synthase, which mediates the final step of glycogen synthesis. Deactivation of GSK-3β activity through phosphorylation has been shown to play a critical role in the acquisition of radiation resistance in cancer cells [47]. Although a direct role for glycogen in conferring protection from radiation has not been established, the availability of increased glycogen reserves could provide cancer cells with glucose through glycogenolysis during radiation-induced oxidative stress. Glucose utilization through the pentose phosphate pathway can lead to the generation of glutathione, which is an important scavenger of radiation-induced free radicals. Our findings expand on these determinations by providing the first direct comparisons of glycogen levels in matched models of resistant and sensitive tumors. However, the smaller effect sizes observed in the lung tumors coupled with its absence in the HN tumors suggest that further investigations are necessary to evaluate the clinical utility of glycogen as a marker of treatment response.
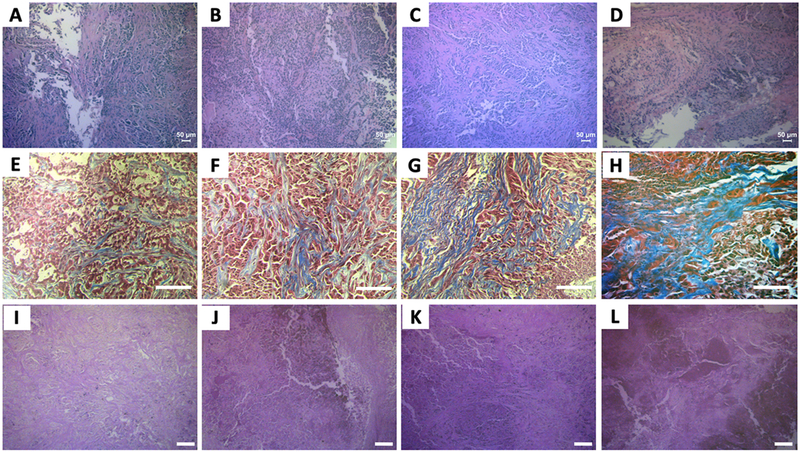
To examine the histological basis of the Raman spectroscopic determinations, tumor sections were stained with hematoxylin and eosin (H&E), Masson’s trichrome, periodic acid-Schiff (PAS) and oil red O (ORO) – for cellular morphology, collagen, glycogen and lipid, respectively (Fig. 5A-L and SF4). While tumor morphology was found to be largely similar across all tumor groups, H&E-stained images identified high levels of necrosis as well as fibrosis in the tumor groups subjected to radiation therapy (XTs) compared with the untreated control tumors (NTs). Specifically, necrosis and fibrosis were found in all groups, and were correlated, with an increase in necrosis associated with an increase in fibrosis. Specifically, within the lung tumor group, the A549-NT and rA549-NT tumors demonstrated higher tumor burdens and lower levels of necrosis and fibrosis. Within the HN tumor group, the lowest levels of necrosis were observed in the 22B-NT tumors while the highest levels were observed in the 47-XT group. Both 22B-XT and 47-NT had similar and intermediate levels of necrosis and fibrosis. This is likely due to the bulkier tumors observed in the 47-NT group. This tumor overgrowth is often associated with a degenerative type of necrosis secondary to ischemia. These histopathological results were largely consistent with the findings from Raman spectral analysis. Furthermore, using bright field images of the Masson’s trichrome and PAS stained slides, we observed increased levels of collagen and glycogen after radiation in both the sensitive and resistant tumors. In addition, collagen content was noticeably higher in the resistant tumors prior to radiation compared with the sensitive tumors. The histological images for the HN tumors have been provided in Supporting Information Fig. SF4.
Figure 5. Histologic assessment of radiation sensitivity and resistance.
Top (A–D), middle (E–H) bottom (I-L) panels display representative microscopic images of H&E, Masson’s trichrome and PAS stained slides, respectively. The columns of panels from left to right - (A, E and I), (B, F and J), (C, G and K) and (D, H and L), respectively, represent fields of view from tumors belonging to the treatment groups - A549-NT, A549-XT, rA549-NT and rA549-XT. The scale bars in panels A-D represent 50 μm and panels E-L represent 100 μm.
In summary, we have used Raman spectroscopic mapping for quantitative assessment of the molecular composition of lung and HN tumors subjected to radiation therapy, and shown that such measurements offer a reliable, non-perturbative method to probe radiation-induced alterations. These findings represent, to the best of our knowledge, the first report comparing the microenvironmental response to radiation in tumor xenografts from different organ sites using optical spectroscopy. Together, our results provide promising evidence for the clinical translation of Raman spectroscopy to discern molecular markers of radiation response either prior to or during the early stages of treatment using fiber optic probes in accessible tumors.
Towards that goal, there are two major focal points of our future investigations. First, the clinical radiation dose of 2 Gy that was used in this study will be delivered on successive days as is usually performed in the clinic. The treatment regimen used here is similar to previous approaches used to establish radiation sensitivity and resistance in tumor xenograft models [28]. Second, radiation-induced microenvironmental changes were evaluated ex vivo from excised tumors. Our evaluation of differences related to intrinsic radiation resistance were conducted on the untreated control tumors. While we observed appreciable differences between the A549-NT and rA549-NT tumors that can be attributed to radiation resistance, these results do not exactly predict if these tumors would go on to respond or fail treatment. The results presented here provide an opportunity to further explore the prediction of long-term treatment response based on measurements made prior to commencing treatment in radiation-naïve tumors in vivo. Our next study would involve in vivo pre-treatment measurements on tumors as well continuous measurements during treatment administered on successive days to enable longitudinal treatment monitoring. Our recent work using diffuse reflectance spectroscopy identified changes in tumor oxygenation in the A549 and rA549 tumors within 48 hours post-radiation; however, minimal or no differences in oxygenation were observed at the time of tumor excision [48]. Therefore, it is possible that the magnitude of radiation-induced biomolecular changes, as sensed by in vivo Raman measurements, will be greater immediately after radiation therapy. Moreover, the fabrication of appropriate probes [49] as well as the emergence of vibrational spectroscopic imaging systems that are already being adopted in clinical studies [50] indicate that translation of our proposed approach is feasible. Hence, based on our current findings as well as these technological developments, we envision that Raman measurements will be employed in the near future to guide treatment planning based on the inclusion of vibrational spectral profiles of a patient’s tumor.
Supplementary Material
Statement of Significance.
Findings highlight the sensitivity of label-free Raman spectroscopy to changes induced by radiation therapy and indicate the potential to predict radiation resistance prior to commencing therapy.
Acknowledgements
I. Barman acknowledges the support from the National Institute of Biomedical Imaging and Bioengineering (2-P41-EB015871-31), National Institute of General Medical Sciences (DP2GM128198) and Johns Hopkins University Catalyst Award. N. Rajaram received funding from the Arkansas Biosciences Institute. R.P.M. Dings acknowledges funding from the Medical Research Endowment Fund, the Winthrop P. Rockefeller Cancer Institute, and the Center for Microbial Pathogenesis and Host Inflammatory Responses (P20GM103625).
Footnotes
Conflict of interest: The authors disclose no potential conflicts of interest.
References
- 1.Fowler J: The rationale of dose fractionation In: The Relationship of Time and Dose in the Radiation Therapy of Cancer. edn.: Karger Publishers; 1969: 6–23. [Google Scholar]
- 2.Withers HR: Biologic basis for altered fractionation schemes. Cancer 1985, 55(S9):2086–2095. [DOI] [PubMed] [Google Scholar]
- 3.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML: Improved survival of patients with human papillomavirus–positive head and neck squamous cell carcinoma in a prospective clinical trial. Journal of the National Cancer Institute 2008, 100(4):261–269. [DOI] [PubMed] [Google Scholar]
- 4.Lindel K, Beer KT, Laissue J, Greiner RH, Aebersold DM: Human papillomavirus positive squamous cell carcinoma of the oropharynx: a radiosensitive subgroup of head and neck carcinoma. Cancer 2001, 92(4):805–813. [DOI] [PubMed] [Google Scholar]
- 5.Brizel DM, Dodge RK, Clough RW, Dewhirst MW: Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiotherapy and Oncology 1999, 53(2):113–117. [DOI] [PubMed] [Google Scholar]
- 6.Brizel D, Sibley G, Prosnitz L, Scher R, Dewhirst M: Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. International Journal of Radiation Oncology* Biology* Physics 1997, 38(2):285–289. [DOI] [PubMed] [Google Scholar]
- 7.Nordsmark M, Bentzen S, Rudat V, Brizel D, Lartigau E, Stadler P, Becker A, Adam M, Molls M, Dunst J: Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiotherapy and Oncology 2005, 77(1):18–24. [DOI] [PubMed] [Google Scholar]
- 8.Baumann M, Herrmann T, Koch R, Matthiessen W, Appold S, Wahlers B, Kepka L, Marschke G, Feltl D, Fietkau R: Final results of the randomized phase III CHARTWEL-trial (ARO 97-1) comparing hyperfractionated-accelerated versus conventionally fractionated radiotherapy in non-small cell lung cancer (NSCLC). Radiotherapy and Oncology 2011, 100(1):76–85. [DOI] [PubMed] [Google Scholar]
- 9.Turrisi AT, Kim K, Blum R, Sause WT, Livingston RB, Komaki R, Wagner H, Aisner S, Johnson DH: Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. New England Journal of Medicine 1999, 340(4):265–271. [DOI] [PubMed] [Google Scholar]
- 10.Saunders M, Dische S, Barrett A, Harvey A, Gibson D, Parmar M, Committee CS: Continuous hyperfractionated accelerated radiotherapy (CHART) versus conventional radiotherapy in non-small-cell lung cancer: a randomised multicentre trial. The Lancet 1997, 350(9072):161–165. [DOI] [PubMed] [Google Scholar]
- 11.Wei Q, Cheng L, Hong WK, Spitz MR: Reduced DNA repair capacity in lung cancer patients. Cancer research 1996, 56(18):4103–4107. [PubMed] [Google Scholar]
- 12.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS: Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322(5909):1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matousek P, Stone N: Development of deep subsurface Raman spectroscopy for medical diagnosis and disease monitoring. Chem Soc Rev 2016, 45(7):1794–1802. [DOI] [PubMed] [Google Scholar]
- 14.Matthaus C, Krafft C, Dietzek B, Brehm BR, Lorkowski S, Popp J: Noninvasive imaging of intracellular lipid metabolism in macrophages by Raman microscopy in combination with stable isotopic labeling. Analytical chemistry 2012, 84(20):8549–8556. [DOI] [PubMed] [Google Scholar]
- 15.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS: Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322(5909):1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paidi SK, Rizwan A, Zheng C, Cheng M, Glunde K, Barman I: Label-Free Raman Spectroscopy Detects Stromal Adaptations in Premetastatic Lungs Primed by Breast Cancer. Cancer research 2017, 77(2):247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winnard PT Jr, Zhang C, Vesuna F, Kang JW, Garry J, Dasari RR, Barman I, Raman V: Organ-specific isogenic metastatic breast cancer cell lines exhibit distinct Raman spectral signatures and metabolomes. Oncotarget 2017, 8(12):20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barman I, Dingari NC, Saha A, McGee S, Galindo LH, Liu W, Plecha D, Klein N, Dasari RR, Fitzmaurice M: Application of Raman spectroscopy to identify microcalcifications and underlying breast lesions at stereotactic core needle biopsy. Cancer research 2013, 73(11):3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sathyavathi R, Saha A, Soares JS, Spegazzini N, McGee S, Dasari RR, Fitzmaurice M, Barman I: Raman spectroscopic sensing of carbonate intercalation in breast microcalcifications at stereotactic biopsy. Scientific reports 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holton S, Walsh M, Kajdacsy-Balla A, Bhargava R: Label-free characterization of cancer-activated fibroblasts using infrared spectroscopic imaging. Biophysical journal 2011, 101(6):1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak JT, Kajdacsy-Balla A, Macias V, Walsh M, Sinha S, Bhargava R: Improving prediction of prostate cancer recurrence using chemical imaging. Scientific reports 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barroso EM, Smits RW, van Lanschot CG, Caspers PJ, ten Hove I, Mast H, Sewnaik A, Hardillo JA, Meeuwis CA, Verdijk R: Water concentration analysis by Raman spectroscopy to determine the location of the tumor border in oral cancer surgery. Cancer research 2016, 76(20):5945–5953. [DOI] [PubMed] [Google Scholar]
- 23.Matthews Q, Isabelle M, Harder SJ, Smazynski J, Beckham W, Brolo AG, Jirasek A, Lum JJ: Radiation-induced glycogen accumulation detected by single cell Raman spectroscopy is associated with radioresistance that can be reversed by metformin. PloS one 2015, 10(8):e0135356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harder SJ, Matthews Q, Isabelle M, Brolo AG, Lum JJ, Jirasek A: A Raman spectroscopic study of cell response to clinical doses of ionizing radiation. Applied spectroscopy 2015, 69(2):193–204. [DOI] [PubMed] [Google Scholar]
- 25.Vidyasagar MS, Maheedhar K, Vadhiraja BM, Fernendes DJ, Kartha VB, Krishna CM: Prediction of radiotherapy response in cervix cancer by Raman spectroscopy: a pilot study. Biopolymers 2008, 89(6):530–537. [DOI] [PubMed] [Google Scholar]
- 26.Harder SJ, Isabelle M, DeVorkin L, Smazynski J, Beckham W, Brolo AG, Lum JJ, Jirasek A: Raman spectroscopy identifies radiation response in human non-small cell lung cancer xenografts. Scientific Reports 2016, 6:21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alhallak K, Jenkins SV, Lee DE, Greene NP, Quinn KP, Griffin RJ, Dings RPM, Rajaram N: Optical imaging of radiation-induced metabolic changes in radiation-sensitive and resistant cancer cells. Journal of Biomedical Optics 2017, 22(6):060502–060502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein AP, Swick AD, Smith MA, Blitzer GC, Yang RZ, Saha S, Harari PM, Lambert PF, Liu CZ, Kimple RJ: Xenograft assessment of predictive biomarkers for standard head and neck cancer therapies. Cancer medicine 2015, 4(5):699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paidi SK, Siddhanta S, Strouse R, McGivney JB, Larkin C, Barman I: Rapid Identification of Biotherapeutics with Label-Free Raman Spectroscopy. Anal Chem 2016, 88(8):4361–4368. [DOI] [PubMed] [Google Scholar]
- 30.Felten J, Hall H, Jaumot J, Tauler R, De Juan A, Gorzsás A: Vibrational spectroscopic image analysis of biological material using multivariate curve resolution-alternating least squares (MCR-ALS). Nature protocols 2015, 10(2):217. [DOI] [PubMed] [Google Scholar]
- 31.Sacré P-Y, Lebrun P, Chavez P-F, Bleye CD, Netchacovitch L, Rozet E, Klinkenberg R, Streel B, Hubert P, Ziemons E: A new criterion to assess distributional homogeneity in hyperspectral images of solid pharmaceutical dosage forms. Analytica Chimica Acta 2014, 818:7–14. [DOI] [PubMed] [Google Scholar]
- 32.WH W: Dealing with a common problem in Social science: A simplified rank-biserial coefficient of correlation based on the U statistic. European Journal of Social Psychology 1972, 2(4):463–465. [Google Scholar]
- 33.Thissen U, Üstün B, Melssen WJ, Buydens LM: Multivariate calibration with least-squares support vector machines. Anal Chem 2004, 76(11):3099–3105. [DOI] [PubMed] [Google Scholar]
- 34.Chang C-C, Lin C-J: LIBSVM: a library for support vector machines. ACM transactions on intelligent systems and technology (TIST) 2011, 2(3):27. [Google Scholar]
- 35.Lyng F, Gazi E, Gardner P: Preparation of tissues and cells for infrared and Raman spectroscopy and imaging. Biomedical Applications of Synchrotron Infrared Microspectroscopy: A Practical Approach Royal Society of Chemistry, Cambridge 2010:147–191. [Google Scholar]
- 36.Arnold MA, Burmeister JJ, Small GW: Phantom glucose calibration models from simulated noninvasive human near-infrared spectra. Anal Chem 1998, 70(9):1773–1781. [DOI] [PubMed] [Google Scholar]
- 37.Halberg N, Khan T, Trujillo ME, Wernstedt-Asterholm I, Attie AD, Sherwani S, Wang ZV, Landskroner-Eiger S, Dineen S, Magalang UJ: Hypoxia-inducible factor 1α induces fibrosis and insulin resistance in white adipose tissue. Molecular and cellular biology 2009, 29(16):4467–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon J-O, Welch TP, Gonzalez FJ, Copple BL: Reduced liver fibrosis in hypoxia-inducible factor-1α-deficient mice. American Journal of Physiology-Gastrointestinal and Liver Physiology 2009, 296(3):G582–G592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL: Hypoxia-inducible Factor 1 (HIF-1) Promotes Extracellular Matrix Remodeling under Hypoxic Conditions by Inducing P4HA1, P4HA2, and PLOD2 Expression in Fibroblasts. Journal of Biological Chemistry 2013, 288(15):10819–10829. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Hofbauer KH, Gess B, Lohaus C, Meyer HE, Katschinski D, Kurtz A: Oxygen tension regulates the expression of a group of procollagen hydroxylases. The FEBS Journal 2003, 270(22):4515–4522. [DOI] [PubMed] [Google Scholar]
- 41.Lee DE, Alhallak K, Jenkins SV, Vargas I, Greene NP, Quinn KP, Griffin RJ, Dings RPM, Rajaram N: A Radiosensitizing Inhibitor of HIF-1 alters the Optical Redox State of Human Lung Cancer Cells In Vitro. Scientific Reports 2018, 8(1):8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn TA, Barron L: Macrophages: master regulators of inflammation and fibrosis In: Seminars in liver disease: 2010: © Thieme Medical Publishers; 2010: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straub JM, New J, Hamilton CD, Lominska C, Shnayder Y, Thomas SM: Radiation-induced fibrosis: mechanisms and implications for therapy. Journal of Cancer Research and Clinical Oncology 2015, 141(11):1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniëls VW, Machiels J: De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer research 2010, 70(20):8117–8126. [DOI] [PubMed] [Google Scholar]
- 45.Mims J, Bansal N, Bharadwaj MS, Chen X, Molina AJ, Tsang AW, Furdui CM: Energy metabolism in a matched model of radiation resistance for head and neck squamous cell cancer. Radiation research 2015, 183(3):291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao Y-C, Lee S-W, Lin L-C, Chen L-T, Hsing C-H, Hsu H-P, Huang H-Y, Shiue Y-L, Chen T-J, Li C-F: Fatty acid synthase overexpression confers an independent prognosticator and associates with radiation resistance in nasopharyngeal carcinoma. Tumor Biology 2013, 34(2):759–768. [DOI] [PubMed] [Google Scholar]
- 47. Shimura T: Acquired Radioresistance of Cancer and the AKT/GSK3β/cyclin D1 Overexpression Cycle. Journal of Radiation Research 2011, 52(5):539–544. [DOI] [PubMed] [Google Scholar]
- 48.Diaz PM, Jenkins SV, Alhallak K, Semeniak D, Griffin RJ, Dings RPM, Rajaram N: Quantitative diffuse reflectance spectroscopy of short-term changes in tumor oxygenation after radiation in a matched model of radiation resistance. Biomedical Optics Express 2018, 9(8):3794–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot M-C, Petrecca K, Leblond F: Intraoperative brain cancer detection with Raman spectroscopy in humans. Science translational medicine 2015, 7(274):274ra219–274ra219. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J-X, Xie XS: Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350(6264):aaa8870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.