Abstract
One of the distinguishing features of the 1918 pandemic is the occurrence of massive, potentially detrimental, activation of the innate immune system in critically ill patients. Whether this reflects an intrinsic capacity of the virus to induce an exaggerated inflammatory responses or its remarkable ability to reproduce in vivo is still open to debate. Tremendous progress has recently been made in our understanding of innate immune responses to influenza infection and it is now time to translate this knowledge into therapeutic strategies, particularly in view of the possible occurrence of future outbreaks caused by virulent strains.
Keywords: Toll-like receptors, RIG-I-like receptors, Interferon, Inflammasome, Inflammation
We summarize here current knowledge of the mechanisms whereby the human body detects the presence of influenza virus infection, controls viral replication and repairs damaged tissue. We discuss the possibility of using this knowledge to treat patients with severe forms of the disease.
Influenza and the innate immune system
The functions of the innate immune system, which are crucial for life, depend on its ability to detect the presence of microorganisms or of tissue damage by means of dedicated receptors. The genes encoding for these receptors are present in the germline and are not recombined in somatic cells. This is at variance with the genes encoding for immunoglobulins or T cell receptors, which are formed by rearrangements occurring in lymphocyte precursor cells and function as receptors in adaptative immune responses [1]. Activation of the innate immune system can have both protective and detrimental effects during influenza. Innate responses are the only weapons that the host can use to prevent or slow down viral replication early during infection, because influenza selectively affects people lacking protective antibodies as a result of mutations of the infecting viral strain [2]. At least 5 days are required after exposure to the virus before adaptative immunity comes into place by producing specific T lymphocytes and antibodies [3]. These first few days are critical in the development of the disease and, without a vigilant system capable of immediately detecting the presence of the pathogen, the virus would spread from the initial focus in the upper respiratory tract to the lower airways and deeper tissues, thereby directly impairing respiratory functions and threatening the life of the host. A vigorous innate response is required not only to mount immediate defensive responses, but also to trigger the subsequent production of antibodies and T cells. Despite their fundamentally protective role, innate responses account for many of the pathological manifestations of influenza and can induce collateral damage to the host. This is particularly evident in life-threatening infections with highly virulent viral strains, such as some pandemic or avian strains, which are often associated with high-level cytokine production and pneumonia [4], [5]. Hypervirulent strains are able to overcome host defences in the upper airways and reach the alveolar space, where inflammatory reactions can disrupt alveolar surface tension causing fluid accumulation and respiratory failure. The resulting clinical condition, which is designated as primary influenza virus pneumonia, can show the clinical, radiologic and pathologic features of the acute respiratory distress syndrome [6].
A primary objective of current influenza research is to understand the mechanisms underlying this dual role of the innate immune system and to ascertain whether the defensive responses can be separated from the excessive inflammatory reactions causing morbidity and lethality. One fundamental question in immunology, which goes beyond the influenza field, is whether pathogen clearance can be accomplished without collateral damage in the presence of substantial infection. This question has of course important practical implications, including the possible use of immunosuppressive or anti-inflammatory measures in influenza therapy [7]. In the case of influenza, one needs to consider not only the innate responses induced by the causative agent itself, but also those directed against the bacterial pathogens that often act as opportunistic invaders and cause secondary bacterial pneumonia. These bacteria are the main proximal cause of the severe illness and the high lethality rates observed during some influenza pandemics, such as the 1918 “Spanish” flu. Priming of the innate immune system, as it occurs during infection with the influenza virus, can deeply alter subsequent response to different stimuli, such as those represented by secondary pathogens. This can occur through a variety of mechanisms and both desensitization (tolerance) and sensitization (training) of the system have been described [8]. Therefore, the extent to which influenza virus infection alters host responses against secondary bacterial invaders must also be carefully considered. This topic has been recently reviewed [9] and will not be further discussed here.
Innate immunity modules
The innate immune responses occurring during influenza can be described as sequentially involving interactions between virions and mucosal secretions, infection of epithelial cells and activation of other resident cell types located in the epithelial or sub-epithelial layers. These include “innate” lymphoid cells, resident macrophages (including alveolar macrophages) and dendritic cells. At later times, other cell types, such as polymorphonuclear leukocytes and monocytes come into play, after being recruited to the infection sites from the blood.
Each of these cell types displays a different set of receptors, which are capable of sensing the presence of the virus and of activating specific defensive functions. Moreover, the receptors are strategically located in various subcellular compartments, such as the cell membrane, the endosomal system, the cytosol and mitochondria. Such an arrangement enables the host to mount defences that are tailored to the invading pathogen and take into account its tissue tropism, intracellular life style and reproductive strategy [10]. Thus, in the case of influenza and other infections, innate responses can be conveniently divided into modules, each involving different cell types, receptors, effector molecules and intracellular compartments [11]. Module components relevant to influenza infection include: 1) soluble extracellular proteins present body fluids; 2) the interferon system; 3) different types of cytokines and chemokines capable of orchestrating the innate response; 4) phagocytosis by macrophages and neutrophils; 5) antigen presentation by dendritic cells. Responses occurring in epithelial cells during influenza infection [12] and the role of extracellular components (including collectins, acute phase proteins and complement components) have been recently reviewed [13] and will not be considered here in depth.
Recognition of influenza virus
The innate immune system is the product of an ancient evolutionary strategy based on the detection of highly conserved microbial components, often called pathogen-associated molecular patterns (PAMPs). Viral RNA, in various forms, is the main PAMP targeted by the system to detect the presence of influenza virus. An important role is also played by the recognition of host products designated as damage-associated molecular patterns (DAMPs) released as a consequence of virus-induced cell lysis. Receptors capable of recognizing the presence of viral products are in the category of pattern recognition receptors (PRRs), which overlap partially with damage recognition receptors (DRR). Once activated, PRRs lead to the secretion of a number of effector molecules that trigger inflammation. These mediators include lipids, such as leukotrienes and prostaglandins, pro-inflammatory cytokines, chemokines and type I interferons [14], [15]. Lipid mediators and pro-inflammatory cytokines, which are produced locally and can also act systemically, cause inflammation, fever and malaise, while chemokines recruit additional cells involved in host defences including myeloid cells (e.g neutrophils and monocytes) and lymphoid cells (e.g. natural killer, or NK cells, T and B cells). A prominent role in host defences is played by NK cells, which kill virus-infected cells, thereby blocking viral replication [16]. Phagocytic inflammatory cells, as well as resident alveolar macrophages, are also important in viral clearance, due to their ability to ingest and destroy infected cells [17].
Three groups of PRRs mediate recognition of PAMPs or DAMPs relevant to influenza infection: 1) Toll-like receptors (TLRs); 2) RIG-I-like receptors (RLRs); 3) NOD-like receptors (NLRs). TLR3 detects the presence of double-stranded RNA (dsRNA), while TLR7 and TLR8 sense single-stranded RNA (ssRNA) and RIG-I specifically recognizes 5′-triphosphate RNA. NLRs, such as NLRP3 (standing for NOD-, LRR- and pyrin domain-containing 3), are also able to directly recognize viral products and DAMPs, leading to the formation of large multiprotein complexes called inflammasomes. The availability of mice lacking single receptors or downstream signal transduction components has greatly contributed to our understanding of innate immune sensing and signalling. By using these models, it is possible to analyse the severity of infection induced by murine-adapted strains of influenza A virus (IAV), as well as disease outcome and pathology, in the absence of specific host factors. Further information can be gained by observing the outcome of influenza in patients with genetic defects in these factors.
TLRs
TLR3, 7 and 8 are predominantly located in internal cell membranes, with the exception of respiratory and intestinal epithelial cells that can express TLR3 also on the plasma membrane [Fig. 1]. These TLRs have a crucial role in the recognition of RNA viruses that, like IAV, enter cells through receptor-mediated endocytosis. In resting cells, these receptors are present in the endoplasmic reticulum, bound to the chaperone protein UNC93B. Upon cell activation by a variety of stimuli, including receptor-mediated endocytosis, nucleic acid-sensing TLRs are moved to the endosomes [18]. This occurs in different steps, involving trafficking to the Golgi apparatus and to early endosomes, which mature into late endosomes and can fuse with lysosomes. Nucleic acid recognition likely occurs in acidified late endosomes or endolysosomes, where full-length, inactive TLRs are cleaved by acid proteases into active and more stable forms [19]. Thus, endosomal acidification and proteolytic cleavage are essential for TLR signaling.
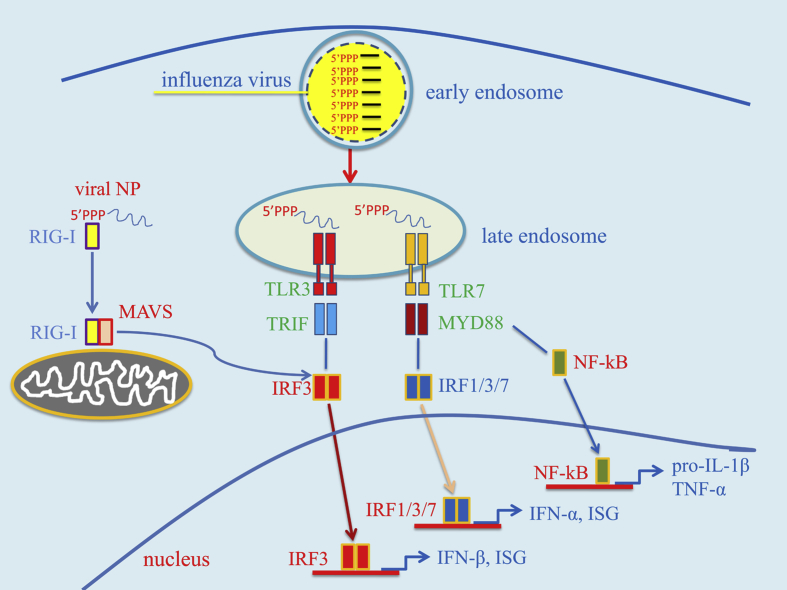
Fig. 1.
Condensed summary of signal transduction pathways activated by TLR3 and TLR7 in cells infected with the influenza virus. After cell entry, the influenza virus is initially found inside endosomes where its genome, consisting of 5′ tri-phosphorylated ssRNA segments, can be recognized by Toll-like receptors 3 and 7 (TLR3/7). TLR7 signalling induces, via the myeloid differentiation primary response 88 (MyD88) adaptor protein and other signalling intermediates, the activation of the transcription factors nuclear factor-κB (NF-κB) and interferon regulatory factors 1, 3 and 7 (IRF1/3/7). NF-κB activates the transcription of pro-inflammatory cytokine genes, such as pro-interleukin1-beta (IL-1β) and tumor necrosis factor-alpha (TNF-α), while the IRFs induce type I interferons (IFN-α and IFN-β) and some interferon-stimulated genes (ISG). TLR3 activates IRF3 via the adaptor TRIF (TIR-domain-containing adapter-inducing interferon-β). Viral RNA is recognized in the cytosol by retinoic acid-inducible gene-I (RIG-I), which, leads to the induction of IFN-β and ISG via the mitochondrial antiviral signaling (MAVS) protein and IRF3. Abbreviation: Viral NP: viral nucleoprotein.
TLR3 is expressed in the endosomes of all cells of the innate immune system, with the exceptions of plasmacytoid dendritic cells (pDCs) and neutrophils [20]. Moreover, TLR3 is expressed both on the surface and in the endosomes of bronchial and alveolar epithelial cells. TLR3 differs from other TLRs in using only TRIF (TIR-domain-containing adaptor protein-inducing IFN-β) as an adaptor for signal transduction [Fig. 1]. The TRIF-dependent pathway leads to the induction of proinflammatory cytokines and type I IFN mainly through the activation of, respectively, NF-κB and IRF3 (or IFN regulatory factor 3) [21]. TLR3 recognizes the dsRNA synthetic analog poly I:C, and, in addition, various forms of double-stranded RNA (dsRNA) that can form during viral infection, including: 1) genomic material of dsRNA virus, such as reovirus; 2) dsRNA transcription intermediates of complex DNA virus, such as herpes viruses; 3) dsRNA replication intermediates of positive strand RNA viruses, such as picornavirus and flavivirus; 4) self dsRNA structures that form in damaged cells; 5) miRNA and siRNA complexes. Although TLR3 is involved in host responses to IAV, the exact structure(s) recognized by this receptor are not entirely clear. TLR3 should be unable to recognize the bulk of influenza virus genomic RNA, which consists of segmented ssRNA [22], [23], [24], [25]. Moreover, dsRNA replication intermediates are believed to be rapidly destroyed by cellular RNA helicases, such as UAP 56, in IAV-infected cells [23], [24]. Accordingly, Weber et al. were unable to detect dsRNA by immunofluorescence in IAV-infected cells [25]. However, recent studies using more sensitive monoclonal antibodies have shown that dsRNA is indeed present in both the cytoplasm and the nucleus of IAV-infected cells [22]. Interestingly, protease pretreatment was necessary to detect dsRNA in the nucleus, suggesting that dsRNA binding proteins (viral nucleocapsid proteins?) block dsRNA exposure in this compartment. In addition, in vitro infection of epithelial cells with IAV readily triggers TLR-3 dependent proinflammatory cytokine production [26]. It is conceivable that TLR3 recognizes the double stranded structures formed by short complementary sequences at the 5′- and 3′-ends of each segment. Moreover, self dsRNA structures forming in damaged cells may also come into contact with endosomal TLR3 in resident or recruited macrophages, which actively phagocytose dying and apoptotic cells [27]. TLR3 is likely to play an important role in anti-IAV innate host defences, since Tlr3−/− mice display higher viral burden in experimental infections, compared with wild-type mice [28], [29]. However, TLR3 signalling may also contribute to pathology since Tlr3−/− mice survive longer under these conditions [28], [30]. These mice generate normal CD8+ and CD4+ T cell responses, suggesting that TLR3 is not required for the initiation of adaptative immune responses to IAV [31].
Human TLR7, which is expressed in the endosomes of bronchial epithelial cells, can sense the presence of the IAV genome in these structures after viral entry through receptor-mediated endocytosis [32]. Endosome acidification is required for both maturation of the receptor and for freeing the genome from the viral core. Specifically, the low endosomal pH activates the M2 viral protein, which functions as a proton channel and acidifies the viral core, promoting dissociation of the M1-nucleoprotein complex [33], [34]. Signals originating from TLR7 and 8 are transduced by the adaptor MyD88, resulting in the activation NF-κB and of IFN-regulatory factors (IRF), which differ in different cell types [Fig. 1]. Stimulation of TLR7 in primary bronchial, but not alveolar, epithelial cells resulted in the release of IL-6 and type III IFN [32]. Plasmacytoid dendritic cells (pDCs), which express TLR7 and TLR8, are recruited to the lung during influenza and produce high amounts of type I IFN by virtue of their high-level, constitutive expression of IRF-7 [35].
TLR7/TLR3 double-deficient or MyD88-deficient mice showed limited ability to restrict IAV replication and were highly susceptible to infection [29]. However, in a different study, TLR7 or MyD88 deficiency was not associated with increased susceptibility to infection [28]. The apparent discrepancies of these studies can be reconciled by taking into account the fact that most of the standard laboratory mouse strains (including the C57BL/6 mice commonly used in studies of this kind) lack functional myxovirus resistance protein 1 (MX1), an important IFN-induced anti-viral factor. Indeed, in an elegant study using MX1 sufficient mice (i.e. strain B6.A2G-Mx1), TLR7 and MyD88 had an essential role in promoting host defences against the highly virulent SC35M H7N7IAV strain and this effect was mediated by pDC, which produce high levels of type I IFN after TLR7 stimulation [36]. TLR7 is also required for generation of B cell germinal centers and of robust memory antibody responses [37]. TLR8 is expressed by human monocytes, macrophages and conventional dendritic cells, but its functional role during influenza infection is presently unclear.
In summary, RNA-sensing TLRs play a crucial role in host defences against IAV by inducing an IFN-dependent antiviral state. Moreover, TLR7 activation promotes the induction of memory B cell responses. Although their protective role seems clearly predominant, these TLRs might also have the potential of inducing detrimental hyper-inflammatory reactions in the context of rapidly lethal, overwhelming infections.
RLRs
The presence of influenza virus in the cytosol of infected cells can be detected by specialized RNA receptors belonging to the RLR family. Among these, RIG-I is perhaps the most important in terms of IAV recognition. This receptor is constitutively expressed by a number of cells, including epithelial cells, macrophages and conventional dendritic cells and is upregulated during viral infection. RIG-I recognizes the 5′-triphosphate RNA promoter region of intact genomic segments or shorter fragments [24], [38], [39], [40]. The “panhandle” double stranded structure formed in vitro by complementary sequences of the promoter is apparently sufficient for RIG activation. Moreover, the degree of complementarity in this region is proportional to its ability to activate the sensor. In infected cells, RIG-I interacts with genomic ssRNA bearing 5′-triphosphates, but not with non-genomic viral transcripts, replication intermediates or cleaved self-RNA [39]. Moreover, RIG-I preferentially associates with shorter viral RNA molecules or subgenomic defective interfering particles [40]. Interactions between RIG-I and viral nucleoprotein were recently found to occur at the level of antiviral stress granules [Fig. 2], in which protein kinase R (PKR), an interferon stimulated gene (ISG), also co-localizes [41]. Notably, PKR promotes granule formation and the viral NS1 protein can interfere with this activity. The RIG-I helicase domain binds to ATP upon interaction with viral RNA, and RIG-I then forms a complex with the mitochondrial antiviral signalling protein (MAVS) through its caspase-recruitment domains [42], [43]. MAVS signalling leads to efficient induction of type I IFN and of pro-inflammatory cytokines via activation of, respectively, IRF3 and NF-κB. Mice that are deficient in MAVS have similar viral loads and survival rates to wild-type mice when challenged with a lethal dose of influenza virus [44]. Moreover MAVS-deficient mice are not impaired in their ability to restrict viral growth and to activate the adaptative immune systems. Again, the results of these studies should be interpreted with caution since they were conducted in MX1-defective mouse strains. Mice lacking RIG-I are unable to clear IAV infection as efficiently as wild-type mice [45]. Melanoma Differentiation-Associated protein 5 (MDA5), another member of the RLR family of dsRNA helicases, might also be involved in IAV sensing and play a role in host defences [46].
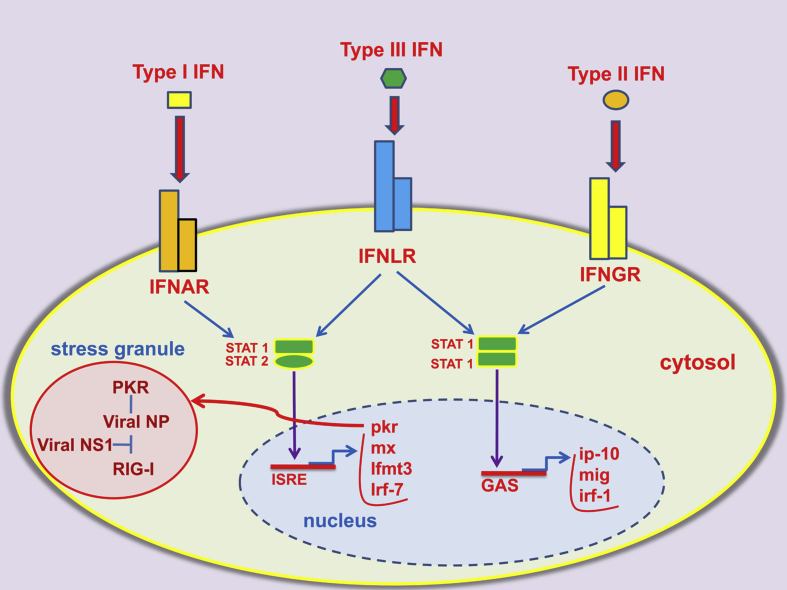
Fig. 2.
Intracellular pathways activated by the interferon system. The three types of interferon (IFN) are recognized by different receptors and activate partially overlapping signalling pathways. Abbreviations: IFNAR: IFN-α/β receptor; IFNLR: IFN-λ receptor; IFNGR: IFN−γ receptor; STAT: signal transducer and activator of transcription protein; ISRE: interferon-stimulated response element; GAS: gamma interferon activation site; PKR: serine–threonine kinase protein kinase R; mx: myxovirus resistance protein; IRF-7: interferon regulator factor-7; IFITM3: interferon-induced transmembrane protein 3; IP-10: interferon gamma-induced protein 10; MIG: monokine induced by interferon gamma, also known as CXCL9; viral NS1: viral non-structural protein 1 (NS1); RIG-I: retinoic acid-inducible gene-I; viral NP: viral nucleoprotein.
The interferon system
The interferon system is perhaps the most important weapon in host defences against influenza virus (for a review see ref. n. [13]). This system includes three types of interferons (IFNs), namely type I (which consists of many subtypes, including IFN-α and β), type II (IFN-γ) and type III (IFN-λ) IFNs. Members of each IFN type bind to a specific receptor (IFNAR, IFNGR and IFNLR, respectively) and this accounts for their different functional properties [Fig. 2]. Indeed, each IFN type induces a partially overlapping, but distinctive, set of genes. The biological activities of the various IFN types have been recently reviewed [47], [48]. In epithelial cells, similar genes are activated by type I and III IFNs, and lack of IFNAR or IFNLR only can be compensated for by the other receptor [49]. Type I and III IFNs apparently play an equally important role in the induction of anti-viral genes in epithelial cells, but not in other cell types, in which the effects of type I IFN predominate. IFN-α and IFN-β are secreted by a large variety of cells, including epithelial cells, macrophages, dendritic cells (DC) and plasmacytoid DCs [14], [15], [50]. Respiratory epithelial cells are important sources of IFN-β early during influenza infection, while pDC come into action at later times by releasing high amounts of IFN-α and IFN-β [15]. Virus-infected cells secrete IFN-α and IFN-β which activate IFNAR receptors present on the surface of the producing cells (autocrinous activity) or nearby cells (paracrinous activity), in which they induce the expression of hundreds IFN-stimulated genes (ISGs). ISGs products are ultimately responsible for the induction of a cell-intrinsic anti-viral state that dramatically reduces viral spread from the initial site of infection. In addition, type I IFNs potently activate natural killer cells (NK), which kill virus-infected cells and produce IFN-γ. Absence of IFN signalling results in systemic viral dissemination and in accelerated lethality in mouse models of influenza infection [51].
When given prophylactically, type I IFN reduces IAV replication and disease severity in both animals [52] and humans [53]. Therapeutic administration, however, is ineffective in animal models and may actually increase lethality despite a reduction in lung IAV titers [52]. This effect is likely related to the induction of epithelial cell death, secondary to the pro-apoptotic effects of type I IFN. Apoptosis is at least in part mediated by IFN-induced upregulation of death receptor 5 (DR5), which functions as a receptor for TNF-related apoptosis-inducing ligand (TRAIL) in lung pneumocytes [54].
Not surprisingly, influenza viruses have developed efficient mechanisms to reduce IFN production and to counteract its effects. The NS1 protein of influenza viruses inhibits IFN-β production by blocking RIG-I mediated viral recognition and maturation of IFN-β gene and ISG transcripts in the nucleus [55]. Genetic deletion of NS1 results in increased IFN production, increased expression of ISGs and decreased viral growth [15]. Interestingly, the NS1 protein produced by the virus responsible for the 1918 pandemic is particularly potent at inhibiting IFN production, which may explain the extreme virulence of this strain. Microscopic observations show that RIG-I co-localizes in cytoplasmic microdomains with genomic and subgenomic 5′-triphosphate ssRNA, which represents its cognate PAMP [38], and with NS1 [38], [41].
Studies in mice have indicated an important role of the serine–threonine kinase protein kinase R (PKR), an ISG product, in controlling viral replication [56]. PKR is present in the cytosol in an inactive state at low levels, which increase dramatically after IFN signalling. Binding to viral nucleic acids triggers the latent serine/threonine kinase activity of the protein and results in phosphorylation of the alpha subunit of the eukaryotic translation initiation factor 2 (eIF2a). This event blocks the translation of viral and cellular mRNAs and markedly inhibits viral growth [57]. In addition, PKR inhibits viral growth by promoting autophagy and apoptosis of infected cells [58]. PKR is activated by the 5′-triphosphate-dsRNA structures present in viral ribonuclear protein complexes exported from the nucleus into the cytosol at late stages during cell infection [57].
In addition to PKR, a number of other ISG products, such as the IFITM and MX proteins, are believed to mediate the antiviral activities of the IFN system. For example, the IFITM proteins block virus entry into the cytosol by inhibiting fusion between viral and host membranes in the endosomes that form after virus attachment and endocytosis [59]. A crucial role for IFITM3 in anti-IAV defences is suggested by observations that mice lacking this protein are hypersusceptible to infection [60]. In addition, human MXA and mouse MX1 can potently inhibit IAV replication [61]. MX1 (or myxovirus resistance protein 1) is a dynamin-like nuclear guanosine triphosphatase that impairs primary transcription of the IAV genome. It should be noted that most standard inbred mouse strains, including C57BL6, lack functional MX1 and/or MX2 proteins [62] and are much more susceptible to influenza infection than MX1-sufficient mice. As mentioned above, this feature should be taken into account when interpreting the results of studies involving targeted mutations, which are commonly performed in mouse strains with spontaneous mutations in the MX proteins. Mx1-sufficient mice, such as MX1 congenic mice on the C57BL/6 background, are highly resistant to IAV infection. These mice provide a useful model to study the role of the main innate immune receptors in anti-influenza host defences. For example, Mx1-sufficient, Tlr7−/−× Mavs−/− double-deficient, mice were unable to mount robust IFN-β responses, lost MX1 expression and rapidly died after influenza infection [63]. These mice might mimic the situation observed in elderly humans showing a selective impairment in IFN-β responses to influenza and other viral infections [63]. Moreover, similar to older human adults, these double-deficient mice undergo severe endogenous bacterial infections secondary to IAV-induced tissue damage. Collectively these data suggest that an age-related impairment in type I IFN responses could predispose the elderly to secondary bacterial pneumonitis after influenza infection.
Proinflammatory cytokines and inflammasomes
Activation of the transcription factor NF-κB by various classes of PRRs induces the expression of proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α), and interleukin 1 beta (IL-1β), and of chemokines, which mediate the influx of inflammatory cells and proteins into the infection site. Signalling by TNF-α and IL-1 family members is involved in an amplification loop, which results in further NF-κB activation and production of proinflammatory mediators. The inflammasomes are cytosolic multimeric protein complexes that play a crucial role in these processes by triggering the release of mature forms of IL-1β and IL-18 and by inducing an inflammatory form of cell death named pyroptosis [64], [65]. These phenomena are mediated by activation of the caspase-1/8 and gasdermin D proteins after two different types of signals are transduced inside the cell. Signal 1 leads to NF-κB-mediated expression of inflammasome components and inactive precursors of proinflammatory cytokines, such as pro-IL-1β and pro-IL-18. Signal 2 involves the activation of cytosolic sensors, such as NLRP3 (NACHT, LRR and PYD domains-containing Protein 3), AIM2 (Absent In Melanoma 2) and RIG-I [65], in response to microbial PAMPs or to products of host cell damage. This results in the formation of multimolecular complexes, caspase-1/8/11 activation, cytokine maturation and pyroptosis. Various inflammasomes contain the adaptor protein ASC that functions as scaffold for caspase-1 recruitment.
An important role of the NLRP3 inflammasome in anti-IAV host defences is suggested by studies in mice with genetic defects in NLRP3, ASC or caspase-1, which all show increased susceptibility to infection with the PR8 IAV strain in association with reduced lung inflammation, cell influx and cytokine production [23], [66]. In contrast, in a other study, NLRP3 or caspase 1/11-deficient mice were protected from lethal infection with the highly virulent H7N9 avian virus, compared with wild-type mice [67]. These apparent discrepancies can be reconciled by taking into account differences in the viral strains used for challenge and in the accompanying severity of infection. In fact, these studies indicate that the NLRP3 inflammasome has a protective role during mild IAV infection, while, at the same time, mediates the severe pathological manifestations observed after challenge with highly virulent strains. Several stimuli can activate NLRP3 in myeloid cells during IAV infection. Changes in mitochondrial membrane potential, such as those occurring after the PRR-induced “respiratory burst”, favour direct interactions of NLRP3 with mitofusin 2, a mitochondrial outer membrane protein, resulting in inflammasome assembly [68], [69]. Moreover, the large molecular weight complexes of the viral protein PB-F2 that can be found in virus-infected cells can readily activate NLRP3 inflammasome formation [70]. The PB1-F2 protein is a virulence factor expressed by pathogenic IAV strains, including the pandemic 1918 H1N1, 1957 H2N2 and 1968 H3N2 strains, as well as the PR8 H1N1 and avian strains. Therefore, it is possible that PB1-F2 is at least partially responsible for the hyperinflammatory response observed during infection with virulent strains. Inflammasome activation can also be induced by the proton fluxes generated in the trans-Golgi network by the viral matrix 2 (M2) protein and can be inhibited by amantadine, an M2 blocker [71].
In addition to NLRP3, other viral sensors such as RIG-I, AIM2 and the Z-DNA binding protein-1 (ZBP1, also known as DAI) have also been reported of being capable of inflammasome activation [11], [27], [72]. The ability of RIG-I to recognize the promoter region of the influenza virus has been discussed above. In infected bronchial epithelial cells, activated RIG-I directly binds to ASC to promote inflammasome formation [73]. AIM-2 can also induce ASC-dependent inflammasome activation after becoming activated by the presence of dsDNA fragments in dying infected cells. The ZBP1 sensor is capable of inducing NLRP3 inflammasome activation and cell death by activating RIP kinase 1 (RIPK1), RIPK3 and caspase-8 after binding to the IAV proteins NP and PB1 [74], [75].
Innate lymphoid cells and γδ T cells
The category of innate lymphoid cells (ILCs) includes cells of lymphoid origin (i.e. those derived from a common lymphoid progenitor and sharing similar morphology) that lack the variable, recombined, antigen-specific receptors expressed by B and T cells (e.g. immunoglobulins and T cell receptors; [76]). ILCs can be further classified into the ILC1, ILC2 and ILC3 subsets, according to their major cytokine products (type 1, type 2 and type 3 cytokines, respectively) and include, in addition, conventional natural killer cells (NK), which are cytotoxic ILCs. Thus, ILCs mirror the phenotypes of T cells since NK cells, ILC1s, ILC2s, and ILC3s can be viewed as the innate counterpart (and perhaps the evolutionary precursors) of, respectively, cytotoxic CD8 T cells, Th1, Th2, and Th17 cells [77]. By producing different sets of mediators, ILCs can shape the host response into one that is adapted to the initial insult. ILCs should not be confused with invariant NKT cells and γδ T cells, which express T cell receptors, but resemble ILCs in their ability to respond to innate stimuli (such as damage signals and PAMPs).
Conventional NK cells have a prominent role in anti-influenza defences by virtue of their ability to recognize virus-infected cells and kill them. Genetic deletion of NKp46, a sialylated NK recognition receptor that can bind viral HA, results in increased susceptibility to influenza in mice [16], [78]. The NKp44 receptor was also shown to bind viral HA [79]. However, influenza viruses may escape NKp44- and NKp46-mediated recognition by expressing heavily glycosylated HA molecules [79]. In addition, NK cells and ILC1s produce the anti-viral chemokine IFNγ, in response to primary cytokines, including type I IFN, IL-12 and IL-18, released in infected tissues. In the early phases of infection, ILC1s may represent an even more important source of IFN-γ compared with NK cells [80]. IFN-γ may inhibit viral replication by inducing a subset of ISGs, such as IP-10, and potentiate the type I IFN response by inducing IRF1 [Fig. 2]. In addition, type II, as well as type I IFN, prostaglandin PGl2, corticosteroids, and testosterone, inhibit the production of IL-5 and other type 2 cytokines by ILC2s [81]. Conversely, ILC2s are activated for increased mediator production by the epithelial cytokines IL-25, IL-33 and thymic stromal lymphopoietin [82].
Type 2 cytokine production by activated ILC2s must be tightly controlled, since IL-5 and IL-13 can induce eosinophil recruitment, mucus overproduction and bronchoconstriction, which can be detrimental during influenza, particularly in asthma patients [83]. On the other hand, ILC2s play an important role in promoting repair of the respiratory epithelium during recovery from influenza by producing amphiregulin, an epidermal growth factor-like growth factor [83]. Thus, ILC2s potentially play a dual role by inducing detrimental inflammatory airway hyperresponsiveness early during infection and tissue repair in the recovery phase. Early during infection with IAV, γδT cells present in the lung produce IL-17A and IL-17F, which activate a common receptor (IL-17R). Genetic deletion of IL-17R results in amelioration of disease manifestations in influenza-infected mice, including reduced weight loss, pulmonary damage and neutrophil infiltration, without affecting viral replication [84]. Similar effects are observed in mice deficient in the TNF receptor TNFR1 [85]. Therefore blocking the activities of IL-17 or TNF may represent an alternative strategy to treat influenza-infected patients with the acute respiratory distress syndrome.
Neutrophils, macrophages and dendritic cells
Neutrophils are the predominant inflammatory cell type during IAV infection and play a crucial role in host defences. Moreover, the severity of the disease correlates with the abundance of neutrophil infiltration. For example, a frequent feature of infections caused by the 1918 pandemic virus was massive neutrophil recruitment to the lungs. Neutrophils have been described as having opposite roles in influenza. The protective effects of this cell type can be shown in experimental animal models using low viral doses as a challenge. Under these conditions, neutrophil ablation is associated with increased viral burden and disease severity [86]. In contrast, neutrophils may play a detrimental role during overwhelming IAV infection and may contribute to the pathophysiological manifestation of the disease [87]. The protective effects of neutrophils have been ascribed to their ability to ingest and destroy dying or apoptotic virus-infected cells. Neutrophils are ideally suited to accomplish this task since like macrophages, they do not support productive influenza virus infection. However, phagocytosis of influenza virus by neutrophils is associated with the generation of a respiratory burst [88], [89] and with the release of neutrophil extracellular traps (NET; [87]), both of which can induce tissue damage [90]. Overall, available data point to a predominantly beneficial role of neutrophils during influenza infection, due to their ability to efficiently reduce viral loads. It is conceivable, however, that, in response to rapid viral growth, high numbers of these cells are recruited to the lung were they release a plethora of extracellular factors capable of damaging surrounding tissues, including the alveolar wall. Therefore, targeting NET components or reactive oxygen species may represent a viable strategy to reduce alveolar epithelial cell leakage in cases of severe IAV infection.
Macrophages residing in the respiratory mucosa or in the alveoli come in contact with influenza viruses once they have established productive infection in the epithelial cells of the upper airways. These resident macrophages, together with epithelial and dendritic cells, produce type I IFNs, proinflammatory cytokines and chemokines capable of attracting neutrophils, monocytes and NK cells [5], [91]. Recruited monocytes differentiate into inflammatory macrophages, which greatly amplify cytokine production. Both macrophages and neutrophils can ingest IAV-infected cells, particularly when they are damaged or apoptotic, and have a crucial role in promoting viral clearance [92]. Depletion of macrophages has been shown to result in increased viral growth in various animal models of influenza infection [93].
Macrophages, neutrophils and dendritic cells are permissive to infection with IAV, resulting in the accumulation of viral RNAs and proteins in various cell compartments where these products are recognized by cytosolic and membrane PRR. Viral replication, however, is abortive and fails to result in the release of virions, due to a defect in the assembly stage [94]. A possible exception is represented by avian influenza strains, which have been shown to reproduce in monocytes. Influenza viruses have developed mechanisms to impair the anti-viral activities of phagocytes and other cells of the innate immunity system. One of such mechanisms is the induction of apoptosis in macrophages by virtue of a protein, named PB1-F2, which is encoded by the PB1 polymerase gene and is produced using an alternative reading frame [95]. Interestingly, the PB1-F2 protein is produced during infection with many pandemic strains and functions in vivo as a virulence factor [95].
The main function of dendritic cells (DCs) during influenza infection is the presentation of viral antigens to naïve CD4+ and CD8+ T lymphocytes. Airway-resident DCs include CD103−CD11bhi and CD103+CD11blowas well as CD11clowB220+ plasmacytoid DC. Both resident and monocyte-derived DC can present viral antigens after they become directly infected or after ingestion of infected cells. Antigen-loaded DCs are recruited to lymph nodes by the activity of specific chemokines (CCL19/21) acting on the CCR7 receptor expressed during maturation of DCs [51], [96]. Antigen can be presented to T cells in lymph nodes either directly by migrated DC or indirectly by lymph-node resident CD8α+DCs after transfer of MHC-peptide complexes to the latter [97]. In addition to their antigen-presenting capabilities, pDC can produce high levels of type I IFN in response to viruses or endosomal TLR agonists [98]. However, in experimental models of IAV infection, viral clearance was not affected by ablation of pDC [99]. Moreover, these cells were not the predominant source of type I IFN, at least at early time points during infection [15].
Again, as discussed above, the results of these studies should be considered with caution since they have been conducted using inbred strains lacking functional MX1, an important IFN-induced antiviral factor [63].
Conclusions
One hundred years ago the “Spanish Flu” pandemic produced devastating consequences and killed an estimated 50 million people. Although better preventive, diagnostic and therapeutic tools have been developed since then, the emergence of new and potentially highly virulent pandemic strains is a continuous and realistic threat. This is clearly documented by studies on the 2009 pandemic precursor virus, which, like the 1918 virus, persisted in swine for sufficient time to allow the rapid reassortment of viruses of human, swine and avian origin. The last two decades have witnessed dramatic progress in our understanding of innate immune responses directed against the influenza viruses. Such responses limit viral growth, promote viral clearance, induce tissue repair and trigger long-lasting adaptive immunity against the infecting strain. At the same time, most of the pathological manifestations of influenza can be explained by localized and generalized inflammatory responses that are triggered by the innate immune system after virus recognition. While control of influenza will ultimately depend on the development of a broadly protective, universal vaccine, there is presently a strong need to translate the accumulated knowledge of innate immunity into effective therapeutic measures. Drugs that target the immune response would circumvent several shortcomings of available anti-viral therapies, such as their narrow treatment windows and the development of drug resistance. For example, currently recommended M2 protein and neuraminidase inhibitors are effective only in the first few days after disease onset and are of little use in patients with severe complications occurring at later times, such as acute lung injury. In these patients, in contrast, it would seem logical to target the cytokine and inflammatory responses that are responsible for the accumulation of cells and fluids in the alveolar space. Currently, however, this strategy remains challenging since we have not yet been able to clearly identify features in the immune responses that can be suppressed without compromising their beneficial, host-protective effect. Accomplishing this task, however, is not impossible and promising results have been obtained in dissecting out detrimental versus protective mechanisms. For example, completely blocking proximal events in the immune response (such as the activation of the pathogen recognition receptors involved in the IFN response) seems unwise, given their general role in orchestrating host defences. In contrast, more limited and specific effector arms, such as the production of oxygen radicals, NET formation and IL-17 production may represent viable targets. Finally, more research is needed to understand the temporal features of the pro-inflammatory response to influenza in relation to the timing of therapeutic interventions.
Funding
The authors have no funding to report.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Olive C. Pattern recognition receptors: sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev Vaccines. 2012;11:237–256. doi: 10.1586/erv.11.189. [DOI] [PubMed] [Google Scholar]
- 2.Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C. Influenza. Nat Rev Dis Prim. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermans D., Webby R.J., Wong S.S. Atypical antibody responses to influenza. J Thorac Dis. 2018;10:S2238–S2247. doi: 10.21037/jtd.2017.12.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N., Chan P.K., Wong C.K., Wong K.T., Choi K.W., Joynt G.M. Viral clearance and inflammatory response patterns in adults hospitalized for pandemic 2009 influenza A(H1N1) virus pneumonia. Antivir Ther. 2011;16:237–247. doi: 10.3851/IMP1722. [DOI] [PubMed] [Google Scholar]
- 5.de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J.D., Chau T.N.B. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Short K.R., Kroeze E.J.B.V., Fouchier R.A.M., Kuiken T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis. 2014;14:57–69. doi: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 7.Li Z.T., Li L., Zhao S., Li J., Zhou H.X., Zhang Y.H. Re-understanding anti-influenza strategy: attach equal importance to antiviral and anti-inflammatory therapies. J Thorac Dis. 2018;10:S2248–S2259. doi: 10.21037/jtd.2018.03.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourbal B., Pinaud S., Beckers G.J.M., Van Der Meer J.W.M., Conrath U., Netea M.G. Innate immune memory: an evolutionary perspective. Immunol Rev. 2018;283:21–40. doi: 10.1111/imr.12647. [DOI] [PubMed] [Google Scholar]
- 9.McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 10.Beutler B. Innate immunity and the new forward genetics. Best Pract Res Clin Haematol. 2016;29:379–387. doi: 10.1016/j.beha.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 12.Denney L., Ho L.P. The role of respiratory epithelium in host defence against influenza virus infection. Biomed J. 2018;41:218–233. doi: 10.1016/j.bj.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White M.R., Doss M., Boland P., Tecle T., Hartshorn K.L. Innate immunity to influenza virus: implications for future therapy. Expert Rev Clin Immunol. 2008;4:497–514. doi: 10.1586/1744666X.4.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogner K., Wolff T., Pleschka S., Plog S., Gruber A.D., Kalinke U. Correction: macrophage-expressed IFN-beta Contributes to Apoptotic Alveolar Epithelial Cell Injury in Severe Influenza Virus Pneumonia. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewell N.A., Vaghefi N., Mertz S.E., Akter P., Peebles R.S., Bakaletz L.O. Differential type I interferon induction by respiratory syncytial virus and influenza A virus in vivo. J Virol. 2007;81:9790–9800. doi: 10.1128/JVI.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazit R., Gruda R., Elboim M., Arnon T.I., Katz G., Achdout H. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto Y., Moki T., Takizawa T., Shiratsuchi A., Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol. 2007;178:2448–2457. doi: 10.4049/jimmunol.178.4.2448. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y.M., Brinkmann M.M., Paquet M.E., Ploegh H.L. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Cattaneo A., Gobert F.X., Muller M., Toscano F., Flores M., Lescure A. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. P Natl Acad Sci USA. 2012;109:9053–9058. doi: 10.1073/pnas.1115091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi F., Means T.K., Luster A.D. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 22.Son K.N., Liang Z.G., Lipton H.L. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J Virol. 2015;89:9383–9392. doi: 10.1128/JVI.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisskirchen C., Ludersdorfer T.H., Muller D.A., Moritz E., Pavlovic J. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza a virus infection. J Virol. 2011;85:8646–8655. doi: 10.1128/JVI.02559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 25.Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Goffic R., Pothlichet J., Vitour D., Fujita T., Meurs E., Chignard M. Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-1-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 27.Schulz O., Diebold S.S., Chen M., Naslund T.I., Nolte M.A., Alexopoulou L. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 28.Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:526–535. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo S.U., Kwon H.J., Song J.H., Byun Y.H., Seong B.L., Kawai T. MyD88 signaling is indispensable for primary influenza a virus infection but dispensable for secondary infection. J Virol. 2010;84:12713–12722. doi: 10.1128/JVI.01675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung Y.H.C., Nicholls J.M., Ho C.K., Sia S.F., Mok C.K.P., Valkenburg S.A. Highly pathogenic avian influenza A H5N1 and pandemic H1N1 virus infections have different phenotypes in Toll-like receptor 3 knockout mice. J Gen Virol. 2014;95:1870–1879. doi: 10.1099/vir.0.066258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heer A.K., Shamshiev A., Donda A., Uematsu S., Akira S., Kopf M. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J Immunol. 2007;178:2182–2191. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis I., Ye F., McNally B., Willette M., Flano E. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J Virol. 2013;87:3261–3270. doi: 10.1128/JVI.01956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diebold S.S., Kaisho T., Hemmi H., Akira S., Sousa C.R.E. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 34.Wang J.P., Liu P., Latz E., Golenbock D.T., Finberg R.W., Libraty D.H. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 2006;177:7114–7121. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 35.Dai J., Megjugorac N.J., Amrute S.B., Fitzgerald-Bocarsly P. Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells. J Immunol. 2004;173:1535–1548. doi: 10.4049/jimmunol.173.3.1535. [DOI] [PubMed] [Google Scholar]
- 36.Kaminski M.M., Ohnemus A., Cornitescu M., Staeheli P. Plasmacytoid dendritic cells and Toll-like receptor 7-dependent signalling promote efficient protection of mice against highly virulent influenza A virus. J Gen Virol. 2012;93:555–559. doi: 10.1099/vir.0.039065-0. [DOI] [PubMed] [Google Scholar]
- 37.Jeisy-Scott V., Kim J.H., Davis W.G., Cao W.P., Katz J.M., Sambhara S. TLR7 recognition is dispensable for influenza virus a infection but important for the induction of hemagglutinin-specific antibodies in response to the 2009 pandemic split vaccine in mice. J Virol. 2012;86:10988–10998. doi: 10.1128/JVI.01064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H. 5 '-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 39.Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Baum A., Sachidanandam R., Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. P Natl Acad Sci USA. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onomoto K., Jogi M., Yoo J.S., Narita R., Morimoto S., Takemura A. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang F.G., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Patel S.S. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Koyama S., Ishii K.J., Kumar H., Tanimoto T., Coban C., Uematsu S. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179:4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 45.Kandasamy M., Suryawanshi A., Tundup S., Perez J.T., Schmolke M., Manicassamy S. RIG-I Signaling Is Critical for Efficient Polyfunctional T Cell Responses during Influenza Virus Infection. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benitez A.A., Panis M., Xue J., Varble A., Shim J.V., Frick A.L. In vivo RNAi screening identifies MDA5 as a significant contributor to the cellular defense against influenza a virus. Cell Rep. 2015;11:1714–1726. doi: 10.1016/j.celrep.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee A.J., Ashkar A.A. The dual nature of type I and type II interferons. Front Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells A.I., Coyne C.B. Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kallfass C., Lienenklaus S., Weiss S., Staeheli P. Visualizing the beta interferon response in mice during infection with influenza a viruses expressing or lacking nonstructural protein 1. J Virol. 2013;87:6925–6930. doi: 10.1128/JVI.00283-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X.Y., Liu S.S., Goraya M.U., Maarouf M., Huang S.L., Chen J.L. Host immune response to influenza a virus infection. Front Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson S., Maini M.K., Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res. 2015;35:252–264. doi: 10.1089/jir.2014.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett A.L., Smith D.W., Cummins M.J., Jacoby P.A., Cummins J.M., Beilharz M.W. Low-dose oral interferon alpha as prophylaxis against viral respiratory illness: a double-blind, parallel controlled trial during an influenza pandemic year. Influenza Other Resp. 2013;7:854–862. doi: 10.1111/irv.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herold S., Becker C., Ridge K.M., Budinger G.R.S. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur Respir J. 2015;45:1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- 55.Jureka A.S., Kleinpeter A.B., Cornilescu G., Cornilescu C.C., Petit C.M. Structural basis for a novel interaction between the NS1 protein derived from the 1918 influenza virus and RIG-I. Structure. 2015;23:2001–2010. doi: 10.1016/j.str.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balachandran S., Roberts P.C., Brown L.E., Truong H., Pattnaik A.K., Archer D.R. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 57.Dauber B., Martinez-Sobrido L., Schneider J., Hai R., Waibler Z., Kalinke U. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M. The IFITM proteins mediate cellular resistance to influenza a H1N1 virus, west nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hefti H.P., Frese M., Landis H., Di Paolo C., Aguzzi A., Haller O. Human MxA protein protects mice lacking a functional alpha beta interferon system against La Crosse virus and other lethal viral infections. J Virol. 1999;73:6984–6991. doi: 10.1128/jvi.73.8.6984-6991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staeheli P., Grob R., Meier E., Sutcliffe J.G., Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pillai P.S., Molony R.D., Martinod K., Dong H., Pang I.K., Tal M.C. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 2016;352:463–466. doi: 10.1126/science.aaf3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raberg L., Sim D., Read A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 65.Ayres J.S., Schneider D.S. Tolerance of infections. Annu Rev Immunol. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 66.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 67.Ren R., Wu S., Cai J., Yang Y., Ren X., Feng Y. The H7N9 influenza A virus infection results in lethal inflammation in the mammalian host via the NLRP3-caspase-1 inflammasome. Sci Rep. 2017;7:7625. doi: 10.1038/s41598-017-07384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ichinohe T., Lee H.K., Ogura Y., Flavell R., Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tal M.C., Iwasaki A. Mitoxosome: a mitochondrial platform for cross-talk between cellular stress and antiviral signaling. Immunol Rev. 2011;243:215–234. doi: 10.1111/j.1600-065X.2011.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McAuley J.L., Tate M.D., MacKenzie-Kludas C.J., Pinar A., Zeng W.G., Stutz A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernandez M.V., Miller E., Krammer F., Gopal R., Greenbaum B.D., Bhardwaj N. Ion efflux and influenza infection trigger NLRP3 inflammasome signaling in human dendritic cells. J Leukoc Biol. 2016;99:723–734. doi: 10.1189/jlb.3A0614-313RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 73.Pothlichet J., Meunier I., Davis B.K., Ting J.P.Y., Skamene E., von Messling V. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza a virus infected cells. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuriakose T., Kanneganti T.D. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol Immunol. 2017;86:56–64. doi: 10.1016/j.molimm.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuriakose T., Man S.M., Malireddi R.K.S., Karki R., Kesavardhana S., Place D.E. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1 doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mikhak Z., Strassner J.P., Luster A.D. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J Exp Med. 2013;210:1855–1869. doi: 10.1084/jem.20130091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eberl G., Colonna M., Di Santo J.P., McKenzie A.N.J. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348 doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 79.Yuan W.M., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weizman O.E., Adams N.M., Schuster I.S., Krishna C., Pritykin Y., Lau C. ILC1 confer early host protection at initial sites of viral infection. Cell. 2017;171:795–808. doi: 10.1016/j.cell.2017.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cephus J.Y., Stier M.T., Fuseini H., Yung J.A., Toki S., Bloodworth M.H. Testosterone attenuates group 2 innate lymphoid cell-mediated airway inflammation. Cell Rep. 2017;21:2487–2499. doi: 10.1016/j.celrep.2017.10.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doherty T.A., Broide D.H. Pathways to limit group 2 innate lymphoid cell activation. J Allergy Clin Immunol. 2017;139:1465–1467. doi: 10.1016/j.jaci.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim H.Y., Umetsu D.T., Dekruyff R.H. Innate lymphoid cells in asthma: will they take your breath away? Eur J Immunol. 2016;46:795–806. doi: 10.1002/eji.201444557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crowe C.R., Chen K., Pociask D.A., Alcorn J.F., Krivich C., Enelow R.I. Critical role of IL-17RA in immunopathology of influenza infection. J Immunol. 2009;183:5301–5310. doi: 10.4049/jimmunol.0900995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szretter K.J., Gangappa S., Lu X., Smith C., Shieh W.J., Zaki S.R. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tate M.D., Deng Y.M., Jones J.E., Anderson G.P., Brooks A.G., Reading P.C. Neutrophils ameliorate lung injury and the development of severe disease during influenza infection. J Immunol. 2009;183:7441–7450. doi: 10.4049/jimmunol.0902497. [DOI] [PubMed] [Google Scholar]
- 87.Narasaraju T., Yang E., Samy R.P., Ng H.H., Poh W.P., Liew A.A. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol. 2011;179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hartshorn K.L., Collamer M., White M.R., Schwartz J.H., Tauber A.I. Characterization of influenza A virus activation of the human neutrophil. Blood. 1990;75:218–226. [PubMed] [Google Scholar]
- 89.Hartshorn K.L., Daigneault D.E., White M.R., Tauber A.I. Anomalous features of human neutrophil activation by influenza A virus are shared by related viruses and sialic acid-binding lectins. J Leukoc Biol. 1992;51:230–236. doi: 10.1002/jlb.51.3.230. [DOI] [PubMed] [Google Scholar]
- 90.Saffarzadeh M., Juenemann C., Queisser M.A., Lochnit G., Barreto G., Galuska S.P. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jayasekera J.P., Vinuesa C.G., Karupiah G., King N.J. Enhanced antiviral antibody secretion and attenuated immunopathology during influenza virus infection in nitric oxide synthase-2-deficient mice. J Gen Virol. 2006;87:3361–3371. doi: 10.1099/vir.0.82131-0. [DOI] [PubMed] [Google Scholar]
- 92.Watanabe Y., Hashimoto Y., Shiratsuchi A., Takizawa T., Nakanishi Y. Augmentation of fatality of influenza in mice by inhibition of phagocytosis. Biochem Biophys Res Commun. 2005;337:881–886. doi: 10.1016/j.bbrc.2005.09.133. [DOI] [PubMed] [Google Scholar]
- 93.Tumpey T.M., Garcia-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Pantin-Jackwood M.J. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Londrigan S.L., Short K.R., Ma J., Gillespie L., Rockman S.P., Brooks A.G. Infection of mouse macrophages by seasonal influenza viruses can Be restricted at the level of virus entry and at a late stage in the virus life cycle. J Virol. 2015;89:12319–12329. doi: 10.1128/JVI.01455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conenello G.M., Zamarin D., Perrone L.A., Tumpey T., Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seth S., Oberdorfer L., Hyde R., Hoff K., Thies V., Worbs T. CCR7 essentially contributes to the homing of plasmacytoid dendritic cells to lymph nodes under steady-state as well as inflammatory conditions. J Immunol. 2011;186:3364–3372. doi: 10.4049/jimmunol.1002598. [DOI] [PubMed] [Google Scholar]
- 97.Qu C., Nguyen V.A., Merad M., Randolph G.J. MHC class I/peptide transfer between dendritic cells overcomes poor cross-presentation by monocyte-derived APCs that engulf dying cells. J Immunol. 2009;182:3650–3659. doi: 10.4049/jimmunol.0801532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.GeurtsvanKessel C.H., Willart M.A., van Rijt L.S., Muskens F., Kool M., Baas C. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]