Highlights
-
•
CYP81Q1 encoding a phenylpropanoid pathway enzyme from sesame was expressed and purified in bacterial heterologous systems.
-
•
Corresponding cytochrome P450 reductase (CPR1) also from sesame was expressed in E. coli for the first time.
-
•
Enhanced activity of CYP81Q1 enzyme by CPR1in the conversion of pinoresinol to sesamin is demonstrated.
-
•
Methodology developed paves way for crystallization of CYP enzymes involved in secondary metabolism.
Keywords: Sesamin, Cytochrome P450, Sesamin synthase, Heterologous expression, CYPR1
Abstract
Members of Cytochromes P450 super family of enzymes catalyse important biochemical reactions in plants. Some of these reactions are so important that they contribute to enormous chemical diversity seen in plants. Many unique secondary metabolites formed by mediation of these enzymes play key role in plant defence and often contribute to maintenance of human health. In oilseed crop Sesamum indicum, the reaction leading to the formation of clinically important sesamin is catalyzed by a unique methylene-di-oxy bridge forming Cytochrome P450 enzyme sesamin synthase. It is encoded by the gene CYP81Q1. In order to elucidate the structure – function relationship of this enzyme and to apply biotechnological tools for enhancing the production of sesamin in the crop, it was intended to clone and express the enzyme in a heterologous system. In this paper we present our results on synthesis of cDNA, cloning, expression and purification of CYP81Q1 from the developing seeds of sesame crop. Following the same procedure we have also cloned a CYP reductase1 (CPR1) gene (CPR1) to facilitate transfer of electron from NADPH to CYP81Q1 enzyme from the same crop. Functional characterization was performed by expressing the recombinant proteins in E. coli (pET28a/BL21-DE3 codon plus) and its activity was evaluated in vitro by HPLC. We demonstrate that purified CYP81Q1 enzyme, on its own, has limited level of activity in the conversion of pinoresinol to sesamin. Its activity gets considerably enhanced in the presence of CPR1.
1. Introduction
Sesamum indicum is an important ancient oilseed crop that serves as major source of the antioxidant lignan sesamin [1]. The lignans, in turn, are dimers of phenyl propane units formed by a covalent linkage between the central carbons of their propyl side chains. Sesamin exhibits clinically significant properties such as lowering of cholesterol, control of hypertension and induction of apoptosis in cancer cells [[2], [3], [4]]. The compound also protects liver from the damaging effects of ethanol and carbon tetrachloride, exhibits anti-inflammatory effect, inhibits vascular superoxide production and induces nitric oxide production in vasculatures [[5], [6], [7], [8], [9], [10]]. In view of these properties there is a great demand for Sesamin [11]. Chemical synthesis of the compound has not been successful [12]. Even though there are many natural sources identified for sesamin, the oil and seed from S. indicum alone continues to be the major source of the compound [13]. Sesamin merely ranges from 0.2% to 0.8% of by the dry weight of seeds in the existing varieties of the sesame crop [[14], [15], [16]]. Genetics of sesamin production indicates that it could be a polygenic trait [17,18]. One of the options available is to enhance sesamin synthesis in live plants of S. indicum by metabolic engineering [13]. Sesamin follows a complex biochemical pathway [19]. In Sesamum, lignan biosynthesis starts with phenylalanine which is sequentially converted to sesamin and related lignans. Pinoresinol is one of the intermediate in the pathway synthesized from coniferyl alcohol by a reaction assisted by a ‘dirigent’ protein [20]. Piperitol synthase converts pinoresinol to piperitol which is then converted to sesamin by sesamin synthase [21,22]. Recently, following a reverse genetics approach, it was shown that the two step conversion of pinoresinol to sesamin was catalysed by a single enzyme that has dual catalytic activity and the enzyme accordingly was named as piperitol-sesamin synthase (PSS). The PSS belongs to cytochrome P450 (CYP) group of enzymes and in Sesamum it is encoded by the gene CYP81Q1, consequently the enzyme was also termed as CYP81Q1 enzyme [23]. The level of expression of CYP81Q1 often differed with genotype of the plant. For example, the PSS from the sesame variety Gomazou exhibited a higher expression than Kin-goma with concomitant increase in sesamin content. Therefore it was inferred that genotypic difference of CYP81Q1 is one of the important factor affecting sesamin content in sesame [17]. CYP81Q3, a homolog of the gene from S. alatum lacked sesamin biosynthesis activity and accordingly the species was designated as a sesamin deficient one. CYP enzymes constitute a super family of enzymes that are involved in diverse primary and secondary metabolic pathways in plants [24]. The divergent CYP enzymes in fact are responsible for much of the phytochemical diversity seen in almost all organisms including prokaryotes, eukaryotes and other higher animals. Many of the CYP enzymes are also involved in degradation of xenobiotics in mammals including human [25]. In order to apply biotechnology for enhancing sesamin content in S. indicum, as a preliminary prerequisite the present study was undertaken to clone, express and characterize CYP81Q1 enzyme in a heterologous system. Earlier, Hemati and Nayeri [26] described amplification and cloning of CYP81Q1 gene from genomic DNA of S. indicum. But its expression and activity was not reported. However, using bioinformatics tools, the authors predicted only the structure but not function of the protein. Gene if amplified from genomic DNA, the CYP81Q genes e.g., CYP81Q1, CYP81Q2, CYP81Q3 and CYP81D2in general contained an intron and therefore remain non expressive in prokaryotes [23]. Therefore, we intended to follow the cDNA approach to enable isolation and expression of the gene in prokaryotic system. CYP81Q1 is amonooxygenase and therefore requires a reduction reaction to proceed prior to its function. PSS meets this requirement by pairing with the membrane bound cytochrome P450 reductase1 (CPR1) that helps transfer of electrons efficiently from the co-factor NADPH to heme unit of the CYP of the CYP81Q1 [27,28]. CPR shaveflavin groups that mediate electron transfer in a highly regulated manner [29]. Herewe present our observation on cDNA cloning, expression and characterization of CYP81Q1 and sesamin CPR1 genes from an Indian variety of S. indicum.
2. Material and method
2.1. Plant material
Seed of Sesamum indicum varietyTMV3, a black seeded one, used in the present study was obtained from National Bureau of Plant Genetic Resource (NBPGR), New Delhi. The crop was maintained by cultivation in the experimental garden of our Department. For cultivation of sesame the procedure as described in Handbook of Agriculture (1996), ICAR, New Delhi was followed. Sesame seeds were sown in rows by hand during the Rabi season (December) and harvested in April. A row to row distance of 50 cm and a plant to plant distance of 20 cm were maintained. Weeds were physically removed and the crop was irrigated on daily routine till seed germination and at regular interval during flowering till pod formation. Fruits of the plant, known as pods, were harvested at different stages of development. Based on the size and colour, the pods were grouped into five classes to correspond to their five developmental stages (Table 1). Seeds were collected separately from each of the stages and split into two sets. One set of the seed was utilized for lignan analysis by HPLC and the other set was used for RNA isolation.
Table 1.
Estimate of certain traits at different stages of development of pod of Sesamum indicum.
| Name of the Trait | Developmental Stage and Characteristic Feature |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Pod colour | Light Green | Light Green | Light Green | Dark Green | Brown |
| Pod length (cm) | 1.2 ± 0.20 | 1.7 ± 0.15 | 2.2 ± 0.10 | 2.2 ± 0.15 | 2.2 ± 0.15 |
| Lignans (mg/g seed) | |||||
| Pinoresinol | 2.4 ± 0.30 | 4.6 ± 0.50 | 3.8 ± 0.40 | 0.38 ± 0.20 | 0.29 ± 0.10 |
| Sesamin | 0 | 0 | 0 | 0.92 ± 0.30 | 0.85 ± 0.10 |
| Sesamolin | 0 | 0 | 0 | 0.43 ± 0.10 | 0.27 ± 0.04 |
2.2. Lignan analysis in developing seeds
200 mg of seeds harvested from different stages of pods were pulverized in liquid nitrogen and the powder obtained was extracted in 1 ml of 80% methanol in an eppendorf tube. The mixture was centrifuged at 5585xg for 3 min and the supernatant was collected to serve as crude lignan extract. The crude samples thus obtained were analysed using Shimadzu HPLC system equipped with C-18 reverse phase column and UV-VIS detector at 290 nm. The mobile phase consisted of 65:35 acetonitrile - water with a flow rate of 0.8 ml/min [30]. Authentic standards of Sesamin (Cat No. SMB00705) and Pinoresinol (Cat No. 40674) were purchased from Sigma-aldrich, USA. HPLC grade acetonitrile (Cat No. AS029) and Water (Cat No. AS077) were purchased from Himedia, India. The lignan content of the harvested sesame seed were quantified following the method described in Dar et al. [16].
2.3. Isolation of RNA, first strand synthesis, cDNA amplification and cloning of CYP81Q1 and CPR1
RNA was isolated from developing seeds collected from stage-4 pods, using RNeasy® Plant Mini Kit (Qiagen) and converted to cDNA using the First strand cDNA synthesis kit (Invitrogen) following the manufacturer’s protocol. Prior to RNA isolation about 100 mg of seeds were crushed using liquid nitrogen into powder in a pestle and mortor. The powder was solubilized in RLC buffer given in the kit. Based on the published reports specific primers were designed and synthesized for amplification of CYP81Q1 and CPR1 genes from the cDNA [23,31]. Gene specific forward and reverse primers were designed in such a way as to include BamHI and XhoI restriction sites respectively, at 5’ and 3’ end of the genes to facilitate their cloning in right direction in the expression vector. The restriction sites are indicated by an underline in the primer sequence. The forward primer for amplification of CYP81Q1 gene consisted of 5’-ATCGGGATCCATGGAAGCTGAAATGCTATATTC-3’ and the reverse primer consisted of 5’-CTAGCTCGAGAACGTTGGAAACCTGACGAAG-3’. Similarly for amplification of cDNA of CPR1, the forward primer consisted of 5’-ATCGGGATCCATGGAACCCACATCGGAAAAGC-3’ and the reverse primers consisted of 5’-CTAGCTCGAGTCACCACACATCACGCAAGTAC-3’. Independent PCRs were performed for the two genes using Q5 high fidelity Taq polymerase (NEB) for 30 cycles of 98 °C for 30 s (denaturation), 60 °C for 15 s (annealing) and 72 °C for 1 min (extension). PCR product was purified using QIA quick PCR Purification Kit (QIAGEN) and quantified using a nanodrop. In two independent experiments the PCR products were cloned into the pET28a (Novogen) expression vector to obtain pET28a-CYP81Q1and pET28a-CPR1 constructs. While CPR1 construct was designed to have only N-terminal His tag, the CYP81Q1 was designed to have both N- as well as C- terminal His tags. They were then independently transformed into the expression host BL21-(DE3) codon plus (Stratagene) following manufacturer’s protocol. pET28a vectors have been designed to have 6xHis tags that get co-expressed with the target protein to facilitate purification and identification of the target proteins (Novagen, TB055 8th Edition 02/99).
2.4. Culture of recombinant bacterium and protein induction
A Single colony of the transformed BL21-(DE3) codon plus harbouring recombinant plasmids for CYP81Q1 or CPR1were grown overnight at 37 °Cin 10 ml LB broth containing 50 μg/l kanamycin in a 50 ml test tube at 200 rpm. The cell suspension of the overnight grown cultures was inoculated in to 1 L fresh LB broth containing the antibiotic at the same concentration taken in a 2 L conical flask and incubated at 37 °C on a shaker set at 200 rpm. The OD600nm, was monitored until it reached a value of 0.8 OD. To this culture 1 mM IPTG (Himedia Cat No. RM2578) was added and incubated for 6 h at 22 °C for induction of the protein production. Cells were then harvested by centrifugation at 3000×g for 15 min at 4 °C.
2.5. Isolation of recombinant protein and purification
The harvested cells were lysed by sonication in a buffer of pH 7.5containing 50 mM Tris, 500 mM NaCl, 20 mM imidazole, 0.5 mM DTT and 5% glycerol with 0.5 mM PMSF. The lysed suspension was centrifuged at 16,000×g for 30 min. Supernatant was passed through 1 ml Ni-NTA packed affinity column (GE healthcare) and washed with 100 volumes of washing buffer of same composition as sonication buffer but with 40 mM imidazole. The bound protein was eluted using 20 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol, 300 mM imidazole, 1 mM DTT and 1 mM PMSF. Purified protein was dialysed in 20 mM HEPES buffer of same composition as that of elution buffer but devoid of imidazole [32].
2.6. Western blotting
SDS-PAGE of the purified protein was performed along with appropriate controls and protein bands were transferred onto Nitrocellulose membrane using semi dry transfer unit with 80 mV for 1 h. The membrane was then washed with the buffer for 30 min, transferred to a tray containing anti-His antibody (Cat No. 2365 T) procured from Cell Signaling Technology (CST) was used at 1:1000 dilution and incubated for 2 h. Subsequently the membrane was washed for 1 h in TBST buffer [33], transferred to a tray containing Anti-rabbit secondary antibody (CST 7074P2) and incubated for 2 h in shaking condition. The membrane was then washed in TBST for 15 min 4 times to remove traces of secondary antibody. The bands were visualized using ECL reagent (Biorad) and photographed.
2.7. Enzyme assay for PSS activity
The enzyme assay was performed by estimating the conversion of pinoresinol to sesamin by HPLC. The reaction mixture consisted of 200 μl of pH7.5 PBS buffer (10 mM), 5 μl each of Pinoresinol (1 mg/ml) and NADPH (Cat No. RM576, 50 mM), and 30 μl each of purified enzymes of CPR1and / or CYP81Q1or water instead. The mixture was incubated at 30 °C for 4 h with intermittent shaking. The reaction was terminated by adding an equal volume of ethylacetate. Following a modified protocol described earlier [30], 10 μl of the resulting mixture was injected into HPLC for lignan analysis.
3. Results
3.1. HPLC of seed lignans at different stages of seed development
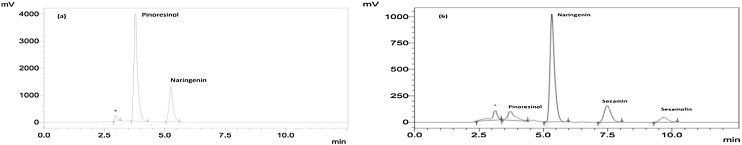
Expression of many of the plant genes is developmentally regulated. In order to identify asuitable stage of pod for harvesting seeds for isolation of target mRNA, an indirect method of analysis was followed. The pod colour and pod length were first measured followed by lignan content in the seeds from five different stages of pod development analysed by HPLC (Table 1). Pods of the first three stages differed in their length but remained light green in colour. Pod of stages 3–5 showed no change in pod length but their colour changed from light green to dark green and then to brown. Seeds harvested from stages 1 to 4 were white and those of stage5 were black in colour (Fig. 1). Lignan analysis of methanol extract of seeds from different stages of development by HPLC revealed that there is no trace of sesamin and sesamolin in seeds upto stage III, but showed a prominent peak with a RT (retention time) of 3.7 min which corresponded to the RT of pinoresinol standard indicating its synthesis during these stages (Fig. 2a).
Fig. 1.
Pods (upper panel) and corresponding seeds (lower panel) of Sesamum indicum harvested at different Stages of development. Further details are given in Table1.
Fig. 2.
HPLC profile of methanol extract of Sesamum indicum seeds harvested at different stages of development. (a) Stage-2 showing a prominent peak of Pinoresinol (RT 3.7) indicating its presence in the seeds. No peaks could be seen for sesamin and sesamolin at this stage. (b) Stage-4 showing additional peaks corresponding to sesamin (RT 7.5) and sesamolin (RT 9.7) Note the peak for Pinoresinol (RT 3.7) is greatly reduced. Internal standard Naringenin is indicated.*represents peak of unknown compound.
HPLC of the extract from seeds of stages 4 and 5 showed prominent peaks for sesamin (RT 7.5 min) and sesamolin (RT 9.7) with considerable reduction in the area of the peak corresponding to pinoresinol (RT 3.7 min) indicating biosynthesis of the sesamin and sesamolin taking place during these stages (Fig. 2b). The lignan content of the seeds at different developmental stages coincided with the difference in HPLC peak area observed (Table1).
3.2. RNA isolation, cDNA synthesis, amplification and sequencing of CYP81Q1 and CPR1
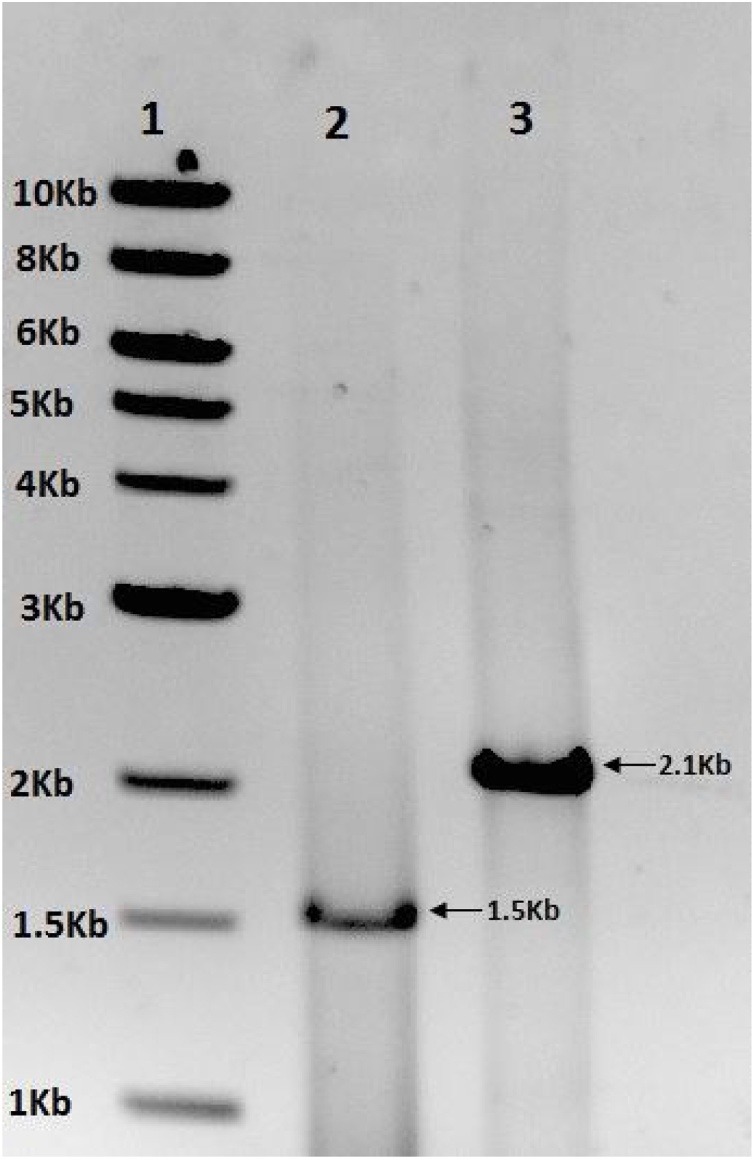
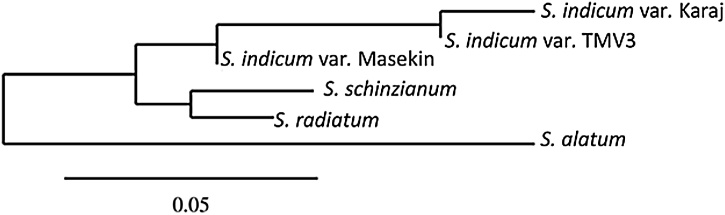
Since sesamin appeared in stage-4, the seeds harvested from stage-4 pods were used for RNA isolation. The total RNA yield was 640 ng/mg of seeds with aA260/A280 of 2.0 as recorded using nanodrop. First strand cDNA was synthesized from the isolated RNA using Invitrogen kit. Gene specific primers enabled amplification of CYP81Q1and CPR1 from cDNA by PCR. Electrophoresis of the PCR products revealed that the size of CYP81Q1 was 1.5 kb and that of CPR1 was 2.1 kb (Fig. 3). Sequencing of the PCR products of the two genes confirmed their identity. Analysis of nucleotide phylogeny of the CYP81Q1 (TMV3) that we have cloned revealed that the sequence had 100% similarity to Sesamum indicum cultivar Karaj cytochrome p450 81Q1 (cyp81q1) gene (the NCBI accession number KP771974.1 Fig. 4) and CPR1sequence available in NCBI data base (data not shown). The PCR products thus obtained from cDNA were cloned and expressed in heterologous system.
Fig. 3.
A gel picture AGE showing molecular sizes of thePCR products amplifiedfrom cDNA using gene specific primers. Lane 1: 1Kb DNA ladder, Lane 2:CYP81Q1(1.5Kb) and Lane 3: CPR1(2.1Kb).
Fig. 4.
Phylogenetic tree based on similarity of sequences of CYP81Q genes sequenced from different accessions of Sesamum.
3.3. Expression and purification of recombinant enzymes
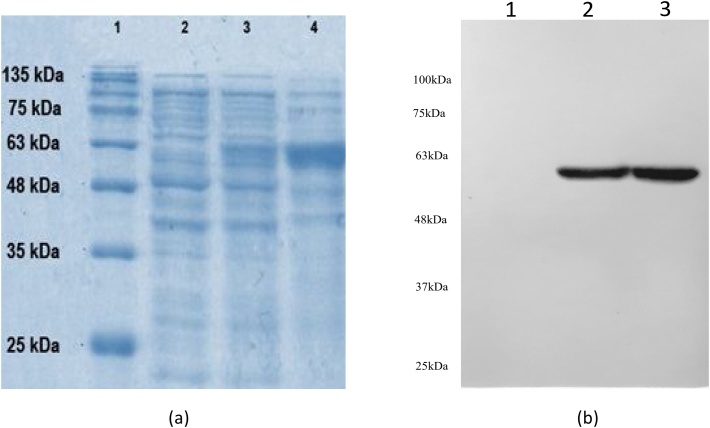
Transcriptional induction of pET28a vector harbouringCYP81Q1transformed into BL21-(DE3) codon plus yielded 4.8 mg of PSS protein per litre of the bacterial culture. SDS-PAGE of the crude protein revealed presence of 55 kDa band corresponding to the molecular weight of PSS protein. An enriched band of protein was observed on purification with His-tag column (Fig. 5). A similar procedure followed for putative CPR1 yielded 3.9 mg of CPR1 protein (MW 72 kDa) per litre of bacterial culture (Supplementary Fig. 1). Thus the cloning and expression of CYP81Q1 and CPR1 genes from S. indicum was confirmed. Expression of the recombinant genes were further confirmed by western blot analysis of the fractions of bacterial lysates from un-induced, induced cultures and the affinity purified protein fraction. The result showed there is no expression of the recombinant CPR1 protein in the un-induced lysates but confirmed its presence in both induced and purified fractions (Supplementary Fig. 1). Thus the cloning of two genes in fusion with 6×His, their over expression and purification of the recombinant protein was further confirmed.
Fig. 5.
(a) Gel pictures of 12% SDS-PAGE of recombinant PSS protein expressed in BL21-DE3 codon strain of E.coli. Lane 1:Pre-stained protein ladder; Lane 2: Cell lysate from un-induced cells; Lane 3: Cell lysate from IPTG induced cells showing 55 kDa PSS enzyme and Lane 4: His-Tag purified protein fraction showing a thick and clear band at size of PSS. (b) Western blot of recombinant protein of CYP81Q1 hybridized with his-tag antibody. Lane 1: un-induced cell lysate, Lane 2: induced cell lysate and Lane 3 his-tag purified protein. (A 10% PAGE was used for preparing the western blot).
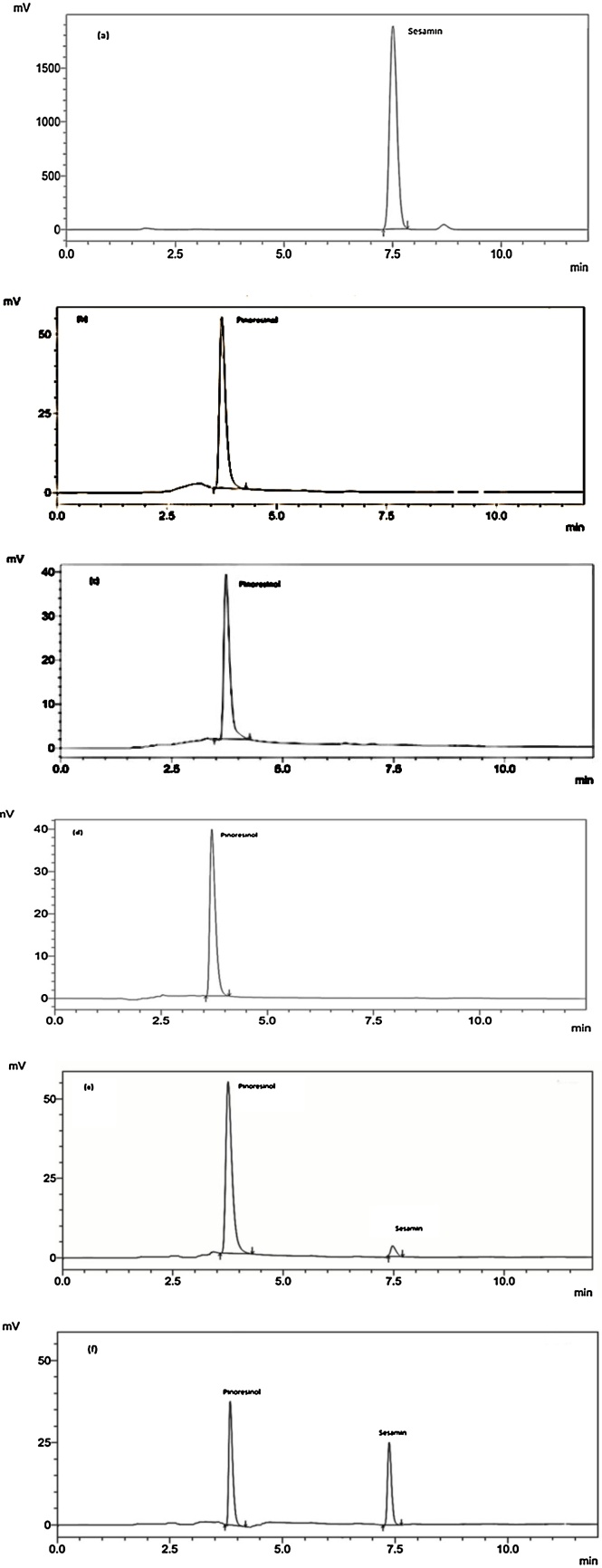
3.4. In vitro assay for CYP81Q1 enzyme activity
HPLC of the standard pinoresinol and sesamin showed a RT of 3.7 min and 7.5 min, respectively (Fig. 6a and b). HPLC of the reaction mixture without the enzyme showed no trace of sesamin (Fig. 6c). The sesamin was also undetectable in the presence of CPR1 alone (Fig. 6d). The same is the case in the presence of NADPH alone or NADPH + CPR1but in the absence of CYP81Q1. However, when CYP81Q1 enzyme (PSS) alonewas supplemented along with NADPH and pinoresinol there appeared a small peak corresponding to sesamin indicating occurrence of some base level activity in the synthesis of sesamin (Fig. 6e). This showed that the purified recombinant protein of PSS, obtained from bacteria is capable of converting substrate pinoresinol to the product sesamin. When both CYP81Q1and CPR1is present together in the reaction mixture, there was considerable increase in CYP81Q1 activity leading to a sharp increase in sesamin produced (Fig. 6f). Thus cloning and expression of functional CYP81Q1 and CPR1 enzymes of sesame is confirmed.
Fig. 6.
HPLC of sesamin standard, pinoresinol standard and PSS assayed in vitrolignans extract. (a) Sesamin standard (RT 7.5); (b) pinoresinol standard (RT 3.7); (c) pinoresinol + NADPH, (d) CPR1 + pinoresinol + NADPH; (e) CYP81Q1 + pinoresinol + NADPH; (f) CYP81Q1 + CPR1 + pinoresinol + NADPH. Note (b), (c) and (d) showing a peak at RT 3.7 for the presence of pinoresinol. (e) Showing a small peak for sesamin. (f) Showing a reduction in pinoresinol with increase in sesamin peak (RT 7.5).
4. Discussion
Sesamin belongs to a group of bioactive compounds known as lignans that are synthesized through phenylpropanoid pathway. There are several reports available on isolation of sesamin and about fifteen other lignans from S. indicum [16,[34], [35], [36]]. Most of these lignans are fat soluble aglycons that on extraction elute in the oil and the remaining is glycosylated that can be isolated mainly from the oil free meal. However, isolation and characterization of enzymes involved in sesamin biosynthesis for biotechnological application has been very few. CYP81Q1 was earlier expressed in heterologous yeast system or in Baculovirus / Sf9 cell system with the enzyme activity assayed using microsomal fractions from the recombinant cells [23,37]. Recently Hemati and Nayeri [26] reported amplification and cloning of CYP81Q1 gene from the genomic DNA isolated from an Iranian S. indicum variety Karaj-1. But its expression, either in vitro or in vivo, has not been demonstrated. It may be relevant to mention that we had also used genomic DNA for amplification CYP81Q1 and obtained a 1.7 kb size PCR product that contained a 171bp intron. Expression of genes in bacterial system would be more preferred approach for industrial production of enzymes for which one needs an intron free gene construct. Following cDNA approach we showed here cloning and expression of the intron free CYP81Q1 gene in E. coli and demonstrated the functioning of the purified recombinant enzyme by in vitro enzyme assay. We also demonstrated here requirement of a reductase for enhanced functioning of the enzyme. Novelty of our paper lies in the fact that we have established a system for sustainable production of both CYP81Q1 (PSS) and CPR1 (reductase) proteins in a bacterial system for conversion of pinoresinol to sesamin. The system paves the way for production of piperitol-sesamin synthase (PSS) and its product(s) thereof at industrial scale. It is relevant to point out here that CYP enzymes have recently been identified as targets for directed evolution by mutation to impart new and/or enhanced catalytic activities that may be followed bysimple expression in E. coli [38].
Recombinant CYP enzymes (monoxygenases and hydroxylases) that are CPR dependant have been assayed in different ways. One of them is to use a mixture of soluble fraction of the lysate from two different recombinant cells, for example in case of catalysis when CYP73A25 (from cotton) and CamCPR (CPR from Camptotheca acuminata) were brought together [27]. In cases where the recombinant proteins are expressed as inclusion body, the insoluble fraction of the lysate was solubilized and used as shown for CYP97C27 and CPR1 from Croton stellatopilosus [28]. Alternately the recombinant chimeric CYP-CPR is immobilized on nanodisc and assayed as in heterologous expression of chimeric CYP725A4-17α-L-CPR of Taxus cuspidata [39]. Earlier the enzyme obtained using heterologous expression of CYP81Q1 of sesame was assayed with the help of yeast microsomal fraction in which some membrane bound native reductase might have served the purpose [23,31]. Here we showed expression of the two plant proteins in bacterial system and followed a procedure modified from Qu et al. [27] and Sintupachee et al. [28] to obtain reproducible results on enzyme activity. We also showed that the presence of reductase is a must for enhanced catalytic conversion of pinoresinol to sesamin in vitro.
In S. indicum var. TMV3, sesamin appears to be synthesized from stage 3 to 5 of the pod development. This correlates with recent report on S. indicum wherein it was shown, by RT-PCR, that maximum amount of CYP81Q1 transcripts accumulated by 25th day of flowering [40]. Unlike the earlier report we found that there is synthesis of pinoresinol in the first three stages as well and there is lack of appearance of piperitol (by HPLC) when sesamin was synthesized from pinoresinol in vitro [23]. We attribute such phenomenon to genotype difference between the varieties as has been suggested earlier [17]. Methylenedioxy bridge formation is an important step in the biosynthesis of several important secondary metabolites [41]. There can be either one (berberine, safrol, yatein) or at the maximum two (sesamin, sangunarine, podophyllotoxin, stylopine) such bridges that are formed in the molecules with their formation catalyzed by a P450 dependent monooxygenases [42]. If there is more than one such bridge, formation of each bridge is catalyzed by different P450 enzymes in successive reactions [43,44]. The PSS enzyme encoded by CYP81Q1 dealt with herein is the first case where the same enzyme catalyzes the formation of two methylenedioxy bridges in two successive reactions leading to synthesis of sesamin in S. indicum [23]. Further the domestication of the crop appears favour selection of sesame crop against sesamin biosynthesis as there are many wild relatives of the only cultivated S. indicum such as S. malabaricum and S. mulayanam characterized by higher sesamin content [40]. Therefore it should be possible to augment sesamin content in cultivated sesame by breeding and further enhanced by metabolic engineering.
Recently by homology modelling and site directed mutagenesis it was shown that residue A308 is important for catalysis leading to formation of characteristic methylenedioxy bridge and a mutation in this residue resulted in inhibition of PSS [37]. Precise catalytic role of the amino acid residues is yet to be known. For the enzymes whose structures have already been elucidated there is a possibility that they can be developed into molecules having clinical or agricultural significance. Such details for the one’s involved in phenylpropanoid pathway are yet to be elucidated [45]. Further analyses such as computational alanine scanning along with comprehensive in vitro studies are required for understanding a complete picture of the mode of action of this enzyme. This may facilitate application of modern methods of genetic engineering such as Crisper/cas9for increasing sesamin content in the crop plants [46]. In conclusion, it may be stated that the CYP81Q1 and CPR1 genes from S. indium have been successfully cloned, expressed in E. coli, functional recombinant enzyme successfully isolated from the heterologous system and its activity demonstrated. Through metabolic engineering it should be possible to modify these genes for optimal expression of the desired secondary metabolite for application towards human welfare [44,38,47].
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
NA acknowledges DBT project BUILDER No. BT/PR14554/INF/22/125/2010 and DST-FIST, for infrastructure. KC acknowledges UGC for NFO Research Fellowship.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2019.e00336.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Ma Y., Xu K., Wang S., Han Y. Simultaneous determination of two epimeric furofuran lignans (Sesamin and Asarinin) of Asarum heterotropoides extract in rat plasma by LC/MS/MS: application to pharmacokinetic study. J. Chromatogr. Sci. 2014;52:793–798. doi: 10.1093/chromsci/bmt114. [DOI] [PubMed] [Google Scholar]
- 2.Penalvo J.L., Hopia A., Adlercreutz H. Effect of sesamin on serum cholesterol and triglycerides levels in LDL receptor-deficient mice. Eur. J. Nutr. 2006;45:439–444. doi: 10.1007/s00394-006-0617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakano D., Takaoka M., Kiso Y., Matsumura Y. Antihypertensive effect of sesamin. Vasc. Dis. Prev. 2004;1:233–241. [Google Scholar]
- 4.Deng P., Wang C., Chen L., Wang C., Du Y., Yan X., Chen M., Yang G., He G. Sesamin induces cell cycle arrest and apoptosis through the inhibition of signal transducer and activator of transcription 3 signalling in human hepatocellular carcinoma cell line HepG2. Biol. Pharm. Bull. 2013;36:1540–1548. doi: 10.1248/bpb.b13-00235. [DOI] [PubMed] [Google Scholar]
- 5.Akimoto K., Kitagawa Y., Akamatsu T., Hirose N., Sugano M., Shimizu S., Yamada H. Protective effects of sesamin against liver damage caused by alcohol or carbon tetrachloride in rodents. Ann. Nutr. Metab. 1993;37:218–224. doi: 10.1159/000177771. [DOI] [PubMed] [Google Scholar]
- 6.Nakai M., Harada M., Nakahara K., Akimoto K., Shibata H., Miki W., Kiso Y. Novel antioxidative metabolites in rat liver with ingested sesamin. J. Agric. Food Chem. 2003;51:1666–1670. doi: 10.1021/jf0258961. [DOI] [PubMed] [Google Scholar]
- 7.Utsunomiya T., Chavali S.R., Zhong W.W., Forse R.A. Effects of sesamin-supplemented dietary fat emulsions on the ex vivo production of lipopolysaccharide induced prostanoids and tumor necrosis factor α in rats. Am. J. Clin. Nutr. 2000;72:804–808. doi: 10.1093/ajcn/72.3.804. [DOI] [PubMed] [Google Scholar]
- 8.Nakano D., Itoh C., Takaoka M., Kiso Y., Tanaka T., Matsumura Y. Antihypertensive effect of sesamin. IV. Inhibition of vascular superoxide production by sesamin. Biol. Pharm. Bull. 2002;25:1247–1249. doi: 10.1248/bpb.25.1247. [DOI] [PubMed] [Google Scholar]
- 9.Nakano D., Itoh C., Ishii F., Kawanishi H., Takaoka M., Kiso Y., Tsuruoka N., Tanaka T., Matsumura Y. Sesamin metabolites induce an endothelial nitric oxide-dependent vasorelaxation through their antioxidative property-independent mechanisms: possible involvement of the metabolites in the antihypertensive effect of sesamin. Biol. Pharm. Bull. 2003;26:1701–1705. [Google Scholar]
- 10.Lee C.C., Chen P.R., Lin S., Tsai S.C., Wang B.W., Chen W.W., Tsai C.E., Shyu K.G. Sesamin induces nitric oxide and decreases endothelin-1 production in HUVECs: Possible implications for its antihypertensive effect. J. Hypertens. 2004;22:2329–2338. doi: 10.1097/00004872-200412000-00015. [DOI] [PubMed] [Google Scholar]
- 11.Beroza M., Milton S.S. The synthesis of dl-sesamin and dl-asarinin. J. Am. Chem. Soc. 1956;78:1242–1247. [Google Scholar]
- 12.Urata H., Nishioka Y., Tobashi T., Matsumura Y., Tomimori N., Ono Y., Kiso Y., Wada S. First chemical synthesis of antioxidative metabolites of sesamin. Chem. Pharm. Bull. 2008;56:1611–1612. doi: 10.1248/cpb.56.1611. [DOI] [PubMed] [Google Scholar]
- 13.Dar A.A., Arumugam N. Lignans of sesame: purification methods, biological activities and biosynthesis – a review. Bioorg. Chem. 2013;50:1–10. doi: 10.1016/j.bioorg.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Hemalatha S., Ghafoorunissa Lignans and tocopherols in Indian sesame cultivars. J. Am. Oil Chem. Soc. 2004;81:467–470. [Google Scholar]
- 15.Williamson K.S., Morris J.B., Pye Q.N., Kamat C.D., Hensley K. A survey of sesamin and composition of tocopherol variability from seeds of eleven diverse sesame (Sesamum indicum L.) genotypes using HPLC-PAD-ECD. Phytochem. Anal. 2008;19:311–322. doi: 10.1002/pca.1050. [DOI] [PubMed] [Google Scholar]
- 16.Dar A.A., Verma N.K., Arumugam N. An updated method for isolation, purification and characterization of clinically important antioxidant lignans - sesamin and sesamolin, from sesame oil. Ind. Crops Prod. 2015;64:201–208. [Google Scholar]
- 17.Hata N., Hayashi Y., Okazawa A., Ono E., Satake H., Kobayashi A. Comparison of sesamin contents and CYP81Q1 gene expressions in above ground vegetative organs between two Japanese sesame (Sesamum indicum L.) varieties differing in seed sesamin contents. Plant Sci. 2010;178:510–516. [Google Scholar]
- 18.Ke T., Dong C., Mao H., Zhao Y., Chen H., Liu H., Dong X., Tong C., Liu S. Analysis of expression sequence tags from a full-length-enriched cDNA library of developing sesame seeds (Sesamum indicum) BMC Plant Biol. 2011;11:180. doi: 10.1186/1471-2229-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umezawa T. Diversity in lignan biosynthesis. Phytochem. Rev. 2003;2:371–390. [Google Scholar]
- 20.Halls S.C., Davin L.B., Kramer D.M., Lewis N.G. Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry. 2004;43:2587–2595. doi: 10.1021/bi035959o. [DOI] [PubMed] [Google Scholar]
- 21.Kato M.J., Chu A., Davin L.B., Lewis N.G. Biosynthesis of antioxidant lignans in Sesamum indicum seeds. Phytochemistry. 1998;47:583–591. [Google Scholar]
- 22.Jiao Y., Davin L.B., Lewis N.G. vol. 49. Pergamon; 1998. pp. 387–394. (Furanofuran Lignan Metabolism as a Function of Seed). [Google Scholar]
- 23.Ono E., Nakai M., Fukui Y., Tmimori N., Fukuchi-Mizutani M., Saito M., Satake H., Tanaka T., Katsuta M., Umezawa T., Tanaka Y. Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. PNAS. 2006;103:10116–10121. doi: 10.1073/pnas.0603865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizutani M. Impacts of diversification of cytochrome P450 on plant metabolism. Biol. Pharm. Bull. 2012;35:824–832. doi: 10.1248/bpb.35.824. [DOI] [PubMed] [Google Scholar]
- 25.Anzenbacher P., Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemati s., Nayeri F.D. Cloning, sequencing, and bioinformatics study of CYP81Q1 gene of iranian sesame (Sesamum indicum L.) cultivar. Modares J. Biotechnol. 2018;9:277–284. [Google Scholar]
- 27.Qu X., Pu X., Chen F., Yang Y., Yang L., Zhang G., Luo Y. Molecular cloning, heterologous expression and functional characterization of an NADPH-cytochrome P450 reductase gene from Camptotheca acuminata, a camptothecin-producing plant. PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0135397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sintupachee S., Ngamrojanavanichc N., Sitthithawornd W., Eknamkul W.D. Molecular cloning, bacterial expression and functional characterisation of cytochrome P450 monooxygenase, CYP97C27 and NADPH-cytochrome P450 reductase CPRI from Croton stellatopilosus Ohba. Plant Sci. 2014;229:131–141. doi: 10.1016/j.plantsci.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Etzerodt T., Wetterhorn K., Dionisio G., Rayment I. Functional characterization of a soluble NADPH-cytochrome P450 reductase from Fusarium graminearum. Protein Expr. Purif. 2017;138:69–75. doi: 10.1016/j.pep.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Moazzami A.A., Andersson R.E., Kamal-Eldin A. Characterization and analysis of sesamolinol diglucoside in sesame seeds. Biosci. Biotechnol. Biochem. 2006;70:1478–1481. doi: 10.1271/bbb.60013. [DOI] [PubMed] [Google Scholar]
- 31.Murata J., Ono E., Yoroizuka S., Toyonaga H., Shiraishi A., Mori S., Tera M., Azuma T., Nagano A.J., Nakayasu M., Mizutani M., Wakasugi T., Yamamoto M.P., Horikawa M. Oxidative rearrangement of (+)-sesamin by CYP92B14 co-generates twin dietary lignans in sesame. Nat. Commun. 2017;8:2155. doi: 10.1038/s41467-017-02053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L., Davidson A.R. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016;169:1829–1838. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmood T., Yang P. Western blot: technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012;4:429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsuzaki H., Kawakishi S., Osawa T. Sesaminol glucosides in sesame seeds. Phytochemistry. 1994;35:773–776. doi: 10.1016/s0031-9422(00)90603-4. [DOI] [PubMed] [Google Scholar]
- 35.Amarowicz R. Application of semipreparative RP-18 HPLC for the purification of sesamin and sesamolin. J. Food Lipids. 2001;8:85–94. [Google Scholar]
- 36.Reshma M.V., Balachandran C., Arumughan C., Sunderasan A., Sukumaran D., Thomas S., Saritha S.S. Extraction, separation and characterisation of sesame oil lignan for nutraceutical applications. Food Chem. 2010;120:1041–1046. [Google Scholar]
- 37.Noguchi A., Horikawa M., Murata J., Tera M., Kawai Y., Ishiguro M., Umezawa T., Mizutani M., Ono E. Mode-of-action and evolution of methylenedioxy bridge forming P450s in plant specialized metabolism. Plant Biotechnol. 2014;31:493–503. [Google Scholar]
- 38.Arnold F.H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 2018;57:4143–4148. doi: 10.1002/anie.201708408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouck J.E., Biggs B.W., Kambalyal A., Arnold W.R., Mey M.D., Ajikumar P.K., Das A. Heterologous expression and characterization of plant Taxadiene-5a- Hydroxylase (CYP725A4) in Escherichia coli. Protein Expr. Purif. 2017;132:60–67. doi: 10.1016/j.pep.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pathak N., Bhaduri A., Bhat K.V., Rai A.K. Tracking sesamin synthase gene expression through seed maturity in wild and cultivated sesame species--a domestication footprint. Plant Biol. 2015;17:1039–1046. doi: 10.1111/plb.12327. [DOI] [PubMed] [Google Scholar]
- 41.Simmonds N.W., Stevens R. Occurrence of the methylene-dioxy bridge in the phenolic components of plants. Nature. 1956;178:752–753. [Google Scholar]
- 42.Atta-ur-Rahman . Elsevier; 2013. Studies in Natural Products Chemistry; p. 29. [Google Scholar]
- 43.Chavez M.L.D., Rolfa M., Gesell A., Kutchan T.M. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 2011;507:186–193. doi: 10.1016/j.abb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Bauer W., Zenk M.H. Two methylenedioxy bridge forming cytochrome P-450 dependent enzymes are involved in (S)-stylopine biosynthesis. Phytochemistry. 1991;30 2953-296. [Google Scholar]
- 45.Ferrer J.L., Austin M.B., Stewart C., Jr., Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian S., Jiang L., Cui X., Zhang J., Guo S., Li M., Zhang H., Ren Y., Gong G., Zong M., Liu F., Chen Q., Xu Y. Engineering herbicide-resistant water melon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018;37:1353. doi: 10.1007/s00299-018-2299-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., Kawar P.G., Hill L., Santino A., Fernie A.R., Martin C. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.