Summary
Viruses, when used as vectors for vaccine antigen delivery, can induce strong cellular and humoral responses against target epitopes. Recent work by Hansen et al. describes the use of a cytomegalovirus‐vectored vaccine, which is able to generate a stable effector‐memory T cell population at the sites of vaccination in rhesus macaques. This vaccine, targeted towards multiple epitopes in simian immunodeficiency virus (SIV), did not induce classical CD8+ T cells. However, non‐canonical CD8+ T cell induction occurred via major histocompatibility complex (MHC) class II and MHC‐E. The MHC‐E‐restricted T cells could recognize broad epitopes across the SIV peptides, and conferred protection against viral challenge to 55% of vaccinated macaques. The human homologue, human leucocyte antigen (HLA)‐E, is now being targeted as a new avenue for vaccine development. In humans, HLA‐E is an unusually oligomorphic class Ib MHC molecule, in comparison to highly polymorphic MHC class Ia. Whereas MHC class Ia presents peptides derived from pathogens to T cells, HLA‐E classically binds defined leader peptides from class Ia MHC peptides and down‐regulates NK cell cytolytic activity when presented on the cell surface. HLA‐E can also restrict non‐canonical CD8+ T cells during natural infection with various pathogens, although the extent to which they are involved in pathogen control is mostly unknown. In this review, an overview is provided of HLA‐E and its ability to interact with NK cells and non‐canonical T cells. Also discussed are the unforeseen beneficial effects of vaccination, including trained immunity of NK cells from bacille Calmette–Guérin (BCG) vaccination, and the broad restriction of non‐canonical CD8+ T cells by cytomegalovirus (CMV)‐vectored vaccines in pre‐clinical trials.
Keywords: cytomegalovirus, HLA‐E, trained immunity, vaccines
Introduction to human leucocyte antigen‐E (HLA‐E)
Sequence, structure and function
HLA‐E is a non‐classical, class Ib major histocompatibility complex (MHC) molecule located on chromosome 6, and is expressed throughout the majority of nucleated tissues in humans 1. Evolutionarily older than the HLA class Ia molecules (HLA‐A, ‐B and ‐C), HLA‐E distorts the traditional boundaries between fast‐acting innate and memory‐driven adaptive immunity 2. In contrast to the highly polymorphic HLA class Ia 3, HLA‐E is relatively more conserved, reducing the capacity for highly varied antigen recognition and peptide binding 4. This reduced polymorphism is also thought to confer tight constraints on binding of specific, hydrophobic epitopes 5. This alone contravenes the dogma that the high polymorphism of the classical major histocompatibility complex facilitates the recognition of (theoretically) 1015 epitopes, which forms the basis of adaptive immunity in vertebrates 6.
Twenty‐seven HLA‐E alleles have been reported to date, but most are found infrequently or as non‐functional proteins 7, 8, 9. HLA‐E exists predominantly in the human population as two alleles, HLA‐E*0101 and HLA‐E*0103, varying by an arginine or a glycine at amino acid position 107, respectively 10, 11. These alleles are found at near‐equal frequencies within the population 12, and probably evolved prior to divergence of humans and primates and the emergence of HLA class Ia genes 13, 14. Both HLA‐E*0101 and HLA‐E*0103 alleles have a relatively low expression on the cell surface 15. Although there is no structural difference of the peptide binding groove between these alleles, HLA‐E*0103 has a higher affinity for peptides than HLA‐E*0101, and is able to stabilize and up‐regulate at the cell surface much more efficiently 10, 16, 17. There is also variation in the peptide‐binding affinities of HLA‐E*0101 and HLA‐E*0103 in the absence of HLA class Ia and tapasin. In the absence of HLA class Ia peptides, for example due to viral down‐regulation, both alleles bind a diverse and mutually exclusive range of cellular‐derived peptides, which promote cell surface stability 18.
Peptide binding and cell surface presentation
Unlike HLA class Ia molecules, the rigidity of the HLA‐E binding groove confers preferential avidity for a limited range of peptides. In a healthy cell setting, HLA‐E binds conserved leader peptides from HLA class Ia molecules 10, 19. The highly hydrophobic binding groove is ideally suited to bind a 9 amino acid ‘leader peptide’, typically VMAPRTL(L/V/I)L (VL9, Table 1) 20. Although VL9 is the optimal peptide for binding to HLA‐E, the affinity of the HLA‐E : peptide complex is defined by specific amino acid anchors; methionine (M) at position 2 and leucine (L) at position 9 21, with ancillary anchors at P3, P6 and P7 22. Higher‐affinity epitopes bind further into the HLA‐E binding groove, improving stability at the cell surface for longer 23. There are also several reported non‐MHC derived self‐peptides that contain a leader sequence able to up‐regulate HLA‐E on the cell surface (Table 2). HSP60 and adenosine triphosphate (ATP)‐binding cassette protein multi‐drug‐resistance protein (MRP)7 both contain putative leader peptides which demonstrate HLA‐E cell‐surface stabilization. However, the immunological function that these peptides exert from HLA‐E up‐regulation is currently unknown 18.
Table 1.
Example human leucocyte antigen (HLA)‐E VL9 epitopes from HLA class Ia peptides 4, 19, 22, 25, 35, 84
| Allele | Sequence |
|---|---|
| HLA‐A*0201 | VMAPRTLVL |
| HLA‐A*01 | VMAPRTLLL |
| HLA‐B*07 | VMAPRTVLL |
| HLA‐B*27 | VTAPRTVLL |
| HLA‐C*07 | VMAPRALLL |
| HLA‐G*01 | VMAPRTLFL |
Table 2.
| Pathogen | Gene product | HLA‐E leader peptide |
|---|---|---|
| HCMV | UL40 | VMAPRTL(I/V/L)L |
| Hepatitis C virus | Core | YLLPRRGPRL |
| Epstein–Barr virus | BZLF1 | SQAPLPCVL |
| HIV | P24 | AISPRTLNA |
| Mycobacterium tuberculosis | Mtb14, P49, Mtb44 | RMAATAQVL, RMPPLGHEL, RLPAKAPLL |
| Salmonella typhimurium serovar Typhi | GroEL | GMQFDRGYL |
| Self‐peptides | ||
| n.a. | HSP60 | QMRPVSRVL |
| n.a. | ATP‐binding cassette protein MRP7 | ALALVRMLI |
HLA = human leucocyte antigen; HCMV = human cytomegalovirus; HSP = heat shock protein; ATP = adenosine triphosphate; MRP = multi‐drug‐resistance protein; n.a. = not available.
HLA‐E orthologues
HLA‐E has defined orthologues within mammals, and is unusually conserved in function across the evolution of adaptive immune genes 24. In mice and rats, the corresponding HLA‐E orthologues are Qa‐1b and RT‐BM1, respectively 25, and Mamu‐E is the HLA‐E homologue in the rhesus macaque (Macaca mulatta) 24. Primate MHC is more polymorphic than human MHC, and Mamu‐E itself exhibits further polymorphism than human HLA‐E, with at least 33 functional alleles identified in rhesus macaque populations 24, 26. However, Mamu‐E is still the most conserved of all the class I MHC loci in macaques. HLA‐E and Mamu‐E share 88% amino acid identity, especially within the peptide binding region of the protein 27, and there is conservation in function between rhesus macaques, cynomolgus macaques (M. fascicularis) and humans 26, 27. Mamu‐E is also able to bind a wider range of peptides than HLA‐E, although it still preferentially binds the canonical VL9 peptide 5.
HLA‐E function in a healthy cell – the fringe of innate and adaptive immunity
HLA class Ia nascent peptides are cleaved by signal peptidase, and are assembled and translocated in the endoplasmic reticulum (ER) of a cell via the peptide‐loading complex consisting of transporter associated with antigen processing (TAP), tapasin and calreticulin 28. HLA‐E epitopes from these peptides are cleaved by signal peptide peptidase, and bind to nascent HLA‐E within the ER, also via interaction with TAP and tapasin 25. On the cell surface, HLA‐E is stabilized by association with β2‐microglobulin 28, and predominantly interacts with CD94/NKG2 receptors on natural killer (NK) cells 15.
HLA‐E and NK cells
Although classified as part of the innate immune system, NK cells span the traditional boundaries of innate and adaptive immunity through generation of memory‐like phenotypes and adaptation to infection 29, 30. They are defined through expression of the cell‐surface molecule CD56 31, and are vital for protection against viral pathogens, especially herpesviruses 19. NK cells target infected cells directly through the up‐regulation of inflammatory markers on the cell surface, or indirectly through their down‐regulation of MHC class Ia 32. HLA‐E, when stabilized with HLA class Ia‐derived peptides, exerts a regulatory function upon NK cells expressing the dimeric receptors CD94 and NKG2A or NKG2C 22, 33, 34. This interaction is dependent on the amino acid residues at positions 5 and 8 of the HLA‐E‐bound leader peptide 35. NKG2A is a C‐type lectin‐like receptor containing an inhibitory immunoreceptor tyrosine‐based inhibition motif (ITIM) sequence 36, which recruits Src homology 2 domain‐containing protein tyrosine phosphatase (SHP)‐1/2 tyrosine phosphatases 37 and prevents a release of cytotoxic granules containing interferon (IFN)‐γ and tumour necrosis factor (TNF)‐α 38. The HLA‐E : class Ia peptide complex can also bind the activating NK cell receptor CD94/NKG2C 39, albeit with an approximately sixfold lower affinity 36, 40. One exception is the HLA class Ib molecule HLA‐G, the VL9 peptide of which has phenylalanine at position 8, and interacts with a much higher affinity with CD94/NKG2C molecules 35. HLA‐G*01 is expressed during development of the placental trophoblast and has the ability to activate cytolytic CD94/NKG2C NK cells, potentially acting to maintain balance of tissue growth 35. CD94/NKG2 NK cell receptors have homologues in macaques and homoplasious molecules in mice 36, all of which interact with the corresponding orthologue of HLA‐E. Up‐regulation of HLA‐E by MHC class Ia peptides facilitates the passive monitoring of epitope presentation and TAP function within the cell; pathogen‐induced cessation of HLA class Ia synthesis prevents HLA‐E cell‐surface expression, and leads to destruction of the cell through ‘missing self’ activation of NKG2C+ NK cells 41.
HLA‐E and infection
Herpesvirus infection
Herpesviruses exhibit convergently evolved mechanisms that alter MHC presentation of viral antigens to the host adaptive immune system. Cytomegalovirus (CMV) is a highly species tropic β‐herpesvirus, and has evolved and diversified in tandem with its mammalian hosts 42. Human CMV (HCMV) is prevalent in 60–100% of a given population 43. HCMV manifests a lifelong latent infection, with subclinical presentation and immune control in immunocompetent individuals 1, 44. Infection is established within salivary gland epithelial cells 45, and disseminates throughout the host during latency 46.
HCMV has evolved the ability to evade the host adaptive immune system through manipulation of HLA expression. The HCMV US2‐11 protein(s) down‐regulate HLA class Ia molecules on the cell surface 47, preventing presentation of viral epitopes to canonical CD8+ T cells (reviewed in Table 3). US2 and US11 induce translocation of HLA class Ia towards the proteasome 48, 49, US6 binds and changes TAP conformation to prohibit peptide binding to HLA class Ia 50 and US3 prevents peptide stabilization in the binding groove of HLA class Ia through direct binding to tapasin 51. Furthermore, UL18 is a homoplasious protein with similar function to HLA class Ia molecules. UL18 complexes with β2‐microglobulin on the cell surface and binds with high affinity to inhibitory leucocyte immunoglobulin‐like receptor 1 on T cells, down‐regulating their cytotoxic activity 52 (Fig. 1).
Table 3.
| Gene product in HCMV | Gene product in RhCMV | Effect on MHC expression |
|---|---|---|
| Down‐regulation of MHC class Ia on the cell surface | ||
| US2, US11 | Rh182, Rh189 | Retrotranslocation of MHC class Ia from endoplasmic reticulum to cytoplasm, for degradation in the proteasome |
| US6 | Rh185 | Alters TAP conformation and peptide binding to MHC class Ia groove |
| US3 | Rh184 | Interacts with tapasin and prevents peptide binding to MHC class Ia groove |
| Up‐regulation of MHC class Ib/prevention of cytotoxic responses | ||
| UL18 | Not present | HLA class Ia functional homologue that can bind inhibitory LIR1 T‐cell receptor |
| UL40 | Rh67 | Stabilizes and up‐regulates MHC‐E at the cell surface |
HLA = human leucocyte antigen; HCMV = human cytomegalovirus; LIR1 = leucocyte immunoglobulin‐like receptor 1; MHC = major histocompatibility complex; RhCMV = rhesus cytomegalovirus; TAP = transporter associated with antigen processing.
Figure 1.
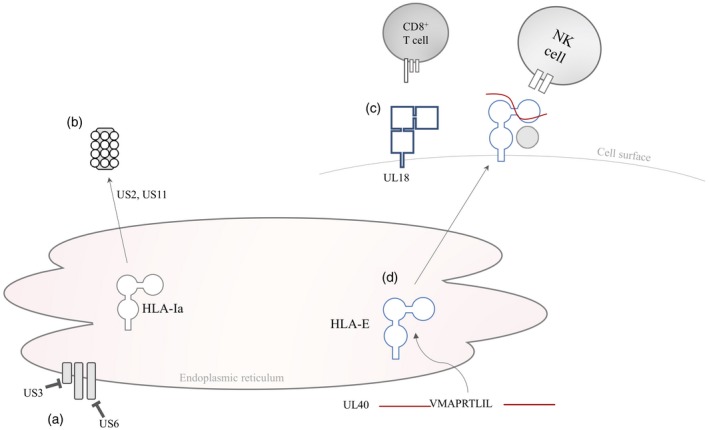
Manipulation of human leucocyte antigen (HLA) molecules by human cytomegalovirus (HCMV). (a) US3 and US6 prevent peptide binding to the HLA class Ia groove via interaction with tapasin and transporter associated with antigen processing (TAP), respectively. (b) US2 and US11 direct nascent HLA class Ia to the proteasome. (c) UL18 acts as a functional homologue of HLA‐E, and binds to the inhibitory leucocyte immunoglobulin‐like receptor 1 (LIR1) on T cells. (d) UL40 contains a VL9 leader peptide, which binds and stabilizes HLA‐E on the cell surface to interact with inhibitory CD94/NKG2A receptors on natural killer (NK) cells 1, 47, 48, 49, 50, 51, 52.
Of most relevance to HLA‐E is UL40, a 221 amino acid glycoprotein containing a 37 amino acid signal sequence with an HLA‐E binding VL9 leader peptide that is identical to the leader peptide of HLA‐C*03 (Tables 1 and 2) 1, 19, 47. Although the function of the UL40 protein is unknown, the UL40 leader peptide binds to nascent HLA‐E in the endoplasmic reticulum in a TAP‐independent manner, working synergistically with the US6 family of HCMV gene products that inhibit TAP function 1, 53. Crucially, the UL40‐VL9 leader peptide is sufficient to up‐regulate HLA‐E expression on the cell surface 54, whereby it prevents NK cell cytolysis of infected cells through interaction with the inhibitory CD94/NKG2A molecule 1. This viral‐mediated up‐regulation of HLA‐E by UL40 can overcome the reduction of the VL9 leader peptide from down‐regulation and proteolysis of HLA class Ia peptides by HCMV. This, in turn, prevents CD94/NKG2A NK cell‐mediated killing of HCMV‐infected cells, despite the lack of HLA class Ia‐derived leader peptide. Loss of UL40 in the CMV genome leads to CD94/NKG2C NK cell‐mediated cytolysis of infected cells 54.
During the course of CMV infection, a subset of NK cells with low CD56 and high NKG2C expression (CD56dimNKG2Cbright) are vastly expanded in approximately 50% individuals, and do not contract during latent infection. This NK subset has the potential to control the virus through increased cytotoxic activity 55. CD56dimNKG2Cbright NK cells may also express CD57, a marker of maturation and terminally differentiated NK cells 56, 57, which potentially induce a form of innate ‘memory’ towards persistent infections such as HCMV.
HIV
Similarly, human immunodeficiency virus (HIV) causes a lifelong and latent infection, which is fatal if left untreated. HIV contains a putative HLA‐E binding leader peptide in the p24 gene product (AA9, Table 2). Although AA9 can stabilize HLA‐E on the cell surface, it is not sufficient to initiate interaction with CD94/NKG2 molecules on NK cells 58. Therefore, CD94/NKG2A+ NK cells are not inhibited by HLA‐E expression, and facilitate cytolytic killing of HIV‐infected cells 59. In HIV infection, there is an expansion of the NKG2Cbright NK compartment, although this has been correlated with HCMV co‐infection and not as a response to HIV 60.
Mycobacterium tuberculosis
Tuberculosis afflicts roughly one‐third of the world’s population, and is caused by persistent latent infection by the bacterium Mycobacterium tuberculosis (Mtb) 61. The current licensed vaccine against Mtb infection is bacillus Calmette–Guérin (BCG), a live‐attenuated vaccine derived from M. bovis 62. NK cells form a substantial part of the innate immune response generated from this vaccine, and produce inflammatory cytokines in response to infection 63. Although understanding of NK cell function during Mycobacteria infection is limited, NK cells in the Mtb granuloma exert cytotoxic pressure through production of granulysin and perforin 64, as well as restriction of bacterial growth through direct contact with infected cells via cytotoxic NKG2D+ NK cells 32. HLA‐E : Mtb peptide complexes are not recognized by CD94/NKG2 molecules, so do not control activation or inhibition of NK cells through these receptors 65.
NK cells and trained immunity
NK cells (and other innate lymphoid cells such as macrophages and monocytes) display ‘trained immunity’ in response to BCG vaccination 66. Trained immunity induces a lasting anti‐pathogen response to secondary, unrelated antigen exposure 62, which in NK cells correlates with increased proinflammatory cytokine production against new pathogens 67, 68. In infants, BCG vaccination correlates with increased weight gain, reduced mortality and reduced infection ability of other Mycobacteria species 66. In immunodeficient SCID mice lacking T and B cells, similar protection is observed after BCG vaccination towards Schistosoma and Candida infection 66, 69. The proinflammatory effect of BCG is also commonly used to treat urothelial cell carcinoma 70. Trained immunity differs from innate memory, as the heightened response is not specific to the original pathogen, although is stronger compared to antigen‐naive innate cells 62 and occurs as a result of histone methylation at the H3K4me1 locus of innate immune cells, inducing a lasting enhanced level of NK cell activation and cytokine production 56, 63, 71.
HLA‐E and T cell restriction
HLA‐E‐mediated presentation of pathogen‐derived peptides to T cells has been observed during infection with CMV, Mtb and Salmonella enterica 72, 73, 74, and recently in CMV‐vectored vaccines against simian immunodeficiency virus (SIV) 5.
Mycobacterium tuberculosis
In humans and mice, CD4+ and CD8+ T cells are vital for control of Mtb infection 75. Unusually, for any known pathogen, there is a large population of CD8+ T cells restricted by HLA‐E induced by infection 76, possibly enhanced by the up‐regulation of HLA‐E on the surface of Mtb‐infected phagosomes 65, 70. Multiple peptides within the Mtb genome can be presented (including peptides from p49 and Mtb44 proteins, Table 2) 61, 72, 74. Their varying amino acid length implies that HLA‐E has higher peptide binding plasticity than solely the VL9 peptide 75. These non‐canonical T cells contribute to the majority of T cells present during active Mtb infection 77, and overshadow T cells restricted by canonical HLA class Ia epitope presentation 78, 79. Mtb antigens presented by HLA‐E to CD8+ T cells can induce either a cytotoxic or regulatory phenotype, consequently inhibiting Mtb pathogenesis and growth in infected macrophages 7, 78. HLA‐E : Mtb‐restricted T cells from active TB infection express a type 2 cytokine profile with increased interleukin (IL)‐4 and IL‐10 production, and assist B cells with antibody and cytokine production to inhibit Mtb growth 65, 76, 77.
Cytomegalovirus
Classically restricted HCMV‐targeting CD8+ T cells are critical in the control of HCMV infection, and constitute up to 10% of the circulating T cell population during active infection 46, 48, 80, typically directed towards epitopes in the pp65 and IE peptides 81. Infection with HCMV also establishes a ‘memory inflation’ population of CD8+ effector memory T cells (TEM), which are defined by their large expansion after infection, terminally differentiated phenotype (CD57+) and their expression of CX3CR1 81, 82, 83. In CMV‐seropositive individuals, a proportion of CD8+ T cells are HLA‐E‐restricted, long‐lasting and express a TEM phenotype 83, although interact with the T cell receptor (TCR) with much lower affinity than HLA class Ia‐bound peptides. These T cells recognize the UL40‐VL9 epitope presented by HLA‐E, and may arise from permanent exposure to HCMV epitopes, which perpetuate a stronger response comparable to canonically restricted CD8+ T cells 83, 84.
Utilizing unusual immune phenotypes for vaccine development
The role of HLA‐E in NK cell activation has been well studied; however, the unforeseen property of HLA‐E to restrict non‐canonical T cells has revealed interesting potential for developments in vaccine design and efficacy.
CMV‐vectored vaccines for SIV
Cytomegalovirus, when used as a vaccine vector in rhesus macaques, induces the restriction of non‐classical T cells by Mamu‐E. This has been demonstrated in a rhesus macaque CMV‐vectored vaccine against SIV, the primate homologue of HIV 85. SIV, like HIV, is a lentivirus that permanently infects the host through integration into the genome. SIV has been used as a close model for HIV, and causes similar pathogenesis including loss of CD4+ T cells, AIDS‐like illness and eventually death in infected macaques 86.
CMV as a virus, and as a vaccine vector, can generate swift TEM cell responses at the site of exposure 87, 88. In contrast, many non‐viral‐vectored vaccines activate a slower‐acting T central memory (TCM) response in the secondary lymphoid organs 89. In these studies, rhesus macaque cytomegalovirus strain 68‐1 (RhCMV 68‐1) was used as a vaccine vector, in accordance with the highly species tropic nature of cytomegaloviruses. The SIV genes gag+rev/nef/ tat+env+pol were expressed in this RhCMV vector 87, 90. In all vaccinated macaques, this vaccine generated robust, long‐lasting CD4+ and CD8+ TEM responses to all SIV peptides, contrasting with predominantly TCM responses from adenovirus‐vectored vaccines containing the same peptides. Fifty‐five per cent of RhCMV : SIV vaccinated macaques generated completely protective immune responses when challenged 59 weeks later with intrarectal infection of highly pathogenic SIVmac239 88. These atypical CD8+ T cell responses were able to recognize broad SIV epitopes, alongside occasional ‘supertopes’ (epitopes recognized by all macaques) within the vaccine, regardless of the MHC haplotype of the macaques 90. Furthermore, classically defined SIV epitopes were not recognized by the T cells of any macaque. Protection was also conferred irrespective of the route of SIV infection, and could be transferred to SIV‐seronegative macaques after adoptive transfer of haematolymphoid cells prior to SIVmac239 challenge 86.
The RhCMV : SIV vaccine restricts non‐canonical CD8+ T cells
In macaques vaccinated with the RhCMV : SIV vaccine, MHC class II‐restricted T cells made up 65% of this non‐canonical response, and the remaining 35% were restricted by Mamu‐E. No canonically restricted CD8+ T cells were produced in response to this vaccine 5, 90. The Mamu‐E‐restricted SIVgag‐specific response was so broad that an average of 20 SIVgag epitopes were recognized per vaccinated macaque, in contrast to ~13 SIVgag epitopes from canonical CD8+ T cell restriction 90. This broad response in the non‐canonical CD8+ T cell compartment has not been previously observed in any preclinical vaccine trial. It may be due to the deletion of Rh157.4 and Rh157.5, which encode an orthologue of the HCMV pentameric glycoprotein receptor complex encoded by UL128 and UL130, which enables non‐fibroblast tropism 5, 90. Similarly to HCMV, normal RhCMV infection only activates canonically restricted T cells 91, and due to the differences between the vaccine and wild‐type RhCMV there is no pre‐existing immunity against this RhCMV‐vectored vaccine in macaques 92.
RhCMV contains an MHC‐E‐binding VL9 sequence
Through this RhCMV‐vectored SIV vaccine, Mamu‐E exhibits the ability to overcome the classical paradigm of MHC class Ia T cell restriction, and in doing so generates an enormous breadth of non‐canonical T cell responses. This, in theory, can offer equal protection to all vaccinated individuals, regardless of MHC genotype, and suggests that Mamu‐E can bind a much broader range of peptides beyond the canonical VL9 leader peptide 5. The VL9 peptide in this vaccine is provided by the RhCMV gene Rh67, which is loaded into Mamu‐E in a TAP‐independent manner. Rh67 initiates Mamu‐E up‐regulation at the cell surface in a convergent fashion to HCMV‐UL40. It is hypothesized that the Rh67‐VL9 peptide can bind and stabilize Mamu‐E deep in the rigid, hydrophobic binding groove, acting as a chaperone and allowing a broader range of SIVgag epitopes to bind higher up in the binding groove and interact with Mamu‐E‐restricted T cells. Although cytomegaloviruses have high species tropism, Rh67 can stabilize and up‐regulate HLA‐E in human cells, suggesting that the function of the ancestral MHC‐E gene has remained conserved 5. This suggests that in the RhCMV : SIV vaccine, Rh67 is facilitating the broad peptide presentation by stabilization of Mamu‐E on the cell surface, allowing presentation of antigen peptides to non‐classical CD8+ T cells. Furthermore, several MHC‐altering genes in RhCMV and HCMV share surprising functional similarity through preventing cell‐surface presentation of MHC class Ia molecules 48 (Table 3). Overall, the convergent evolution of RhCMV‐Rh67 and HCMV‐UL40, and other genes involved in MHC down‐regulation, suggests that the mechanism of HLA‐E stabilization is conserved between cytomegalovirus species, and therefore a HCMV‐vectored vaccine in humans may function in the same way.
CMV‐vectored vaccines for tuberculosis
Recently, Hansen et al. 93 published a preclinical trial testing three RhCMV strain 68‐1‐vectored vaccines expressing six or nine proteins from Mtb. All vaccines induced IFN‐γ‐ and TNF‐producing TEM cells that are vital in protection against Mtb infection, and induced complete or partial protection in > 40% vaccinated macaques. One vaccine, using the original RhCMV68‐1 vector from the SIV vaccine challenge, elicited unconventionally restricted MHC class II‐ and MHC‐E‐restricted T cells. However, two new ‘68‐1.2 RhCMV’‐vectored vaccines only exhibited a canonical T cell response in vaccinated macaques. All three vaccines, however, obtained the same levels of efficacy during challenge trials, suggesting that, contrary to earlier reports 61, 65, Mamu‐E‐restricted T cell responses are not vital for control of Mtb infection 93. Natural Mtb infection induces both cytotoxic and regulatory‐like HLA‐E‐restricted T cells in humans. HLA‐E‐restricted regulatory T cells are likely to be beneficial in containing the Mycobacterium and preventing dissemination 61, 76. However, HLA‐E‐mediated peptide presentation may not be as beneficial to Mtb vaccine function if the induction of regulatory CD8+ T cells reduces the progression of disease, but does not establish a cytolytic environment targeted to Mtb infection. It will also be interesting to see if, in future studies, this Mtb vaccine can generate similar trained immunity in innate and NK cells, as is seen in the current BCG vaccine.
CMV vaccines in human clinical trials
Phase I clinical trials of CMV vaccines in humans were not able to elicit the same broad epitope response as RhCMV‐vectored vaccines. The vaccine, created from chimeric Towne and Toledo fibroblast‐adapted strains, was aimed at inducing immunity against HCMV infection, rather than for use as a viral vector. This vaccine did not contain the pentameric glycoprotein complex, facilitating non‐fibroblast trophism in a similar fashion to the RhCMV vector. However, vaccination did not elicit any non‐canonically restricted T cells in the human participants 94, 95, 96. There is also reasonable concern regarding the use of CMV as a vaccine in CMV‐seropositive individuals, thus creating a ‘superinfection’ serostatus, and possibly negating the immunogenic effect of the vaccine. Although this proved unproblematic in macaques, which have a RhCMV seropositivity of > 90% in captive populations 97, it is still uncertain what effect this will have in human clinical trials, given the ability of HCMV to reduce vaccine efficacy 98.
Future directions for utilizing pathogens to enhance vaccine efficacy
The dual functionality of HLA‐E, through its ability to inhibit NK cells and activate non‐classical CD8+ T cells, has made it an intriguing target of research for both understanding the immunology behind pathogen infection and improving vaccine design. Although it is attractive to think that the results obtained by Hansen et al. 5, 87, 88 in the SIV field could lead to the creation of a non‐conventional T cell stimulating vaccine in humans, there are still fundamental questions that must be answered to assess the potential for and safety of developing a vaccine that can induce HLA‐E restricted T‐cells. It is also important to acknowledge the existence of self‐peptides, including HSP60, that contain potential HLA‐E leader peptides 99. These peptides are able to up‐regulate HLA‐E, although their immunological function is not well understood, and therefore may have an impact on vaccine efficacy.
However, as HLA‐E is emerging as an important aspect of the host response to several pathogens, understanding how it can restrict T cells via non‐conventional mechanisms will continue to be an important avenue of vaccine development, and also improve fundamental understanding of how HLA‐E borders innate and adaptive immunity. Furthermore, the important contribution of NK cells towards vaccine efficacy has still to be fully elucidated.
In conclusion, trained immunity and the restriction of non‐canonical CD8+ T cells by CMV‐vectored vaccines are just two examples of unexpected effects caused by vaccination, which are impacting future vaccine design. If viral‐vectored vaccines can be developed to induce HLA‐E‐restricted T cells in human patients, it may pave the way for the development of vaccines with broad, fast‐acting and best‐placed immunogenicity against many pathogens.
Disclosure
The authors declare no competing interests.
Author contributions
H. R. S. and T. L. conceptualized and wrote the review; G. B. and S. B. contributed to the review structure, contents and proofreading.
Acknowledgements
We would like to thank Ciaran Gilbride, Jyothi Purushotham and Sarah Gilbert for proofreading the manuscript. The authors acknowledge funding support from the Wellcome trust (H. R. S.). The funding sources had no involvement in the writing of this manuscript or decision to submit this manuscript for publication.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
Vaccines for emerging pathogens: from research to the clinic. Clinical and Experimental Immunology 2019, 196: 155–156.
Emerging viruses and current strategies for vaccine intervention. Clinical and Experimental Immunology 2019, 196: 157–166.
Novel multi‐component vaccine approaches for Burkholderia pseudomallei. Clinical and Experimental Immunology 2019, 196: 178–188.
Novel approaches for the design, delivery and administration of vaccine technologies. Clinical and Experimental Immunology 2019, 196: 189–204.
Mucosal vaccines and technology. Clinical and Experimental Immunology 2019, 196: 205–214.
Vaccines for emerging pathogens: prospects for licensure. Clinical and Experimental Immunology 2019, doi: 10.1111/cei.13284
Contributor Information
H. R. Sharpe, Email: hannah.sharpe@msdtc.ox.ac.uk.
T. Lambe, Email: teresa.lambe@ndm.ox.ac.uk.
References
- 1. Tomasec P, Braud VM, Rickards C et al Surface expression of HLA‐E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287:1031. [DOI] [PubMed] [Google Scholar]
- 2. Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol 2005; 5:459–71. [DOI] [PubMed] [Google Scholar]
- 3. Little AM, Parham P. Polymorphism and evolution of HLA class I and II genes and molecules. Rev Immunogenet 1999; 1:105–23. [PubMed] [Google Scholar]
- 4. Braud V, Jones EV, McMichael A. The human major histocompatibility complex class Ib molecule in HLA‐E binds signal sequence‐derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol 1997; 27:1164–9. [DOI] [PubMed] [Google Scholar]
- 5. Hansen SG, Wu HL, Burwitz BJ et al Broadly targeted CD8⁺ T cell responses restricted by major histocompatibility complex E. Science 2016; 351:714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wooldridge L, Ekeruche‐Makinde J, van den Berg HA et al A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 2012; 287:1168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prezzemolo T, van Meijgaarden KE, Franken KLMC et al Detailed characterization of human Mycobacterium tuberculosis specific HLA‐E restricted CD8. Eur J Immunol 2018; 48:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grimsley C, Kawasaki A, Gassner C et al Definitive high resolution typing of HLA‐E allelic polymorphisms: identifying potential errors in existing allele data. Tissue Antigens 2002; 60:206–12. [DOI] [PubMed] [Google Scholar]
- 9. Felício LP, Porto IO, Mendes‐Junior CT et al Worldwide HLA‐E nucleotide and haplotype variability reveals a conserved gene for coding and 3’ untranslated regions. Tissue Antigens 2014; 83:82–93. [DOI] [PubMed] [Google Scholar]
- 10. Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA‐E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem 2003; 278:5082–90. [DOI] [PubMed] [Google Scholar]
- 11. Veiga‐Castelli LC, Castelli EC, Mendes CT et al Non‐classical HLA‐E gene variability in Brazilians: a nearly invariable locus surrounded by the most variable genes in the human genome. Tissue Antigens 2012; 79:15–24. [DOI] [PubMed] [Google Scholar]
- 12. Grimsley C, Ober C. Population genetic studies of HLA‐E: evidence for selection. Hum Immunol 1997; 52:33–40. [DOI] [PubMed] [Google Scholar]
- 13. Sawai H, Kawamoto Y, Takahata N, Satta Y. Evolutionary relationships of major histocompatibility complex class I genes in simian primates. Genetics 2004; 166:1897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geraghty DE, Stockschleader M, Ishitani A, Hansen JA. Polymorphism at the HLA‐E locus predates most HLA‐A and ‐B polymorphism. Hum Immunol 1992; 33:174–84. [DOI] [PubMed] [Google Scholar]
- 15. Braud VM, Allan DS, O’Callaghan CA et al HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998; 391:795–9. [DOI] [PubMed] [Google Scholar]
- 16. Tripathi P, Agrawal S. The role of human leukocyte antigen E and G in HIV infection. AIDS 2007; 21:1395–404. [DOI] [PubMed] [Google Scholar]
- 17. Lauterbach N, Wieten L, Popeijus HE et al Peptide‐induced HLA‐E expression in human PBMCs is dependent on peptide sequence and the HLA‐E genotype. Tissue Antigens 2015; 85:242–51. [DOI] [PubMed] [Google Scholar]
- 18. Celik AA, Kraemer T, Huyton T, Blasczyk R, Bade‐Döding C. The diversity of the HLA‐E‐restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 2016; 68:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ulbrecht M, Martinozzi S, Grzeschik M et al Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA‐E and prevents NK cell‐mediated lysis. J Immunol 2000; 164:5019–22. [DOI] [PubMed] [Google Scholar]
- 20. O'Callaghan CA, Tormo J, Willcox BE et al Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA‐E. Mol Cell 1998; 1:531–41. [DOI] [PubMed] [Google Scholar]
- 21. Joosten SA, Sullivan LC, Ottenhoff TH. Characteristics of HLA‐E restricted T‐cell responses and their role in infectious diseases. J Immunol Res 2016; 2016:2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoare HL, Sullivan LC, Clements CS et al Subtle changes in peptide conformation profoundly affect recognition of the non‐classical MHC class I molecule HLA‐E by the CD94‐NKG2 natural killer cell receptors. J Mol Biol 2008; 377:1297–303. [DOI] [PubMed] [Google Scholar]
- 23. Lampen MH, Hassan C, Sluijter M et al Alternative peptide repertoire of HLA‐E reveals a binding motif that is strikingly similar to HLA‐A2. Mol Immunol 2013; 53:126–31. [DOI] [PubMed] [Google Scholar]
- 24. Dambaeva SV, Bondarenko GI, Grendell RL, Kravitz RH, Durning M, Golos TG. Non‐classical MHC‐E (Mamu‐E) expression in the rhesus monkey placenta. Placenta 2008; 29:58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Hall T, Oliveira CC, Joosten SA, Ottenhoff TH. The other Janus face of Qa‐1 and HLA‐E: diverse peptide repertoires in times of stress. Microbes Infect 2010; 12:910–8. [DOI] [PubMed] [Google Scholar]
- 26. Wu HL, Wiseman RW, Hughes CM et al The role of MHC‐E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J Immunol 2018; 200:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyson JE, McAdam SN, Gallimore A et al The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics 1995; 41:59–68. [DOI] [PubMed] [Google Scholar]
- 28. Braud VM, Allan DS, Wilson D, McMichael AJ. TAP‐ and tapasin‐dependent HLA‐E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol 1998; 8:1–10. [DOI] [PubMed] [Google Scholar]
- 29. Romee R, Foley B, Lenvik T et al NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease‐17 (ADAM17). Blood 2013; 121:3599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 1989; 320:1731–5. [DOI] [PubMed] [Google Scholar]
- 31. Cook KD, Waggoner SN, Whitmire JK. NK cells and their ability to modulate T cells during virus infections. Crit Rev Immunol 2014; 34:359–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esin S, Batoni G. Natural killer cells: a coherent model for their functional role in Mycobacterium tuberculosis infection. J Innate Immun 2015; 7:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA‐E. Proc Natl Acad Sci USA 2008; 105:6696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrie EJ, Clements CS, Lin J et al CD94‐NKG2A recognition of human leukocyte antigen (HLA)‐E bound to an HLA class I leader sequence. J Exp Med 2008; 205:725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Llano M, Lee N, Navarro F et al HLA‐E‐bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA‐G‐derived nonamer. Eur J Immunol 1998; 28:2854–63. [DOI] [PubMed] [Google Scholar]
- 36. Cheent KS, Jamil KM, Cassidy S et al Synergistic inhibition of natural killer cells by the nonsignaling molecule CD94. Proc Natl Acad Sci USA 2013; 110:16981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. García P, Llano M, de Heredia AB et al Human T cell receptor‐mediated recognition of HLA‐E. Eur J Immunol 2002; 32:936–44. [DOI] [PubMed] [Google Scholar]
- 38. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 2013; 31:227–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lauterbach N, Wieten L, Popeijus HE, Voorter CE, Tilanus MG. HLA‐E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum Immunol 2015; 76:578–86. [DOI] [PubMed] [Google Scholar]
- 40. Sullivan LC, Clements CS, Beddoe T et al The heterodimeric assembly of the CD94‐NKG2 receptor family and implications for human leukocyte antigen‐E recognition. Immunity 2007; 27:900–11. [DOI] [PubMed] [Google Scholar]
- 41. López‐Botet M, Bellón T, Llano M, Navarro F, García P, de Miguel M. Paired inhibitory and triggering NK cell receptors for HLA class I molecules. Hum Immunol 2000; 61:7–17. [DOI] [PubMed] [Google Scholar]
- 42. Yan G, Zhang G, Fang X et al Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 2011; 29:1019–23. [DOI] [PubMed] [Google Scholar]
- 43. Powers C, Früh K. Rhesus CMV: an emerging animal model for human CMV. Med Microbiol Immunol 2008; 197:109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilkinson GW, Tomasec P, Stanton RJ et al Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 2008; 41:206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sathiyamoorthy K, Chen J, Longnecker R, Jardetzky TS. The COMPLEXity in herpesvirus entry. Curr Opin Virol 2017; 24:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gillespie GM, Wills MR, Appay V et al Functional heterogeneity and high frequencies of cytomegalovirus‐specific CD8(+) T lymphocytes in healthy seropositive donors. J Virol 2000; 74:8140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mazzarino P, Pietra G, Vacca P et al Identification of effector‐memory CMV‐specific T lymphocytes that kill CMV‐infected target cells in an HLA‐E‐restricted fashion. Eur J Immunol 2005; 35:3240–7. [DOI] [PubMed] [Google Scholar]
- 48. Pande NT, Powers C, Ahn K, Früh K. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol 2005; 79:5786–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 1996; 84:769–79. [DOI] [PubMed] [Google Scholar]
- 50. Hengel H, Koopmann JO, Flohr T et al A viral ER‐resident glycoprotein inactivates the MHC‐encoded peptide transporter. Immunity 1997; 6:623–32. [DOI] [PubMed] [Google Scholar]
- 51. Park B, Kim Y, Shin J et al Human cytomegalovirus inhibits tapasin‐dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 2004; 20:71–85. [DOI] [PubMed] [Google Scholar]
- 52. Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR‐1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 1999; 11:603–13. [DOI] [PubMed] [Google Scholar]
- 53. Heatley SL, Pietra G, Lin J et al Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen‐E (HLA‐E) by natural killer cells. J Biol Chem 2013; 288:8679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang EC, McSharry B, Retiere C et al UL40‐mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci USA 2002; 99:7570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muntasell A, Vilches C, Angulo A, López‐Botet M. Adaptive reconfiguration of the human NK‐cell compartment in response to cytomegalovirus: a different perspective of the host‐pathogen interaction. Eur J Immunol 2013; 43:1133–41. [DOI] [PubMed] [Google Scholar]
- 56. Netea MG. Training innate immunity: the changing concept of immunological memory in innate host defence. Eur J Clin Invest 2013; 43:881–4. [DOI] [PubMed] [Google Scholar]
- 57. Min‐Oo G, Lanier LL. Cytomegalovirus generates long‐lived antigen‐specific NK cells with diminished bystander activation to heterologous infection. J Exp Med 2014; 211:2669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nattermann J, Nischalke HD, Hofmeister V et al HIV‐1 infection leads to increased HLA‐E expression resulting in impaired function of natural killer cells. Antivir Ther 2005; 10:95–107. [DOI] [PubMed] [Google Scholar]
- 59. Davis ZB, Cogswell A, Scott H et al A Conserved HIV‐1‐derived peptide presented by HLA‐E renders infected T‐cells highly susceptible to attack by NKG2A/CD94‐bearing natural killer cells. PLOS Pathog 2016; 12:e1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brunetta E, Fogli M, Varchetta S et al Chronic HIV‐1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co‐infection. AIDS 2010; 24:27–34. [DOI] [PubMed] [Google Scholar]
- 61. Joosten SA, van Meijgaarden KE, van Weeren PC et al Mycobacterium tuberculosis peptides presented by HLA‐E molecules are targets for human CD8 T‐cells with cytotoxic as well as regulatory activity. PLOS Pathog 2010; 6:e1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gardiner CM, Mills KH. The cells that mediate innate immune memory and their functional significance in inflammatory and infectious diseases. Semin Immunol 2016; 28:343–50. [DOI] [PubMed] [Google Scholar]
- 63. Della Chiesa M, Marcenaro E, Sivori S, Carlomagno S, Pesce S, Moretta A. Human NK cell response to pathogens. Semin Immunol 2014; 26:152–60. [DOI] [PubMed] [Google Scholar]
- 64. Garand M, Goodier M, Owolabi O, Donkor S, Kampmann B, Sutherland JS. Functional and phenotypic changes of natural killer cells in whole blood during Mycobacterium tuberculosis infection and disease. Front Immunol 2018; 9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Caccamo N, Pietra G, Sullivan LC et al Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA‐E during active tuberculosis and express type 2 cytokines. Eur J Immunol 2015; 45:1069–81. [DOI] [PubMed] [Google Scholar]
- 66. Blok BA, Arts RJ, van Crevel R, Benn CS, Netea MG. Trained innate immunity as underlying mechanism for the long‐term, nonspecific effects of vaccines. J Leukoc Biol 2015; 98:347–56. [DOI] [PubMed] [Google Scholar]
- 67. Rusek P, Wala M, Druszczyńska M, Fol M. Infectious agents as stimuli of trained innate immunity. Int J Mol Sci 2018; 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kleinnijenhuis J, Quintin J, Preijers F et al BCG‐induced trained immunity in NK cells: role for non‐specific protection to infection. Clin Immunol 2014; 155:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe 2011; 9:355–61. [DOI] [PubMed] [Google Scholar]
- 70. Buffen K, Oosting M, Quintin J et al Autophagy controls BCG‐induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLOS Pathog 2014; 10:e1004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fanucchi S, Fok ET, Dalla E et al Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat Genet 2019; 51:138–50. [DOI] [PubMed] [Google Scholar]
- 72. Heinzel AS, Grotzke JE, Lines RA et al HLA‐E‐dependent presentation of Mtb‐derived antigen to human CD8+ T cells. J Exp Med 2002; 196:1473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Salerno‐Gonçalves R, Fernandez‐Viña M, Lewinsohn DM, Sztein MB. Identification of a human HLA‐E‐restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol 2004; 173:5852–62. [DOI] [PubMed] [Google Scholar]
- 74. Walters LC, Harlos K, Brackenridge S et al Pathogen‐derived HLA‐E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat Commun 2018; 9:3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McMurtrey C, Harriff MJ, Swarbrick GM et al T cell recognition of Mycobacterium tuberculosis peptides presented by HLA‐E derived from infected human cells. PLOS ONE 2017; 12:e0188288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH, Joosten SA. Human CD8+ T‐cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA‐E have an unorthodox Th2‐like, multifunctional, Mtb inhibitory phenotype and represent a novel human T‐cell subset. PLOS Pathog 2015; 11:e1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Harriff MJ, Wolfe LM, Swarbrick G et al HLA‐E presents glycopeptides from the Mycobacterium tuberculosis protein MPT32 to human CD8. Sci Rep 2017; 7:4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bian Y, Shang S, Siddiqui S et al MHC Ib molecule Qa‐1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLOS Pathog 2017; 13:e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McMichael AJ, Picker LJ. Unusual antigen presentation offers new insight into HIV vaccine design. Curr Opin Immunol 2017; 46:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sylwester AW, Mitchell BL, Edgar JB et al Broadly targeted human cytomegalovirus‐specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 2005; 202:673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Klenerman P. The (gradual) rise of memory inflation. Immunol Rev 2018; 283:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gordon CL, Lee LN, Swadling L et al Induction and maintenance of CX3CR82‐intermediate peripheral memory CD8. Cell Rep 2018; 23:768–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jouand N, Bressollette‐Bodin C, Gérard N et al HCMV triggers frequent and persistent UL40‐specific unconventional HLA‐E‐restricted CD8 T‐cell responses with potential autologous and allogeneic peptide recognition. PLOS Pathog 2018; 14:e1007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hoare HL, Sullivan LC, Pietra G et al Structural basis for a major histocompatibility complex class Ib‐restricted T cell response. Nat Immunol 2006; 7:256–64. [DOI] [PubMed] [Google Scholar]
- 85. Sharp PM, Shaw GM, Hahn BH. Simian immunodeficiency virus infection of chimpanzees. J Virol 2005; 79:3891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hansen SG, Piatak M, Ventura AB et al Immune clearance of highly pathogenic SIV infection. Nature 2013; 502:100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hansen SG, Vieville C, Whizin N et al Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 2009; 15:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hansen SG, Ford JC, Lewis MS et al Profound early control of highly pathogenic SIV by an effector memory T‐cell vaccine. Nature 2011; 473:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Small JC, Haut LH, Bian A, Ertl HC. The effect of adenovirus‐specific antibodies on adenoviral vector‐induced, transgene product‐specific T cell responses. J Leukoc Biol 2014; 96:821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hansen SG, Sacha JB, Hughes CM et al Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013; 340:1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hansen SG, Powers CJ, Richards R et al Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 2010; 328:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Früh K, Picker L. CD8+ T cell programming by cytomegalovirus vectors: applications in prophylactic and therapeutic vaccination. Curr Opin Immunol 2017; 47:52–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hansen SG, Zak DE, Xu G et al Prevention of tuberculosis in rhesus macaques by a cytomegalovirus‐based vaccine. Nat Med 2018; 24:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Murray SE, Nesterenko PA, Vanarsdall AL et al Fibroblast‐adapted human CMV vaccines elicit predominantly conventional CD8 T cell responses in humans. J Exp Med 2017; 214:1889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Adler SP, Manganello AM, Lee R et al A Phase 1 study of 4 live, recombinant human cytomegalovirus Towne/Toledo chimera vaccines in cytomegalovirus‐seronegative men. J Infect Dis 2016; 214:1341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Suárez NM, Lau B, Kemble GM et al Genomic analysis of chimeric human cytomegalovirus vaccine candidates derived from strains Towne and Toledo. Virus Genes 2017; 53:650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol 2003; 77:6620–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Falconer O, Newell ML, Jones CE. The effect of human immunodeficiency virus and cytomegalovirus infection on infant responses to vaccines: a review. Front Immunol 2018; 9:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Michaëlsson J, Teixeira de Matos C, Achour A, Lanier LL, Kärre K, Söderström K. A signal peptide derived from hsp60 binds HLA‐E and interferes with CD94/NKG2A recognition. J Exp Med 2002; 196:1403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jensen PE, Sullivan BA, Reed‐Loisel LM, Weber DA. Qa‐1, a nonclassical class I histocompatibility molecule with roles in innate and adaptive immunity. Immunol Res 2004; 29:81–92. [DOI] [PubMed] [Google Scholar]
- 101. Araújo RC, Dias FC, Bertol BC et al Liver HLA‐E expression is associated with severity of liver disease in chronic hepatitis C. J Immunol Res 2018; 2018:2563563. [DOI] [PMC free article] [PubMed] [Google Scholar]


