Abstract
Background & Aims
Fibrolamellar carcinoma (FLC) is a rare liver cancer that primarily affects adolescents and young adults. It is characterized by a heterozygous approximately 400-kb deletion on chromosome 19 that results in a unique fusion between DnaJ heat shock protein family member B1 (DNAJB1) and the alpha catalytic subunit of protein kinase A (PRKACA). The role of microRNAs (miRNAs) in FLC remains unclear. We identified dysregulated miRNAs in FLC and investigated whether dysregulation of 1 key miRNA contributes to FLC pathogenesis.
Methods
We analyzed small RNA sequencing (smRNA-seq) data from The Cancer Genome Atlas to identify dysregulated miRNAs in primary FLC tumors and validated the findings in 3 independent FLC cohorts. smRNA-seq also was performed on a FLC patient-derived xenograft model as well as purified cell populations of the liver to determine whether key miRNA changes were tumor cell–intrinsic. We then used clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (Cas9) technology and transposon-mediated gene transfer in mice to determine if the presence of DNAJB1-PRKACA is sufficient to suppress miR-375 expression. Finally, we established a new FLC cell line and performed colony formation and scratch wound assays to determine the functional consequences of miR-375 overexpression.
Results
We identified miR-375 as the most dysregulated miRNA in primary FLC tumors (27-fold down-regulation; P = .009). miR-375 expression also was decreased significantly in a FLC patient-derived xenograft model compared to 4 different cell populations of the liver. Introduction of DNAJB1-PRKACA by clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 engineering and transposon-mediated somatic gene transfer in mice was sufficient to induce significant loss of miR-375 expression (P < .05). Overexpression of miR-375 in FLC cells inhibited Hippo signaling pathway proteins, including yes-associated protein 1 and connective tissue growth factor, and suppressed cell proliferation and migration (P < .05).
Conclusions
We identified miR-375 as the most down-regulated miRNA in FLC tumors and showed that overexpression of miR-375 mitigated tumor cell growth and invasive potential. These findings open a potentially new molecular therapeutic approach. Further studies are necessary to determine how DNAJB1-PRKACA suppresses miR-375 expression and whether miR-375 has additional important targets in this tumor. Transcript profiling: GEO accession numbers: GSE114974 and GSE125602.
Keywords: Fibrolamellar Carcinoma, Pediatric Cancer, miRNA, Cancer Genomics
Abbreviations used in this paper: AML12, alpha mouse liver 12; cAMP, cyclic adenosine monophosphate; Cas9, CRISPR-associated protein 9; CHOL, cholangiocarcinoma; CRISPR, clustered regularly interspaced short palindromic repeats; CTGF, connective tissue growth factor; DNAJB1, DnaJ heat shock protein family member B1; EdU, 5-ethynyl-2'-deoxyuridine; FLC, fibrolamellar carcinoma; GI, gastrointestinal; HCC, hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; mRNA, messenger RNA; NML, nonmalignant liver; oncomiR, oncogenic microRNA; PCR, polymerase chain reaction; PDX, patient-derived xenograft; PI, phosphatidylinositol; PKA, protein kinase A; prFLC, partially reprogrammed fibrolamellar carcinoma; PRKACA, α catalytic subunit of protein kinase A; smRNA-seq, small RNA sequencing; TCGA, The Cancer Genome Atlas; WT, wild-type; YAP1, yes-associated protein 1
Summary.
Small RNA profiling of primary fibrolamellar carcinoma tumors shows microRNA-375 (miR-375) as the most dysregulated miRNA. Several disease models recapitulate the dramatic suppression of miR-375. Functional interrogation of miR-375 in a novel fibrolamellar carcinoma cell line shows that it suppresses fibrolamellar carcinoma cell growth and migration.
Fibrolamellar carcinoma (FLC) is a rare type of liver cancer that primarily affects teenagers and young adults.1 Unlike typical hepatocellular carcinoma (HCC), FLC generally is not preceded by cirrhosis, fatty liver, or other forms of liver injury.2 Currently, the only effective treatment option for FLC is surgical resection. Unfortunately, many patients have metastatic disease at the time of diagnosis, rendering surgical cures difficult.3 The lack of knowledge of underlying disease mechanisms has hindered our understanding of this cancer and the development of novel therapeutics for FLC patients.
Several studies have identified and validated a single recurrent mutation that occurs in approximately 80%–100% of FLC tumors.4, 5 This mutation, a heterozygous ∼400-kb deletion on chromosome 19, fuses exon 1 of DnaJ heat shock protein family member B1 (DNAJB1) with exons 2–10 of the α catalytic subunit of protein kinase A (PRKACA). This chimeric fusion retains its kinase activity4, 6 and is sufficient to induce liver tumors in wild-type mice.7, 8 Importantly, introduction of DNAJB1-PRKACA by transposon-mediated somatic gene transfer and by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 engineering of the mouse genome induced similar tumors, indicating that formation of DNAJB1-PRKACA, rather than loss of genes in the approximately 400-kb deletion, is central to FLC initiation.8 Although DNAJB1-PRKACA is sufficient to induce tumors in mice, the downstream signaling effectors of this fusion remain unclear.
MicroRNAs (miRNAs) are ∼22 nucleotide noncoding RNA molecules that function as negative regulators of gene expression at the post-transcriptional level. miRNAs typically bind to their targets by sequence complementarity in their seed region (nucleotides 2–8 counting from the 5’ end of the miRNA) to mediate target degradation, messenger RNA (mRNA) destabilization, and/or translational repression.9 They have been implicated as drivers and candidate therapeutic targets in numerous diseases including a wide variety of cancers.10 Recently, 1 group profiled miRNA expression in FLC tumors and identified several dysregulated miRNAs compared with nonmalignant livers (NMLs).11 However, it still remains unclear whether dysregulation of these miRNAs is the result of DNAJB1-PRKACA signaling and if any of these miRNAs have important functions in FLC pathogenesis and disease progression.
Here, we performed small RNA-sequencing (smRNA-seq) in primary FLC tumors from The Cancer Genome Atlas (TCGA) and from an independent cohort to identify dysregulated miRNAs. We identified miR-375 as the most down-regulated miRNA in FLC and validated the massive down-regulation in 3 additional FLC cohorts. MiR-375 also was down-regulated significantly in a patient-derived xenograft (PDX) model of FLC compared with 4 liver cell populations, and was more down-regulated in FLC than in 21 of 22 other tumor types within TCGA. We also showed that introduction of the DNAJB1-PRKACA fusion in cells and in mice was sufficient to suppress miR-375 expression. Finally, we derived a new FLC cell line and showed that treatment with miR-375 mimic inhibits numerous Hippo signaling pathway members, including yes-associated protein 1 (YAP1) and connective tissue growth factor (CTGF), and significantly reduces cell proliferation and migration. Overall, our results show that miR-375 functions as a tumor suppressor in FLC and points toward future therapies based on miR-375 mimics that may provide a viable option for patients.
Results
Identification of Dysregulated miRNAs in FLC
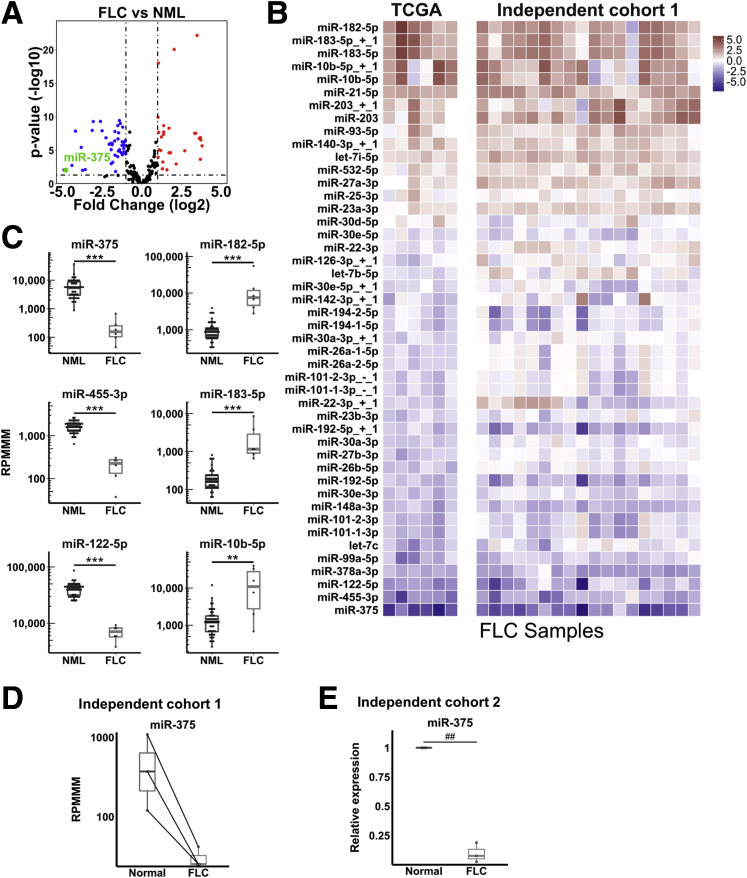
To identify miRNAs dysregulated in FLC, we first analyzed smRNA-seq data from FLC samples (n = 6) in TCGA. We previously confirmed the presence of the DNAJB1-PRKACA fusion transcript as well as classic histologic features of FLC in these samples.12 By using miRquant 2.0, our previously published smRNA-seq analysis pipeline,13 we quantified the expression of canonical mature miRNAs and isomiRs, sequence variants resulting from alternative miRNA processing or postprocessing modifications. We identified 30 significantly up-regulated and 46 significantly down-regulated miRNAs in FLC compared with nonmalignant liver (average expression, >100 reads per million mapped to miRNAs in either FLC or NMLs; ≥2-fold change; P < .05) (Figure 1A). To confirm our findings, we performed smRNA-seq on an independent cohort of FLC samples (n = 18) and observed strong concordance with the TCGA cohort (Figure 1B). Because different smRNA-seq library preparation protocols introduce different biases through adaptor ligation,14, 15 it would not be appropriate to directly compare the 2 cohorts; therefore, we sought to confirm similar trends within each cohort. In both data sets, miR-375, miR-455-3p, and miR-122-5p, all proposed as tumor suppressors,16, 17, 18, 19 were found to be down-regulated significantly. Conversely, miR-182-5p, miR-183-5p, and miR-10b-5p, all previously reported oncogenic miRNAs (oncomiRs),20, 21, 22 were up-regulated significantly in FLC (Figure 1B and C).
Figure 1.
miR-375 is the most dysregulated miRNA in FLC. (A) Volcano plot showing differentially expressed miRNAs from TCGA small RNA-seq data in FLC (n = 6) relative to NML (n = 50). Dashed lines represent fold change of -2 or +2 (vertical) and P = .05 (horizontal). (B) Heatmaps showing miRNA expression in the TCGA cohort (n = 6) and independent validation cohort 1 (n = 18, compared with n = 3 NML). Color intensity shows log2 (fold change) relative to NML within each cohort. miRNAs are included in the heatmap if they had an average expression greater than 1000 reads per million mapped to miRNAs (RPMMM) in FLC or NML and P < .05 in the TCGA cohort. (C) Expression of 3 candidate tumor-suppressor miRNAs (miR-375, miR-455-3p, and miR-122-5p) and 3 candidate oncomiRs (miR-182-5p, miR-183-5p, and miR-10b-5p) from TCGA small RNA-seq data in FLC vs NML. Samples are plotted as individual points. Boxes represent the 25th (bottom), 50th (middle), and 75th (top) percentiles of the data. Whiskers represent data <25th and >75th percentiles. (D) Expression of miR-375 of matched FLC tumor and NML samples (n = 3) from independent cohort 1. (E) Reverse-transcription quantitative PCR validation of loss of miR-375 expression in independent cohort 2 (n = 3) of FLC samples and matched adjacent NML. Data are presented from 3 independent experiments. **P < .01, ***P < .001 (Mann–Whitney U test, 2-sided), ##P < .01 (2-tailed Student paired t test; P > .05; Wilcoxon signed-rank test).
We next focused our attention on miR-375 for 4 reasons. First, miR-375 together with its isomiR miR-375+1 are the 2 most down-regulated miRNAs in FLC in terms of fold change (Figure 1A). Second, miR-375 is down-regulated in numerous cancer types including hepatocellular carcinoma,23, 24 gastric cancer,25, 26 esophageal cancer,27 and head and neck squamous cell carcinomas.28 Third, miR-375 directly targets known oncogenes including JAK2,25 IGF1R,27 and YAP1.16, 26 Fourth, miR-375 has been reported to be suppressed by the cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) signaling axis29 that is thought to be aberrantly activated in FLC.4 To further validate miR-375 suppression in FLC samples, we analyzed the subset of tumors from our independent cohort with matched NMLs (n = 3). As expected, we observed a substantial decrease in miR-375 levels in tumor tissue (Figure 1D). We also measured miR-375 by reverse-transcription quantitative polymerase chain reaction (PCR) in an additional independent set of FLC samples along with matched adjacent nonmalignant tissue (n = 3). Consistent with our previous results, we observed dramatic loss of miR-375 expression in FLC (Figure 1E). As a final validation, we examined recently published smRNA-seq data from FLC tumors11 and confirmed significant loss of miR-375 expression in this data set (∼20-fold down-regulation in FLC compared with NML, FDR < 2.5 × 10-12, DESeq2). Together, these data indicate that miR-375 expression is dramatically suppressed in FLC tumors.
MiR-375 Is Dramatically Suppressed in an FLC Patient-Derived Xenograft Model Compared With Every Major Lineage Stage of the Liver
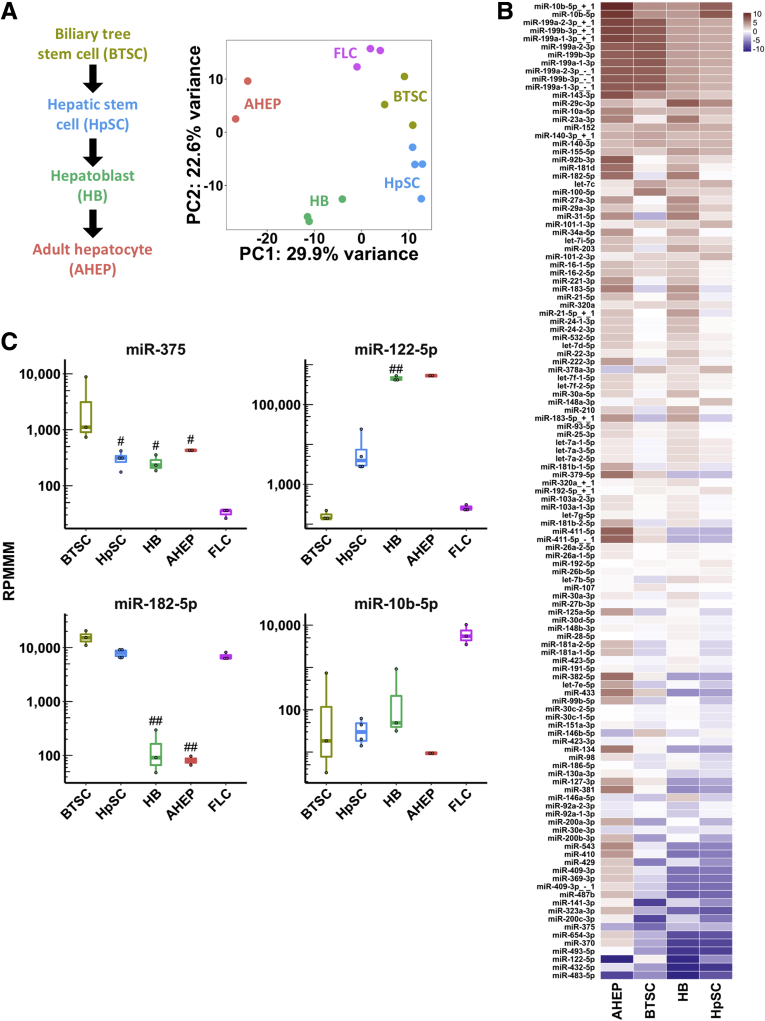
To determine whether miR-375 is specifically suppressed in FLC tumor cells, we performed smRNA-seq on FLC spheroids derived from our previously described PDX model,30 and on 4 different maturational lineage stages of the liver. Because the cell type of origin of FLC remains undetermined, comparison of FLC with multiple different parenchymal cell types within the liver will provide evidence that loss of miR-375 expression occurs in FLC tumor cells as opposed to other cell types (eg, stromal) within primary tumor tissue. The lineage stages we analyzed comprise, from the most primitive to the most mature, biliary tree stem cells, hepatic stem cells, hepatoblasts, and adult hepatocytes, representing a maturational lineage trajectory (Figure 2A). Principal component analysis showed that samples clustered closely by cell type (Figure 2A). Notably, FLC PDX spheroids clustered most closely with biliary tree stem cells (Figure 2A), consistent with our previous analysis of mRNA profiles in the same cell types.30 Importantly, we found that miR-375 is among the most dramatically suppressed miRNAs in FLC PDX tumor cells compared with all 4 maturational lineage stages of the liver (Figure 2B), suggesting that loss of miR-375 does indeed occur in FLC tumor cells. Specifically, miR-375 is more than 13-fold down-regulated in PDX tumor spheroids compared with hepatocytes and more than 105-fold down-regulated compared with biliary tree stem cells (Figure 2C). We also observed up-regulation of miR-182-5p and miR-10b-5p as well as down-regulation of miR-122-5p in PDX spheroids compared with hepatocytes (Figure 2C). Taken together with our findings from primary tumor tissue, our results strongly suggest that miR-375 may function as a tumor suppressor in FLC.
Figure 2.
miR-375 is dramatically suppressed in an FLC patient-derived xenograft model relative to every lineage stage of the liver. (A) Maturational trajectory depicts the lineage stages within the liver (left). Principal component analysis of small RNA-seq data from a FLC PDX model and 4 maturational lineage stages of the liver (right). (B) Heatmap showing miRNA expression in the FLC PDX model. Color intensity shows log2 (fold change) relative to 4 maturational lineage stages of the liver. miRNAs are included in the heatmap if they had an average expression >1000 reads per million mapped to miRNAs (RPMMM) in any cell type and P < .05 in any cell type compared with FLC. (C) Expression of 2 candidate tumor-suppressor miRNAs (miR-375 and miR-122-5p) and 2 candidate oncomiRs (miR-182-5p and miR-10b-5p) from small RNA-seq in 4 maturational lineage stages of the liver and the FLC PDX model. #P < .05, ##P < .01 (2-tailed Student t test; P ≥ .05, Mann–Whitney U test). PC, principal component.
Expression of the DNAJB1-PRKACA Fusion Is Sufficient to Suppress miR-375 Expression
The cAMP/PKA signaling axis has been shown previously to suppress miR-375 expression in pancreatic β cells.29 The ∼400-kb deletion that creates the DNAJB1-PRKACA fusion is the most common genetic lesion in FLC tumors (occurring in ∼80%–100% of patients),4, 5 and the resulting chimeric protein retains PKA activity.4, 6, 31 We hypothesized that DNAJB1-PRKACA is sufficient to suppress miR-375 expression. To test this hypothesis, we used 2 independent approaches.
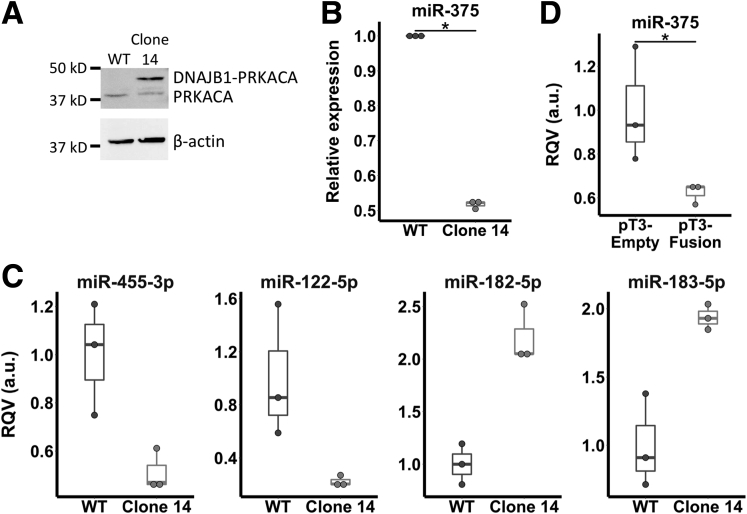
First, we used CRISPR/Cas9 technology to recapitulate the genetic lesion found in human FLC tumors by creating a heterozygous deletion on mouse chromosome 8 in the murine hepatocyte cell line alpha mouse liver 12 (AML12). This region is syntenic to human chromosome 19, allowing us to faithfully re-create the deletion event in mouse cells (Figure 3A). A comparison of cells harboring the deletion (clone 14) with wild-type (WT) AML12 cells showed a significant reduction of miR-375 expression in the mutated cells that express DNAJB1-PRKACA (Figure 3B). We also observed up-regulation of the previously described oncomiRs miR-182-5p and miR-183-5p along with down-regulation of the tumor suppressors miR-122-5p and miR-455-3p in cells expressing DNAJB1-PRKACA compared with WT AML12 cells (Figure 3C).
Figure 3.
Expression of DNAJB1-PRKACA fusion is sufficient to suppress miR-375 and recapitulate the miRNA dysregulation in FLC. (A) Western blot of WT and gene-edited AML12 cells (clone 14) with an antibody against PKA catalytic subunit α. The higher molecular weight band represents the DNAJB1-PRKACA fusion. Data are representative of 2 independent experiments. (B) miR-375 expression in WT (n = 3) and fusion-expressing AML12 cells (clone 14, n = 3). (C) Expression of previously described tumor-suppressor miRNAs (miR-455-3p and miR-122-5p) and oncomiRs (miR-182-5p and miR-183-5p) in AML12 cells with (clone 14, n = 3) and without (n = 3) DNAJB1-PRKACA fusion. Data are presented from 3 independent replicates. (D) miR-375 expression in liver tissue and liver tumors of mice expressing empty (pT3-empty, n = 3) and fusion-containing (pT3-fusion, n = 3) transposon, respectively. Data are presented from 3 independent biological replicates. *P < .05 (Mann–Whitney U test, 1-sided). RQV, relative quantitative value.
We also introduced DNAJB1-PRKACA to C57BL6/N mouse livers by hydrodynamic tail-vein injection of a transposon containing the fusion, as described previously.8 We harvested tumors 4 months after injection from fusion-injected mice and liver tissue from empty vector-injected mice and compared miR-375 expression. Introduction of DNAJB1-PRKACA by transposon resulted in a significant down-regulation of miR-375 compared with control (Figure 3C). Together, these results suggest that DNAJB1-PRKACA suppresses miR-375 expression. However, we note that short-term introduction of DNAJB1-PRKACA in HepG2 cells did not result in a significant loss of miR-375 expression, suggesting that suppression of miR-375 may require prolonged DNAJB1-PRKACA expression and/or a very specific cellular context.
miR-375 Is a Candidate Master Regulator of Cancer Pathways in FLC
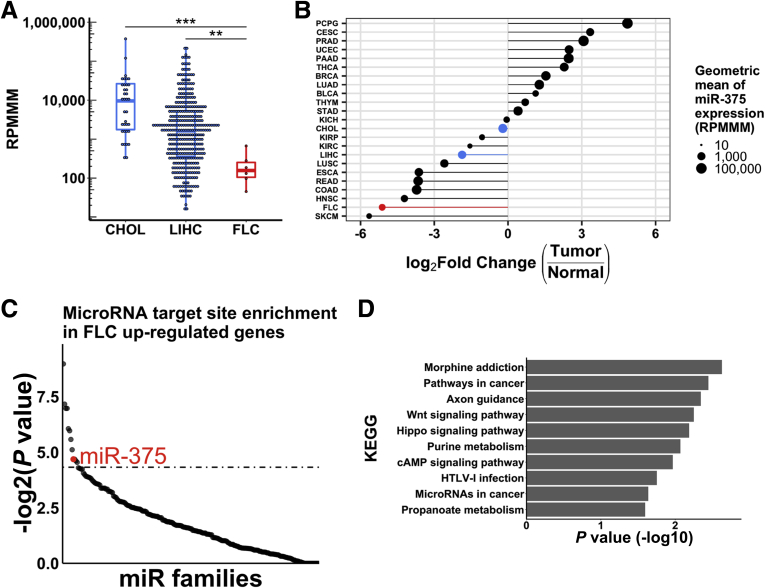
We next compared the levels of miR-375 in FLC with 2 other liver-associated cancers, HCC (denoted as liver hepatocellular carcinoma within TCGA) and cholangiocarcinoma (CHOL), and found that miR-375 expression was decreased significantly in FLC compared with these other liver cancers (Figure 4A). To compare miR-375 expression in FLC with other nonliver cancers, we analyzed smRNA-seq data from 23 tumor types within TCGA. Strikingly, miR-375 expression levels were most reduced in skin cutaneous melanoma and FLC when compared with all other tumor types (Figure 4B). Notably, miR-375 has tumor-suppressive roles in melanoma.32
Figure 4.
miR-375 is a candidate master regulator of cancer pathways in FLC. (A) Expression of miR-375 in CHOL (n = 36), LIHC (n = 366), and FLC (n = 6) in TCGA smRNA-seq data. (B) Log2 (fold change) of miR-375 expression in different tumor types (n = 23) within TCGA. The size of each circle represents the geometric mean of miR-375 expression in each tumor type. Each tumor type is ranked on the y-axis by the log2 (fold change) of the geometric mean of tumor expression over non–tumor expression of miR-375. Geometric means were used instead of arithmetic means to provide robustness to outliers. Highlighted are FLC (red) as well as CHOL and LIHC (blue). (C) Ranked –log2 (P value) of miRhub Monte Carlo simulation. miRNAs were examined for target site enrichment in genes up-regulated in FLC. Dotted line represents P = .05. (D) KEGG enrichment analysis of up-regulated miR-375 target genes in FLC. **P < .01, ***P < .001 (Mann–Whitney U test, 2-sided). BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal papillary cell carcinoma; KIRP, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma.
To determine if miR-375 may function as a potential master regulator of gene expression in FLC, we implemented our previously described Monte Carlo simulation approach called miRhub.33 Because loss of miR-375 is expected to lead to the de-repression of its targets, we examined the set of genes up-regulated in FLC compared with HCC. We chose to compare FLC with HCC as a conservative approach because miR-375 is more highly expressed in HCC than FLC, but less than NML. However, it is important to note that comparison of FLC with both HCC and NML yielded similar results. We found that miR-375 is predicted to target genes up-regulated in FLC significantly more than expected by chance (Figure 4C), suggesting that the loss of miR-375 is a major driver of FLC gene expression. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the set of up-regulated miR-375 target genes showed enrichment in numerous oncogenic pathways including the Wnt, Hippo, and cAMP signaling pathways (Figure 4D). The Wnt signaling pathway has been shown to promote DNAJB1-PRKACA–induced tumor progression,8 cAMP is an established regulator of the canonical PKA signaling axis,34, 35 and Hippo signaling is overactive in FLC.36
miR-375 Regulates Hippo Signaling
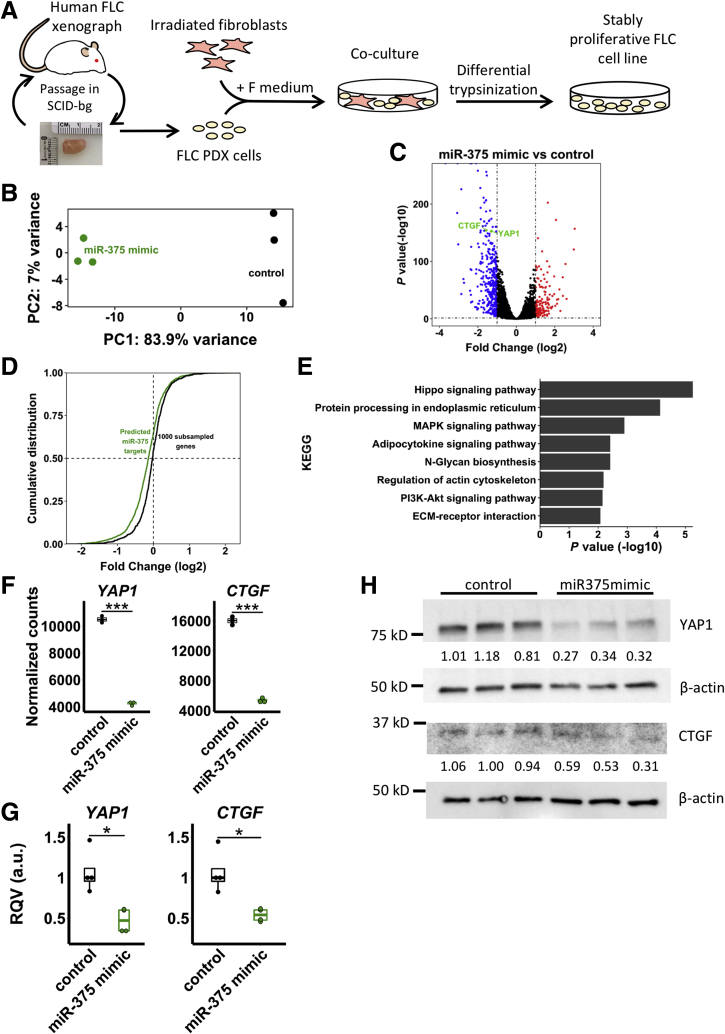
To further investigate the role of miR-375 in FLC, we adapted a patient-derived xenograft model of FLC for use as a stable cell line (Figure 5A), as previously described,37 and performed RNA sequencing after miR-375 overexpression. miR-375 mimic-treated samples cluster separately from samples treated with a scramble control (Figure 5B). We identified 353 significantly down-regulated and 178 significantly up-regulated genes (Figure 5C) (average normalized count, ≥100 across all samples; adjusted P < .05; fold change, >2) after miR-375 overexpression. As expected, down-regulated genes were enriched in predicted miR-375 targets (P = 1.217 × 10-12, Fisher exact test). We also observed greater down-regulation in predicted miR-375 targets compared with a randomly sampled set of genes (Figure 5D) (P < 2.2 ×10-16, Kolmogorov–Smirnov test). To identify important candidate miR-375 targets, we intersected genes that were down-regulated on miR-375 overexpression with genes up-regulated in primary FLC samples, where miR-375 expression is lost. KEGG pathway analysis of these genes (n = 296; average normalized count, ≥100; adjusted P < .05; log2 fold change, >0.5) showed enrichment in several pathways including extracellular matrix-receptor interaction (P = 2.134 × 10-4), phosphatidylinositol (PI)3K-Akt signaling (P = .01310), Hippo signaling (P = .007852), and Notch signaling (P = .005485). Examination of the genes that are down-regulated after miR-375 overexpression and also are predicted miR-375 targets (n = 428) showed the greatest enrichment in the Hippo signaling pathway (Figure 5E), providing support that miR-375 regulates Hippo signaling in FLC.
Figure 5.
miR-375 regulates the Hippo signaling pathway. (A) Diagram of FLC cell line derivation. (B) Principal components analysis of RNA-seq data from miR-375 mimic and control-treated FLC cells. The 500 most variable genes were used in the analysis. (C) Volcano plot showing differentially expressed genes in miR-375 mimic relative to control-treated FLC cells. Dashed lines represent fold change of -2 or +2 (vertical) and adjusted P value = .05 (horizontal). (D) Cumulative density distribution of expression fold change (log2, miR-375 mimic compared with control) for miR-375 targets and 1000 randomly selected genes. (E) KEGG enrichment analysis of miR-375 targets that are down-regulated (average normalized count, ≥100 across all samples; adjusted P value < .05; log2 fold change, <-0.5; n = 428). (F and G) RNA expression of CTGF and YAP1 after treatment of miR-375 mimic or control by RNA-seq (n = 3 per condition in panel F) and reverse-transcription quantitative PCR (RT-qPCR) (n = 4 per condition in panel G). Gene expression was normalized to that of GAPDH. RT-qPCR data are representative of 3 independent experiments. Statistical significance was determined by the Wald test (DESeq2) and the Mann–Whitney U test (1-sided) for RNA-seq and RT-qPCR data, respectively. (H) Protein expression of CTGF and YAP1 after treatment of miR-375 mimic or control. Western blot densitometry is normalized to β-actin. Data are representative of 2 independent experiments. *P < .05, ***P < .001. ECM, extracellular matrix; PC, principal component; SCID, severe combined immunodeficiency.
miR-375 has been shown to directly target the Hippo signaling members YAP1 and CTGF in other cellular contexts.16, 24, 26, 38, 39, 40 Indeed, we observed a significant reduction in both CTGF and YAP1 mRNA expression by both RNA-seq (Figure 5F) and reverse-transcription quantitative PCR (Figure 5G). Concomitant with the decrease in mRNA levels, we also observed a reduction in protein levels of YAP1 and CTGF after miR-375 overexpression (Figure 5H). To confirm these findings, we analyzed previously published argonaute high-throughput sequencing and cross-linking immunoprecipitation data in the human hepatoma cell line Huh741 and observed enrichment of signal at the miR-375 target sites in YAP1 and CTGF.
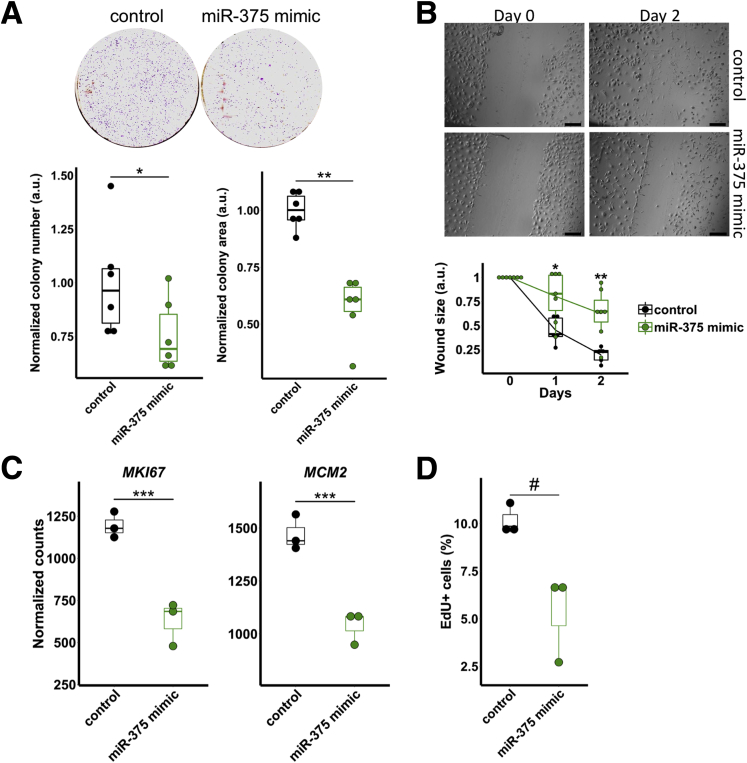
Importantly, miR-375 overexpression caused a significant decrease in colony formation (Figure 6A) as well as in cell migration (Figure 6B), which are indicative of reduced survival and invasive potential, respectively. The decrease in colony formation was consistent with reduced proliferation after miR-375 overexpression, as measured by classic markers of proliferation, KI67 and MCM2 (Figure 6C), as well as 5-ethynyl-2’-deoxyuridine (EdU) incorporation (Figure 6D). Collectively, our results show that miR-375 regulates members of the Hippo signaling pathway and functions as a tumor suppressor in FLC.
Figure 6.
miR-375 inhibits growth and migration of FLC. (A) Colony formation assays after treatment of control (n = 6) or miR-375 mimic (n = 6). Colony number (bottom left) and colony area (bottom right) were quantified. Data are presented from 2 independent experiments. (B) Scratch wound assays after treatment of control (n = 5) or miR-375 mimic (n = 7). Scale bar: 100 μm. Data are presented from 2 independent experiments. (C) RNA expression quantified by RNA-seq of the proliferation markers MKI67 and MCM2 after treatment of miR-375 mimic. Statistical significance was determined by the Wald test (DESeq2). (D) EdU incorporation assays after treatment of control (n = 3) or miR-375 mimic (n = 3). Data are representative of 2 independent experiments. *P < .05, **P < .01, ***P < .001 (Mann–Whitney U test, 1-sided, unless otherwise indicated), #P < .05 (Student 1-tailed t test, P ≥ .05 Mann–Whitney U test, 1-sided).
Discussion
MiR-375 has been characterized previously as a tumor suppressor in numerous gastrointestinal (GI) tissues including liver, stomach, esophagus, and colon.23, 24, 27, 42 In addition, we have shown that miR-375 has antiproliferative effects in intestinal stem cells,43 suggesting that miR-375 might have broad tumor-suppressive functions in the GI tract. Using smRNA-seq, we identified miR-375 as the most down-regulated miRNA in FLC tumors. By comparing miR-375 expression in FLC with 22 additional tumor types in TCGA, including HCC and CHOL, we found that FLC tumors showed the second greatest loss of miR-375 expression, behind only melanoma. MiR-375 has been shown to have antiproliferative and anti-invasive properties in melanoma,32 indicating that miR-375 also functions as a tumor suppressor in non-GI contexts.
In addition to miR-375, we showed down-regulation of additional tumor suppressors (miR-455-3p and miR-122-5p) and up-regulation of oncomiRs (miR-182-5p, miR-183-5p, and miR-10b-5p) in FLC relative to NML. However, it remains to be seen if these miRNAs function as bona fide regulators of FLC.
We also report the characterization of a miRNA profile of maturational lineage stages of the liver. Small RNA profiling in 4 cell types (biliary tree stem cells, hepatic stem cells, hepatoblasts, and adult hepatocytes) and an FLC PDX model facilitated the comparison of miRNA expression across purified cell populations rather than the mixtures of cell types present in primary tumors. We found that FLC cells grouped most closely with biliary tree stem cells, matching our previous results from the analysis of gene expression profiles.30 Importantly, miR-375 expression was decreased significantly in FLC cells compared with all other normal parenchymal cell types in the liver, suggesting that miR-375 expression is indeed lost in FLC tumor cells. Because we do not have data in nonparenchymal tumor cells (eg, stromal, immune), it remains to be seen whether miR-375 also has functional roles in these cell types. Although we have analyzed these data in the context of FLC, this data set provides a rich resource for further understanding liver development and other liver-related disorders.
To gain a more mechanistic understanding of DNAJB1-PRKACA function, it is necessary to identify the genes and pathways downstream of this fusion. MiR-375 has been reported to be transcriptionally repressed by the cAMP/PKA signaling axis.29 To determine whether miR-375 is downstream of DNAJB1-PRKACA, we used 2 independent approaches. First, we used CRISPR/Cas9 technology to engineer the heterozygous deletion creating Dnajb1-Prkaca into the murine cell line AML12. Simultaneously, we introduced DNAJB1-PRKACA to the livers of WT C57BL6/N mice using transposon-mediated somatic gene transfer. Both methods showed significant suppression of miR-375 upon introduction of DNAJB1-PRKACA, suggesting that expression of DNAJB1-PRKACA, rather than loss of genes in the approximately 400-kb deletion, is responsible for the loss of miR-375 expression. In addition, loss of miR-375 expression is not dependent on the process of tumor formation, which occurred in mice injected with DNAJB1-PRKACA expressing transposons, because suppression of miR-375 expression also was observed in cultured AML12 cells engineered with the deletion leading to DNAJB1-PRKACA fusion. We did note, however, that introduction of DNAJB1-PRKACA into HepG2 cells, a human hepatoma cell line, did not result in suppression of miR-375, suggesting that cellular context might be important for miR-375 regulation. It remains to be seen how DNAJB1-PRKACA causes loss of miR-375 expression.
Computational prediction of miR-375 targets in genes up-regulated in FLC showed enrichment in genes involved in a number of biological processes. Notably, predicted miR-375 targets were enriched in Wnt, cAMP, and Hippo signaling. Wnt signaling has been previously connected to a genetically engineered mouse model of FLC. Expression of DNAJB1-PRKACA together with an activated form of β-catenin (CTNNB1T41A) significantly accelerated tumor formation and progression compared with DNAJB1-PRKACA alone.8 Wnt signaling also has been implicated in stem cell renewal in the liver and biliary tree, in agreement with the finding that FLC is rich in cancer stem cells.30 Frizzled 8, a member of the canonical Wnt signaling pathway and validated target of miR-375,44 is overexpressed in FLC. Loss of miR-375 in colorectal cancer promotes metastasis through Frizzled 8 and Wnt pathway activation,42 raising the possibility that Wnt signaling may play an important role in FLC pathogenesis and metastasis.
Our finding that miR-375 targets are enriched in the cAMP signaling pathway is notable because DNAJB1-PRKACA is a cAMP-responsive kinase.31 Loss of miR-375 would enhance cAMP signaling and promote PKA activity. Interestingly, increased cAMP/PKA activity has been reported to repress miR-37529 in other cellular contexts, and if this is occurring in FLC, it would be expected to increase cAMP/PKA activity even further.
miR-375 has been shown to repress Hippo signaling by directly targeting YAP1 and CTGF as confirmed by 3’ untranslated region luciferase reporter assays.16, 24, 26, 38, 39, 40 Furthermore, dysregulation of Hippo signaling has been observed previously in FLC tumors.36 In agreement with this, introduction of a miR-375 mimic in a newly developed FLC cell line led to the suppression of a number of Hippo pathway members, most notably YAP1 and CTGF, for which mRNA and protein levels were reduced. Importantly, introduction of a miR-375 mimic significantly reduced colony formation, EdU incorporation, and migration, indicative of reduced survival, proliferation, and metastatic potential, respectively. Previous studies have reported interactions between YAP1 and CREB,45, 46 a key mediator of cAMP/PKA signaling, including in liver cancer. CREB transcriptionally activates YAP1, whereas YAP1 promotes protein stability of CREB. These findings suggest that multiple pathways regulated by miR-375 also may interact in a miR-375–independent manner to promote tumor growth.
We also identified genes that were down-regulated upon miR-375 overexpression and up-regulated in primary FLC samples, where miR-375 expression is drastically reduced. These genes were enriched in multiple pathways including extracellular matrix-receptor interaction, PI3K-Akt signaling, and Hippo signaling. FLC tumors have characteristic lamellar fibrosis from which the cancer derives its name. Despite this obvious feature, its roles in FLC biology remain unclear. Collagen and cell adhesion genes that we identified as up-regulated in primary FLC tumors include COL1A1, COL4A1, COL4A2, COL4A5, ITGA6, and ITGB4. One previous study found the fibrotic bands in FLC contained collagen I and IV, as well as additional matrix molecules.47 Loss of miR-375 may promote the up-regulation of multiple genes encoding for extracellular matrix molecules, thereby contributing to the fibrotic phenotype observed in FLC.
Dysregulated PI3K-Akt signaling has been linked to many cancers. Interestingly, miR-375 suppresses PI3K-Akt signaling in osteosarcoma48 and colorectal cancer,49 suggesting that loss of miR-375 may activate PI3K-Akt signaling in FLC. Overall, miR-375 represents a regulatory hub in FLC with loss of miR-375 expression activating multiple signaling cascades that promote FLC pathogenesis.
In addition to miR-375, other miRNAs may play a role in activating the Hippo signaling pathway. Notably, miR-455-3p is among the most down-regulated miRNA in FLC (Figure 1B) and is known to target WW Domain Containing Transcription Regulator 1 (WWTR1), a YAP1 paralog and validated oncogene.50 Loss of miR-375 and miR-455-3p expression in FLC may function synergistically to overactivate the Hippo signaling pathway, resulting in increased proliferation and stemness. Interestingly, activated Hippo signaling has been implicated in globally impaired miRNA processing by sequestering DEAD-box helicase 17, a component of the microprocessor complex important in miRNA maturation.51 This impairment ultimately could result in further decreasing miR-375/miR-455-3p levels and activating Hippo signaling.
Our results provide mechanistic insight into the development of FLC outside of DNAJB1-PRKACA fusion. Our model proposes that DNAJB1-PRKACA suppresses miR-375, resulting in enhanced Hippo signaling, likely in part through increased expression of YAP1 and CTGF, ultimately promoting pro-oncogenic phenotypes including proliferation, survival, and metastasis. With RNA-based therapies showing increasing promise,10 miR-375–based therapies merit future consideration for FLC therapeutics.
Materials and Methods
Small RNA Sequencing
Total RNA was isolated using the Total RNA Purification Kit (Norgen Biotek, Thorold, Ontario, Canada) per the manufacturer’s instructions. RNA purity was quantified with the Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA), and RNA integrity was quantified with the Agilent 2100 Bioanalyzer or 4200 Tapestation (Agilent Technologies, Santa Clara, CA). Libraries were prepared using the CleanTag Small RNA Library Prep kit (TriLink Biotechnologies, San Diego, CA). Sequencing was performed on the HiSeq2000 platform (Illumina, San Diego, CA) at the Genome Sequencing Facility of the Greehey Children’s Cancer Research Institute (University of Texas Health Science Center, San Antonio, TX).
RNA Sequencing
Total RNA was isolated using the Total RNA Purification Kit (Norgen Biotek) per the manufacturer’s instructions. RNA purity was quantified with the Nanodrop 2000 (Thermo Fisher Scientific), and RNA integrity was quantified with the Agilent 4200 Tapestation (Agilent Technologies). Libraries were prepared using the NEBNext Ultra II Directional Library Prep Kit (New England Biolabs, Ipswich, MA). Sequencing was performed on the NextSeq500 platform (Illumina) at the Biotechnology Research Center (Cornell University, Ithaca, NY).
Bioinformatic Analyses
Read quality was assessed using FastQC. For smRNA-seq, reads were trimmed, mapped, and quantified to the hg19 genome using miRquant 2.0, our previously described smRNA-seq analysis pipeline.13 Briefly, reads were trimmed using Cutadapt and reads were mapped to the genome using Bowtie.52 Perfectly aligned reads represented miRNA loci and imperfectly mapped reads (from isomiRs) were re-aligned to these loci using SHRiMP.53 Aligned reads were quantified and normalized using reads per million mapped to miRNAs. TCGA smRNA-seq data for cholangiocarcinoma (TCGA CHOL), FLC, and HCC (TCGA liver hepatocellular carcinoma) were downloaded from the Cancer Genomics Hub and mapped using the miRquant 2.0 pipeline. TCGA smRNA-seq data for 23 tumor types were downloaded using TCGA-assembler 2.54 Tumor types without normal samples were discarded and the geometric mean of miR-375 expression for tumor and normal samples across the remaining tumor types was calculated. The geometric mean was used instead of the arithmetic mean to provide increased robustness toward outliers. TCGA mRNA-seq was downloaded from the Cancer Genomics Hub. RNA-seq data were mapped to the hg38 genome with STAR.55 Transcripts were quantified with Salmon56 using GENCODE release 25 transcript annotations. Normalization and differential analysis was performed using DESeq2.57 miRhub analysis was performed using our previously described Monte Carlo simulation tool.33 KEGG pathway analysis was performed using Enrichr.58 MiR-375 targets were obtained from TargetScan release 7.2.
Primary Samples
Informed consent was obtained from all human subjects. Samples were collected according to Institutional Review Board protocols 1802007780 (Cornell University) and 31281 (University of Washington).
Animals
Mice were treated as previously described.8 Briefly, female 6- to 10-week-old C57BL6/N mice were subjected to hydrodynamic tail-vein injection with sterile 0.9% NaCl solution containing 20 μg transposon and CMV-SB13 transposase (1:5 molar ratio). 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC, 0.1%) diet was administered after tail-vein injection. Animal experiments were approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (protocol 11-06-011).
Cell Line Generation
The FLC xenograft model established by the Reid Lab (University of North Carolina, Chapel Hill, NC) from patient-derived ascite tumor cells30 was maintained at Duke in severe combined immunodeficiency (SCID) bg immunocompromised mice. For passage, tumors were minced in Kubota’s medium (PhoenixSongs Biologicals, Branford, CT) supplemented with 1% hyaluronic acid (Sigma, St. Louis, MO) and 50 ng/mL each of hepatocyte growth factor and vascular endothelial growth factor (Peprotech, Rocky Hill, NJ) and serially transplanted approximately every 3 months as previously described.30 The partially reprogrammed FLC line (prFLC) was generated in the Duke induced Pluripotent Stem Cell (iPSC) Shared Resource Facility and maintained at 37°C and 5% CO2 in complete F medium according to a published protocol.37 The presence of transcripts for DNAJB1, PRKACA, and DNAJB1-PRKACA fusion in both the xenograft tumor and the cell line were validated by PCR using the following primer set to simultaneously detect wild-type DNAJB1 (forward: GAGGAGATCAAGCGGGCCTAC, reverse: AGCCAGAGAATGGGTCATCAATG), wild-type PRKACA (forward: ACGCCGCCGCCGCCAAGAA, reverse: GAGGAACGGAAAGTTGACAGCT), and their exon 1–exon 2 fusion (DNAJB1 forward-PRKACA reverse), and product bands confirmed by Sanger sequencing.
HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA). DNAJB1-PRKACA was cloned by PCR from a previously described FLC model30 using forward and reverse primers to DNAJB1 (5’-GAGGATCCATGGGTAAAGACTACTACCAGACGTTGGGCC) and PRKACA (5’-GAGCGGCCGCCTAAAACTCAGAAAACTCCTTGCCACACTTCTCATTG), respectively. Enhanced green fluorescent protein (EGFP) was PCR-cloned using forward and reverse primers (5’-AAGGATCCATGGTGAGCAAGG and 5’-CCCGAATTCTTACTTGTACAGCTCG). PCR products were cloned into the pCR-Blunt II-TOPO vector (Thermo Fisher Scientific) and subcloned into the pLV-EF1a-IRES-Puro vector (Addgene plasmid 85132; a gift from Tobias Meyer). HEK293/T17 cells (American Type Culture Collection) were transfected with DNAJB1-PRKACA and EGFP independently along with psPAX2 (Addgene plasmid 12260; a gift from Didier Trono) and pMD2.G (Addgene plasmid 12259; a gift from Didier Trono) to produce lentiviral particles, which were concentrated using Lentiviral-X Concentration (Takara Bio USA, Mountain View, CA) per the manufacturer’s protocol. HepG2 cells were transduced with varying concentrations of lentivirus for 24 hours and selected with 2 μg/mL puromycin (Thermo Fisher Scientific) for 4 days. Selectable cells treated with the lowest concentration of virus were used for further passaging and experiments to obtain a majority of cells with 1 viral integration.
WT AML12 cells were a gift from the Fausto laboratory.59 Guide RNA sequences were chosen in the first intron of the mouse Dnajb1 gene and the first intron of the mouse Prkaca gene. The sequences of the guide RNAs used were as follows: 5’-GCATTCCGGGGATCTAGCGG (Dnajb1) and 5’- TAGTGCTGAGGAGAGTGGGG (Prkaca). These guide RNAs are expected to induce double-strand breaks at each site; when they occur on the same allele, nonhomologous end-joining will fuse the genomic regions, deleting the intervening approximately 400 kb, and generating a chimeric fusion between Dnajb1 and Prkaca. AML12 cells were transfected with a modified PX458 plasmid encoding Streptococcus pyogenes CRISPR associated protein 9 (SpCas9) and the guide RNAs driven from individual U6 promoters. Transfected cells were plated as individual cells and grown to individual colonies. These clones then were screened by immunoblot and PCR analysis.
Quantitative PCR
Total RNA was isolated from AML12 cells, pT3 transposon-injected samples, and Huh7 cells using the Total RNA Purification Kit (Norgen Biotek) per the manufacturer’s instructions. RNA from AML12 cells and liver tissue from FLC and paired NML was extracted using TRIzol (Life Technologies, Carlsbad, CA) and the rNeasy Mini Kit (Qiagen, Germantown, MD) as described by the manufacturer. Reverse-transcription was performed using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific) or the Iscript Complementary DNA synthesis kit (Bio-Rad, Hercules, CA). Gene expression was quantified with TaqMan Gene Expression Assays (Thermo Fisher Scientific) on a CFX96 Touch Real-Time System (Bio-Rad) or the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). miRNA expression levels were normalized to U6. The TaqMan assays used were miR-375 (assay ID 000564), miR-122 (assay ID 002245), miR-455 (assay ID 002455), miR-182 (assay ID 002599), miR-183 (assay ID 002269), and miR-10b (assay ID 002218).
Total RNA was extracted from prFLC cells using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to the manufacturer’s instructions. Complementary DNA was synthesized using the Superscript First Strand Synthesis System (Invitrogen, Carlsbad, CA) and amplified with SYBR Green PCR Master Mix (Applied Biosystems) using gene-specific primers (glyceraldehyde-3-phosphate dehydrogenase: 5'-TGGGTGTGAACCATGAGAAG, 5'-GCTAAGCAGTTGGTGGTGC; YAP1: 5’-CCGACTCCTTCTTCAAGCC, 5’-CGAACATGCTGTGGAGTCA; and CTGF: 5’-GAAATGCTGCGAGGAGTG, 5’-GCTCTAATCATAGTTGGGTCTG). All targets were amplified on a Step One Plus Real Time PCR System (Applied Biosystems), and expression was normalized to the housekeeping gene GAPDH.
Western Blot
AML12 cells were lysed at 4°C in RIPA buffer (Sigma) supplemented with Complete Protease Inhibitor Cocktail (Sigma), Pierce Phosphatase Inhibitor (Thermo Fisher Scientific), 1 mmol/L phenylmethanesulfonyl fluoride, 0.1% β-mercaptoethanol, and 1 mmol/L dithiothreitol. Lysates were flash-frozen, thawed, and centrifuged for 10 minutes at 14,000 × g at 4°C. Protein concentration was quantified using the Pierce Microplate BCA Protein Assay Kit–Reducing Agent Compatible (Thermo Fisher Scientific). Samples were diluted 3:1 in NuPAGE LDS Sample Buffer (Thermo Fisher Scientific), heated at 70°C for 10 minutes, loaded into NuPAGE 10% Bis-Tris Protein Gels (Thermo Fisher Scientific), and run in 1× MOPS buffer (Thermo Fisher Scientific). Transfer to polyvinylidene difluoride membranes was performed with the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked with 5% bovine serum albumin for 1 hour at room temperature, probed with rabbit anti-PRKACA (1:200, sc-903; Santa Cruz Biotechnology, Dallas, TX) and mouse anti–β-actin (1:1000, 3700; Cell Signaling Technology, Danvers, MA) overnight at 4°C, and then incubated with donkey anti-rabbit 680RD (1:10,000; LI-COR Biosciences, Lincoln, NE) and donkey anti-mouse 800CW (1:10,000; LI-COR Biosciences) for 1 hour at room temperature. Membranes were visualized using the ChemiDoc MP (Bio-Rad).
prFLC cell lysates were prepared using RIPA buffer (Sigma) supplemented with 1 mmol/L sodium orthovanadate and protease inhibitor cocktail (Sigma) and spun at 16,000 × g. Supernatants were quantified, resuspended in sodium dodecyl sulfate sample buffer, and stored at -80°C. Proteins were separated on 4%–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose. Membranes were blocked with 5% bovine serum albumin and exposed overnight to the following antibodies: YAP1 (1:1000, 14074; Cell Signaling Technology), CTGF (1:1000, 86641; Cell Signaling Technology), and developed with anti-rabbit horseradish peroxidase secondary antibody (GE Healthcare, Chicago, IL) using SuperSignal West Femto (Thermo Fisher Scientific) on a Bio-Rad ChemiDoc system. Protein loading was normalized using signal from β-actin–horseradish peroxidase (GenScript, Piscataway, NJ).
Colony Formation and Cell Migration Assays
miR-375 miRCURY LNA mimic and negative control miRCURY LNA (Qiagen) were transfected at a 50 nmol/L final concentration using RNAiMAX Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For cell migration assays, prFLC cells were transfected in F medium and replated 4 days later in fresh F medium in 24-well, flat-bottom, tissue culture plates at 50,000 cells/well to achieve confluence. The next day, scratch wounding was initiated with a P200 pipette tip and cells were imaged at 0, 24, 48, and 72 hours. For colony formation assessment in monolayer cultures, prFLC cells were plated in 6-well plates at a concentration of 15,000 or 36,000 cells/well and transfected. After 4 days, cells were stained with 0.1% crystal violet in 0.1 mol/L borate, pH 9.0, and 2% ethanol for 20 minutes and colonies were quantified in ImageJ (FIJI).
EdU Incorporation
prFLC cells were plated in F medium in 24-well plates at a density of 10,000 cells/well. After overnight incubation, cells were transfected with miR-375 or negative control miRCURY LNA mimic (Qiagen) at a 50 nmol/L final concentration using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were incubated for 2 hours with 10 μmol/L EdU. Cells were washed with phosphate-buffered saline, fixed in 4% paraformaldehyde for 20 minutes, and permeabilized with Triton X-100 (Fisher Scientific, Hampton, NH) for 20 minutes. EdU was detected using the Click-iT Plus EdU Alexa Fluor 594 Imaging kit (Invitrogen) according to the manufacturer’s instructions with reactions scaled appropriately for 24-well plates. Cells were counterstained with Hoechst 33342 (Life Technologies). Images were acquired on a ZOE Fluorescent Cell Image (Bio-Rad). For each experiment, at least 5 independent fields were imaged per well. Images were analyzed in ImageJ (National Institutes of Health, Bethesda, MD) and cells were manually and independently counted by 2 researchers.
Statistics
Statistical significance was determined primarily by the Mann–Whitney U test for unpaired samples and the Wilcoxon signed-rank test for paired samples as indicated in the figure legends. Any alternative statistical tests that were used are noted in the text and figure legends. Statistical comparisons were performed in R (3.3.1). P < .05 was considered statistically significant.
Acknowledgments
The authors would like to thank members of the Anna M. Diehl and Praveen Sethupathy laboratories for helpful discussions regarding this study, and Zhao Lai from the Genome Sequencing Facility of the Greehey Children’s Cancer Research Institute of the University of Texas Health Science Center at San Antonio for assistance with small RNA sequencing. The authors also thank Tom Stockwell, John Hopper, Dr John Craig, Marna Davis, and the entire Fibrolamellar Cancer Foundation for their support of the patients and the research network.
Footnotes
Author contributions Timothy A. Dinh designed the study, acquired, analyzed, and interpreted the data, prepared figures, and drafted the manuscript; Mark L. Jewell acquired, analyzed, and interpreted the data, provided resources, and prepared figures; Matt Kanke analyzed and interpreted the data and prepared figures; Ramja Sritharan, Seona Lee, and Adam Francisco acquired, analyzed, and interpreted data; Rigney E. Turnham acquired, analyzed, and interpreted data and provided resources; Edward R. Kastenhuber, Eliane Wauthier, Cynthia D. Guy, Raymond S. Yeung, Scott W. Lowe, Lola M. Reid, and John D. Scott provided resources; Anna M. Diehl supervised the study; and Praveen Sethupathy designed the study, analyzed and interpreted data, drafted the manuscript, and supervised the study.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants DK105542 and 5R01DK077794-10 from the National Institute of Diabetes and Digestive and Kidney Diseases (J.D.S. and A.M.D., respectively); grant 5R37AA010154-22 from the National Institute on Alcohol Abuse and Alcoholism (A.M.D.); and grants from the Fibrolamellar Cancer Foundation (A.M.D. and P.S.).
Contributor Information
Anna M. Diehl, Email: annamae.diehl@duke.edu.
Praveen Sethupathy, Email: pr46@cornell.edu.
References
- 1.Craig J.R., Peters R.L., Edmondson H.A., Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46:372–379. doi: 10.1002/1097-0142(19800715)46:2<372::aid-cncr2820460227>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica. 2012;2012:1–15. doi: 10.6064/2012/743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stipa F., Yoon S.S., Liau K.H., Fong Y., Jarnagin W.R., D’Angelica M., Abou-Alfa G., Blumgart L.H., DeMatteo R.P. Outcome of patients with fibrolamellar hepatocellular carcinoma. Cancer. 2006;106:1331–1338. doi: 10.1002/cncr.21703. [DOI] [PubMed] [Google Scholar]
- 4.Honeyman J.N., Simon E.P., Robine N., Chiaroni-Clarke R., Darcy D.G., Lim I.I.P., Gleason C.E., Murphy J.M., Rosenberg B.R., Teegan L., Takacs C.N., Botero S., Belote R., Germer S., Emde A.-K., Vacic V., Bhanot U., LaQuaglia M.P., Simon S.M. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornella H., Alsinet C., Sayols S., Zhang Z., Hao K., Cabellos L., Hoshida Y., Villanueva A., Thung S., Ward S.C., Rodriguez-Carunchio L., Vila-Casadesús M., Imbeaud S., Lachenmayer A., Quaglia A., Nagorney D.M., Minguez B., Carrilho F., Roberts L.R., Waxman S., Mazzaferro V., Schwartz M., Esteller M., Heaton N.D., Zucman-Rossi J., Llovet J.M. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology. 2014;148:806–818. doi: 10.1053/j.gastro.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung J., Ginter C., Cassidy M., Franklin M.C., Rudolph M.J., Robine N., Darnell R.B., Hendrickson W.A. Structural insights into mis-regulation of protein kinase A in human tumors. Proc Natl Acad Sci U S A. 2015;112:1374–1379. doi: 10.1073/pnas.1424206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelholm L.H., Riaz A., Serra D., Dagnæs-Hansen F., Johansen J.V., Santoni-Rugiu E., Hansen S.H., Niola F., Frödin M. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1–Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology. 2017;153:1662–1673. doi: 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kastenhuber E.R., Lalazar G., Tschaharganeh D.F., Houlihan S.L., Baslan T., Chen C.-C., Requena D., Tian S., Bosbach B., Wilkinson J.E., Simon S.M., Lowe S.W. DNAJB1-PRKACA fusion kinase drives tumorigenesis and interacts with β-catenin and the liver regenerative response. Proc Natl Acad Sci U S A. 2017;114:13076–13084. doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–221. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 11.Farber B.A., Lalazar G., Simon E.P., Hammond W.J., Requena D., Bhanot U.K., La Quaglia M.P., Simon S.M. Non coding RNA analysis in fibrolamellar hepatocellular carcinoma. Oncotarget. 2018;9:10211–10227. doi: 10.18632/oncotarget.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinh T.A., Vitucci E.C.M., Wauthier E., Graham R.P., Pitman W.A., Oikawa T., Chen M., Silva G.O., Greene K.G., Torbenson M.S., Reid L.M., Sethupathy P. Comprehensive analysis of The Cancer Genome Atlas reveals a unique gene and non-coding RNA signature of fibrolamellar carcinoma. Sci Rep. 2017;7:44653. doi: 10.1038/srep44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanke M., Baran-Gale J., Villanueva J., Sethupathy P. miRquant 2.0 : an expanded tool for accurate annotation and quantification of microRNAs and their isomiRs from small RNA-sequencing data. J Integr Bioinform. 2016;13:307. doi: 10.2390/biecoll-jib-2016-307. [DOI] [PubMed] [Google Scholar]
- 14.Hafner M., Renwick N., Brown M., Mihailović A., Holoch D., Lin C., Pena J.T.G., Nusbaum J.D., Morozov P., Ludwig J., Ojo T., Luo S., Schroth G., Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baran-Gale J., Lisa Kurtz C., Erdos M.R., Sison C., Young A., Fannin E.E., Chines P.S., Sethupathy P. Addressing bias in small RNA library preparation for sequencing: a new protocol recovers microRNAs that evade capture by current methods. Front Genet. 2015;6:1–9. doi: 10.3389/fgene.2015.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selth L.A., Das R., Townley S.L., Coutinho I., Hanson A.R., Centenera M.M., Stylianou N., Sweeney K., Soekmadji C., Jovanovic L., Nelson C.C., Zoubeidi A., Butler L.M., Goodall G.J., Hollier B.G., Gregory P.A., Tilley W.D. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2017;36:24–34. doi: 10.1038/onc.2016.185. [DOI] [PubMed] [Google Scholar]
- 17.Yan J.W., Lin J.S., He X.X. The emerging role of miR-375 in cancer. Int J Cancer. 2014;135:1011–1018. doi: 10.1002/ijc.28563. [DOI] [PubMed] [Google Scholar]
- 18.Wang B., Zou A., Ma L., Chen X., Wang L., Zeng X., Tan T. miR-455 inhibits breast cancer cell proliferation through targeting CDK14. Eur J Pharmacol. 2017;807:138–143. doi: 10.1016/j.ejphar.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Tsai W.C., Hsu P.W.C., Lai T.C., Chau G.Y., Lin C.W., Chen C.M., Lin C Der, Liao Y.L., Wang J.L., Chau Y.P., Hsu M.T., Hsiao M., Huang H Da, Tsou A.P. MicroRNA-122, a tumor suppressor MicroRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 20.Cao M.-Q., You A.-B., Zhu X.-D., Zhang W., Zhang Y.-Y., Zhang S.-Z., Zhang K., Cai H., Shi W.-K., Li X.-L., Li K.-S., Gao D.-M., Ma D.-N., Ye B.-G., Wang C.-H., Qin C.-D., Sun H.-C., Zhang T., Tang Z.-Y. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. 2018;11:12. doi: 10.1186/s13045-018-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung W.K.C., He M., Chan A.W.H., Law P.T.Y., Wong N. Wnt/β-catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362:97–105. doi: 10.1016/j.canlet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 23.He X.X., Chang Y., Meng F.Y., Wang M.Y., Xie Q.H., Tang F., Li P.Y., Song Y.H., Lin J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 24.Liu A.M., Poon R.T.P., Luk J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 25.Ding L., Xu Y., Zhang W., Deng Y., Si M., Du Y., Yao H., Liu X., Ke Y., Si J., Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 26.Kang W., Huang T., Zhou Y., Zhang J., Lung R.W.M., Tong J.H.M., Chan A.W.H., Zhang B., Wong C.C., Wu F., Dong Y., Wang S., Yang W., Pan Y., Chak W.P., Cheung A.H.K., Pang J.C.S., Yu J., Cheng A.S.L., To K.F. MiR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis article. Cell Death Dis. 2018;9:92. doi: 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong K.L., Kwong D.L.W., Chan T.H.-M., Law S.Y.-K., Chen L., Li Y., Qin Y.-R., Guan X.-Y. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 28.Harris T., Jimenez L., Kawachi N., Fan J.B., Chen J., Belbin T., Ramnauth A., Loudig O., Keller C.E., Smith R., Prystowsky M.B., Schlecht N.F., Segall J.E., Childs G. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am J Pathol. 2012;180:917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller D.M., Clark E.A., Goodman R.H. Regulation of microRNA-375 by cAMP in pancreatic β-cells. Mol Endocrinol. 2012;26:989–999. doi: 10.1210/me.2011-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikawa T., Wauthier E., Dinh T.A., Selitsky S.R., Reyna-Neyra A., Carpino G., Levine R., Cardinale V., Klimstra D., Gaudio E., Alvaro D., Carrasco N., Sethupathy P., Reid L.M. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat Commun. 2015;6:8070. doi: 10.1038/ncomms9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riggle K.M., Riehle K.J., Kenerson H.L., Turnham R., Homma M.K., Kazami M., Samelson B., Bauer R., McKnight G.S., Scott J.D., Yeung R.S. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr Res. 2016;80:110–118. doi: 10.1038/pr.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazar J., DeBlasio D., Govindarajan S.S., Zhang S., Perera R.J. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011;585:2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Baran-Gale J., Fannin E.E., Kurtz C.L., Sethupathy P. Beta cell 5′-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One. 2013;8:e73240. doi: 10.1371/journal.pone.0073240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor S.S., Ilouz R., Zhang P., Kornev A.P. Assembly of allosteric macromolecular switches: lessons from PKA. Nat Rev Mol Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langeberg L.K., Scott J.D. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol. 2015;16:232–244. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaQuaglia M.J., Grijalva J.L., Mueller K.A., Perez-Atayde A.R., Kim H.B., Sadri-Vakili G., Vakili K. YAP subcellular localization and Hippo pathway transcriptome analysis in pediatric hepatocellular carcinoma. Sci Rep. 2016;6:1–12. doi: 10.1038/srep30238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Krawczyk E., Suprynowicz F.A., Palechor-Ceron N., Yuan H., Dakic A., Simic V., Zheng Y., Sripadhan P., Chen C., Lu J., Hou T., Choudhury S., Kallakury B., Tang D.G., Darling T., Thangapazham R., Timofeeva O., Dritschilo A., Randell S.H., Albanese C., Agarwal S., Schlegel R. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc. 2017;12:439–451. doi: 10.1038/nprot.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikawa E., Osada H., Okazaki Y., Arima C., Tomida S., Tatematsu Y., Taguchi A., Shimada Y., Yanagisawa K., Yatabe Y., Toyokuni S., Sekido Y., Takahashi T. MiR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71:6165–6173. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 39.Ou J., Kou L., Liang L., Tang C. MiR-375 attenuates injury of cerebral ischemia/reperfusion via targeting Ctgf. Biosci Rep. 2017;37 doi: 10.1042/BSR20171242. BSR20171242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam K.J., Mo J.-S., Han S.-H., Park W.-C., Kim H.-S., Yun K.-J., Chae S.-C. MicroRNA 375 regulates proliferation and migration of colon cancer cells by suppressing the CTGF-EGFR signaling pathway. Int J Cancer. 2017;141:1614–1629. doi: 10.1002/ijc.30861. [DOI] [PubMed] [Google Scholar]
- 41.Luna J.M., Scheel T.K.H., Danino T., Shaw K.S., Mele A., Fak J.J., Nishiuchi E., Takacs C.N., Catanese M.T., de Jong Y.P., Jacobson I.M., Rice C.M., Darnell R.B. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L., Wen T., Liu Z., Xu F., Yang L., Liu J., Feng G., An G. MicroRNA-375 suppresses human colorectal cancer metastasis by targeting Frizzled 8. Oncotarget. 2016;7:40644–40656. doi: 10.18632/oncotarget.9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peck B.C.E., Mah A.T., Pitman W.A., Ding S., Lund P.K., Sethupathy P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust MicroRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem. 2017;292:2586–2600. doi: 10.1074/jbc.M116.770099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao C., Shi W., Xiong Y., Yu X., Zhang X., Qin M., Du C., Song T., Li J. MiR-375 regulates the canonical Wnt pathway through FZD8 silencing in arthritis synovial fibroblasts. Immunol Lett. 2015;164:1–10. doi: 10.1016/j.imlet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Chen L., Feng P., Peng A., Qiu X., Zhu X., He S., Zhou D. cAMP response element-binding protein and Yes-associated protein form a feedback loop that promotes neurite outgrowth. J Cell Mol Med. 2018;22:374–381. doi: 10.1111/jcmm.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., Ma L., Weng W., Qiao Y., Zhang Y., He J., Wang H., Xiao W., Li L., Chu Q., Pan Q., Yu Y., Sun F. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58:1011–1020. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- 47.Scoazec J.Y., Flejou J.F., D’Errico A., Fiorentino M., Zamparelli A., Bringuier A.F., Feldmann G., Grigioni W.F. Fibrolamellar carcinoma of the liver: composition of the extracellular matrix and expression of cell-matrix and cell-cell adhesion molecules. Hepatology. 1996;24:1128–1136. doi: 10.1002/hep.510240525. [DOI] [PubMed] [Google Scholar]
- 48.Zhou H., Zhang M., Yuan H., Zheng W., Meng C., Zhao D. MicroRNA-375 functions as a tumor suppressor in osteosarcoma by targeting PIK3CA. Tumor Biol. 2015;36:8579–8584. doi: 10.1007/s13277-015-3614-9. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Tang Q., Li M., Jiang S., Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444:199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 50.Zhan T., Huang X., Tian X., Chen X., Ding Y., Luo H., Zhang Y. Downregulation of microRNA-455-3p links to proliferation and drug resistance of pancreatic cancer cells via targeting TAZ. Mol Ther Nucleic Acids. 2018;10:215–226. doi: 10.1016/j.omtn.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D., Gregory R.I. Hippo signaling regulates microprocessor and links cell-density-dependent mirna biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rumble S.M., Lacroute P., Dalca A.V., Fiume M., Sidow A., Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei L., Jin Z., Yang S., Xu Y., Zhu Y., Ji Y. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34:1615–1617. doi: 10.1093/bioinformatics/btx812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J.C., Merlino G., Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]