Abstract
Background & Aims
Intratumor heterogeneity is a common feature of colorectal cancer (CRC). Here, we analyzed whether mesenchymal-like CRC cells promote the progression of epithelial-like CRC cells via paracrine mechanisms.
Methods
Six CRC cell lines that show an epithelial phenotype were treated with conditioned media (CM) from CRC cell lines that show a mesenchymal phenotype, and effects on epithelial-mesenchymal transition (EMT), migration, invasion, and chemoresistance were determined. Secreted factors potentially mediating these effects were identified by using cytokine arrays. Associations of these factors with tumor progression and patient survival were determined.
Results
CM obtained from mesenchymal-like CRC cells induced EMT associated with increased migration, invasion, and chemoresistance in epithelial-like CRC cell lines. Notably, activation of p53 in mesenchymal-like CRC cells prevented these effects of CM. Increased concentrations of several cytokines were identified in CM from mesenchymal-like CRC cell lines and a subset of these cytokines showed repression by p53. The down-regulation of nidogen-1 (NID1) was particularly significant and was owing to p53-mediated induction of microRNA-192 and microRNA-215, which directly target the NID1 messenger RNA. NID1 was found to be required and sufficient for inducing EMT, invasion, and migration in epithelial-like CRC cells. In primary CRCs, increased NID1 expression was associated with p53 mutation and microRNA-192/215 down-regulation. Importantly, increased NID1 expression in CRCs correlated with enhanced tumor progression and poor patient survival.
Conclusions
Taken together, our results show that CRC cells promote tumor progression via secreting NID1, which induces EMT in neighboring tumor cells. Importantly, the interference of p53 with this paracrine signaling between tumor cells may critically contribute to tumor suppression.
Keywords: Metastasis, Nidogen1, microRNAs, Tumor Progression
Abbreviations used in this paper: cDNA, complementary DNA; CM, conditioned medium; CMS, consensus molecular subtype; COAD, colonic adenocarcinomas; CRC, colorectal cancer; DOX, doxycycline; EMT, epithelial-mesenchymal transition; ERK, extracellular signal-regulated kinase; FBS, fetal bovine serum; MAPK, mitogen-activated protein kinase; miR, microRNA; miRNA, microRNA; mRNA, messenger RNA; NID1, nidogen-1; qPCR, quantitative polymerase chain reaction; READ, rectal adenocarcinomas; siRNA, small interfering RNA; TCGA, The Cancer Genome Atlas; TGFβ, transforming growth factor β; VIM, vimentin; VSV, Vesicular Stomatitis Virus; wt, wild-type; ZEB1, Zinc Finger E-Box Binding Homeobox 1
Graphical abstract
Summary.
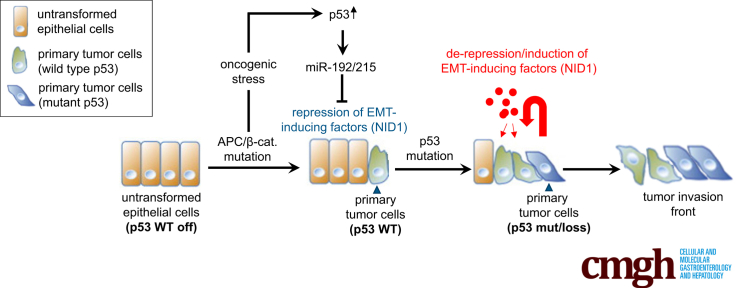
We show that factors secreted by mesenchymal-like, invasive colorectal cancer cells induce epithelial-mesenchymal transition, migration, and invasion of epithelial-like, noninvasive tumor cells. This effect was abrogated by the p53 tumor suppressor via microRNA-192/215–mediated repression of secreted nidogen1.
During tumor progression, cancer cells acquire a mesenchymal phenotype that is associated with an increased invasive and migratory capacity, which allows them to leave the site of the primary tumor, invade surrounding tissues, and migrate to distant organs. Once these migrating cancer cells reach their new niche, they switch back to an epithelial phenotype and proliferate to form metastases.1 The understanding of these processes is clinically relevant because metastasis accounts for more than 90% of cancer mortality. The processes by which cells switch between epithelial and mesenchymal phenotypes are known as the epithelial-mesenchymal transition (EMT) and its counterpart, the mesenchymal-epithelial transition.2 Through EMT tumor cells also acquire features of stem cells3 and a multidrug-resistant phenotype.4
Tumors represent a heterogeneous mix of cells because they are composed of numerous subclones.5, 6, 7 This intratumor heterogeneity and the interaction of tumor cells with the surrounding microenvironment critically contributes to metastasis.8, 9, 10, 11 Invasive colorectal cancer cells penetrate through the muscularis mucosae and subsequently enter the bloodstream.12 It is thought that the invasive front of colorectal tumors contains cancer cells that have undergone EMT as a result of signals emanating from the tumor microenvironment and therefore have mesenchymal properties.12 Notably, the loss of p53 function has been associated with EMT, invasion, and metastasis in colorectal cancer.13, 14, 15
Although reciprocal cross-talk between numerous intracellular signaling pathways is known to regulate and maintain EMT, it now is emerging that extracellular factors provided by the tumor microenvironment also can influence the tumor cell state and invasive potential. Indeed, it has been shown that cells undergoing EMT enhance the secretion of various factors, especially growth factors (transforming growth factor β [TGFβ], hepatocyte growth factor [HGF])16 and proinflammatory cytokines, such as interleukins 6 and 8, which induce EMT.17, 18 Therefore, factors secreted from single invasive tumor cells might be responsible for the generation and maintenance of a tumor invasion front.
The p53 transcription factor is encoded by the most commonly mutated tumor-suppressor gene.19 The p53 network responds to a variety of intrinsic and extrinsic stress signals by inducing genes that encode factors that mediate cell-cycle arrest, senescence, or apoptosis.20, 21, 22 More recently, a paracrine role for p53 was established, because p53 also induces the production of secreted proteins that influence the proliferation and viability of neighboring but also more distantly localized cells.23, 24, 25, 26, 27
In this study, we identified nidogen-1 (NID1) as a p53-repressed, secreted factor that mediates the inhibitory effect of p53 on paracrine EMT and presumably on metastasis. Besides shedding light on the mechanisms of tumor suppression by p53, components of this regulatory circuit may be exploited for tumor therapeutic and diagnostic/prognostic purposes in the future.
Results
Paracrine Signaling Between Colorectal Cancer Cells Induces EMT
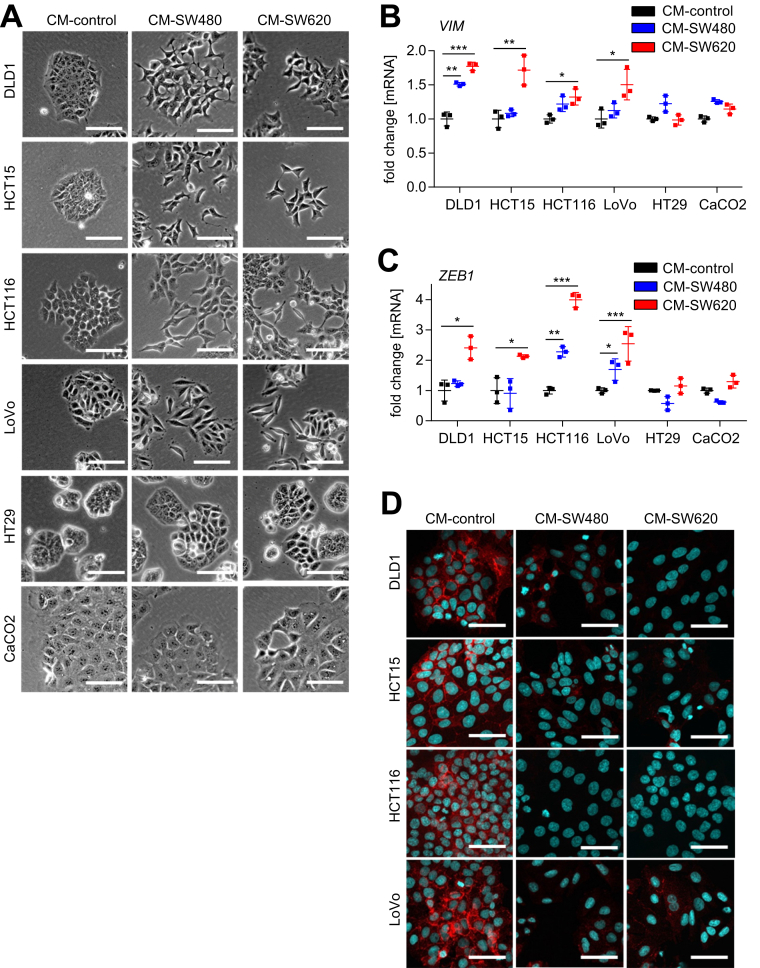
Here, we asked whether mesenchymal-like tumor cells secrete factors that may induce EMT in epithelial-like tumor cells that reside in their vicinity. Therefore, we harvested conditioned medium (CM) derived from the mesenchymal-like colorectal cancer (CRC) cells SW480 and SW620 and applied it to the epithelial-like CRC cell lines DLD1, HCT15, HCT116, LoVo, Caco2, and HT29. After incubation with the CM for 96 hours, epithelial-like cells DLD1, HCT15, HCT116, and LoVo changed from a cobblestone-like shape and island-like growth pattern to a spindle-shaped mesenchymal morphology with scattered cell distribution and decreased cell–cell contacts, whereas Caco2 and HT29 cells did not show changes in morphology (Figure 1A). The mesenchymal markers vimentin (VIM) and ZEB1 were up-regulated on the level of messenger RNA (mRNA) expression in DLD1, HCT15, HCT116, and LoVo cells after the addition of CM from mesenchymal-like CRC cell lines (Figure 1B and 1C). In addition, the cytoplasmic and membrane-bound E-cadherin localization typical for DLD1, HCT15, HCT116, and LoVo cells was lost after the addition of CM from SW480 and SW620 cells (Figure 1D). Therefore, CM from mesenchymal-like CRC cells induces EMT in epithelial-like CRC cells.
Figure 1.
Paracrine signaling from mesenchymal- to epithelial-like CRC cells induces EMT. (A) Phase-contrast images of DLD1, HCT15, HCT116, LoVo, HT29, and Caco2 cells cultured for 96 hours in CM collected from SW480 or SW620 CRC cells after 72 hours of cultivation. (B and C) qPCR analyses of EMT-related genes VIM and ZEB1 in DLD1, HCT15, HCT116, LoVo, HT29, and Caco2 cells cultured for 96 hours in CM obtained from SW480 or SW620 cells. Mean values ± SD (n = 3 biological replicates) are provided. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001. (D) Immunofluorescence detection of E-cadherin in DLD1, HCT15, HCT116, and LoVo cells cultured in CM obtained from SW480 or SW620 cells. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm.
Paracrine Enhancement of Migration, Invasion, and Chemoresistance
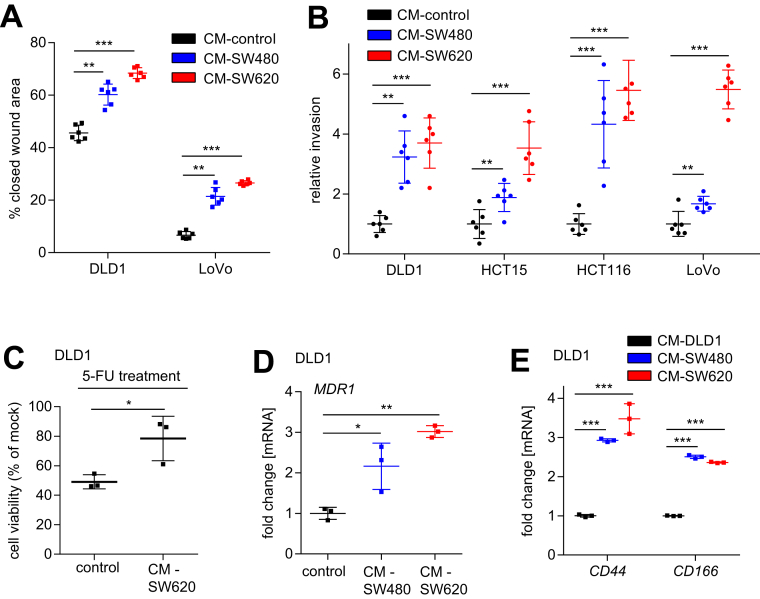
EMT is known to increase the invasiveness of cancer cells.28 In line with the induction of EMT in epithelial-like CRC cells exposed to CM derived from SW480 and SW620, these cells also showed increased migration (Figure 2A) and invasion (Figure 2B) in wound healing and modified Boyden chamber assays, respectively. Moreover, recent studies have shown that EMT promotes chemoresistance and stemness in cancer cells.3, 4 Here, we observed that pretreatment of epithelial-like DLD1 cells with CM from mesenchymal-like SW620 cells increased their viability after exposure to the chemotherapeutic agent 5-fluorouracil (Figure 2C), suggesting that SW620-derived CM enhances chemoresistance. In line with these results, CM from mesenchymal-like CRC cells SW480 or SW620 induced expression of ABCB1/MDR1 in DLD1 cells (Figure 2D). Because MDR1 mediates the adenosine triphosphate–dependent export of numerous anticancer drugs,29 its increased expression may explain the observed increase in chemoresistance. In addition, cultivation of DLD1 cells in SW480/SW620-derived CM induced the expression of the stem cell markers CD44 and CD166, indicating that stemness of these cells presumably also was enhanced by exposure to CM (Figure 2E). Taken together, these results show that the EMT induced by CM from mesenchymal CRC cell lines is associated with functional consequences, such as increased migration, invasion, and chemoresistance.
Figure 2.
Paracrine enhancement of migration, invasion, and chemoresistance in epithelial-like CRC cells. (A) Wound healing assay of DLD1 and LoVo cells treated with SW480 or SW620 CM for 30 hours. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (B) Invasion assay in modified Boyden chambers. DLD1, HCT15, HCT116, and LoVo cells were seeded on Matrigel-coated filters with control or SW480/SW620 CM used as a chemoattractant in the lower well. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. (C) Cell viability was determined by a 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromidefor assay. DLD1 cells were treated with control or SW620 CM for 72 hours before treatment with 5-fluorouracil (5-FU) (50 μg/mL) for 48 hours. (D and E) qPCR analyses of MDR1, CD44, and CD166 expression in DLD1 cells cultured in CM obtained from SW480 and SW620 cells. (A and B) Mean values ± SD (n = 2 biological replicates; each n = 3 technical replicates), and (C–E) mean values ± SD (n = 3 biological replicates) are provided. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001
p53 Suppresses Paracrine Induction of EMT
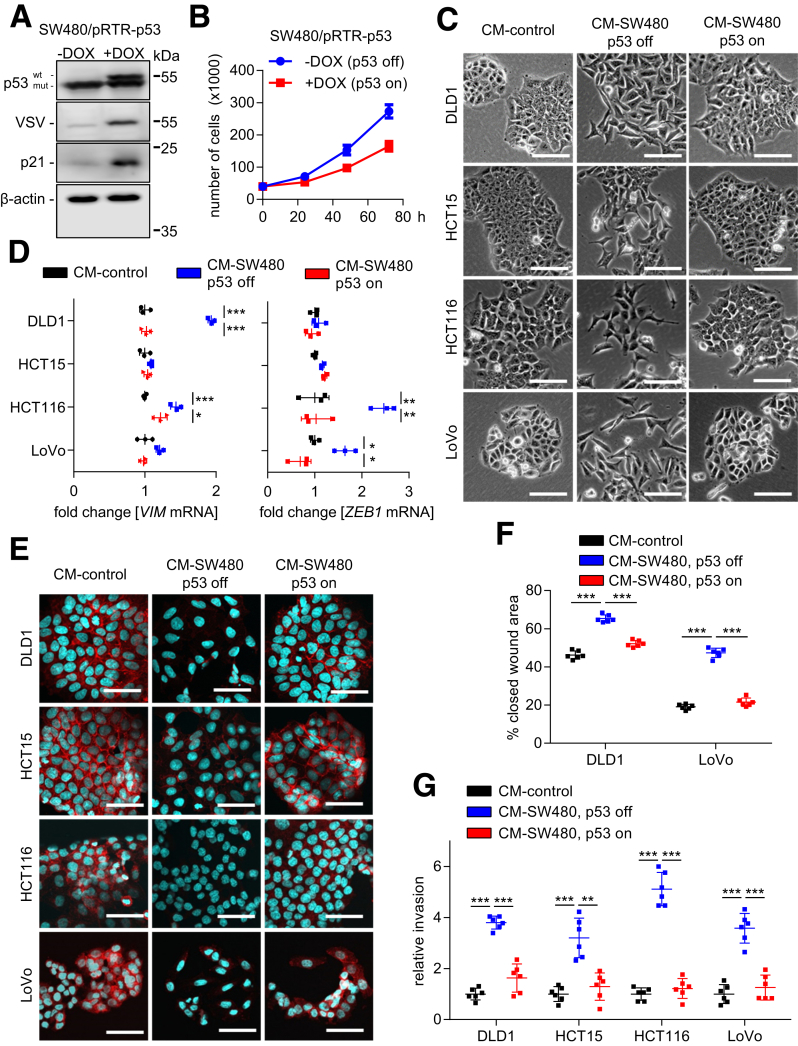
We hypothesized that p53 may inhibit the paracrine induction of EMT observed here. To test this hypothesis, we used SW480 cells ectopically expressing p53 under control of a doxycycline (DOX)-inducible promoter (SW480/pRTR-p53-VSV).30 SW480 cells harbor mutant p53 protein because the remaining p53 allele has R273H and P309S mutations.31 After addition of DOX for 48 hours, SW480/pRTR-p53-VSV cells also expressed the tagged wild-type (wt) p53 protein at similar levels as the mutant p53 protein (Figure 3A). Because treatment with DOX resulted in the induction of the p53 target gene p21, the ectopic wild-type (wt) p53 protein is transcriptionally active (Figure 3A). Ectopic expression of wt p53 decreased the proliferation of SW480 cells (approximately 2-fold after 72 hours) (Figure 3B). Therefore, we seeded 2 times more DOX-treated than untreated SW480/pRTR-p53-VSV cells and collected CM 72 hours later. CM harvested from SW480/pRTR-p53-VSV cells treated with DOX did not induce EMT in DLD1, HCT15, HCT116, and LoVo cells as evidenced by the morphology and the expression of EMT markers when compared with CM collected from SW480/pRTR-p53-VSV cells not exposed to DOX (Figure 3C–E). In addition, ectopic expression of p53 in SW480 cells prevented the enhancement of migration and invasion in epithelial-like CRC cells, which was observed after exposure to SW480-derived CM (Figure 3F and G). Altogether, these results show that activation of p53 suppresses the expression and/or secretion of EMT-inducing factors in mesenchymal-like SW480 cells.
Figure 3.
p53 activation suppresses paracrine induction of EMT. (A) Western blot analyses of indicated proteins in SW480/pRTR-p53-VSV cells, treated with vehicle (p53 off) or with DOX (p53 on) for 48 hours. (B) Cell number of SW480/pRTR-p53-VSV cells, treated with vehicle (p53 off) or with DOX (p53 on) at the indicated time points. (C) Phase-contrast images showing cell morphology of DLD1, HCT15, HCT116, and LoVo cells treated with CM from SW480/pRTR-p53-VSV cells for 96 hours, which were treated with vehicle (p53 off) or DOX (p53 on) for 72 hours. (D) qPCR analyses of indicated EMT-related genes in DLD1, HCT15, HCT116, and LoVo cells treated with CM from control or SW480/pRTR-p53-VSV cells for 96 hours, which were treated with vehicle (p53 off) or DOX (p53 on) for 72 hours. (E) Immunofluorescence detections of E-cadherin in DLD1, HCT15, HCT116, and LoVo cells treated with CM from SW480/pRTR-p53-VSV cells for 96 hours, which were treated with vehicle (p53 off) or DOX (p53 on) for 72 hours. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm. (F) Wound healing assay of DLD1 and LoVo cells treated for 30 hours with CM collected from SW480/pRTR-p53-VSV cells, which were treated with vehicle (p53 off) or DOX (p53 on) for 72 hours. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (F) Invasion assay in modified Boyden chambers. DLD1, HCT15, HCT116, and LoVo cells were seeded on Matrigel-coated filters with CM from SW480/pRTR-p53-VSV cells, which were treated with vehicle (p53 off) or DOX (p53 on), used as a chemoattractant in the lower well. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. (B and D) Means ± SD (n = 3 biological replicates), and (F and G) means ± SD (n = 2 biological replicates; each n = 3 technical replicates) are provided. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001.
Identification of Secreted EMT Regulators Within CM of CRC Cell Lines
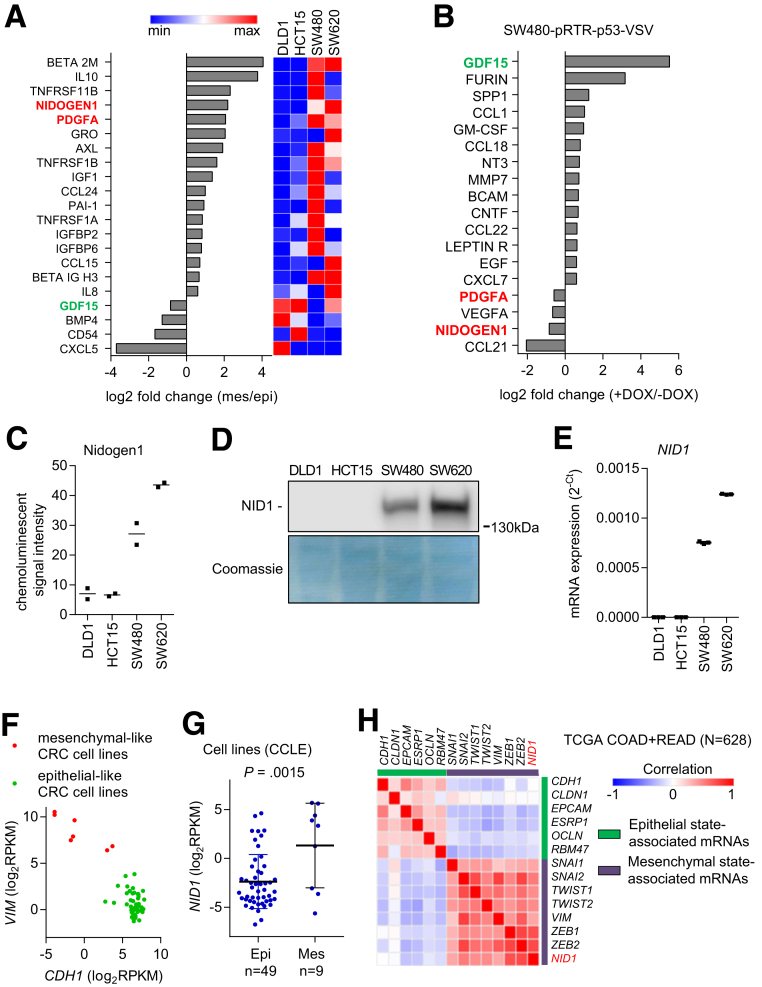
Next, we aimed to identify EMT-inducing factors preferentially secreted by SW480 and SW620 cells. Therefore, we used an array that detects 274 cytokines to compare cytokine expression levels in conditioned media obtained from epithelial-like DLD1/HCT15 and mesenchymal-like SW480/SW620 cells. Seventeen proteins were present at increased levels and 4 proteins were present at decreased levels in mesenchymal-like vs epithelial-like CRC cells (changes >1.5-fold) (Figure 4A). Because the activation of p53 in mesenchymal-like cancer cells had suppressed the paracrine induction of EMT in DLD1 cells (Figure 3), we also analyzed the CM of SW480 cells after activation of p53. Fourteen proteins were present at increased levels and 4 proteins were present at decreased levels in SW480 cells after activation of p53 (changes > 1.5-fold) (Figure 4B). By comparing both cytokine expression analyses, we found that NID1 and platelet-derived growth factor A were present at increased levels in the CM from mesenchymal-like cells and at decreased levels in SW480 cells after activation of p53. In contrast, growth/differentiation factor 15 (GDF15) was present at decreased levels in the CM from mesenchymal-like cells and at increased levels in SW480 cells after activation of p53. In the subsequent analyses we focused on NID1 because it was present at the highest levels in the CM from SW620 cells, which induced the most pronounced EMT in epithelial-like CRC cells (Figure 4C and D). In addition, on the mRNA level, NID1 showed the highest expression in SW480 and SW620 cells, and very low expression in the epithelial-like DLD1 and HCT15 cells (Figure 4E). Next, we analyzed whether the expression of NID1 generally is associated with mesenchymal-like cell states of established CRC cell lines. Therefore, we used expression data of CRC cell lines deposited within the Cancer Cell Line Encyclopedia. First, we classified colorectal cancer cell lines as epithelial- or mesenchymal-like based on their expression of CDH1 and vimentin, respectively32 (Figure 4F). NID1 expression was significantly higher in mesenchymal-like CRC cell lines (Figure 4G). Furthermore, the expression of NID1 correlated positively with mesenchymal-state–associated genes and negatively with epithelial-state–associated genes in expression profiles of primary CRCs derived from 456 cases of colonic adenocarcinomas (COAD) and 172 cases of rectal adenocarcinomas (READ) deposited in The Cancer Genome Atlas (TCGA) database33 (Figure 4H).
Figure 4.
Identification of EMT regulators within CM of CRC cell lines. (A) Cytokine array analyses of CM from epithelial (DLD1 and HCT15) and mesenchymal (SW480 and SW620) CRC cell lines. Secreted factors that showed at least 1.5-fold difference between epithelial-like and mesenchymal-like CRC cell lines are shown. (B) Cytokine array analyses of CM from SW480-pRTR-p53-VSV cells. Secreted factors that showed at least 1.5-fold difference between SW480-pRTR-p53-VSV cells treated with vehicle (p53 off) or DOX (p53 on) are shown. (C) NID1 levels in CM of indicated cell lines assessed by densitometric quantification of NID1-specific signals from cytokine array analyses. (D) Western blot analysis of NID1 protein in concentrated CM from indicated cell lines. Coomassie Blue staining was used as a loading control. (E) qPCR analyses of NID1 expression in indicated cell lines. (F) Classification of colorectal cancer cell lines as epithelial- or mesenchymal-like based on the distribution of vimentin (VIM) and E-cadherin (CDH1) mRNA expression. Data are from the Cancer Cell Line Encyclopedia database (CCLE). (G) NID1 expression in epithelial- and mesenchymal-like CRC cell lines represented in the CCLE database. Individual data points and means ± SD are provided. (H) Correlation of NID1 expression with epithelial- and mesenchymal-state–associated mRNAs in primary CRC tumors. Expression data are from the TCGA collection of human colorectal adenocarcinomas (COAD + READ; N = 628). (C) Individual values and the means (n = 2 technical replicates/1 biological sample), and (E) means ± SD (n = 3 biological replicates) are provided. Epi, epithelial-like CRC cell lines; Mes, mesenchymal-like CRC cell lines.
Expression of NID1 Is Suppressed by p53 via Induction of microRNA-192 and microRNA-215
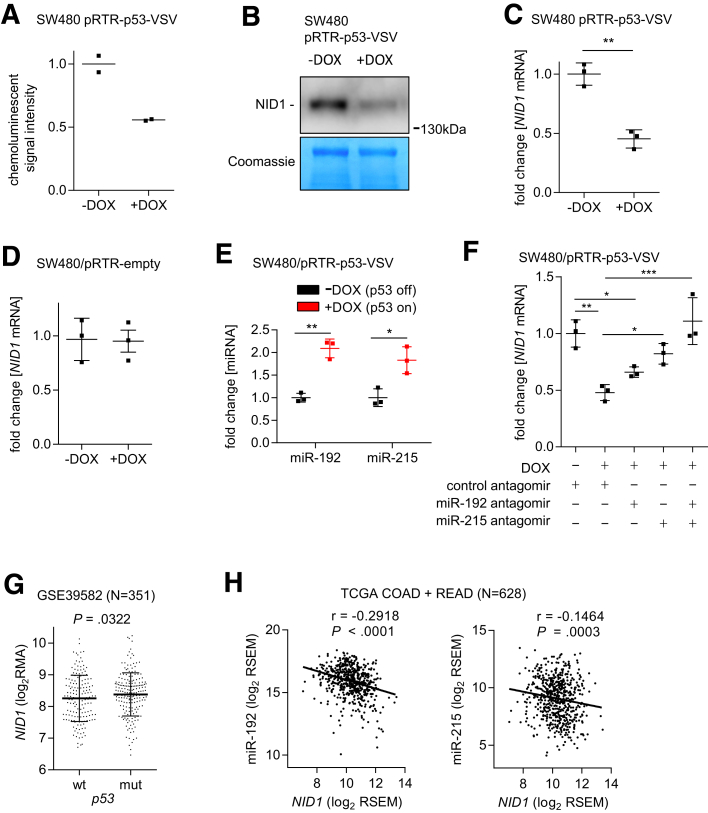
The decreased levels of NID1 protein in CM from SW480 cells after activation of p53 assessed by cytokine arrays (Figure 5A) was validated by Western blot analysis of concentrated CM from these cells with a NID1-specific antibody (Figure 5B). Similarly, NID1 mRNA expression was repressed after activation of p53 in SW480 cells (Figure 5C). To exclude the possibility that DOX modulates NID1 expression, we analyzed SW480 cells stably transfected with an empty pRTR vector. Treatment with DOX did not cause a change in NID1 expression (Figure 5D). p53 does not directly repress its targets, but suppresses gene expression through p53-inducible microRNAs (miRNAs) or the dimerization partner, RB-like, E2F and multi-vulval class B complexes.21 Our bioinformatic analyses showed that the NID1 3’-untranslated region contains miR-192 and miR-215 seed-matching sites and therefore represent a putative target of these miRNAs, which we previously identified as being induced more than 2-fold by p53 activation in SW480 cells.34 Therefore, we hypothesized that the repression of NID1 by p53 is mediated by miR-192 and miR-215. Indeed, quantitative polymerase chain reaction (qPCR) analyses confirmed that expression of mature miR-192 and miR-215 is induced by ectopic p53 in SW480 cells (Figure 5E). Furthermore, antagomir-mediated inhibition of either miR-192 or miR-215 partially prevented NID1 repression by p53, while combined inhibition of miR-192 and miR-215 completely prevented NID1 repression by p53 (Figure 5F). In line with the observed repression of NID1 by ectopic p53 in CRC cell lines, NID1 expression also was increased in primary CRCs harboring mutant p53 (Figure 5G). Furthermore, expression of miR-192 and miR-215 showed a significantly inverse correlation with NID1 expression in 628 primary CRCs from the TCGA database (Figure 5H). Therefore, the observations made in cell lines are presumably also relevant for primary CRCs. Taken together, these results show that NID1 represents an attractive candidate for mediating the effects of CM described earlier and that NID1 is controlled negatively by p53 via the microRNAs miR-192 or miR-215.
Figure 5.
Expression of NID1 is suppressed by p53 via induction of miR-192 and miR-215. (A) NID1 protein levels in CM from SW480-pRTR-p53-VSV cells treated with vehicle (p53 off) or DOX (p53 on) assessed by densitometric quantification of NID1-specific signals from cytokine array analyses. (B) Western blot analysis of NID1 protein in concentrated CM from SW480-pRTR-p53-VSV cells treated with vehicle (p53 off) or DOX (p53 on). Coomassie Blue staining was used as a loading control. (C) qPCR analyses of NID1 expression in SW480/pRTR-p53-VSV cells treated with vehicle or DOX. (D) qPCR analyses of NID1 expression in SW480/pRTR empty cells treated with vehicle or DOX. (E) qPCR analyses of mature miR-192 and miR-215 expression in SW480/pRTR-p53-VSV cells treated with vehicle (p53 off) or DOX (p53 on). (F) qPCR analyzes of NID1 expression in SW480/pRTR-p53-VSV cells transfected with miR-192 or miR-215 antagomirs and treated with DOX. (G) NID1 expression in p53 wt and mutant primary colorectal tumors (data from GSE39582; Student t test). Individual data points and means ± SD are provided. (H) Correlation of NID1 with miR-192 (left) and miR-215 (right) expression in primary CRC tumors. Expression data are from the TCGA collection of human colorectal adenocarcinomas (COAD + READ; N = 628). Spearman correlation coefficient (r) is provided. (A) Individual values and the means (n = 2 technical replicates/1 biological sample), and (C–F) means ± SD (n = 3 biological replicates) are provided. (C) Significance was determined using the Student t test, and (E and F) by 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001. RSEM, RNA-Seq by Expectation Maximization.
Expression of NID1 Is Suppressed by DNA Methylation
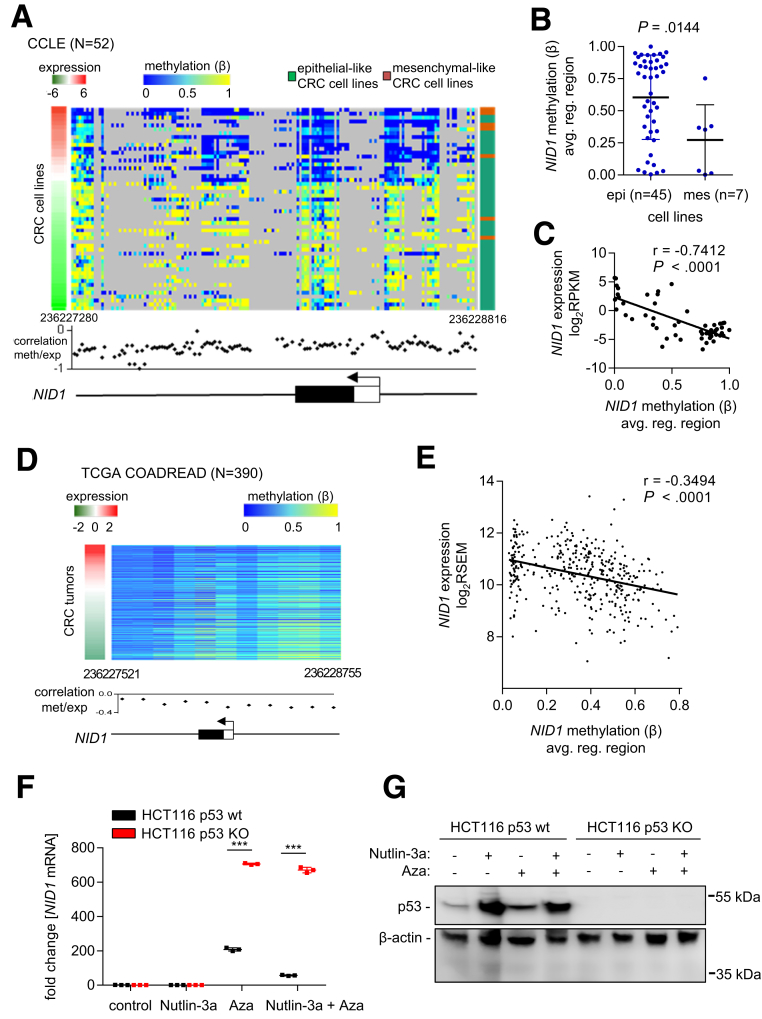
Our results suggest that upon loss of functional p53, CRC cells may express NID1 at increased levels. To test this hypothesis, we first analyzed NID1 expression in 3 pairs of epithelial-like p53 wt CRC cell lines (HCT116, RKO, and SW48) and their derivatives with a homozygous deletion of p53. However, none of the cell lines, neither p53 wt nor p53 deficient, expressed NID1 (qPCR crossing point values > 35; data not shown). By analyzing the expression and DNA methylation data from 52 CRC cell lines from the Cancer Cell Line Encyclopedia, we noticed that in most mesenchymal-like cell lines (including SW480 and SW620), the NID1 gene is not methylated, whereas in epithelial-like cell lines (including DLD, HCT15, LoVo, HCT116, RKO, and SW48), the NID1 gene is highly methylated (Figure 6A and B). Furthermore, NID1 expression and DNA methylation show a significantly inverse correlation (Figure 6C). In line with these results, a negative correlation between NID1 expression and DNA methylation also was observed in primary colorectal tumors from the TCGA COAD + READ data set (Figure 6D and E). Treatment of HCT116 p53 wt and p53-deficient cells with the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine resulted in a pronounced induction of NID1 expression (Figure 6F). However, the induction of NID1 was approximately 3 times lower in p53 wt compared with p53-deficient cells. Moreover, treatment with the p53 activator Nutlin-3a further prevented the induction of NID1 by 5-aza-2’-deoxycytidine in p53 wt cells, whereas in p53-deficient cells Nutlin-3a showed no effect (Figure 6F and G). Thus, in CRC cells with methylated NID1 gene, loss of functional p53 is not sufficient to induce NID1 expression, but additional demethylation together with loss of p53 is required for complete de-repression of NID1.
Figure 6.
Expression of NID1 is suppressed by DNA methylation. (A) DNA methylation and expression of NID1, as well as epithelial/mesenchymal status of 52 CRC cell lines (data from the Cancer Cell Line Encyclopedia). Left: red/green colored bar: NID1 expression; middle: blue/yellow heatmap: DNA methylation (β value) for each CG site in the NID1 regulatory genomic region; right: green/orange bar: epithelial/mesenchymal status of cell lines. The graph below the heatmap shows the correlation of each CG site in the NID1 regulatory genomic region with NID1 expression. hg19 genomic coordinates and the NID1 gene structure with indicated first exon (white box: untranslated region; black box: protein encoding region) and transcription start site (arrow) is shown at the bottom. (B) Average DNA methylation of all CG sites in the NID1 regulatory genomic region in epithelial-like and mesenchymal-like CRC cell lines (data from the Cancer Cell Line Encyclopedia; Student t test). Individual data points and means ± SD are provided. (C) Correlation between average DNA methylation of all CG sites in the NID1 regulatory gene region and NID1 expression in 52 CRC cell lines (data from the Cancer Cell Line Encyclopedia). Spearman correlation coefficient is provided. (D) DNA methylation and expression of NID1 in 390 primary colorectal tumors (data from the TCGA COAD + READ cohort). Left: red/green colored bar: NID1 expression; middle: blue/yellow heatmap: DNA methylation (β value) for each CG site in the NID1 regulatory genomic region. The graph bellow the heatmap shows the correlation of each CG site in the NID1 regulatory genomic region with NID1 expression. hg19 genomic coordinates and the NID1 gene structure with indicated first exon (white box: untranslated region; black box: protein encoding region) and transcription start site (arrow) is shown at the bottom. (E) Correlation between average DNA methylation of all CG sites in the NID1 regulatory genomic region and NID1 expression in primary colorectal tumors (data from the TCGA COAD + READ cohort). Spearman correlation coefficient is provided. (F) NID1 mRNA expression in HCT116 p53 wt and p53 knockout cells. Cells were treated with the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine for 96 hours and with Nutlin-3a for the last 48 hours. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; ***P < .001. (G) Western blot analysis of p53 protein in HCT116 p53 wt and p53 knockout cells treated as described in panel F.
Requirement of NID1 for CM-Induced EMT, Migration, and Invasion
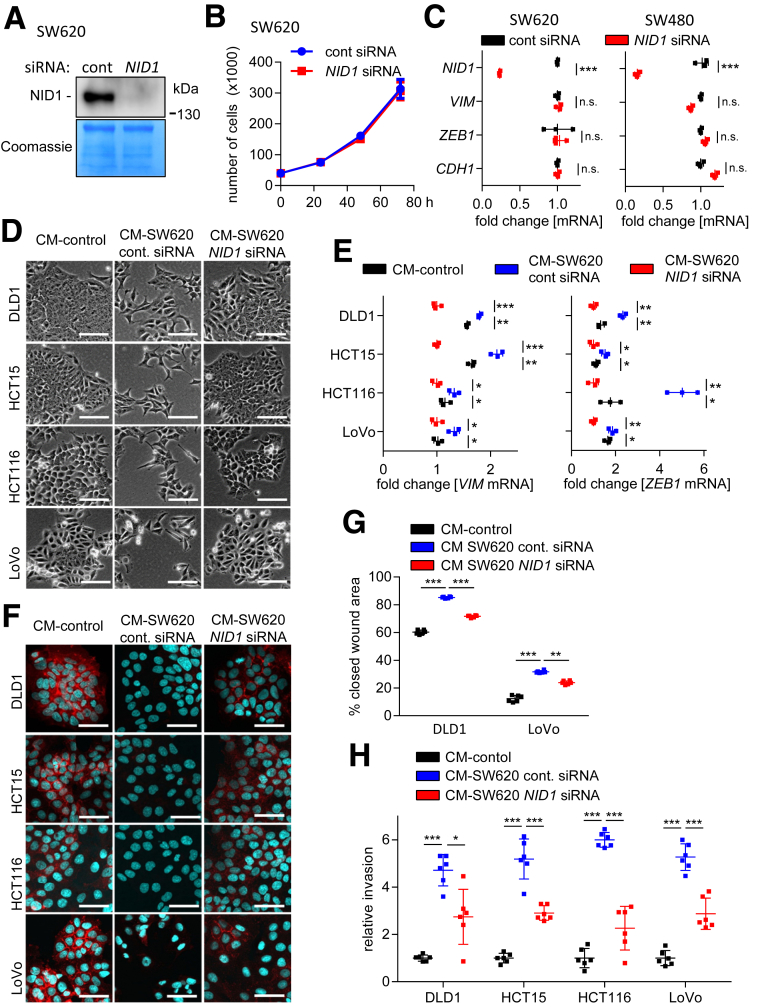
Next, we determined the relevance of NID1 for the induction of EMT by CM obtained from SW480/SW620 cells. First, we confirmed that NID1 protein expression is effectively repressed after transfection of NID1-specific small interfering RNA (siRNA) in SW620 cells (Figure 7A). NID1 knockdown did not affect proliferation of SW620 cells (Figure 7B). Furthermore, suppression of NID1 in SW480 and SW620 cells did not modulate the expression of the EMT-related genes ZEB1, VIM, and CDH1 in these cells (Figure 7C). Therefore, depletion of NID1 is not sufficient to induce mesenchymal-epithelial transition in established mesenchymal-like CRC cells. However, CM harvested from SW620 cells treated with NID1-specific siRNA was less potent in inducing EMT in DLD1, HCT15, HCT116, and LoVo cells as evidenced by the morphology and the expression of EMT markers when compared with treatment with CM from SW620 cells transfected with control siRNAs (Figure 7D–F). In addition, silencing of NID1 in SW620 cells significantly suppressed the ability of CM from these cells to enhance the migration and invasion of epithelial-like CRC cells (Figure 7G and H). Therefore, NID1 is a required mediator of the paracrine induction of EMT, invasion, and migration by mesenchymal-like CRC cells.
Figure 7.
Requirement of NID1 for CM-induced EMT, migration, and invasion. (A) Western blot analysis of NID1 protein in concentrated CM from SW620 cells transfected with control or NID1-specific siRNAs for 72 hours. Coomassie Blue staining was used as loading control. (B) Cell number of SW620 cells transfected with control or NID1-specific siRNAs at the indicated time points. (C) qPCR analysis of NID1 and EMT-related mRNAs ZEB1, VIM, and CDH1 in SW480 and SW620 cells transfected with control or NID1-specific siRNAs for 72 hours. (D) Phase-contrast images showing cell morphology of DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM from SW620 cells 72 hours after transfection with control or NID1-specific siRNAs. (E) qPCR analyses of VIM and ZEB1 in DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM collected from SW620 cells 72 hours after transfection with control or NID1 siRNA. (F) Immunofluorescence detections of E-cadherin in DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM collected from SW620 cells 72 hours after transfection with control or NID1 siRNA. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm. (G) Wound healing assay of DLD1 and LoVo cells treated with indicated CMs for 30 hours. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (H) Invasion assay in modified Boyden chambers: DLD1, HCT15, HCT116, and LoVo cells were seeded on Matrigel-coated filters with indicated CMs used as a chemoattractant in the lower well. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. Means ± SD of (B, C, and E) n = 3 biological replicates, and (G and H) n = 2 biological replicates (each n = 3 technical replicates) are provided. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001. cont, control.
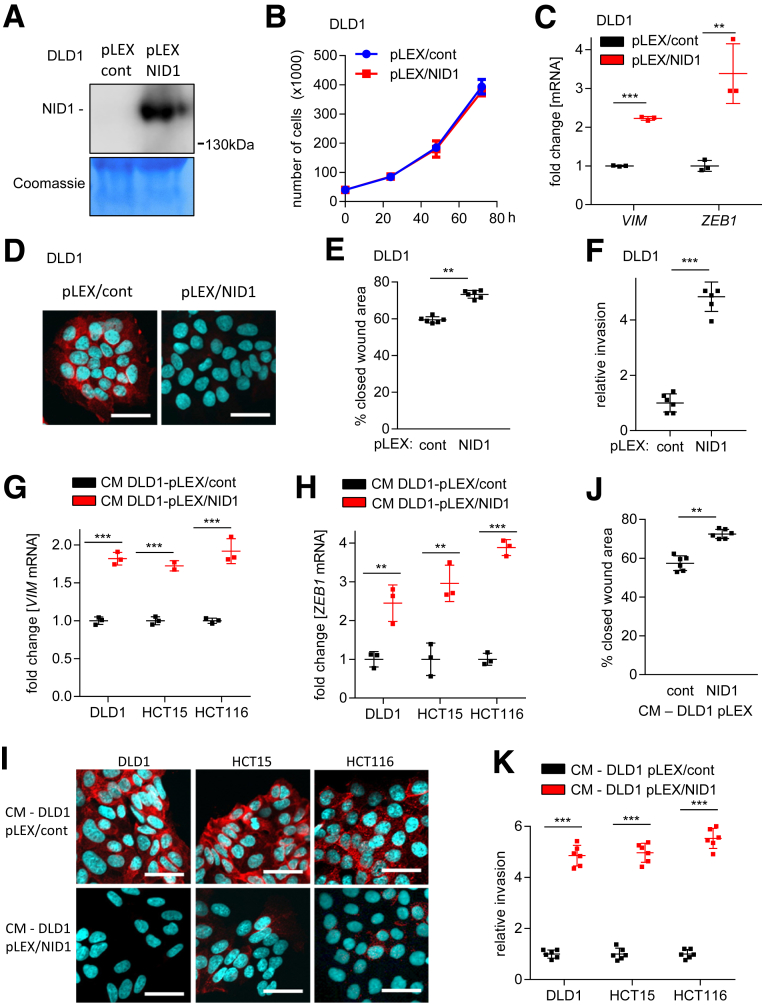
Ectopic NID1 Expression Induces EMT, Migration, and Invasion
Next, we analyzed whether NID1 is sufficient for induction of EMT, migration, and invasion in epithelial-like CRC cells. Therefore, we generated DLD1 cells (DLD1-pLEX/NID1) that stably express and secrete NID1 (Figure 8A). As a control, DLD1 harboring the empty pLEX vector were generated. Ectopic expression of NID1 did not affect proliferation of DLD1 cells (Figure 8B). However, ectopic NID1 expression resulted in increased expression of the VIM and ZEB1 mRNAs, and decreased expression of E-cadherin protein (Figure 8C and D). Moreover, DLD1-pLEX/NID1 cells showed increased migration and invasion (Figure 8E and F). Therefore, the ectopic NID1 produced by DLD1 cells presumably had autocrine effects. Importantly, CM from DLD1-pLEX/NID1 cells induced EMT in the epithelial-like CRC cells DLD1, HCT15, and HCT116 as evidenced by the expression of EMT markers (Figure 8G-I). Finally, epithelial-like CRC cells showed increased migration and invasion after treatment with CM from DLD1-pLEX/NID1 cells (Figure 8J and K). Therefore, secreted NID1 is sufficient to confer the paracrine induction of EMT.
Figure 8.
Ectopic NID1 expression induces EMT, migration, and invasion in an autocrine and paracrine manner. (A) Western blot analysis of NID1 protein in concentrated CM from DLD1 cells stably transfected with control (pLEX/cont) or NID1 expression (pLEX/NID1) vector. Coomassie Blue staining was used as a loading control. (B) Cell number of DLD1-pLEX/cont and DLD1-pLEX/NID1 cells at indicated time points. (C) qPCR analysis of EMT-related mRNAs VIM and ZEB1 in DLD1-pLEX/cont and DLD1-pLEX/NID1 cells. (D) Immunofluorescence detection of E-cadherin in DLD1-pLEX/cont and DLD1-pLEX/NID1 cells. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm. (E) Wound healing assay of DLD1-pLEX/cont and DLD1-pLEX/NID1 cells. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (F) Invasion assay in modified Boyden chambers: DLD1-pLEX/cont and DLD1-pLEX/NID1 cells were seeded on Matrigel-coated filters. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. (G and H) qPCR analysis of VIM and ZEB1 in DLD1, HCT15, and HCT116 cells treated with CM from DLD1-pLEX/cont and DLD1-pLEX/NID1 cells for 96 hours. (I) Immunofluorescence detection of E-cadherin in DLD1, HCT15, and HCT116 cells treated with CM from DLD1-pLEX/cont and DLD1-pLEX/NID1 cells for 96 hours. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm. (J) Wound healing assay of DLD1 cells treated with CM from DLD1-pLEX/cont and DLD1-pLEX/NID1 cells for 30 hours. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (K) Invasion assay in modified Boyden chambers: DLD1, HCT15, and HCT116 cells were seeded on Matrigel-coated filters with indicated CMs used as a chemoattractant in the lower well. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. Means ± SD of (B, C, G, and H) n = 3 biological replicates, and (E, F, J, and K) n = 2 biological replicates (each n = 3 technical replicates) are shown. (E, F, and J) Significance was determined using the Student t test and (C, G, H, and K) by 1-way analysis of variance with the Tukey multiple comparison post-test; **P < .01; ***P < .001. cont, control.
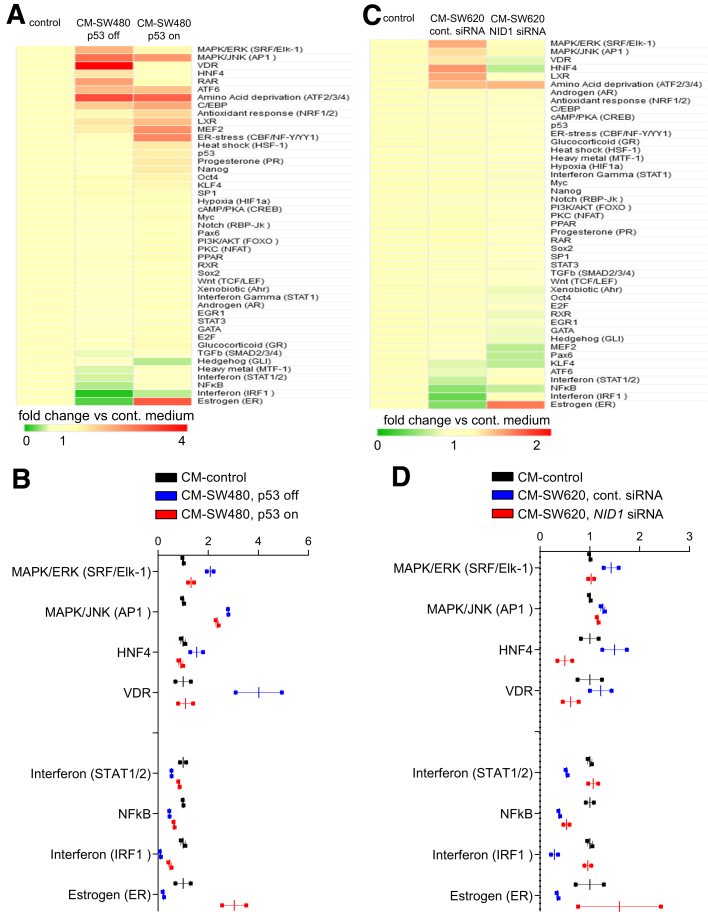
Changes in Signaling Pathways Induced by CM-Induced EMT
To determine which signaling pathways were affected in epithelial-like CRC cells exposed to CM from mesenchymal-like CRC cells and therefore presumably mediate the induction of EMT, we determined the activity of 45 signal transduction pathways using a reporter assay platform. First, we analyzed DLD1 cells treated with CM from SW480 with and without activation of ectopic p53 expression (Figure 9A and B). Next, we analyzed pathway activity in DLD1 cells treated with CM from SW620 cells transfected with control or NID1 siRNA (Figure 9C and D). Thereby, we found that mitogen-activated protein kinase (MAPK), HNF4, and vitamin D receptor signaling was induced, whereas interferon, nuclear factor-κB, and estrogen receptor signaling was repressed in DLD1 cells after exposure to CM collected from SW480 and SW620 cells. Ectopic expression of p53 in SW480 cells or knockdown of NID1 in SW620 cells prevented these CM-induced changes in DLD1 cells. Numerous studies have shown that the MAPK/extracellular signal-regulated kinase (ERK) signaling pathway induces EMT and promotes cancer progression.35 For example, activation of ERK2, which is an effector of MAPK-ERK signaling, increased the expression of the EMT-inducing transcription factors ZEB1 and ZEB2. Consequently, induction of ZEB1/2 by ERK2 resulted in repression of E-cadherin and induction of VIM in mammary epithelial cells MCF-10a.36, 37 Furthermore, it has been shown that in primary colorectal cancers, tumor cells with high MAPK activity reside specifically at the leading tumor edge, undergo EMT, and express markers related to CRC stem cells.38
Figure 9.
Signaling pathway activity in DLD1 cells treated with CM from mesenchymal-like cancer cells. (A and B) Analysis of 45 signal transduction pathways in DLD1 cells cultured for 48 hours in CM collected from SW480/pRTR-p53-VSV cells treated with DOX (p53 on) or without DOX (p53 off) for 72 hours. (A) Complete results of the assay are shown and (B) pathways showing significant changes are shown (means ± SEM, n = 2 technical replicates). (C and D) Analysis of 45 signal transduction pathways in DLD1 cells treated with CM from SW620 cells transfected with control or NID1 siRNA. (C) Complete results of the assay are shown and (D) pathways showing significant changes are shown (means ± SEM, n = 2 technical replicates). cont, control.
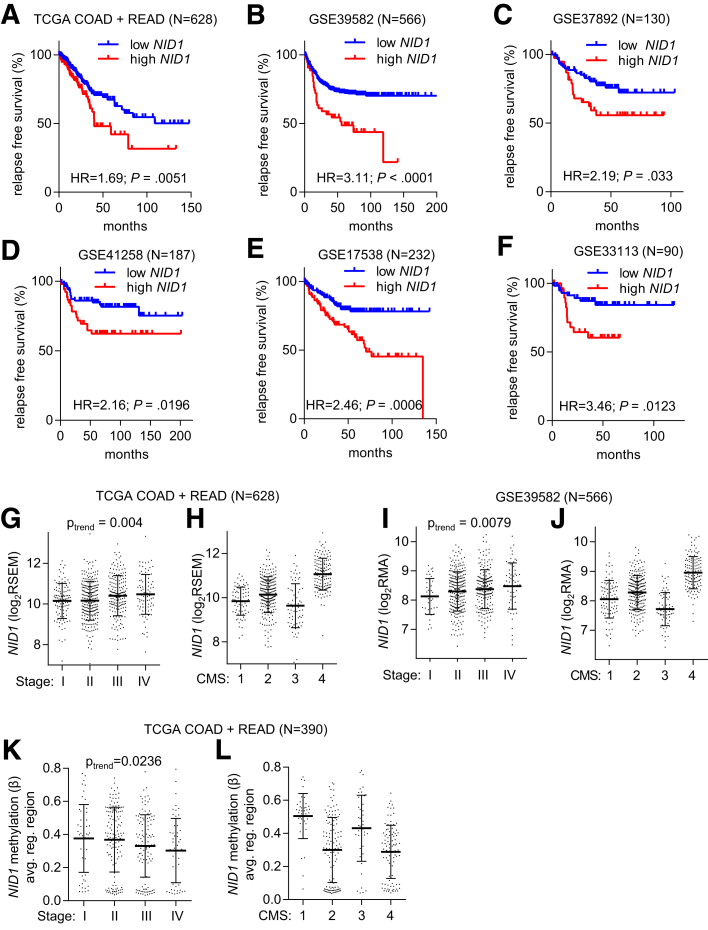
High NID1 Expression Is Associated With Poor Prognosis in CRC Patients
Finally, we analyzed, whether expression of NID1 is associated with clinical parameters in CRC patients. The expression of NID1 showed a positive correlation with poor relapse-free survival in 6 independent cohorts of CRC patients (Figure 10A–F). Moreover, NID1 expression was increased significantly from stage I to stage IV CRC tumors in a linear trend (Figure 10G and I). Recently, a consensus molecular subtype (CMS) classification was introduced that groups CRCs into 4 main subtypes.39 The expression of NID1 was highest in the CMS4 subtype, which represents CRCs with mesenchymal features and patients who show the poorest prognosis when compared with the other subtypes (Figure 10H and J). Finally, NID1 DNA methylation was highest in stage I and lowest in stage IV, as well as CMS4 colorectal tumors within the TCGA data set (Figure 10K and L). Therefore, the association of increased NID1 expression with a mesenchymal phenotype that promotes invasion and metastasis also is found in primary CRCs and therefore presumably is of prognostic and clinical relevance.
Figure 10.
Increased NID1 expression is associated with poor prognosis in CRC patients. (A–F) Associations of NID1 expression with relapse-free survival in the indicated CRC patient cohorts. The statistics were calculated using the log-rank test. (G–J) Associations of NID1 expression with (G and I) tumor stage and the (H and J) CMS of CRC in the (G and H) TCGA COAD + READ and the (I and J) GSE39582 CRC patient cohorts. (K and L) Associations of NID1 DNA methylation with (K) tumor stage and the (L) CMS in the TCGA COAD + READ patient cohort. (G–L) Individual data points and means ± SD are provided. (G, I, and K) Significance was determined using 1-way analysis of variance with a post-test for linear trend. RSEM, RNA-Seq by Expectation Maximization.
Discussion
CRCs originate from normal epithelial cells lining the colon. Therefore, early stage CRC cells show an epithelial-like phenotype. However, during tumor progression, single tumor cells acquire mesenchymal-like properties, which increase their migratory and invasive capacities. Here, we show that these mesenchymal-like CRC cells may act in a paracrine fashion by secreting NID1, which enhances the malignant properties of less progressed, epithelial-like tumor cells (for a schematic summary see the graphical abstract). The paracrine process described here might be involved in the accumulation of tumor cells with an increased capacity to invade and migrate at the invasion front. Importantly, we showed that p53 suppresses this paracrine signaling between tumor cells by negatively regulating the expression of NID1. Interestingly, NID1 was reported to have protumorigenic effects in other tumor entities previously: for example, NID1 induces EMT and promotes chemoresistance in ovarian cancer.40 Furthermore, NID1 promotes the formation of lung metastasis by breast cancer cells.41 NID1 is associated with the extracellular matrix and basement membranes, wherein it functions as a linker between laminins, collagens, and proteoglycans, and cell surface receptors to control cell polarization, migration, and invasion.42 NID1-deficient mice show impaired wound healing43 and NID1 is produced mainly by mesenchymal cells.44 It has been shown that inhibition of NID1 reduces tumor growth and metastasis in an endometrial cancer model45 and NID1 levels are increased in plasma samples from ovarian cancer patients.42, 46 Our findings suggest that NID1 may have similar functions and properties in CRC.
It has been reported that besides having cell-autonomous functions, p53 also affects the cellular secretome: activation of p53 in fibroblasts selectively induced apoptosis in p53-deficient cancer cells via secretion of the tumor-suppressor Par-4.25 Furthermore, p53-expressing senescent stellate cells released factors that skew macrophage polarization toward a tumor-suppressive M1 state.26 Thereby, p53 expressed in stromal cells had a paracrine, tumor-suppressive effect on tumor cells. Our data suggest that p53 activation within tumor cells also modulates the tumor microenvironment and that the loss of p53 function creates a protumorigenic environment, which enhances the malignant properties of neighboring tumor cells. The tumor-promoting effects of p53 loss are mediated by p53-regulated secreted factors, which are up-regulated after inactivation of p53 by mutation. Besides NID1, our cytokine array data suggest that platelet-derived growth factor A also might represent such a factor. Numerous studies showed that members of the platelet-derived growth factor family promote cancer progression.47, 48, 49, 50 On the other hand, wt p53 might repress tumor progression by inducing the expression and secretion of factors that suppress EMT and metastasis. Our results indicate that GDF15, which is a member of the TGFβ superfamily, might represent such a factor because it was present at decreased levels in CM from mesenchymal-like CRC cells and at increased levels in CM from SW480 cells after activation of p53.
It has been suggested that secreted factors, such as TGFβ, function as autocrine or paracrine signals that maintain the mesenchymal status of breast and kidney cells.51, 52 However, colorectal tumors frequently have impaired TGFβ signaling. Moreover, we did not observe differential expression of TGFB1, TGFB2, and TGFB3 ligands between epithelial- and mesenchymal-like CRC cells (data not shown). Therefore, other factors, such as NID1, presumably play a more important role in CRC-associated EMT.
CM derived from SW480 and SW620 cells induced EMT in DLD1, HCT15, HCT116, and LoVo, but not in the HT29 and Caco2 cell lines. Because HT29 and Caco2 are the most differentiated among the studied cell lines,53, 54 it is conceivable that a certain level of de-differentiation is necessary to make cells permissive for EMT induced by NID1. Alternatively, the receptors for NID1 or their downstream signaling cascades might be altered in HT29 and Caco2 cells. Further analyses are required to clarify these issues in the future.
Here, we showed that the expression of NID1 is repressed by DNA methylation on the transcriptional level, and by p53/miR-192/215 on the post-transcriptional level. Our results suggest that during tumor progression CRC cells secrete NID1 after mutational inactivation of p53 combined with demethylation of the NID1 gene. The secreted NID1 then induces EMT in neighboring tumor cells and thereby contributes to the progression of CRCs. The conversion of noninvasive epithelial cells to invasive, mesenchymal-like CRC cells mediated by factors present in conditioned medium of more malignant, mesenchymal-like CRC cells described here represents an experimental system that may facilitate the identification and characterization of additional secreted factors, which critically contribute to tumor progression.
Material and Methods
Cell Lines, Cell Culture, and Reagents
The colorectal cancer cell lines DLD1, HCT15, HCT116, LoVo, Caco2, HT29, SW480, and SW620 were maintained in McCoy’s 5A Medium (16600082; Thermo Fisher, Waltham, MA) containing 10% fetal bovine serum (FBS) (10270106; Thermo Fisher). All cells were cultivated in the presence of 100 U/mL penicillin and 0.1 mg/mL streptomycin. siRNAs (Thermo Fisher silencer siRNA: negative control [ID 4611] and NID1 [ID s9554]) were transfected at a final concentration of 10 nmol/L using Lipofectamine 2000 (11668027; Thermo Fisher) or HiPerfect transfection reagent (301705; Qiagen). DLD1 cells were stably transfected with pLEX/NID1 and pLEX/control plasmids (kindly provided by Yibin Kang, Princeton University, Princeton, NJ) by using the Lipofectamine LTX transfection reagent (15338100; Thermo Fisher) followed by selection with puromycin. CM was harvested from mesenchymal-like CRC cell lines and filtered through 0.22-μm filters to ensure that no cells were present in the CM. CM from parental epithelial cells was used as control. CMs were diluted 1:1 with fresh McCoy’s 5A medium containing 10% FBS to ensure adequate nutrients. Nutlin-3a (SML0580; Sigma, St. Louis, MO) was used at a final concentration of 10 μmol/L. 5-Aza-2’-deoxycytidine (A3656; Sigma) was used at a final concentration of 5 μmol/L.
RNA Isolation and qPCR
Total RNA was isolated using the Total RNA Isolation Kit (11828665001; Roche, Basel, Switzerland) according to the manufacturer’s instructions. For mRNA analyses, complementary DNA (cDNA) was generated from 1 μg total RNA per sample using the Verso cDNA synthesis kit (AB1453A; Thermo Fisher). Real-time qPCR was performed using the LightCycler 480 (Roche) and the Fast SYBR Green Master Mix (4309155; Thermo Fischer). The sequences of oligonucleotides used as qPCR primers are listed in Table 1. For analyses of mature miRNA expression, cDNA was generated and qPCR was performed using the miRCURY LNA RT kit (339340; Qiagen), the miRCURY LNA SYBR Green PCR kit (339345; Qiagen), and the following commercially available primers: SNORD48 (YP00203903; Qiagen), hsa-miR-192-5p (YP00204099; Qiagen, Hilden, Germany), and hsa-miR-215-5p (YP00204398; Qiagen). Expression was normalized to GAPDH for mRNAs and SNORD48 for miRNAs using the ΔΔCt method.55 Results are represented as fold induction of the treated/transfected condition compared with the control condition.
Table 1.
Oligonucleotides Used for qPCR
| Gene | Forward (5’–3’) | Reverse (5’–3’) |
|---|---|---|
| GAPDH | TGTTGCCATCAATGACCCCTT | CTCCACGACGTACTCAGCG |
| NID1 | ATCAGCAATCCTTGGCTCAC | CCTTGGGATTCCTCTGTTCA |
| CDH1 | CCCGGGACAACGTTTATTAC | GCTGGCTCAAGTCAAAGTCC |
| VIM | TACAGGAAGCTGCTGGAAGG | ACCAGAGGGAGTGAATCCAG |
| ZEB1 | TCAAAAGGAAGTCAATGGACAA | GTGCAGGAGGGACCTCTTTA |
| MDR1 | CAGGAAATCTATTTCAAGGTCTGC | CATCACCAAAGGACTCAGCA |
| CD44 | GCCTACTGCAAATCCAAACAC | GAAGCTCTGAGAATTACTCTGCTG |
| CD166 | CGTCTGCTCTTCTGCCTCTT | GTACGTCAAGTCGGCAAGGT |
Western Blot Analysis and Antibodies
Cell-lysates were collected in RIPA lysis buffer (50 mmol/L Tris/HCl, pH 8.0, 250 mmol/L NaCl, 1% NP40, 0.5% [wt/vol] sodium deoxycholate, 0.1% sodium dodecyl sulfate, complete mini protease [05892970001; Roche]) and phosphatase inhibitors (04906837001; Roche). Lysates were sonicated and centrifuged for 15 minutes at 4°C. For analyses of CM, cells were incubated with serum-free medium for 24 hours. CM was filtered through 0.22-μm filters to ensure that no cells were present. CM was concentrated approximately 50-fold using Amicon Ultra columns (50 kilodaltons, UFC205024; Merck Millipore, Burlington, MA). Proteins from whole-cell lysate or CM were separated on sodium dodecyl sulfate–acrylamide gels and transferred to Immobilon polyvinylidene difluoride membranes (IPVH00010; Merck Millipore). For immunodetection, membranes were incubated with the antibodies listed in Table 2. Signals from horseradish-peroxidase–coupled secondary antibodies were generated by Immobilon Western horseradish peroxidase Substrate (WBKLS0500; Merck Millipore) and recorded with a Charged Coupled Device camera (Odyssey Fc; LI-COR, Lincoln, NE). In Western blots from CM, membranes were stained with Coomassie Blue for loading control.
Table 2.
List of Antibodies
| Antibodies | Catalog number | Company | Use | Dilution | Source |
|---|---|---|---|---|---|
| Primary | |||||
| Epitope | |||||
| E-cadherin | 334000 | Thermo Fisher | IF | 1:50 | Mouse |
| Actin | A2066 | Sigma | WB | 1:10,000 | Rabbit |
| p53 | sc-126 | Santa Cruz (Dallas, TX) | WB | 1:1000 | Mouse |
| VSV | V4888 | Sigma | WB | 1:1000 | Rabbit |
| p21 | 556431 | BD | WB | 1:1000 | Mouse |
| NID1 | MAB2570 | R&D (Minneapolis, MN) | WB | 1:1000 | Mouse |
| Secondary or conjugates | |||||
| Anti-mouse HRP | W4021 | Promega | WB | 1:10,000 | Goat |
| Anti-rabbit HRP | A0545 | Sigma | WB | 1:10,000 | Goat |
| Alexa Flour 555, conjugated anti-mouse | A21422 | Thermo Fisher | IF | 1:500 | Goat |
HRP, horseradish peroxidase.
Boyden Chamber Invasion Assay
To analyze invasion, cell inserts (8.0-μm pore size membrane, 15718039; Corning, Corning, NY) were first coated with Matrigel (354230; BD Bioscience, Franklin Lakes, NJ) at a dilution of 3.3 ng/mL in medium without serum. Subsequently, 5 × 104 cells, previously deprived of serum (0.1%) for 24 hours, were seeded on Matrigel in the upper chamber in serum-free medium. As a chemoattractant, CM from mesenchymal-like CRC cells was placed in the lower chamber. After 48 hours, nonmotile cells at the top of the filter were removed and the cells in the bottom chamber were fixed with methanol and stained with 4′,6-diamidino-2-phenylindole (6335.1; Roth, Karlsruhe, Germany) and counted using immunofluorescence microscopy. Results represent the average number of cells in 5 fields per membrane in triplicate inserts.
Wound Healing Assay
Cells were cultured until they reached complete confluence. Mitomycin C (10 μg/mL, 089K0731; Sigma) was added 2 hours before generating a scratch using a pipette tip. After washing twice with Hank’s balanced salt solution to remove Mitomycin C and detached cells, conditioned medium was added. Images were captured on an Axiovert Observer Z.1 microscope connected to an AxioCam MRm camera using the Axiovision software (Zeiss, Oberkochen, Germany) at the respective time points.
Immunofluorescence and Confocal Laser-Scanning Microscopy
Cells cultivated on glass coverslides were fixed in 4% paraformaldehyde/phosphate-buffered saline for 10 minutes, permeabilized in 0.2% Triton X-100 (Sigma) for 20 minutes, and blocked in 100% FBS for 1 hour. Proteins were detected with the antibodies against E-cadherin (334000, 1:50 dilution; Thermo Fisher). Chromatin was stained by 4′,6-diamidino-2-phenylindole (6335.1; Roth). Specimens were covered with ProLong Gold antifade (P36931; Thermo Fisher). Confocal laser scanning microscopy images were captured with a LSM700 microscope using a Plan Apochromat 20×/0.8 M27 objective and ZEN 2009 software (Zeiss) with the following settings: image size, 2048 × 2048; 16 bit; pixel/dwell, 25.2 μs; pixel size, 0.31 μm; laser power, 2%; and master gain, 600–1000. After image capturing the original confocal laser scanning microscopy files were converted into TIFF files.
Cytokine Array
A human cytokine Antibody Array C Series 4000 (AAH-CYT-4000; RayBiotech, Norcross, GA) was used to determine the cytokines present in the media of CRC cell lines. After 48 hours of cell cultivation in 0.2% fetal calf serum medium, the conditioned medium was transferred to the array, and the analysis was performed according to the manufacturer’s instructions. Chemiluminescence signals were recorded with a CCD camera (Odyssey Fc, LI-COR) and quantified using densitometric analysis with LI-COR Image Studio v5.2 software. The assays were performed in 2 technical replicates within 1 biological sample.
Analyses of Signal Transduction Pathway Activity
To simultaneously analyze 45 signal transduction pathways, we used the Cignal 45-pathway reporter array (CCA-901L; Qiagen). DLD1 cells (20,000/well) were transfected with reporter plasmids using the Lipofectamine 2000 transfection reagent (11668027; Thermo Fisher). Twenty-four hours after transfection, medium was replaced with conditioned medium from SW480/pRTR-p53-VSV or SW620 cells. After 48 hours, luminescence intensities were measured with the Dual Luciferase Reporter assay (E1910; Promega, Madison, WI) using an Orion II luminometer (Berthold, Bad Wildbad, Germany).
Analysis of Expression and DNA Methylation Data From Public Databases
Expression, DNA methylation, and clinical data of the TCGA COAD and READ cohort was obtained from the TCGA data portal (gdc.cancer.gov)56 and the cBIO portal (www.cbioportal.org).57 For expression analyzes, the RNA sequencing by expectation-maximization normalized expression values from the Illumina (San Diego, CA) RNASeqV2 (genes) data sets were used, and for methylation analyzes, β methylation values ranging from 0 (nonmethylated) to 1 (100% methylation), determined by 450K BeadChip arrays (Illumina), were used. Expression data of cancer cell lines was obtained from The Cancer Cell line Encyclopedia (www.broadinstitute.org/ccle/home).58 For expression analyzes, the reads per kilobase of transcript, per million mapped reads values obtained by RNA sequencing were used and for methylation analyzes, β methylation values (determined by reduced representation bisulfite sequencing) were used. Expression and clinical data of GSE39582, GSE37892, GSE41258, GSE17538, and GSE33113 data sets were downloaded from NCBI GEO (www.ncbi.nlm.nih.gov/geo). The CMS classification of TCGA and GSE39582 samples was obtained from Synapse (www.synapse.org; syn2623706). Correlations between NID1 and miRNA expression, and between NID1 expression and DNA methylation were calculated using the Spearman correlation coefficient. The statistics for survival curves was calculated by the log-rank test. For binary classification of cases (high/low expression), receiver operated characteristics curve analysis was used to determine optimal cut-off values.59 Associations between expression/methylation and tumor TNM stage were calculated by 1-way analysis of variance with a post-test for linear trend.
Statistical Analysis
qPCR, migration, invasion, and cell growth data are presented as means ± SD. The differences between 2 groups were analyzed by a 2-tailed Student t test. One-way analysis of variance with the Tukey multiple comparison post-test was used to compare more than 2 groups. Calculations were performed using Prism7 (GraphPad Software, Inc, San Diego, CA) and P values ≤ .05 were considered significant.
Acknowledgments
The authors thank Bert Vogelstein (Johns Hopkins Medical School, Baltimore, MD) for providing the HCT116 p53 knockout cell line and Yibin Kang (Princeton University, Princeton, NJ) for providing pLEX/NID1 and pLEX/control plasmids.
Footnotes
Author contributions Matjaz Rokavec and Nassim Bouznad performed the experiments and analyzed data; Heiko Hermeking conceived the concept of the study, and Matjaz Rokavec and Heiko Hermeking designed the analyses and wrote the paper.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grant 2017.050.1 from Wilhelm-Sander Stiftung (H.H.).
References
- 1.Nguyen D.X., Bos P.D., Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 2.Tam W.L., Weinberg R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena M., Stephens M.A., Pathak H., Rangarajan A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011;2:e179. doi: 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., Curtis C. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji T., Ibaragi S., Hu G.F. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135–7139. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calbo J., van Montfort E., Proost N., van Drunen E., Beverloo H.B., Meuwissen R., Berns A. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–256. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Punt C.J., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 11.Joung J.G., Oh B.Y., Hong H.K., Al-Khalidi H., Al-Alem F., Lee H.O., Bae J.S., Kim J., Cha H.U., Alotaibi M., Cho Y.B., Hassanain M., Park W.Y., Lee W.Y. Tumor heterogeneity predicts metastatic potential in colorectal cancer. Clin Cancer Res. 2017;23:7209–7216. doi: 10.1158/1078-0432.CCR-17-0306. [DOI] [PubMed] [Google Scholar]
- 12.Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller P.A., Vousden K.H., Norman J.C. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell E., Piwnica-Worms D., Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4:405–414. doi: 10.1158/2159-8290.CD-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanrion M., Kuperstein I., Barriere C., El Marjou F., Cohen D., Vignjevic D., Stimmer L., Paul-Gilloteaux P., Bieche I., Tavares Sdos R., Boccia G.F., Cacheux W., Meseure D., Fre S., Martignetti L., Legoix-Ne P., Girard E., Fetler L., Barillot E., Louvard D., Zinovyev A., Robine S. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat Commun. 2014;5:5005. doi: 10.1038/ncomms6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Rokavec M., Oner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., Slotta-Huspenina J., Bader F.G., Greten F.R., Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palena C., Hamilton D.H., Fernando R.I. Influence of IL-8 on the epithelial-mesenchymal transition and the tumor microenvironment. Future Oncol. 2012;8:713–722. doi: 10.2217/fon.12.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin S., Levine A.J. The p53 functional circuit. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 20.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 23.Charni M., Molchadsky A., Goldstein I., Solomon H., Tal P., Goldfinger N., Yang P., Porat Z., Lozano G., Rotter V. Novel p53 target genes secreted by the liver are involved in non-cell-autonomous regulation. Cell Death Differ. 2016;23:509–520. doi: 10.1038/cdd.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bar J., Moskovits N., Oren M. Involvement of stromal p53 in tumor-stroma interactions. Semin Cell Dev Biol. 2010;21:47–54. doi: 10.1016/j.semcdb.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burikhanov R., Shrestha-Bhattarai T., Hebbar N., Qiu S., Zhao Y., Zambetti G.P., Rangnekar V.M. Paracrine apoptotic effect of p53 mediated by tumor suppressor Par-4. Cell Rep. 2014;6:271–277. doi: 10.1016/j.celrep.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lujambio A., Akkari L., Simon J., Grace D., Tschaharganeh D.F., Bolden J.E., Zhao Z., Thapar V., Joyce J.A., Krizhanovsky V., Lowe S.W. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khwaja F.W., Svoboda P., Reed M., Pohl J., Pyrzynska B., Van Meir E.G. Proteomic identification of the wt-p53-regulated tumor cell secretome. Oncogene. 2006;25:7650–7661. doi: 10.1038/sj.onc.1209969. [DOI] [PubMed] [Google Scholar]
- 28.Hahn S., Jackstadt R., Siemens H., Hunten S., Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callaghan R., Luk F., Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab Dispos. 2014;42:623–631. doi: 10.1124/dmd.113.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunten S., Siemens H., Kaller M., Hermeking H. The p53/microRNA network in cancer: experimental and bioinformatics approaches. Adv Exp Med Biol. 2013;774:77–101. doi: 10.1007/978-94-007-5590-1_5. [DOI] [PubMed] [Google Scholar]
- 31.Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16:116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rokavec M., Kaller M., Horst D., Hermeking H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci Rep. 2017;7:4687. doi: 10.1038/s41598-017-04234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunten S., Kaller M., Drepper F., Oeljeklaus S., Bonfert T., Erhard F., Dueck A., Eichner N., Friedel C.C., Meister G., Zimmer R., Warscheid B., Hermeking H. p53-regulated networks of protein, mRNA, miRNA, and lncRNA expression revealed by integrated pulsed stable isotope labeling with amino acids in cell culture (pSILAC) and next generation sequencing (NGS) analyses. Mol Cell Proteomics. 2015;14:2609–2629. doi: 10.1074/mcp.M115.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhillon A.S., Hagan S., Rath O., Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 36.Shin S., Dimitri C.A., Yoon S.O., Dowdle W., Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsum I.A., Martin C., Humbert P.O. Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J Cell Sci. 2013;126:3990–3999. doi: 10.1242/jcs.129387. [DOI] [PubMed] [Google Scholar]
- 38.Blaj C., Schmidt E.M., Lamprecht S., Hermeking H., Jung A., Kirchner T., Horst D. Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Cancer Res. 2017;77:1763–1774. doi: 10.1158/0008-5472.CAN-16-2821. [DOI] [PubMed] [Google Scholar]
- 39.Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., Bot B.M., Morris J.S., Simon I.M., Gerster S., Fessler E., De Sousa E.M.F., Missiaglia E., Ramay H., Barras D., Homicsko K., Maru D., Manyam G.C., Broom B., Boige V., Perez-Villamil B., Laderas T., Salazar R., Gray J.W., Hanahan D., Tabernero J., Bernards R., Friend S.H., Laurent-Puig P., Medema J.P., Sadanandam A., Wessels L., Delorenzi M., Kopetz S., Vermeulen L., Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y., Zhu Y., Fan X., Zhang C., Wang Y., Zhang L., Zhang H., Wen T., Zhang K., Huo X., Jiang X., Bu Y., Zhang Y. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget. 2017;8:33110–33121. doi: 10.18632/oncotarget.16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aleckovic M., Wei Y., LeRoy G., Sidoli S., Liu D.D., Garcia B.A., Kang Y. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017;31:1439–1455. doi: 10.1101/gad.301937.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L., Zhang Y., Li N., Feng L., Yao H., Zhang R., Li B., Li X., Han N., Gao Y., Xiao T., Wu L. Nidogen-1: a candidate biomarker for ovarian serous cancer. Jpn J Clin Oncol. 2015;45:176–182. doi: 10.1093/jjco/hyu187. [DOI] [PubMed] [Google Scholar]
- 43.Baranowsky A., Mokkapati S., Bechtel M., Krugel J., Miosge N., Wickenhauser C., Smyth N., Nischt R. Impaired wound healing in mice lacking the basement membrane protein nidogen 1. Matrix Biol. 2010;29:15–21. doi: 10.1016/j.matbio.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Ekblom P., Ekblom M., Fecker L., Klein G., Zhang H.Y., Kadoya Y., Chu M.L., Mayer U., Timpl R. Role of mesenchymal nidogen for epithelial morphogenesis in vitro. Development. 1994;120:2003–2014. doi: 10.1242/dev.120.7.2003. [DOI] [PubMed] [Google Scholar]
- 45.Pedrola N., Devis L., Llaurado M., Campoy I., Martinez-Garcia E., Garcia M., Muinelo-Romay L., Alonso-Alconada L., Abal M., Alameda F., Mancebo G., Carreras R., Castellvi J., Cabrera S., Gil-Moreno A., Matias-Guiu X., Iovanna J.L., Colas E., Reventos J., Ruiz A. Nidogen 1 and nuclear protein 1: novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin Exp Metastasis. 2015;32:467–478. doi: 10.1007/s10585-015-9720-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Xu B., Liu Y., Yao H., Lu N., Li B., Gao J., Guo S., Han N., Qi J., Zhang K., Cheng S., Wang H., Zhang X., Xiao T., Wu L., Gao Y. The ovarian cancer-derived secretory/releasing proteome: a repertoire of tumor markers. Proteomics. 2012;12:1883–1891. doi: 10.1002/pmic.201100654. [DOI] [PubMed] [Google Scholar]
- 47.Chen J., Yuan W., Wu L., Tang Q., Xia Q., Ji J., Liu Z., Ma Z., Zhou Z., Cheng Y., Shu X. PDGF-D promotes cell growth, aggressiveness, angiogenesis and EMT transformation of colorectal cancer by activation of Notch1/Twist1 pathway. Oncotarget. 2017;8:9961–9973. doi: 10.18632/oncotarget.14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H., Sun J.D., Yan L.J., Zhao X.P. PDGF-D/PDGFRbeta promotes tongue squamous carcinoma cell (TSCC) progression via activating p38/AKT/ERK/EMT signal pathway. Biochem Biophys Res Commun. 2016;478:845–851. doi: 10.1016/j.bbrc.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 49.Wang R., Li Y., Hou Y., Yang Q., Chen S., Wang X., Wang Z., Yang Y., Chen C., Wu Q. The PDGF-D/miR-106a/Twist1 pathway orchestrates epithelial-mesenchymal transition in gemcitabine resistance hepatoma cells. Oncotarget. 2015;6:7000–7010. doi: 10.18632/oncotarget.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Q., Hou X., Xia J., Qian X., Miele L., Sarkar F.H., Wang Z. Emerging roles of PDGF-D in EMT progression during tumorigenesis. Cancer Treat Rev. 2013;39:640–646. doi: 10.1016/j.ctrv.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L., Weinberg R.A. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gregory P.A., Bracken C.P., Smith E., Bert A.G., Wright J.A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y.Y., Lindeman G.J., Shannon M.F., Drew P.A., Khew-Goodall Y., Goodall G.J. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–1040. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 54.Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D., Tomlinson I.P., Bodmer W.F., Mariadason J.M., Sieber O.M. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.K., Yu J., Aspesi P., Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Budczies J., Klauschen F., Sinn B.V., Gyorffy B., Schmitt W.D., Darb-Esfahani S., Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]