Abstract
Regulation of rDNA transcription depends on the formation and dissociation of a functional complex between RNA polymerase I (pol I) and transcription initiation factor Rrn3p. We analyzed whether phosphorylation is involved in this molecular switch. Rrn3p is a phosphoprotein that is predominantly phosphorylated in vivo when it is not bound to pol I. In vitro, Rrn3p is able both to associate with pol I and to enter the transcription cycle in its nonphosphorylated form. By contrast, phosphorylation of pol I is required to form a stable pol I-Rrn3p complex for efficient transcription initiation. Furthermore, association of pol I with Rrn3p correlates with a change in the phosphorylation state of pol I in vivo. We suggest that phosphorylation at specific sites of pol I is a prerequisite for proper transcription initiation and that phosphorylation/dephosphorylation of pol I is one possibility to modulate cellular rDNA transcription activity.
Precise regulation of rRNA synthesis according to environmental conditions plays a key role in ribosome biosynthesis, which is tightly controlled through all prokaryotic and eukaryotic organisms (1, 2). The level of rRNA production correlates with the activity of the RNA polymerase I (pol I)-containing transcription machinery that generates the precursor transcript of the mature 18S, 25S, and 5.8S rRNA. Transcription efficiency depends on the formation of active initiation complexes on the rDNA promoter, which requires several transcription initiation factors and the recruitment of initiation competent pol I. In the yeast Saccharomyces cerevisiae, the first pol I-dependent initiation factor identified was the TATA binding protein TBP (3, 4), which could be isolated within a stable complex of 240 kDa (5). TBP contacts polypeptides of two multiprotein complexes, the core factor (CF; refs. 6, 7) and the upstream activating factor (UAF; refs. 6 and 8). One crucial role of TBP in pol I transcription is to recruit the pol I machinery to the promoter in a manner completely dependent on UAF (6, 8, 9). CF is composed of three stably associated proteins encoded by the genes RRN6, RRN7, and RRN11 (7, 10, 11) whereas UAF is a multisubunit complex consisting of at least five different proteins: Rrn5p, Rrn7p, Rrn10p (12), and the histones H3 and H4 (13). It has been proposed that binding of UAF upstream to the promoter is necessary to efficiently recruit CF and, finally, the initiation active form of pol I to the start site of rRNA synthesis (8). Initiation-competent pol I is stably associated with the pol I-specific initiation factor Rrn3p (14, 15). By binding the C-terminal part of Rrn6p and the pol I-specific subunit A43, Rrn3p connects CF to pol I, which results in recruitment of pol I to the promoter and in the formation of an active initiation complex (16). Recently, it was suggested that pol I-Rrn3p, together with CF, cycles on and off the promoter, whereas UAF remains bound (17). One possibility to regulate transcription initiation is the formation and dissociation of the pol I-Rrn3p complex, which represents a molecular switch to regulate rRNA synthesis. This idea was supported by the following previously published data (15). First, stable association of pol I with Rrn3p in solution is a necessary prerequisite to initiate transcription; second, only a minor proportion of both Rrn3p and pol I were found in a stable complex with each other in whole cell extracts, whereas most of both pol I and Rrn3p were uncomplexed and were found to be inactive in transcription initiation; third, the pol I-Rrn3p complex dissociated during one round of transcription in vitro and lost its capacity for subsequent reinitiation; and fourth, stationary cells, which are silent in rDNA transcription, show a strong reduction of pol I-Rrn3p complexes; however, they contain substantial levels of both free Rrn3p and pol I (15). Because both pol I and Rrn3p exist in parallel as uncomplexed and as stably associated molecules, it is conceivable that different posttranslational modifications have to exist in either one or both components to trigger initiation activity.
Recently, the human homolog of yRrn3p has been identified. It is able to replace yeast Rrn3p in vivo (18) and was shown to be identical with TIF-IA, the factor mediating growth-dependent control of mammalian ribosomal RNA synthesis (19). In accordance with its yeast counterpart, human Rrn3p was found to be essential to recruit initiation-competent pol I to the promoter, thereby linking human pol I to the promoter selectivity factor SL1 (20). Similar to the situation in yeast, only a minor proportion of human pol I complexes were found in stable association with hRrn3p to result in an initiation-competent enzyme (20). These findings suggest that the reversible interaction between Rrn3p and pol I by posttranslational modifications might reflect a conserved basal mechanism to regulate rDNA transcription in eukaryotes.
Phosphorylation is a common posttranslational modification that is involved in many regulatory pathways. Because previous results demonstrated that the pol I subunits A190, A43, A34.5, A23, and A19 are phosphorylated in vivo (21, 22), we asked whether phosphorylation of pol I is a possible target that could modulate the interaction between pol I and Rrn3p. In deed, association of pol I with Rrn3p is paralleled by a change in the pol I phosphorylation pattern, suggesting a transition in the phosphorylation state of pol I to reach initiation competence. Dephosphorylation of pol I in vitro reduced initiation activity of yeast pol I, resulted in destabilization of the preformed pol I-Rrn3p complex, and inhibited complex formation with bacterially expressed Rrn3p. By contrast, initiation competence of Rrn3p did not require its phosphorylation. From these results, we propose that phosphorylation of pol I is one possibility to modulate pol I-Rrn3p interaction and, thus, to control activity of the transcription process.
Materials and Methods
Strains and Templates.
Yeast wild-type strain BJ926, strain NOY657 containing hemagglutinin antigen (HA)-tagged Rrn3p (23), and strain RRN3-ProtA were used for preparation of the extracts, subsequent fractionation, and purification of the transcription activities. Strain RRN3-ProtA was constructed by genomic integration of ProteinA-epitope (ProtA) with a preceding TEV cleavage site in frame with RRN3 as described (24).(Oligonucleotides used: Rrn3-Modu5′ and Rrn3-Modu3′.)
Preparation of the Transcriptionally Active Fractions.
Fraction PA600 (protein concentration 2.5–5 mg/ml) and the fractions required for reconstitution of transcription (fraction B600 and TBP-cpl), as well as fraction B2000, were generated on a large scale according to refs. 5 and 25. Homogenous RNA polymerase I that did not contain Rrn3p (pol I-A) was purified according to refs. 36 and 37. Initiation competent pol I (pol I-Rrn3p complex) was purified as described (5).
In Vitro Transcription.
Promoter-dependent transcription reactions were performed as described elsewhere (5). If adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) was used for the transcription reaction, ATP in the transcription buffer was replaced by AMP-PNP (Roche) at a final concentration of 0.2 mM.
Immunoaffinity Purification of pol I-Rrn3p-ProtA Complex.
Protein A purification of Rrn3p was performed as described previously (26) except that 20 ml whole cell extract was applied to a 0.1-ml IgG-Sepharose column. If pol I-Rrn3p-ProtA complex was purified from the transcriptionally active fraction PA600, 0.1 ml IgG Sepharose beads were incubated with 1 ml of fraction PA600 in the presence of 20% glycerol, 20 mM Hepes (pH 7.8), 2 mM MgCl2, 0.02 mM EDTA, 1 mM DTT, 1 mM PMSF, 2 mM Benzamidine, and 600 mM potassium acetate. The beads were washed three times with the same buffer except that 1.5 M potassium acetate was included followed by a wash step of 20 mM Hepes (pH 7.8). To generate fraction PA600 depleted of pol I-Rrn3p-ProtA, two rounds of incubation with 0.1 ml IgG Sepharose beads were performed.
Immunoaffinity Purification of pol I and HA-Tagged Rrn3p.
HA-tagged Rrn3p and pol I-bound Rrn3p were immunoprecipitated via α-HA antibodies and α-pol I antibodies that were cross-linked to ProtA-Sepharose beads according to ref. 27.
In Vivo 33P Labeling of Yeast Cells.
Five hundred millimeter yeast cultures were grown in yeast extract/peptone/dextrose (YPD) medium depleted from inorganic phosphate to an OD600 between 1 and 2. Phosphate-depleted medium was generated as described elsewhere (28) and supplied with 2 mCi of 33PO4 (Amersham Pharmacia). Cells were harvested by centrifugation in a Haereus (Kendro Hanau, Germany) centrifuge at 4000 rpm for 10 min. The pellet was resuspended in the same volume of ice-cold breaking buffer (20% glycerol/400 mM NH4SO4/150 mM Hepes, pH 8/10 mM MgCl2/5 mM β-Mercaptoethanol/1 mM PMSF). Cells were broken with precooled glass beads for 45 min on a IKA-Vibrax (Staufen, Germany) shaker at 1800 rpm at 4°C. Glass beads were pelleted in a tabletop centrifuge for 5 min at 13,000 rpm, and the supernatant was centrifuged in a tabletop centrifuge for 1 h at 40,000 rpm to remove cell remnants.
The resulting whole cell extracts were adjusted to 600 mM potassium acetate, 0.5% Nonidet P-40, 0.05% Triton X-100, and supplied with 50 μl IgG beads or ProtA-Sepharose beads that have been prepared as followed. Beads were preincubated for 15 min with 20 mM Hepes, 0.5% Nonidet P-40, and 10 mg/ml milk powder. After this preincubation, the beads were washed two times with 20 mM Hepes, 0.5% Nonidet P-40, and 10 mg/ml milk powder, two times with 1.5 M potassium acetate, 0.5% Nonidet P-40, 10 mg/ml milk powder, and further preincubated for 1 h with a whole cell extract of wild-type yeast in the presence of 600 mM potassium acetate, 0.5% Nonidet P-40, and 0.05% Triton X-100. The whole cell extracts of the epitope-tagged strains were incubated for 1 h at 4°C with IgG beads. After immunoprecipitation, the beads were washed two times with buffer TST (150 mM NaCl/50 mM Tris, pH 7.5/0.05% Triton X-100/1% PMSF), three times with 5 mM NH4Ac (pH 5), three times with 1.5 M potassium acetate, 0.5% Nonidet P-40 followed by a further wash step with TST. Beads were suspended in SDS sample buffer and analyzed by SDS/PAGE.
Cloning and Purification of Bacterially Expressed Rrn3p.
Primers were constructed that introduced a BspHI cleavage site 5′ in front of the N terminus (Rrn3-ProtA-F1) and a HindIII cleavage site 3′ of the C terminus of yeast ProtA-Rrn3p (Rrn3-ProtA-R1). ProtA-Rrn3p was PCR amplified from genomic DNA of a yeast strain containing a chromosomally integrated TEV-ProtA-7His-Rrn3p by using Rrn3-ProtA-F1 and Rrn3-ProtA-R1 as primers. The PCR product was digested with BspHI, and the resulting N terminus of the protein was introduced in a pQE60 expressing vector (Qiagen, Chatsworth, CA) that was opened with NcoI. To insert the C terminus of ProtA-Rrn3p, the PCR-product was digested with EcoRV and HindIII and cloned into a pQE60-expressing vector containing the previously introduced N terminus of ProtA-Rrn3p. The resulting DNA fragments were sequenced after each cloning step.
After transformation M15 Escherichia coli cells were grown at 16°C in 2× TY medium supplemented with ampicillin (50 μg/ml) and kanamycin (25 μg/ml) to an OD600 of 0.5. Expression of Rrn3p was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 40 min. Cells were harvested at 7,000 rpm for 10 min and resuspended in breaking buffer (20% glycerol/100 mM Tris⋅HCl, pH 8/400 mM KOAc/1 mM EDTA/5 mM Mg(OAc)2/5 mM β-mercaptoethanol/0.1% Tween 20) supplemented with 1 mM PMSF. Cells were lysed by using a cell disrupter. The resulting whole cell extract was cleared by centrifugation (100,000 × g, 60 min) and loaded on a ResourceQ-column equilibrated with Q-buffer (20% glycerol/20 mM Tris⋅HCl, pH 8/5 mM Mg(OAc)2/0.1% Tween 20) supplemented with 400 mM KOAc. After washing with the same buffer, proteins were eluted with 90 ml of a linear gradient from 400 mM to 2 M potassium acetate in Q-buffer at a flow rate of 2 ml/min. Rrn3p-containing fractions were analyzed by Western blotting, pooled, adjusted to 5 mM imidazole, and loaded on a 1.5 ml NTA-Ni-column equilibrated with Ni-buffer (20% glycerol/20 mM Tris⋅HCl, pH 8/500 mM KOAc/5 mM Mg(OAc)2/0.1% Tween 20) containing 5 mM imidazole. The column was subsequently washed with 15 ml Ni-buffer containing 5 mM and 20 mM imidazole, respectively, and proteins were eluted in 0.5-ml steps with Ni-buffer containing 250 mM imidazole. Rrn3p-containing fractions were pooled and loaded on a MonoQ-column equilibrated with Q-buffer containing 400 mM KOAc and 5 mM β-mercaptoethanol. Proteins were eluted with 20 ml of a linear gradient from 400 mM to 2 M KOAc in Q-buffer (flow rate 0.25 ml/min).
Interaction of pol I-A with Bacterially Expressed Rrn3p.
Two micrograms of pol I-A (3-fold molar excess) were incubated for 30 min at room temperature with 75 ng purified bRrn3p-ProtA in a total volume of 17.5 μl 20 mM Hepes (pH 7.8), 1 mM MgCl2, and 50 mM NH4 Cl2. Various amounts of the fraction were directly tested in the in vitro assay. ProtA-tagged Rrn3p was affinity purified with IgG-Sepharose under stringent conditions: 20 μl human IgG-Sepharose (Amersham Pharmacia) were prepared as described in the section about in vivo labeling of Rrn3p. The beads were added to the preincubated fraction, the total volume was adjusted to 0.5 ml of 20 mM Hepes (pH 7.8), 600 mM potassium acetate, and 0.5% Nonidet P-40, and the suspension was incubated at 8°C on a turning wheel. Beads were further processed as described above (in vivo labeling).
Phosphatase Treatment.
Alkaline phosphatase covalently attached to agarose beads (Sigma) was washed 2 times with 20 mM Hepes (pH 7.8) and 1 mM MgCl2. Either 40 μl fraction PA600 were incubated with 20 μl beads (14 U) phosphatase or 20 μl pol I-A (0.4 mg/ml), 25 μl purified yeast pol I-Rrn3p complex or 20 μl bRrn3p (10 ng/μl) were incubated with 10 ml beads (7 units) phosphatase for 30 min at 37°C. The beads were pelletted twice, and the supernatants were carefully removed with capillar pipette tips. If pol I-Rrn3p was attached to IgG-Sepharose, 10-μl beads were incubated with soluble alkaline phosphatase (Roche) 40 units for 15 min or 30 min at 37°C. In control reactions, phosphatase was inhibited either by heat denaturation (15 min 95°C) or by addition of a mixture of phosphatase inhibitors (final concentration 10 mM NaF, 5 mM sodium pyrophosphate, and 0.5 mM EDTA).
Oligonucleotides.
Rrn3-ProtA-F1: 5′-TTTTTTTCATGATGGCTTTTGAGAATACAAGT-3′. Rrn3-ProtA-R1: 5′-TTTTTTAAGCTTAGAAGTGGCGCGCCCTAGTGATG-3′. Rrn3-Modu5′: 5′-TGGAGTGAAGCAAGCGGGGAATATGAAAGTGATGGGTCGGATGACCGTACGCTGCAGGTAGAC-3′. Rrn3-Modu3′: 5′-TAGTTTGTGACGGGCATGTCTCGAAGATACCTATGAAAAAAGACCATCGATGAATTCGAGCTCG-3′.
Results
Phosphorylation Is Required To Maintain a Stable and Transcriptionally Active pol I-Rrn3p Complex.
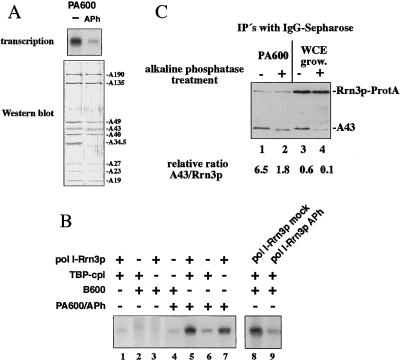
If phosphorylation of certain proteins is involved in the process of accurate transcription, dephosphorylation of pol I and its transcription factors should affect rRNA synthesis in vitro. We have recently reported the isolation of a subnucleolar structure containing all pol I-dependent transcription factors required for efficient transcription initiation in vitro (27). Treatment of this macromolecular complex (fraction PA600) with immobilized alkaline phosphatase resulted in a strong reduction of initiation activity (Fig. 1A). Western blot analysis of pol I before and after treatment with phosphatase showed a similar distribution of pol I-subunits (except subunit A34.5), indicating that same amounts of most pol I subunits were still present after the dephosphorylation procedure. To find out which component of the initiation complex was functionally affected by the dephosphorylation reaction, the different fractions required to reconstitute transcription in vitro were subsequently added back to the phosphatase-treated fraction PA600. We have previously published that efficient reconstitution of transcription depended on three different activities (5, 15): fraction B600, a TBP-containing 240-kDa protein complex, and the pol I-Rrn3p complex. When the purified pol I-Rrn3p complex was added back to the dephosphorylated initiation complex, transcriptional activity was restored (Fig. 1B, lanes 5 and 7). Addition of neither fraction B600 nor TBP-complex alone significantly stimulated the activity (Fig. 1B, lanes 4 and 6). These results indicate that phosphorylation of one or more components in the pol I-Rrn3p complex is needed for efficient accurate transcription. To directly demonstrate that pol I-Rrn3p is the target of the dephosphorylation reaction, the purified pol I-Rrn3p complex was treated with active or inactivated immobilized alkaline phosphatase and tested in the reconstituted transcription assay. As depicted in lanes 8 and 9 of Fig. 1B, initiation activity of the phosphatase-treated pol I-Rrn3p complex was strongly reduced, but not when heat inactivated phosphatase was used. Because formation and dissociation of the pol I-Rrn3p complex was shown to be crucial to trigger rRNA synthesis (15), we wondered whether dephosphorylation of the pol I-Rrn3p complex was a possibility to dissociate Rrn3p from pol I. We took advantage of a yeast strain expressing a chromosomal encoded Rrn3p-ProtA fusion protein and used IgG-Sepharose to purify immobilized pol I-Rrn3p-ProtA complexes from both whole cell extracts and the transcriptionally active fraction PA600 (Fig. 1C, lanes 1, 3). Our coimmunoprecipitation experiments confirmed our previously published results that most of the cellular Rrn3p is not associated to pol I whereas functionable pol I-bound Rrn3p is significantly enriched in fraction PA600 (15). Therefore, immunoprecipitated Rrn3p from fraction PA600 exhibits a 10-fold higher ratio of bound pol I as compared with whole cell extracts (compare Fig. 1C, lanes 1 and 3).
Figure 1.
Phosphatase treatment of pol I-Rrn3p complex inhibits transcription initiation and results in destabilization of the complex. (A) Initation-active fraction PA600 was treated without (lane 1) and with (lane 2) immobilized alkaline phosphatase. Four-microliter aliquots were tested in transcription initiation (Upper) and analyzed by Western blotting by using anti-pol I-antibodies (Lower). Because of the dephosphorylation reaction, the pol I-subunits A43 and A34.5 shift to an apparently smaller molecular mass. (B) One microliter of purified fraction B600, 2 μl pol I-Rrn3p complex, and/or 1.5 μl TBP-complex were added to 4 μl phosphatase inhibited fraction PA600 (lanes 4–7). In the control reactions depicted in lanes 1–3, phosphatase-treated fraction PA600 was omitted. The transcriptional activities of 2.5 μl either mock- (lane 8) or phosphatase-treated (lane 9) purified pol I-Rrn3p complex was tested in the presence of 1.5 μl TBP-complex and 1 μl fraction B600. (C) Rrn3p-ProtA was immunopurified by using IgG-beads either from the transcriptional active fraction PA600 (lanes 1, 2), or from whole cell extracts (lanes 3, 4). Each 10 μl immobilized Rrn3p-ProtA were treated without (lanes 1 and 3) or with (lanes 2 and 4) 40 units of alkaline phosphatase for 15 min at 37°C. Fractions were analyzed by Western blotting by using antibodies directed against the A43 subunit. The relative A43/Rrn3p ratio of intensities is indicated at the bottom. Signal intensities were evaluated from four different Western blots by using the image system NIH Image. Because of phosphatase treatment, the A43 subunit exhibited a shift toward a smaller molecular mass.
However, in both fractions (whole cell extracts and PA600) the amount of Rrn3p-bound pol I dropped significantly after phosphatase treatment (Fig. 1C, lanes 2 and 4), suggesting that dephosphorylation of the pol I-Rrn3p complex results in a destabilization of the interaction between pol I and Rrn3p. We conclude that dephosphorylation of the pol I-Rrn3p complex weakens the physical interaction of the binding partners and impairs the formation of a functional transcription initiation complex.
The Free Form of Rrn3p Is Predominantly Phosphorylated in Vivo.
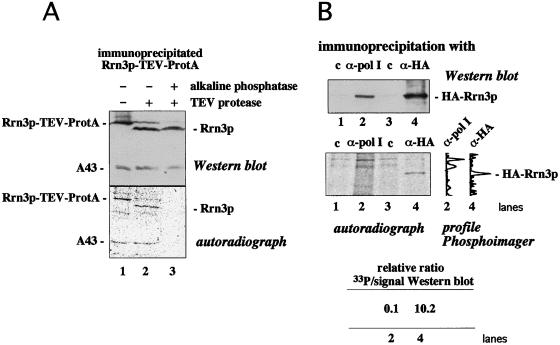
Because Rrn3p is one possible target that could be affected by the dephosphorylation reaction, we investigated whether Rrn3p is a phosphoprotein. We cultivated yeast strains expressing a chromosomal encoded Rrn3p-TEV-ProtA fusion protein in the presence of inorganic [33P]phosphate. Immunoprecipitation of Rrn3p from whole cell extracts by using IgG-Sepharose revealed a radiolabeled protein that comigrated with Rrn3p-TEV-ProtA on SDS/PAGE, whereas no radioactive band in the same area was visible if the immunoprecipitation was performed with control beads (data not shown). After treatment of the immunoprecipitate with TEV-protease, a 33P-labeled polypeptide with the apparent molecular mass of Rrn3p was generated that was immunologically verified as Rrn3p (Fig. 2A). Incubation of the immunoprecipitated Rrn3p with alkaline phosphatase removed the 33P labeling whereas the amount of the precipitated Rrn3p remained unaffected. These results demonstrate that Rrn3p present in whole cell extracts is phosphorylated in vivo.
Figure 2.
The free form of Rrn3p is predominantly phosphorylated in vivo. (A) Logarithmically growing cells were labeled with [33P]phosphate, and Rrn3p-TEV-ProtA fusion protein was immunopurified with IgG-Sepharose. Immobilized Rrn3p-TEV-ProtA fusion protein was treated without (lane 1) or with (lanes 2 and 3) TEV protease, followed by incubation without (lanes 1 and 2) or with (lane 3) 40 units alkaline phosphatase. The fractions were analyzed by Western blotting with antibodies directed against both the N-terminal domain of Rrn3p and the pol I subunit A43 (Upper) and subsequent autoradiography (Lower). Note that subunit A43 of the Rrn3p-bound pol I is also phosphorylated. (B) 33P-labeled pol I and HA-Rrn3p fusion protein were immunoprecipitated from whole cell extracts by using empty ProtA-beads (lanes 1 and 3) or ProtA-beads that had been crosslinked to either anti pol I-antibodies (lane 2) or anti HA-antibodies (lane 4). Fractions were analyzed by Western blotting with anti HA-antibodies (Upper) followed by exposing the membrane to a x-ray film (Lower). The 33P-profiles with background subtracted as established by a PhosphorImager (Molecular Dynamics) are depicted for lanes 2 and 4, and the relative 33P/Western blot intensities are indicated.
Therefore, we wanted to know whether phosphorylation of Rrn3p is a requirement for complex formation between pol I and Rrn3p. If so, one would expect that pol I-bound Rrn3p is phosphorylated in vivo whereas free Rrn3p is phosphorylated at a lower degree—at least at some specific sites. We labeled a yeast strain encoding an HA-tagged Rrn3p with [33P]phosphate. Pol I-bound Rrn3p was coimmunoprecipitated with antibodies directed against pol I, and Rrn3p-specific radioactivity was compared with that of total cellular Rrn3p, which was immunoprecipitated via its HA-tag. Congruent with previous observations (ref. 15; see also Fig. 1C, lanes 1 and 3) we found only a minor proportion (less than 15%) of Rrn3p associated to pol I. Consequently, if exclusively pol I-bound Rrn3p is phosphorylated, its 33P to protein ratio should be significantly higher (at least 7-fold higher) than in free Rrn3p. Quantitative analysis of the Western blot depicted in Fig. 2B excluded this possibility: the 8-fold amount of total Rrn3p was precipitated via its HA-tag (Fig. 2B, lane 4) compared with precipitated pol I-associated Rrn3p (lane 2). Accordingly, if phosphorylation is restricted to pol I-bound Rrn3p, about the same amount of radiolabeled Rrn3p should be detectable in both lanes. However, Rrn3p precipitated by its HA-tag was significantly more radiolabeled than pol I-bound Rrn3p that did not exhibit a radioactive band, which is clearly set off from the background of nonspecific radioactivity (compare Fig. 2B, lanes 2 and 4). These experiments show that uncomplexed Rrn3p is phosphorylated in vivo and that phosphorylation of Rrn3p is not limited to its pol I-bound form. In contrast, the higher 33P to protein ratio of total cellular Rrn3p (see Fig. 2B) suggests that free Rrn3p is predominantly phosphorylated.
Rrn3p Is Able To Enter the Transcription Cycle in Its Nonphosphorylated Form.
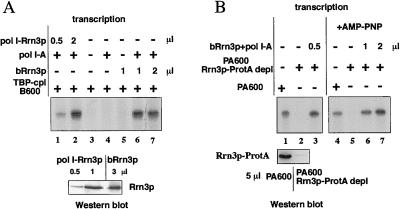
Because phosphorylation of Rrn3p correlated with pol I-free Rrn3p, we asked whether unmodified rather than modified Rrn3p represents the active pol I-interacting molecule. Consequently, bacterially expressed Rrn3p, which should not contain posttranslational modifications of eukaryotes like proper phosphorylation, should be able to functionally interact with purified pol I to result in a similar active complex as the preformed pol I-Rrn3p complex isolated from yeast. It was recently published that purified Rrn3p from bacterial sources associates with pol I and plays an active role in transcription initiation (23). However, in these experiments, it was not addressed whether bacterially expressed Rrn3p exhibits a similar transcriptional activity as the active form isolated from yeast. Therefore, we compared the transcriptional activities of both Rrn3ps and found a high level of transcriptional activity even after short preincubation times (15 min) of purified pol I (pol I-A) with bacterially expressed Rrn3p (bRrn3p). Transcription efficiency was comparable no matter whether isolated yeast pol I-Rrn3p complexes formed in vivo or bacterially expressed Rrn3p incubated with pol I-A was used (Fig. 3A, compare lanes 1, 2 and 6, 7 and the signal intensities of the Western blot underneath). Apparently, unmodified or nonspecific modified Rrn3p from bacterial sources is as efficient as endogenous pol I-bound Rrn3p to establish transcriptional activity.
Figure 3.
Efficient and functional interaction of unmodified Rrn3p with pol I. (A) Bacterially expressed Rrn3p (bRrn3p) was incubated with (lane 6 and 7) or without (lane 5) purified pol I (1 μl, 400 ng/μl pol I-A) for 15 min at room temperature and tested in the reconstituted transcription assay. In control experiments, either bRrn3p alone (lane 4) or bRrn3p and pol I-A (lane 3) were omitted whereas, in the experiment depicted in lanes 1 and 2, isolated yeast pol I-Rrn3p complex was assayed. Aliquots of the Rrn3p-containing fractions used in the transcription assay were tested in their amount of Rrn3p by Western blotting with antibodies directed against the N terminus of Rrn3p (Lower). (B) Bacterially expressed Rrn3p is transcriptionally active in the presence of AMP-PNP. Fraction PA600 was depleted from Rrn3p-ProtA with IgG beads and used to assay functional pol I Rrn3p-complexes (lanes 1–3). Each 1 μl of fraction PA600 before (lanes 1 and 4) or after (lanes 2 and 5) depletion with IgG-beads was tested in transcription initiation. After 15-min preincubation, bRrn3p and pol I-A were added to the Rrn3p-depleted fraction (lanes 3, and 6, and 7). In the experiments depicted in lanes 4–7, ATP in the transcription reaction was replaced by AMP-PNP.
To distinguish whether Rrn3p has to be phosphorylated at specific sites by some putative kinases present in the transcription assay before Rrn3p gains initiation competence, transcription reactions were performed with bacterially expressed Rrn3p in the presence of AMP-PNP (adenylyl-imidodiphosphate). This ATP-analogue with a non-hydrolyzable γ-phosphate bond can be incorporated in the growing RNA chain; however, it is unsuitable for phosphorylation of proteins. We used an immunological Rrn3p-depleted fraction PA600 to monitor Rrn3p-specific activity: adsorption of Rrn3p-ProtA to IgG-Sepharose removed transcriptional activity together with Rrn3p from fraction PA600 (see Fig. 3B, lanes 1 and 2, and Western blot beneath). Transcriptional activity could be restored by adding both purified pol I and bRrn3p (Fig. 3B, lane 3). As depicted in Fig. 3B, lanes 4–7, efficient transcriptional activity was also observed when transcription was performed in the presence of AMP-PNP after bacterially expressed Rrn3p was preincubated with purified pol I. This experiment rules out that phosphorylation at sites specific for S. cerevisiae is a prerequisite for Rrn3p to enter the transcription cycle.
In Vivo Phosphorylation of pol I Changes in Dependency of Its Association with Rrn3p.
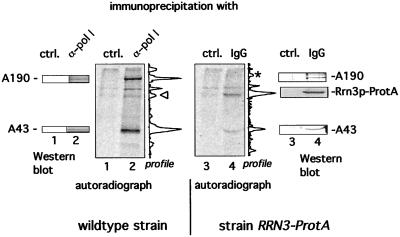
Because establishment of a functional pol I-Rrn3p complex is not dependent on phosphorylation of Rrn3p, we investigated whether Rrn3p-bound pol I is phosphorylated and whether phosphorylation of pol I is a prerequisite to functionally interact with Rrn3p. We labeled equal amounts of wild-type yeast and cells encoding a chromosomal integrated RRN3-ProtA fusion gene with inorganic [33P]phosphate and immunoprecipitated pol I of the wild-type strain with anti-pol I antibodies and Rrn3p-ProtA-bound pol I with IgG beads. Precipitated pol I subunits and Rrn3p-ProtA were verified by Western blotting. As expected, the pol I subunits A190 and A43 were radiolabeled if immunoprecipitation was performed with anti-pol I antibodies (Fig. 4, lane 2). However, if Rrn3p-associated pol I was immunopurified, the ratio in terms of 33P-radioactivity between subunit A43 and A190 changed significantly: the relative value of the A43/A190 radioactivity in total pol I was 0.8, compared with 6 in Rrn3p-associated pol I (Fig. 4, lanes 2 and 4). Apparently, association of pol I with Rrn3p is correlated by a distinct phosphorylation pattern of pol I, which can be monitored by a higher phosphorylation ratio of subunit A43 compared with A190. We conclude that distinct pol I subunits like A43 are phosphorylated in the initiation active enzyme and that pol I undergoes a transition in its phosphorylation state when it becomes associated with Rrn3p.
Figure 4.
Pol I exhibits a different phosphorylation pattern when it is associated with Rrn3p compared with total cellular pol I. Logarithmically growing wild-type cells and strain RRN3-ProtA were labeled with [33P]phosphate. Pol I was precipitated with anti-pol I antibodies that had been crosslinked to ProtA-beads. Rrn3p-TEV-ProtA fusion protein was immunopurified with IgG-Sepharose. The fractions were analyzed by Western blotting with antibodies directed against pol I subunits or ProtA and subsequent autoradiography. The 33P-profiles of lane 2 and 4, as established by a PhosphorImager, are depicted right next to the corresponding autoradiograph. Note that the peaks marked by Δ and * do not correspond to wild-type Rrn3p and A190, respectively.
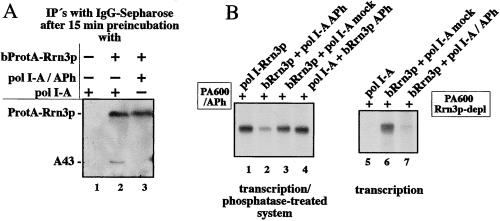
Phosphorylation of pol I Is Required To Form a Functional pol I-Rrn3p Complex.
To find out whether phosphorylation of pol I is required for stable and functional association with Rrn3p, we performed interaction studies with native and dephosphorylated pol I-A and purified bacterially expressed Rrn3p. As depicted in Fig. 5A, lane 3, phosphatase treatment of pol I-A abolished its capability to form a stable complex with Rrn3p. In contrast, a substantial amount of mock-treated pol I-A was capable of interacting with purified bacterial expressed Rrn3p-ProtA and remained associated even under very stringent conditions (compare lanes 2 and 3, Fig. 5, and see figure legend). Consistently, incubation of phosphatase-treated pol I-A with bRrn3p resulted in reduced transcriptional activity (Fig. 5B, lanes 2 and 7) both in a transcription system in which activity had been impaired by phosphatase treatment (lanes 1–4) and in a system from which functional Rrn3p had been immunodepleted (lanes 5–7). In control reactions, combinations of mock-treated pol I-A with bRrn3p, as well as phosphatase-treated bRrn3p with pol I-A showed no significant decline in transcriptional activity (Fig. 5B, lanes 3, 4, and 6), demonstrating that, in these selected states, both components can function with each other. Taken together, these experiments confirm that specific phosphorylation of pol I, but not that of Rrn3p, is essential for formation of the initiation active pol I-Rrn3p complex.
Figure 5.
Phosphorylation of pol I-A is required for the formation of a functional pol I-Rrn3p complex. (A) Rrn3p-ProtA associates only with the phosphorylated form of pol I. Each 10 μg of purified pol I-A was treated for 30 min at 37°C with 14 units active immobilized alkaline phosphatase (lane 3) or without enzyme (lanes 1 and 2). Two micrograms of the resulting pol I (3-fold molar excess) were incubated for 30 min without (lane 1) or with (lanes 2 and 3) 75 ng purified bRrn3p-ProtA in a total volume of 17.5 μl. ProtA-tagged Rrn3p was affinity purified with IgG-Sepharose under very stringent conditions (see Materials and Methods) to prevent nonspecific binding of pol I to the beads. IgG-bound proteins were analyzed by Western blotting by using anti-A43 antibodies. (B) Phosphatase-treated pol I-A but not bRrn3p is impaired in transcription initiation. Each 200 ng of either phosphatase (lane 2) or mock-treated (lane 3) pol I-A were incubated with 1 μl untreated (lane 2 and 3) or phosphatase-treated (lane 4) bRrn3p. After 15 min preincubation, the fractions and isolated yeast pol I-Rrn3p complex (lane 1) were added either to 4 μl of the dephosphorylated fraction PA600 (lanes 1–4) or to 1 μl of the Rrn3p-depleted fraction PA600 (lanes 5–7) to assay them for transcriptional activity.
Discussion
We suggest that phosphorylation at specific pol I sites is a prerequisite for the formation of the initiation competent enzyme and that adjustment of pol I and Rrn3p to different phosphorylation states enables the cell to modulate the activity of its rDNA transcription machinery. In particular, phosphorylation of pol I at specific sites is a prerequisite for the formation of an initiation competent pol I-Rrn3p complex, whereas phosphorylation of Rrn3p is not required for functional complex formation. Because Rrn3p is phosphorylated in its pol I-free form, it is possible that Rrn3p associates with pol I in a low phosphorylated state and becomes more phosphorylated during or after transcription. Our data show that phosphorylation reactions are involved in the regulation of rDNA transcription in yeast.
Phosphorylation/dephosphorylation represents a principal mechanism in the regulation of mammalian rDNA transcription. Mitotic silencing of mammalian rRNA synthesis was described to correlate with phosphorylation of the human promoter selectivity factor SL1 (29), upstream binding factor UBF (30), and transcription termination factor TTF1 (31). On the other hand, phosphorylation of UBF at Ser-484 (32) and at its C terminus (33–35) is necessary for activated transcription.
Our results suggest that, in yeast, the pol I-Rrn3p complex is a key target for phosphorylation/dephosphorylation reactions. The other two fractions required for reconstitution of transcription in vitro (TBP-complex and fraction B600) were both active after phosphatase treatment. This finding could mean that either the phosphorylation states of the factors have no impact on their transcriptional activity or that kinases are present in the rather crude fraction B600 that rephosphorylate the crucial residues by using in vitro transcription conditions.
Although phosphorylation of the pol I-subunits A190, A43, A34.5, A23, and A19 was reported quite a while ago (21, 22), not much is known about the impact of their phosphorylation pattern on the pathway of rRNA synthesis. We provide evidence that a certain degree of phosphorylation at specific pol I-subunits is necessary for its binding to Rrn3p and, thus, for efficient promoter-dependent rRNA synthesis. From previous observations (15) and from our coimmunoprecipitation experiments, we conclude that only a small fraction of pol I is complexed with Rrn3p and that this minor proportion of pol I differs in its phosphorylation state from the majority of cellular pol I. If the distinct phosphorylation state of this pol I population is required to interact with Rrn3p, it is obvious why only such a small amount of pol I is complexed with Rrn3p. A limiting amount of properly phosphorylated pol I would also explain why the in vitro binding efficiency of pol I and Rrn3p was rather low although the interaction was strikingly stable.
One could speculate that phosphorylation/dephosphorylation at specific pol I sites mediates the interaction with Rrn3p, thereby regulating the initiation activity of the enzyme. For instance, Rrn3p might interact with a specifically phosphorylated form of pol I to recruit initiation active pol I to the promoter. After transcription initiation, Rrn3p dissociates from the transcription complex (15). Plausible mechanisms that cause the release of Rrn3p could be the dephosphorylation at specific sites within pol I (for instance A43) or hyperphosphorylation of Rrn3p. However, it is also possible that the hyperphosphorylated form of subunit A190, which is present in Rrn3p-free pol I, triggers dissociation of Rrn3p. Preliminary interaction experiments with pol I and Rrn3p, which were randomly hyperphosphorylated in vitro, showed no altered binding properties, indicating that specific phosphorylation sites are involved in this process. Therefore, it will now be necessary to identify all modified sites in Rrn3p and in the pol I subunits to study the roles of the particular modifications in transcription initiation and elongation. Furthermore, it will be necessary to test whether and how the phosphorylation pattern of both pol I and Rrn3p changes through one transcription cycle and whether in stationary cells both components are incapable of binding each other because of their specific phosphorylation states.
Our findings that free Rrn3p is predominantly phosphorylated and that bacterially expressed, unmodified Rrn3p is able to functionally interact with pol I indicate that dephosphorylated rather than phosphorylated Rrn3p represents the active form of Rrn3p in transcription. On the other hand, phosphatase treatment of uncomplexed and inactive yeast Rrn3p did not restore its transcriptional activity (data not shown), which suggests that either a second posttranslational modification or for instance an inhibiting activity might be additionally involved in regulation of Rrn3p activity.
The determination of all modified residues in both pol I and Rrn3p in dependency of their functional state will provide an important tool to understand the basic mechanism of rDNA transcription and will contribute to the knowledge of how transcriptional regulation is coupled to growth control of the cell.
Acknowledgments
We thank Dr. Nomura for providing us with the strain NOY657, E. Draken for expert technical assistance, I. Eckstein for yeast cultivation, and S. Schütz for assistance to generate the Rrn3p-ProtA strain. We are especially grateful to Dr. I. Haas for helpful discussion and for critical reading of the manuscript. This work was supported by funds from the Deutsche Forschungsgemeinschaft and from the Human Frontier Science Program Organization (HFSPO).
Abbreviations
- pol I
RNA polymerase I
- ProtA
ProteinA-epitope
- HA
hemagglutinin antigen
- UAF
upstream activating factor
- CF
core factor
- TBP
TATA binding protein
- AMP-PNP
adenosine 5′-[β,γ-imido]triphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Thomas G. Nat Cell Biol. 2000;2:E71–E72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- 2.Warner J R. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 3.Cormack B P, Struhl K. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 4.Schultz M C, Reeder R H, Hahn S. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 5.Milkereit P, Schultz P, Tschochner H. Biol Chem. 1997;378:1433–1443. doi: 10.1515/bchm.1997.378.12.1433. [DOI] [PubMed] [Google Scholar]
- 6.Steffan J S, Keys D A, Dodd J A, Nomura M. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 7.Lin C W, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder R H. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steffan J S, Keys D A, Vu L, Nomura M. Mol Cell Biol. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiqi I, Keener J, Vu L, Nomura M. Mol Cell Biol. 2001;21:2292–2297. doi: 10.1128/MCB.21.7.2292-2297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keys D A, Vu L, Steffan J S, Dodd J A, Yamamoto R T, Nogi Y, Nomura M. Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- 11.Lalo D, Steffan J S, Dodd J A, Nomura M. J Biol Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 12.Keys D A, Lee B S, Dodd J A, Nguyen T T, Vu L, Fantino E, Burson L M, Nogi Y, Nomura M. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 13.Keener J, Dodd J A, Lalo D, Nomura M. Proc Natl Acad Sci USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto R T, Nogi Y, Dodd J A, Nomura M. EMBO J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
- 15.Milkereit P, Tschochner H. EMBO J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aprikian P, Moorefield B, Reeder R H. Mol Cell Biol. 2001;21:4847–4855. doi: 10.1128/MCB.21.15.4847-4855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moorefield B, Greene E A, Reeder R H. Proc Natl Acad Sci USA. 2000;97:4724–4729. doi: 10.1073/pnas.080063997. . (First Published April 11, 2000; 10.1073/pnas.080063997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodem J, Dobreva G, Hoffmann-Rohrer U, Iben S, Zentgraf H, Delius H, Vingron M, Grummt I. EMBO Reports. 2000;1:171–175. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller G, Panov K I, Friedrich J K, Trinkle-Mulcahy L, Lamond A I, Zomerdijk J C. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bréant B, Buhler J-M, Sentenac A, Fromageot P. Eur J Biochem. 1983;130:247–251. doi: 10.1111/j.1432-1033.1983.tb07143.x. [DOI] [PubMed] [Google Scholar]
- 22.Buhler J-M, Iborra F, Sentenac A, Fromageot P. FEBS Lett. 1976;71:37–41. doi: 10.1016/0014-5793(76)80893-9. [DOI] [PubMed] [Google Scholar]
- 23.Keener J, Josaitis C, Dodd J, Nomura M. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 24.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 25.Tschochner H. Proc Natl Acad Sci USA. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Cell. 2001;705:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 27.Fath S, Milkereit P, Podtelejnikov A V, Bischler N, Schultz P, Bier M, Mann M, Tschochner H. J Cell Biol. 2000;149:575–590. doi: 10.1083/jcb.149.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 29.Heix J, Vente A, Voit R, Budde A, Michaelidis T M, Grummt I. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein J, Grummt I. Proc Natl Acad Sci USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirri V, Roussel P, Hernandez-Verdun D. J Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- 32.Voit R, Hoffmann M, Grummt I. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuan J C, Zhai W, Comai L. Mol Cell Biol. 1999;19:2872–2879. doi: 10.1128/mcb.19.4.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihm A J, Hershey J C, Haystead T A, Madsen C S, Owens G K. Proc Natl Acad Sci USA. 1998;95:14816–14820. doi: 10.1073/pnas.95.25.14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Mahony D J, Smith S D, Xie W, Rothblum L I. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huet J, Phalente L, Buttin G, Sentenac A, Fromageot P. Embo J. 1982;1:1193–1198. doi: 10.1002/j.1460-2075.1982.tb00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buhler J M, Huet J, Davies K E, Sentenac A, Fromageot P. J Biol Chem. 1980;255:9949–9954. [PubMed] [Google Scholar]