Abstract
Argonaute proteins are present and conserved in all domains of life. Recently characterized prokaryotic Argonaute proteins (pAgos) participates in host defense by DNA interference. Here, we report that the Natronobacterium gregoryi Argonaute (NgAgo) enhances gene insertions or deletions in Pasteurella multocida and Escherichia coli at efficiencies of 80–100%. Additionally, the effects are in a homologous arms-dependent but guide DNA- and potential enzyme activity-independent manner. Interestingly, such effects were also observed in other pAgos fragments including Thermus thermophilus Argonaute (TtAgo), Aquifex aeolicus Argonaute (AaAgo) and Pyrococcus furiosus Argonaute (PfAgo). The underlying mechanism of the NgAgo system is a positive selection process mainly through its PIWI-like domain interacting with recombinase A (recA) to enhance recA-mediated DNA strand exchange. Our study reveals a novel system for enhancing homologous sequence-guided gene editing in bacteria.
INTRODUCTION
Developing an efficient gene editing tool is crucial for the development of vaccines against pathogenic bacteria, the promotion of beneficial products for probiotic microorganisms and industrial microbiology (1–3), and gene-function studies in bacteria (4). During recent years, a few bacterial gene editing tools have been developed. These generally include the following three types: (i) random integration mediated by transposons (5); (ii) the site-specific recombination based on the integrase family and the resolvase/invertase family, which rely on specific sequences in the bacterial genome (6,7) and (iii) bacterial homologous recombination systems, which can concisely manipulate the bacterial genome. However, the efficiency of homologous recombination is low in its natural condition (8,9). How to obtain the mutant at a high efficiency is an essential issue. To overcome such obstacles, two strategies have been developed (10). One strategy is a negative selection system that is based on the killing of non-mutant strains for the isolation of mutant strains, which includes antibiotic selection systems, SacB selection system (11), galk selection system (12), thermo-sensitive system (13), thymidylate synthase A selection system (14), Toxin-Antitoxin Cassette (15), and Restriction-Modification System (16). Actually, this strategy has also been used by the newly developed clustered, regularly interspaced, short palindromic repeats (CRISPR)–associated Cas9 system for bacterial engineering (1,17–19). The CRISPR-Cas9 system relies on guide RNA (gRNA):Cas9-directed cleavage at a targeted genomic site to kill unmutated cells and circumvents the need for selectable markers or counter-selection systems (18). The other strategy is a positive selection system that depends on the promotion efficiency of homologous recombination, which includes the introduction of recombinase A (recA) (20,21), a key enzyme involved in the DNA recombination and repair (22). Although the introduction of recA could significantly promote the efficiency, the recombination efficacy was still quite low (21).
Pasteurella multocida is a gram-negative coccobacillus that infects various hosts and leads to huge economic losses in the poultry, cattle and saiga antelopes industries (23,24). It is also the most common causative organism isolated from dog-bite or cat-bite wounds (25). Additionally, the isolation of a few higher virulent and multiple antibiotic-resistant strains has made the development of novel vaccines and pathogenesis studies more urgent (26). Although a thermo-sensitive system was developed for gene manipulation in P. multocida NADC TT94 (13), it worked at a low efficiency in the highly virulent and multiple antibiotic-resistant strain GX-PM (26). We further attempted various methods for gene editing, such as the galk selection system, the thymidylate synthase A selection system, and the SacB system, but all failed. Therefore, finding a new, highly safe and efficient system for robust gene editing is of high priority.
Argonaute proteins (Agos) were initially discovered in eukaryotes as key proteins in RNA interference systems (27). Both eukaryotic Agos and long prokaryotic Agos (pAgos) form a bi-lobed scaffold, in which one lobe consists of the amino-terminal (N) and PIWI–Argonaute–Zwille (PAZ) domains, whereas the other lobe consists of the middle (MID) and PIWI domains (28–30). The MID and PAZ domains usually form binding pockets that facilitate the anchoring of the 5′ and 3′ ends of an oligonucleotide guide, respectively (30). On target binding, the PIWI domain of catalytically active Agos (that contain the DEDX motif, in which X denotes D, H or N (31)) mediates cleavage of cognate DNA targets (31–34) or RNA targets in vitro (32,35). These suggest a potential for prokaryotic Argonaute proteins as a novel gene-editing tool (30). Recently, DNA-guided genome editing with Natronobacterium gregoryi Argonaute (NgAgo) has been developed for genome editing in eukaryotic cells, but the original report was retracted due to the continued inability of the research community to replicate the key findings (36,37). Interestingly, NgAgo was identified as a genetic tool for gene silencing in zebrafish (38) or for degradation of pregenomic RNA during Hepatitis B virus infection (39). However, no study reports its potential role in bacterial gene editing. Here, we present the successful genetic manipulation of a few bacterial isogenic mutants with the NgAgo and other pAgos fragments systems, demonstrating its potential role in bacterial gene editing as an effective positive selection system independent of its potential enzyme activity.
MATERIALS AND METHODS
Bacterial strains, plasmids and DNA oligos
The bacterial strains and plasmids used in this study are presented in Supplementary Table S1. The Primers for PCR are presented in Supplementary Table S2. The guide DNA (gDNA) sequences for target genes are presented in Supplementary Table S3.
Construction of pSHK5(TS)-NgAgo
A NgAgo gene (GenBank ID: KU899087.1) was amplified by PCR with the NgAgo-L/R primers and a High-Fidelity DNA polymerase (NEB). A ribosome binding sequence (rbs) was amplified by PCR from the avian P. multocida strain GX-PM genome with the RBS-L/R primers (Supplementary Table S2). The NgAgo gene and rbs were connected by fusion PCR with the RBS-RH-L/NgAgo-RH-R primers, and then inserted upstream of the kanamycin resistance gene (kanr) in the temperature-sensitive P. multocida–E. coli shuttle vector pSHK5(TS) to generate the pSHK5(TS)-NgAgo plasmid with the pSHK5(TS)-RBS-RH-F/pSHK5(TS)-NgAgo-RH-R and NgAgo-Kan-RH-F/R primers.
Next-generation sequencing of the bacterial genome
Pasteurella multocida strains (GX-PM and the constructed isogenic lyi gene mutants with or without gDNA) were sequenced at the Shanghai Sangon Biotech using Illumina technology. For all bacteria genomes, we constructed and sequenced an Illumina short-insert paired-end library. The majority of the genomes were assembled using the ALLPATHS and Velvet assembly methods.
Pull-down assay coupled with LC-MS/MS
An E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag, pSHK5(TS)-NgAgo fragments (A, B or D)-Flag, or pSHK5(TS). The bacteria were collected in 2 ml RIPA buffer (Beyotime), and then lysed by two passages through a French pressure cell (Thermo, USA) at 20 000 lb/in.2. After centrifugation at 12 000g for 10 min, the supernatants were collected and further purified with FLAG M Purification Kit (Sigma-Aldrich, CELLMM2). Finally, the interacting proteins were recovered with PBS buffer containing 300 μg/ml of 3× FLAG peptides (Sigma-Aldrich, F4799). The eluted proteins were subjected for analysis by LC-MS/MS with Eksigent nanoLC-Ultra 2D (AB SCIEX) and TripleTOF 5600 (AB SCIEX). The data analysis was performed with the Mascot 2.3 (Matrix Science) and was based on the SwissProt 57.15 database (Taxonomy: E. coli) with the following parameters: Type of search: MS/MS Ion Search; Mass values: Monoisotopic; Fixed modifications: Carbamidomethyl (C); Variable modifications: Acetyl (Protein N-term), Deamidated (NQ), Dioxidation (W) and Oxidation (M); Peptide mass tolerance: ±30 ppm; Fragment mass tolerance: ±0.15 Da; and Max missed cleavages: 2. To ensure the reliability of identified proteins, proteins that more than two unique peptides were successfully matched were recognized as the identified proteins.
Analysis of the interaction of NgAgo and recA
An E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL (the left arm sequences of aste gene), pSHK5(TS)-asteL, pSHK5(TS)-NgAgo-Flag or pSHK5(TS). The bacteria were lysed by two passages through a French pressure cell (Thermo, USA) at 20 000 lb/in.2, and the proteins were collected for analysis with FLAG M Purification Kit (Sigma-Aldrich, CELLMM2). The eluted proteins were collected in 100 μl of TE buffer containing 1% SDS and then were subjected for immunoblot analysis with rabbit polyclonal antibodies against recA (Abcam, ab63797) or rabbit polyclonal antibodies against Bacteriophage phic31 integrase (Abcam, ab93248). Images were obtained on the MF Chem BIS Bio-Imaging System (DNR). All experiments were completed in triplicate. This assay was also performed to detect the interaction between the recA and NgAgo fragments (A, B, D, E and F), Pyrococcus furiosus Argonaute fragment D (PfAgo-D, 316–771 aa, containing MID and PIWI domains), Thermus thermophilus Argonaute fragment D (TtAgo-D, 316–685 aa, containing MID and PIWI domains), or Aquifex aeolicus Argonaute fragment D (AaAgo-D, 314–706 aa, containing MID and PIWI domains).
An E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL, pSHK5(TS)-asteL, pSHK5(TS)-NgAgo-Flag or pSHK5(TS). Proteins were extracted for analysis with 50 μl Protein A agarose (KPL) with or without 5 μl rabbit polyclonal antibodies against recA (Abcam) and incubated for 3 h at 4°C. To confirm the identification of the recA-interacting proteins, the purified proteins were further subjected for immunoblot analysis with rabbit polyclonal antibodies against recA (Abcam, ab63797) and peroxidase-labelled goat anti-rabbit IgG(H+L) (KPL) or anti-DDDDK-Tag FLA-1 purified IgG/Mouse (MBL) and peroxidase-labeled goat anti-mouse IgG(H+L) (KPL), followed by development with the clarity™ western ECL Substrate (Bio-Rad). This assay was also performed to detect the interactions between the NgAgo fragments (A, B, D, E and F) and recA.
The interaction of recA and NgAgo in the presence of Dnase I
An E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL, pSHK5(TS)-asteL, pSHK5(TS)-NgAgo-Flag or pSHK5(TS). The total proteins were extracted as described before and then digested with or without 8 μl Dnase I (10g/ml, Sangon Biotech) at 37°C for 3 h. Subsequently, these proteins were subjected for analysis with FLAG M Purification Kit (Sigma-Aldrich, CELLMM2). The eluted protein were collected in 100 μl of TE buffer containing 1% SDS and then were subjected for immunoblot analysis with rabbit polyclonal antibodies against recA (Abcam, ab63797) and peroxidase-labelled goat anti-rabbit IgG(H+L) (KPL) or anti-DDDDK-Tag FLA-1 purified IgG/Mouse (MBL) and peroxidase-labeled goat anti-mouse IgG(H+L) (KPL), Images were obtained on the MF Chem BIS Bio-Imaging System (DNR). All experiments were completed in triplicate.
Recombinant expression and purification of NgAgo and recA from E. coli and P. multocida
The NgAgo gene was amplified with primers NgAgo-pET28a-RH-F/R or Flag-pET28a-NgAgo-RH-F/R (Table S2). Then both PCR fragments were cloned into the prokaryotic expression vector pET28a. The recombinant NgAgo-His (rNgAgo-His) and NgAgo-FLAG (rNgAgo-FLAG) was expressed in E. coli (BL21), and purified with Ni-NTA agarose (GE Healthcare) or ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) according to the manufacturer's instructions.
The ErecA or PmrecA gene was amplified from E. coli genome or P. multocida GX-PM genome with primers ErecA-pET28a-RH-F/R or PmrecA-pET28a-RH-F/R. Both PCR fragments were cloned into the prokaryotic expression vector pET28a, and then transformed into E. coli (BL21) for expression. Either recombinant ErecA (rErecA) or PmrecA (rPmrecA) was purified with Ni-NTA agarose (GE Healthcare) according to the manufacturer's instructions.
Analysis of the direct interaction of NgAgo and recA
The purified rNgAgo-FLAG and the rErecA or rPmrecA were incubated for 3 h at 4°C. Then the proteins were subjected for co-purification with Ni-NTA agarose (GE Healthcare) or ANTI-FLAG M2 Affinity Gel. The eluted proteins were further subjected for immunoblot analysis with rabbit polyclonal antibodies against recA (Abcam, ab63797) and peroxidase-labelled goat anti-rabbit IgG(H+L) (KPL) or anti-DDDDK-Tag FLA-1 purified IgG/Mouse (MBL) and peroxidase-labelled goat anti-mouse IgG(H+L) (KPL), followed by development with the clarity™ western ECL Substrate (BIO-RAD).
Binding studied by surface plasmon resonance (SPR)
The binding of recA to NgAgo under laminar flow was analyzed by SPR using a BIAcore T200 system (GE Healthcare). The surface of a carboxymethylated dextran (CM5) sensor chip (GE Healthcare) was activated with 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 0.1 M N-hydroxysuccinimide. rNgAgo-His was immobilized by amine coupling to one flow cell. All free reactive surface groups were blocked using 1 M ethanolamine. BSA was immobilized onto the other flow cell as nonspecific binding control. Different concentrations (0–4 μM) of rErecA in PBS buffer containing 0.5% Tween-20 were injected over the flow cells at 60 μL/min (contact time, 2 min). After each injection, any bound protein was stripped with 6.25 mM NaOH (15 s). Data analysis was performed using the BIAcore T200 evaluation software 3.1 (GE Healthcare), which allowed determination of Ka, Kd, and KD values by fitting the derived sensograms to 1:1 Langmuir model. A separate determination of the equilibrium constant KD was performed to validate curve fitting by plotting the maximal responses (RUmax) against substrate concentration and fitting to one-binding site model. The binding of rNgAgo-His to rErecA was also performed similarly. rErecA and BSA were immobilized by amine coupling to each flow cell, and different concentrations (0–2.5 μM) of rNgAgo-His in PBS buffer containing 0.5% Tween-20 was injected over the flow cells for analysis.
Chromatin immunoprecipitation (ChIP)-qPCR
E. coli was transformed with pSHK5(TS)-NgAgo-Flag-asteL or pSHK5(TS)-asteL (Supplementary Table S1) and cultured in 10 ml LB medium for 12 hours. Then the bacteria were cross-linked for 1 h with formaldehyde at a 1% final concentration (Sigma-Aldrich) and quenched by the addition of glycine to 125 mM for 10 min. The bacterial pellets were resuspended in 2 ml RIPA buffer (Beyotime) with 50 μl lysozyme (100 mg/ml, Sangon Biotech) and incubated at 37°C for 30 min. The samples were chilled and sonicated for 5 min in a Uibra cell TM Sonics (500W, Shanghai SpringSci Co., Ltd) with 9 s on/9 s off pulsing at 40% amplitude. After centrifugation at 12 000g for 10 min, 100 μl of supernatants were collected as input controls for qPCR. The other supernatants were incubated with 50 μl Protein A agarose beads (KPL) with 5 μl rabbit polyclonal antibodies against recA (Abcam) for 2 h at 4°C. The agarose beads were resuspended in 100 μl of elution buffer as ChIP samples. Next, 4 μl of 500 mM NaCl was added into the prepared samples, including the ChIP samples and input controls, and then incubated at 65°C for 8 h to reverse the DNA–protein crosslinking. Then, all the samples were purified by using a PCR purification kit (Qiagen). The FastStart Universal SYBRR Green master (ROX) (Roche) was applied for qPCR according to the manufacturer's instructions (Supplementary Table S2).
Electrophoretic mobility shift (EMSA) assay
Reactions contained Tris–HCI (70 mM, pH 7.6), MgCl2 (10 mM), dithiothreitol (5 mM), ATP-γS (2.0 mM), rErecA (variable concentrations), and phiX174-oligo chemically labeled with Cy5 (125 nM) were combined to a final volume of 20 μl and incubated at 37°C for 60 min to form recA-ssDNA filaments in the presence of various concentration of rNgAgo-His. The reaction products were resolved on a 1% agarose gel and imaged on a Fujifilm FLA-5100 (FUJIFILM Life Science, Japan).
DNA Strand exchange assay
A reaction mixture containing Tris–HCl (70 mM, pH 7.6), MgCl2 (10 mM), dithiothreitol (5 mM), ATP-γS (2.0 mM), rErecA (variable concentrations), HPLC-purified phiX174-oligo chemically labeled with Cy5 (125 nM), was incubated at 37°C for 10 min to form recA-ssDNA filaments. D loop formation was initiated with the addition of 1 μg phiX174 RF1 DNA and various concentration of rNgAgo-His to the mixture, and incubated at 37°C. The reaction was stopped via addition of Proteinase K and 1% SDS at room temperature. Reaction products were resolved on a 1% agarose gel and imaged on a Fujifilm FLA-5100 (FUJIFILM Life Science, Japan).
RESULTS
NgAgo-assisted gene editing in P. multocida
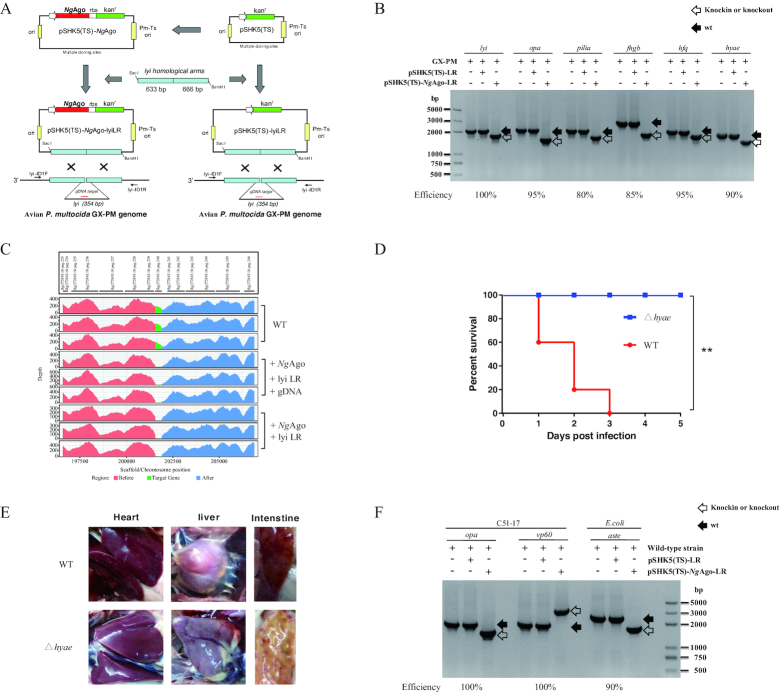
An NgAgo gene was inserted upstream of the kanamycin resistance gene (kanr) in the temperature-sensitive P. multocida–E. coli shuttle vector pSHK5(TS) to generate the pSHK5(TS)–NgAgo plasmid (Figure 1A, Supplementary Table S1-2). The homologous arms of the target gene, e.g. the lyi (lysozyme inhibitor) gene, were inserted into the pSHK5(TS) or pSHK5(TS)-NgAgo plasmid to yield pSHK5(TS)-lyiLR or pSHK5(TS)-NgAgo-lyiLR, respectively. To test whether NgAgo could promote gene editing, either the pSHK5(TS)-lyiLR plasmid or pSHK5(TS)-NgAgo-lyiLR plasmid with guide-DNA (gDNA) were electro-transformed into the P. multocida strain GX-PM (Figure 1A, Supplementary Table S1 and S3). As a result, isogenic mutants could only be detected at 100% efficiency with the NgAgo system by PCR with the lyi-ID1F/R primers (Figure 1B and Supplementary Figure S1), indicating that the efficiency of natural homologous recombination in the P. multocida strain is negligible, while the NgAgo system is robust for gene editing in P. multocida without off-target effect which was confirmed by next-generation sequencing (Figure 1C).
Figure 1.
NgAgo-assisted gene editing in P. multocida and E. coli. (A) Schematic construction of the isogenic lyi (lysozyme inhibitor) gene mutant for P. multocida. An NgAgo gene with a ribosome binding sequence (rbs) was introduced upstream of the kanamycin resistance gene (kanr) in the temperature-sensitive P. multocida-E. coli shuttle vector pSHK5(TS) to generate the pSHK5(TS)-NgAgo plasmid. The homologous arms of the lyi gene were then inserted into the pSHK5(TS) or pSHK5(TS)-NgAgo plasmid to yield pSHK5(TS)-lyiLR or pSHK5(TS)-NgAgo-lyiLR, respectively. Subsequently, the two plasmids were electro-transformed into the P. multocida strain GX-PM with guide DNA (gDNA) to isolate the isogenic mutants by PCR with the lyi-ID1F/R primers. (B) Construction efficiency of the avian P. multocida strain GX-PM with deletion of the lyi, opa (Opacity-associated protein), pilia (type IV fimbrial subunit protein), fhgb (Flavohemoglobin), hfq (RNA chaperone Hfq), or hyae (hydrogenase-1 operon protein) gene with or without NgAgo system. (C) Representatives of whole genome sequencing for the P. multocida strain GX-PM with the deletion of the lyi gene with or without gDNA. (D) Comparison of the virulence of the avian P. multocida strain GX-PM with the constructed isogenic hyae mutant in chickens. Infection of chickens with the avian P. multocida strain GX-PM caused mortality of 100% in the third day of post-infection, while infection of the constructed isogenic hyae mutant could not cause any death during the trial (n = 5). (E) Severe damage to several vital organs in response to infection with the avian P. multocida strain GX-PM was not observed in the chickens infected with the constructed isogenic hyae mutant. (F) Construction efficiency of the rabbit P. multocida strain C51-17 with deletion of the opa (Opacity-associated protein) gene or insertion of a vp60 gene from rabbit haemorrhagic disease virus; and the E. coli strain with the aste (succinylglutamate desuccinylase) gene deletion with or without NgAgo system.
To test whether the NgAgo-based gene knockout in P. multocida is successful only for a lyi gene, we applied this approach to manipulate several additional genes. As shown in Figure 1B and Supplementary Figure S2-S6, opa (Opacity-associated protein), pilia (type IV fimbrial subunit protein), fhgb (Flavohemoglobin), hfq (RNA chaperone Hfq), and hyae (hydrogenase-1 operon protein) genes, were successfully removed from the genome with phenotype efficiencies of 95%, 80%, 85%, 95% and 90%, respectively. The virulence of the constructed isogenic hyae mutant in chickens was significant decreased compared with the wt strain GX-PM, which caused high mortality and severe damage to several vital organs (26), e.g., liver, heart and duodenum (Figure 1D and E), indicating that the NgAgo system could help successfully delete the genes in P. multocida at a high efficiency.
NgAgo-assisted gene editing in E. coli and rabbit P. multocida strain
To further test whether the NgAgo-based system is bacterial strain-specific, we applied it to the rabbit P. multocida strain C51-17 and E. coli. As shown in Figure 1F and Supplementary Figure S7-S8, both representative genes opa and aste (succinylglutamate desuccinylase) were completely removed from the genome of the C51-17 strain and E. coli, respectively, at an efficiency of 100%. Additionally, NgAgo-based specific gene (a vp60 gene from rabbit haemorrhagic disease virus) knock-in editing is also possible, as shown in Figure 1F and Supplementary Figure S9. These data suggest that the NgAgo system is potentially efficient for bacterial gene editing.
NgAgo-assisted gene editing is based on the homologous sequences and is independent of its potential enzymatic activity and gDNA
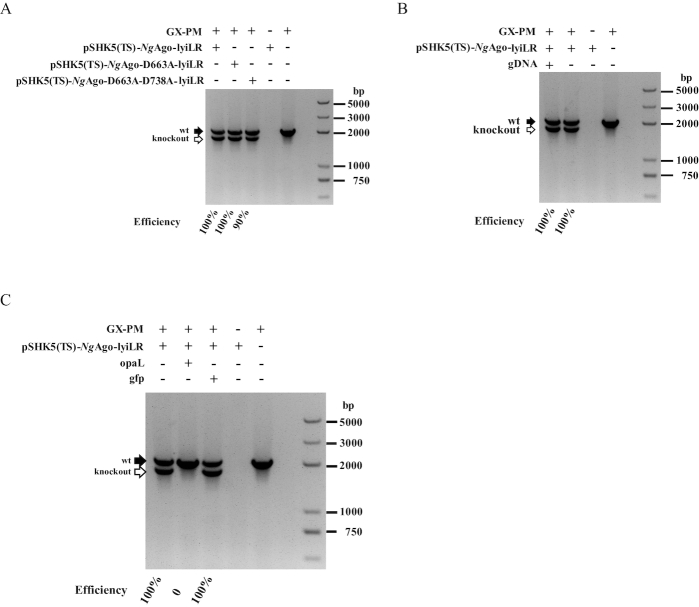
In terms of the high efficiency of NgAgo-based gene editing in bacteria, we aimed to determine the crucial role of NgAgo in bacterial homologous recombination. As both eukaryotic and prokaryotic Agos have conserved DEDX motif with confirmed or predicted catalytic activity (Supplementary Figures S10 and S11), we introduced single or double mutations for Asp663 and Asp738 in NgAgo, obtaining NgAgo-D663A and NgAgo-D663A-D738A (Supplementary Table S1-S2), which are predicted to inhibit the potential catalytic activity of NgAgo (38,40). We found that transformation of each pSHK5(TS)-NgAgo-lyiLR mutant vector efficiently caused lyi knockout, with phenotype efficiencies of 100% and 90% (Figure 2A and Supplementary Figure S12). This indicates that the NgAgo system enhances homologous sequence-guided gene editing in bacteria independent of its potential enzyme activity. Additionally, this was further supported by the finding that no insertion/deletion mutations were detected in the bacterium transformed with gDNA/NgAgo using a T7E1 enzyme assay (Supplementary Figure S13). These results seem to conflict with our previous attempt to use the NgAgo/guide complex to cleave the target sequences of genome for counter-selection. Surprisingly, gDNA is not required in the bacteria as long as the homologous arms are present (Figure 2B and Supplementary Figure S14). Thus, we hypothesized that the homologous arms are required for NgAgo-mediated gene editing in bacteria, while both the potential enzyme activity and gDNA are not necessary.
Figure 2.
NgAgo-assisted gene editing independent of its potential enzymatic activity and guide DNA (gDNA). (A) Construction efficiency of isogenic lyi mutant in the P. multocida strain GX-PM based on NgAgo or the NgAgo mutants NgAgo-D663A and NgAgo- D663A-D738A, which are predicted to lack NgAgo potential catalytic activity. (B) Comparison of the construction efficiency of the isogenic lyi mutant in the P. multocida strain GX-PM based on NgAgo with or without gDNA. (C) Construction of isogenic lyi mutant in the P. multocida strain GX-PM with pSHK5(TS)-NgAgo-lyiLR, which was further inserted with other homologous sequences (opaL) or non-homologous sequences (gfp gene sequence).
Because the homologous arms were enough for NgAgo-assisted gene editing, we further tested whether other homologous sequences had an inhibitory effect of target gene deletion. Indeed, we observed that the phenotype based on the bacterial NgAgo system was significantly disturbed when additional homologous sequences (opal sequence) are introduced (Figure 2C and Supplementary Figure S15). In contrast, an exogenous non-homologous sequence (gfp sequence) could not interfere with the construction of an isogenic lyi mutant based on the NgAgo system (Figure 2C and Supplementary Figure S15). This result further suggested that NgAgo-assisted gene editing is based on homologous sequence-directed recombination.
NgAgo-assisted gene editing through binding to recA
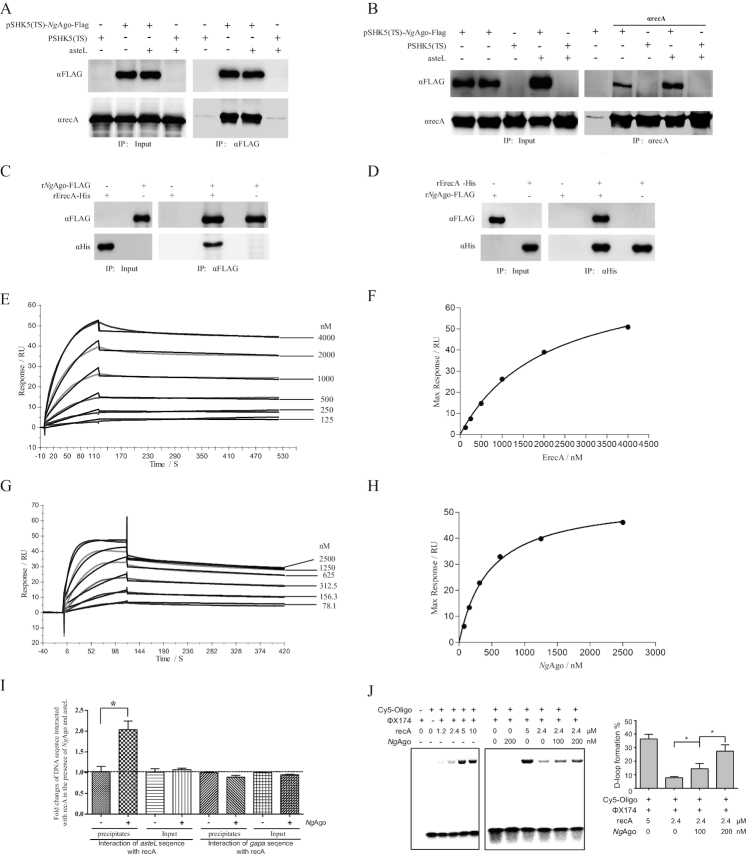
To elucidate the mechanism underlying NgAgo-assisted genetic manipulation, we used pull-down approaches coupled with LC-MS/MS to identify NgAgo-interacting proteins. Interestingly, homologous recombinase recA, an essential protein in bacterial homologous recombination and genome DNA repair (22), was identified as the dominant NgAgo-interacting protein (Supplementary Table S4). This specific interaction between NgAgo and recA was further validated by Co-IP approaches (Figure 3A and B), and phiC31 integrase as a control exhibited no interaction with NgAgo (Supplementary Figure S16). Furthermore, the interaction of NgAgo and recA was not significantly blocked in the presence of Dnase I (Supplementary Figure S17), indicating that the interaction was not mainly through binding to DNA.
Figure 3.
NgAgo interacts with recA and enhances recA-mediated DNA strand exchange in bacteria. (A) Co-IP assay detection of the interaction between NgAgo-FLAG and endogenous recA with αFLAG agarose. The E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL (the left arm sequences of aste gene), pSHK5(TS)-asteL, pSHK5(TS)-NgAgo-Flag or pSHK5(TS) and cultured for 48 hours. Total proteins were extracted for Co-IP assay with αFLAG agarose. (B) Co-IP assay detection of the interaction between NgAgo-FLAG and endogenous recA with SPA/G agarose and αrecA. The E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL, pSHK5(TS)-asteL, pSHK5(TS)-NgAgo-Flag or pSHK5(TS) and cultured for 48 h. Total proteins were extracted for Co-IP assay with SPA/G agarose and αrecA. (C) Direct interaction between the purified recombinant NgAgo-FLAG with the purified recombinant ErecA-His with αFLAG agarose. The eluted proteins were further subject for immunoblot with antibodies against recA or FLAG. (D) Direct interaction between the purified recombinant NgAgo-FLAG with the purified recombinant ErecA-His with Ni-NTA agarose. The eluted proteins were further subject for immunoblot with antibodies against recA or FLAG. (E) The curves representing the binding of purified rErecA-His to rNgAgo-His. rNgAgo-His and BSA were immobilized by amine coupling to each flow cell, and different concentrations (0-4 μM) of rErecA was injected over the flow cells for surface plasmon resonance analysis. Data were obtained as response value of rErecA-His to rNgAgo-His subtracting response value of rErecA-His to BSA. (F) The plotting of the RUmax at each concentration of rErecA for rNgAgo-His. These curves were fitted to the one-binding site model, giving values for KD constants. (G) The curves representing the binding of purified rNgAgo-His to rErecA-His. rErecA-His and BSA were immobilized by amine coupling to each flow cell, and different concentrations (0–2.5 μM) of rNgAgo-His was injected over the flow cells for surface plasmon resonance analysis. Data were obtained as response value of rNgAgo-His to rErecA-His subtracting response value of rNgAgo-His to BSA. (H) The plotting of the RUmax at each concentration of rNgAgo-His for rErecA. These curve was fitted to the one-binding site model, giving values for KD constants. (I) Enhanced binding of recA with the target DNA by NgAgo. The E. coli strain was transformed with pSHK5(TS)-NgAgo-Flag-asteL or pSHK5(TS)-asteL and cultured for 12 h, then cross-linked with formaldehyde. After sonication, the lysate was precipitated with SPA agarose and αrecA. After reverse crosslinking the samples, the samples were used for qPCR detection of asteL with the aste-RT-F/R primers or the irrespective gapa sequence with the gapa-RT-F/R primers. The reca sequence was used as an internal control for qPCR with the reca-RT-F/R primers. (J) NgAgo-assisted gene editing is through enhancing recA-mediated DNA strand exchange. DNA strand exchange assay was performed based on the phiX174 RF1 DNA and HPLC-purified phiX174-oligo chemically labeled with Cy5 in the presence of rErecA and various concentration of rNgAgo-His. The products were resolved on a 1% agarose gel, and the D-loop formation and free of Cy5-labeled oligos were imaged on a Fujifilm FLA-5100 (FUJIFILM Life Science, Japan). The D-loop formation % is the percentage of the signal of the D-loop formation of Cy5-labed oligos in the signal of the D-loop formation and free of Cy5-labeled oligos.
Moreover, the purified rNgAgo-FLAG could be co-precipitated with the purified recombinant His-tagged recA through Ni-NTA agarose. In addition, the purified recombinant His-tagged recA could also be co-precipitated with the purified rNgAgo-FLAG through αFLAG M2 agarose (Figure 3C, D and Supplementary Figure S18). These indicated the direct interaction of NgAgo with recA.
To further analyze the direct interaction of both proteins, the purified recombinant NgAgo was covalently bound to the SPR sensor chip, and a concentration-dependent binding response was observed for the purified recombinant recA (Ka = 5.74 ± 0.23 × 103 M−1 s−1, Kd = 2.4 ± 1.0 × 10−4 S−1, and KD = 42.4 ± 20.3 nM) (Figure 3E and F). Subsequently, the purified recombinant recA was covalently bound to the SPR sensor chip, and a concentration-dependent binding response was also observed for the purified recombinant NgAgo (Ka = 2.32 ± 0.17 × 104 M−1 s−1, kd = 7.07 ± 0.19 × 10−4 S−1, and KD = 30.6 ± 3.2 nM) (Figure 3G and H). These results further confirmed the direct interaction of both proteins.
It is known that the efficiency of homologous sequence-mediated recombination was dependent on the interaction of recA with the target sequences. We further confirmed that NgAgo could enhance interaction of recA with the target sequences, which was exhibited by Chip-qPCR (Figure 3I, Supplementary Table S2). As a result, the enhanced interaction of recA with the target sequences may account for NgAgo-promoted gene editing through homologous recombination.
NgAgo-assisted gene editing through enhancing recA-mediated DNA strand exchange
The recA promotes the central steps of recombination: it binds to single-stranded DNA (ssDNA) in an ATP-dependent manner, aligns and pairs two DNA molecules, and then promoting a strand switch followed by branch migration (22,41,42). The present study indicates NgAgo-assisted gene editing through binding to recA, we conceived NgAgo probably promotes recA-mediated DNA strand exchange, a major experimental model for the recombination activities (41). Interestingly, the purified recombinant NgAgo indeed promoted recA-mediated DNA strand exchange in a concentration-dependent manner (Figure 3J), while it did not significantly promote the interaction of recA with ssDNA (Supplementary Figure S19). Therefore, the present study indicated that NgAgo-assisted gene editing is through enhancing recA-mediated DNA strand exchange.
The PIWI-like domain of NgAgo is required to enhance gene editing in bacteria
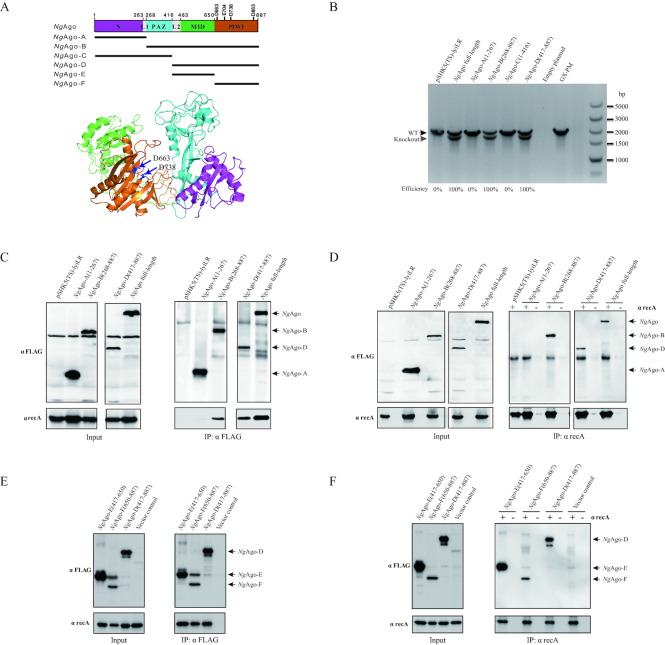
To functionally characterize the responsible domain of NgAgo for enhancement of gene editing, we constructed four NgAgo fragments to generate the pSHK5(TS)-NgAgo A (1-267 aa, containing N domain), B (268–887 aa, containing PAZ, MID and PIWI domains), C (1–416 aa, containing N and PAZ domains) and D (417-887 aa, containing MID and PIWI domains) plasmids (Figure 4A, Supplementary Table S1-S2). The pSHK5(TS)-NgAgo (A, B, C and D)-lyiLR plasmids were individually electro-transformed into P. multocida. As a result, isogenic mutants could only be detected with transformed NgAgo B and D rather than A and C (Figure 4B and Supplementary Figure S20), indicating that the NgAgo D fragment is enough to enhance gene editing in bacteria. Amino acid sequence alignment demonstrated that the NgAgo D fragment (C-terminus of NgAgo) contains the MID and PIWI domains. The MID domain usually forms binding pockets that facilitate the anchoring of the 5′-phosphate of the oligonucleotide guide, while the PIWI domain of catalytically active Agos mediates target-strand cleavage (28,30). Here, the C-terminus of NgAgo is as efficient as full-length NgAgo in enhancement of gene editing in bacteria, with almost 100% efficiency. Interestingly, using pull-down approaches coupled with LC-MS/MS, recA was identified from full-length of NgAgo, and NgAgo fragments B and D, but not from NgAgo A fragment with the highest MASCOT search score and the largest numbers of identified unique peptides (Supplementary Table S4). The interaction between the C-terminus of NgAgo and recA was further confirmed by forward/reverse protein-interaction assay (Figure 4C-D). Thus, we conclude that the C-terminus of NgAgo is responsible for gene editing in bacteria by interaction with recA.
Figure 4.
The PIWI-like domain of NgAgo is required for promotion of gene editing in bacteria. (A) The structural homology modelling of NgAgo major domains aligned to Aquifex aeolicus Argonaute (PDB: 1YVU) and Thermus thermophilus Argonaute (PDB: 5XOU) using the HHPred server and Pymol. N, L, PAZ, MID and PIWI domains are highlighted in purple, grey, blue, green and orange, respectively. The structure of other pAgos were supplied in Figure S11. (B) Comparison of the construction efficiency for the isogenic lyi mutant in the P. multocida strain GX-PM based on various NgAgo fragments: A (1-267), B (268-887), C (1–416) and D (417–887). (C) Co-IP assay detection of interaction between recA and the various NgAgo fragments with αFLAG agarose. The E. coli strain was transformed with pSHK5(TS)-NgAgo (A, B or D)-Flag-asteL or pSHK5(TS)-asteL and cultured for 48 h. Total proteins were extracted for Co-IP assay with αFLAG agarose. (D) Co-IP assay detection of interaction between recA and the various NgAgo fragments with SPA/G agarose and αrecA. The E. coli strain was transformed with pSHK5(TS)-NgAgo (A, B or D)-Flag-asteL or pSHK5(TS)-asteL and cultured for 48 h. Total proteins were extracted for Co-IP assay with SPA/G agarose and αrecA. (E) Co-IP assay detection of interaction between recA and the MID domain (fragment E) or PIWI-like domain (fragment F) of NgAgo with αFLAG agarose. The fragment D of NgAgo and the vector were severed as the positive and negative controls. (F) Co-IP assay detection of interaction between recA and the MID domain (fragment E) or PIWI-like domain (fragment F) of NgAgo with SPA/G agarose and αrecA. The fragment D of NgAgo and the vector were severed as the positive and negative controls.
To further elucidate whether the effect relies on MID and / or PIWI domain, pSHK5(TS)–NgAgo E (417–650 aa, containing MID domain) and F (650–887 aa, containing PIWI-like domain) -opaLR plasmids were individually electro-transformed into P. multocida. As a result, isogenic mutants could be detected with transformed NgAgo E and F with efficiency of 20% and 100% (Supplementary Figure S21), indicating that the PIWI-like domain is enough to enhance gene editing in bacteria. The interaction between the PIWI-like domain of NgAgo and recA was further confirmed by forward/reverse protein-interaction assay (Figure 4E and F). Thus, we conclude that the PIWI-like domain of NgAgo is responsible for gene editing in bacteria by interaction with recA.
The pAgos enhance gene editing in bacteria
NgAgo enhances homologous sequence-guided gene editing in bacteria, which promoted us to further evaluate the effect of other pAgos. Interestingly, the isogenic opa gene mutants could easily be detected at 100% efficiency with PfAgo-D, TtAgo-D and AaAgo-D, while the isogenic mutant based on the natural homologous recombination in the P. multocida strain was still not detected (Supplementary Figure S22). It indicated that enhancement of gene editing in bacteria was not solely for NgAgo, and it might be mediated by other pAgos. Furthermore, the interactions of these pAgos with recA were also detected (Supplementary Figure S23), suggesting that these pAgos enhance gene editing in bacteria in a similar manner by which NgAgo takes effect.
DISCUSSION
The development of a broad and effective genetic manipulation system is the fundamental for vaccine development or the study of pathogenesis. The developed NgAgo-based system was highly efficient for gene editing in various bacterial strains. More importantly, this tool makes the rapid construction of an avirulent strain feasible. For example, the virulence of the constructed isogenic hyae mutant in the highly pathogenic P. multocida strain was significantly decreased (Figure 1D and E), potentiating the construction of a novel generation of avirulent vaccine strain with gene editing for fowl cholera. Additionally, the successful insertion of a vp60 gene from rabbit haemorrhagic disease virus into the rabbit P. multocida strain provided an alternative strategy to construct a novel vaccine strain against two important infectious diseases in rabbit (Figure 1F). This was essential for developing a safe and cheap vaccine against rabbit haemorrhagic disease virus due to a lack of choices in addition to an inactivated vaccine derived from a clarified liver suspension from experimentally infected rabbits (43).
Argonaute proteins in eukaryotes and prokaryotes are evolutionarily conserved (44). The eukaryotic Argonaute proteins play a key role in RNA interference pathways either directly by binding and cleavage of the target RNA, or indirectly by binding to the target RNA followed by the recruitment of the silencing proteins (44–46). In contrast, several pAgos were reported to mediate nucleic acid-guided cleavage of cognate DNA targets (31–34) or RNA targets in vitro (32,35). The present study identified a new way for the PIWI-like domain of some pAgos in enhancing homologous sequence-guided gene editing in bacteria independent of its potential enzymatic activity and a guide DNA or RNA. This will undoubtedly strengthen our understanding of the applications potential of pAgos on gene editing.
The recently developed CRISPR/Cas9 system is clearly a powerful tool for bacterial genome editing (1,17–19,47–49) that relies on gRNA:Cas9-directed cleavage at a targeted genomic site to kill unmutated cells and circumvent the need for selectable markers or counter-selection systems (18). The effect depends on cleavage by Cas9 with a guided RNA, meaning that the efficiency can change with different guide RNA sequences (50) and in different bacterial species; for example, it is ∼100% in S. pneumonia but only 65% in E. coli (18). Although a few improved CRISPR/Cas9 systems have been developed to prevent the off-target effects (51,52), their application in bacteria has not been evaluated to date. More importantly, the CRISPR/Cas9 system requires a large gene (the gene length of Cas9 is ∼ 4 kb) and elements expressing a guide RNA, which increase the load of the tool vector. Thus, it will be difficult to introduce this system into some bacteria with unknown genetic background or lack of genetic technique (e.g. P. multocida) for gene editing.
In contrast, the pAgos system is completely distinct from CRISPR-Cas systems. Apart from its high efficiency for bacterial gene editing, this pAgos system has other additional merits. The PIWI-like domain of NgAgo was only ∼0.7 kb, which makes the system easy to manipulate in only one vector to accomplish recombination at a high efficiency. Additionally, this system is independent of its potential nuclease activity, which could eliminate the concern for potential off-target damage to the genome (Figure 1C). In light of the PIWI-like domain of pAgos-assisted gene editing through the promotion of homologous recombination in bacteria, which was different from the currently developed tool to isolate mutants by killing non-mutant strains, like in the CRISPR-Cas system, the pAgos system could not only be considered an alternative system, but might also be a component in combination with other tools (e.g. CRISPR-Cas system) to further enhance the efficiency of gene editing.
While a DNA-guided NgAgo system is controversial in its application in eukaryotic cells (36,38,53–55), based on the above findings, we conclude that the PIWI-like domain of NgAgo system substantially enhances homologous recombination in bacteria through binding to recA to enhance recA-mediated DNA strand exchange, a major experimental model for the recombination activities (41). As the pAgos system only requires homologous arms and enhances homologous recombination without detectable off-target effects (genome damage), it offers great convenience for gene editing in bacteria, though the role of this system in mammal genome manipulation remains to be further elucidated. The present study also demonstrated a novel aspect of pAgos on gene editing in bacteria.
DATA AVAILABILITY
The NGS data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP135583. The MS data have been submitted to the integrated proteome resources iProX (http://www.iprox.org) under Project ID number IPX0001178000.
More additional materials and methods are provided in the Supplemental materials and methods.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2017YFD0500204, 2016YFD0500801, 2017YFC1700402]; National Natural Science Foundation of China [81822048, 81770256, 34115109]; Agro-scientific Research in the Public Interest [201303044]; Natural Science Foundation of Hubei Province [2015CFA041, 2016CFA015]; Fundamental Research Funds for the Central University [2662018PY044]; Foundation of State Key Laboratory of Veterinary Biotechnology [SKLVBF2019103]. Funding for open access charge: National Key Research and Development Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jiang Y., Qian F.H., Yang J.J., Liu Y.M., Dong F., Xu C.M., Sun B.B., Chen B., Xu X.S., Li Y. et al.. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017; 8:15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li K., Cai D., Wang Z., He Z., Chen S.. Development of an efficient genome editing tool in bacillus licheniformis using CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 2018; 84:e02608-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bravo D., Landete J.M.. Genetic engineering as a powerful tool to improve probiotic strains. Biotechnol. Genet. Eng. Rev. 2017; 1:1–17. [DOI] [PubMed] [Google Scholar]
- 4. Case M.E., Geever R.F., Asch D.K.. Use of gene replacement transformation to elucidate gene function in the qa gene cluster of Neurospora crassa. Genetics. 1992; 130:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnsrud L., Calos M.P., Miller J.H.. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell. 1978; 15:1209–1219. [DOI] [PubMed] [Google Scholar]
- 6. Thorpe H.M., Smith M.C.M.. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:5505–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang P., Wang J., Qi Q.S.. Prophage recombinases-mediated genome engineering in Lactobacillus plantarum. Microb. Cell Factor. 2015; 14:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Staroscik A.M., Hunnicutt D.W., Archibald K.E., Nelson D.R.. Development of methods for the genetic manipulation of Flavobacterium columnare. BMC Microbiol. 2008; 8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wielgoss S., Barrick J.E., Tenaillon O., Wiser M.J., Dittmar W.J., Cruveiller S., Chane-Woon-Ming B., Medigue C., Lenski R.E., Schneider D.. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Court D.L., Sawitzke J.A., Thomason L.C.. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002; 36:361–388. [DOI] [PubMed] [Google Scholar]
- 11. Kamoun S., Tola E., Kamdar H., Kado C.I.. Rapid generation of directed and unmarked deletions in Xanthomonas. Mol. Microbiol. 1992; 6:809–816. [DOI] [PubMed] [Google Scholar]
- 12. Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G.. Simple and highly efficient BAC recombineering using gaIK selection. Nucleic Acids Res. 2005; 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Briggs R.E., Tatum F.M.. Generation and molecular characterization of new temperature-sensitive plasmids intended for genetic engineering of Pasteurellaceae. Appl. Environ. Microbiol. 2005; 71:7187–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong Q.N.Y., Ng V.C.W., Lin M.C.M., Kung H.F., Chan D., Huang J.D.. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Res. 2005; 33:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu J., Deng A., Sun Q., Bai H., Sun Z., Shang X., Zhang Y., Liu Q., Liang Y., Liu S. et al.. Bacterial genome editing via a designed Toxin-Antitoxin cassette. ACS Synth. Biol. 2017; 7:822–831. [DOI] [PubMed] [Google Scholar]
- 16. Bai H., Deng A., Liu S., Cui D., Qiu Q., Wang L., Yang Z., Wu J., Shang X., Zhang Y. et al.. A novel tool for microbial genome editing using the Restriction-Modification system. ACS Synth. Biol. 2018; 7:98–106. [DOI] [PubMed] [Google Scholar]
- 17. Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S.. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015; 81:2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A.. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013; 31:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cobb R.E., Wang Y., Zhao H.. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 2015; 4:723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishag H.Z.A., Xiong Q.Y., Liu M.J., Feng Z.X., Shao G.Q.. E-coli recA gene improves gene targeted homologous recombination in Mycoplasma hyorhinis. J. Microbiol. Methods. 2017; 136:49–56. [DOI] [PubMed] [Google Scholar]
- 21. Allam A.B., Reyes L., Assad-Garcia N., Glass J.I., Brown M.B.. Enhancement of targeted homologous recombination in Mycoplasma mycoides subsp. capri by inclusion of heterologous recA. Appl. Environ. Microbiol. 2010; 76:6951–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Z., Yang H., Pavletich N.P.. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008; 453:489–484. [DOI] [PubMed] [Google Scholar]
- 23. Harper M., Boyce J.D., Adler B.. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 2006; 265:1–10. [DOI] [PubMed] [Google Scholar]
- 24. Kock R.A., Orynbayev M., Robinson S., Zuther S., Singh N.J., Beauvais W., Morgan E.R., Kerimbayev A., Khomenko S., Martineau H.M. et al.. Saigas on the brink: Multidisciplinary analysis of the factors influencing mass mortality events. Sci. Adv. 2018; 4:eaao2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christenson E.S., Ahmed H.M., Durand C.M.. Pasteurella multocida infection in solid organ transplantation. Lancet Infect. Dis. 2015; 15:235–240. [DOI] [PubMed] [Google Scholar]
- 26. Yu C., Sizhu S., Luo Q., Xu X., Fu L., Zhang A.. Genome sequencing of a virulent avian Pasteurella multocida strain GX-Pm reveals the candidate genes involved in the pathogenesis. Res. Vet. Sci. 2016; 105:23–27. [DOI] [PubMed] [Google Scholar]
- 27. Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C.. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998; 17:170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L.. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004; 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 30. Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools. Nat. Rev. Microbiol. 2018; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 31. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J. et al.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D.. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014; 11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swarts D.C., Koehorst J.J., Westra E.R., Schaap P.J., van der Oost J.. Effects of argonaute on gene expression in thermus thermophilus. PLoS One. 2015; 10:e0124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S.H., Turchiano G., Ata H., Nowsheen S., Romito M., Lou Z., Ryu S.M., Ekker S.C., Cathomen T., Kim J.S.. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2016; 35:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cyranoski D. Updated: NgAgo gene-editing controversy escalates in peer-reviewed papers. Nature. 2016; 540:20–21. [DOI] [PubMed] [Google Scholar]
- 38. Qi J., Dong Z., Shi Y., Wang X., Qin Y., Wang Y., Liu D.. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 2016; 26:1349–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Z., Tan S., Xu L., Gao L., Zhu H., Ma C., Liang X.. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017; 145:20–23. [DOI] [PubMed] [Google Scholar]
- 40. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J.. Crystal structure of A-aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox M.M. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007; 42:41–63. [DOI] [PubMed] [Google Scholar]
- 42. Cox M.M. The RecA protein as a recombinational repair system. Mol. Microbiol. 1991; 5:1295–1299. [DOI] [PubMed] [Google Scholar]
- 43. Lavazza A., Capucci L.. Chapter 2.6.2: rabbit hemorrhagic disease. The Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Terrestrial Manual). 2016; 1–18. [Google Scholar]
- 44. Hock J., Meister G.. The Argonaute protein family. Genome Biol. 2008; 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 46. Hannon G.J. RNA interference. Nature. 2002; 418:244–251. [DOI] [PubMed] [Google Scholar]
- 47. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Selle K., Barrangou R.. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015; 23:225–232. [DOI] [PubMed] [Google Scholar]
- 49. Wang Y., Zhang Z.T., Seo S.O., Lynn P., Lu T., Jin Y.S., Blaschek H.P.. Bacterial Genome Editing with CRISPR-Cas9: Deletion,Integration, Single Nucleotide Modification, and Desirable “Clean" Mutant Selection in Clostridium beijerinckii as an Example. ACS Synth Biol. 2016; 5:721–732. [DOI] [PubMed] [Google Scholar]
- 50. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z.L., Joung J.K.. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A.. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017; 550:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao F., Shen X.Z., Jiang F., Wu Y., Han C.. Retraction: DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2017; 35:797. [DOI] [PubMed] [Google Scholar]
- 54. Gao F., Shen X.Z., Jiang F., Wu Y., Han C.. Addendum: Editorial Expression of Concern: DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2017; 35:481. [DOI] [PubMed] [Google Scholar]
- 55. Gao F., Shen X.Z., Jiang F., Wu Y., Han C.. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2016; 34:768–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP135583. The MS data have been submitted to the integrated proteome resources iProX (http://www.iprox.org) under Project ID number IPX0001178000.
More additional materials and methods are provided in the Supplemental materials and methods.