Abstract
Centromere identity is determined by the specific deposition of CENP-A, a histone H3 variant localizing exclusively at centromeres. Increased CENP-A expression, which is a frequent event in cancer, causes mislocalization, ectopic kinetochore assembly and genomic instability. Proteolysis regulates CENP-A expression and prevents its misincorporation across chromatin. How proteolysis restricts CENP-A localization to centromeres is not well understood. Here we report that, in Drosophila, CENP-ACID expression levels are regulated throughout the cell cycle by the combined action of SCFPpa and APC/CCdh1. We show that SCFPpa regulates CENP-ACID expression in G1 and, importantly, in S-phase preventing its promiscuous incorporation across chromatin during replication. In G1, CENP-ACID expression is also regulated by APC/CCdh1. We also show that Cal1, the specific chaperone that deposits CENP-ACID at centromeres, protects CENP-ACID from SCFPpa-mediated degradation but not from APC/CCdh1-mediated degradation. These results suggest that, whereas SCFPpa targets the fraction of CENP-ACID that is not in complex with Cal1, APC/CCdh1 mediates also degradation of the Cal1-CENP-ACID complex and, thus, likely contributes to the regulation of centromeric CENP-ACID deposition.
INTRODUCTION
Centromere identity is determined epigenetically by the specific deposition at centromeres of the histone H3 variant CENP-A (also called CenH3) (reviewed in (1–6)). Several mechanisms ensure that CENP-A is deposited only at centromeres. Centromeric CENP-A deposition is replication-independent (7,8) and, in most studied cases, occurs in G1 (9–14). CENP-A deposition requires the contribution of licensing factors, such as M18BP1 that recognizes centromeric chromatin and modifies it for deposition (15–19), and specific CENP-A chaperones, such as Scm3 in yeasts (20–22), Cal1 in Drosophila (23–28) and HJURP in vertebrates (29–31). In mammals, Cdk1/2 phosphorylates M18BP1 and HJURP inhibiting CENP-A loading outside of G1 (32). These mechanisms are overcome when CENP-A is overexpressed in yeast, Drosophila and mammalian cells, leading to its misincorporation across chromatin (33–42). In mammalian cells, ectopic CENP-A deposition depends on the H3.3 chaperone DAXX (43). Mistargeting of CENP-A to non-centromeric sites has important consequences since it leads to ectopic kinetochore formation, chromosome instability and aneuploidy (37,44–46). In this regard, increased CENP-A expression has been reported in several tumors, correlating with high aggressiveness and invasiveness (44,47–52). Therefore, it is of great importance to better understand the mechanisms that regulate CENP-A expression and stability.
Proteolytic degradation has been shown to regulate CENP-A expression in yeast and Drosophila, acting as a safeguard mechanism that prevents CENP-A misincorporation across chromatin (35,38). In yeast, four different E3-ubiquitin ligases have been reported to be involved in CENP-ACse4 degradation (53–56). In Drosophila, the F-box protein Partner-of-paired (Ppa), which is a variable subunit of the E3-ubiquitin ligase SCF, has been shown to interact with CENP-ACID and down-regulate its expression, preventing its ectopic deposition at non-centromeric sites (57). However, how these proteolytic activities regulate CENP-A stability across the cell cycle and contribute to its timely deposition at centromeres remains largely unknown. Here we analyze these questions in vivo in Drosophila and identify APC/CCdh1 as a second major E3-ubiquitin ligase that, together with SCFPpa, regulates CENP-ACID stability during cell cycle progression.
MATERIALS AND METHODS
Antibodies
Rabbit polyclonal αCENP-ACID is described in (38). Rabbit polyclonal αCal1 is a gift from Dr Erhardt and is described in (73). The rest of antibodies used in these experiments are commercially available: mouse monoclonal αGFP (Roche, 11 814 460 001), rabbit polyclonal αActin (Sigma, A2066), rat monoclonal αElav (DSHB, 7E8A10), rabbit polyclonal αPH3 (Millipore, 06-570), mouse monoclonal αProspero (DSHB, MR1A) and mouse monoclonal αCut (DSHB, 2B10).
Stable S2 cell lines
ppa promoter (nucleotide position +1 to –1000) and ppa cDNA (the 3′UTR included) were obtained from genomic DNA by PCR-amplification using appropriate primers and cloned into pEGFP-C1 (Clontech) to generate plasmid pGFP-Ppa, which expresses GFP::Ppa under the control of the ppa promoter. To obtain stable cell lines, Drosophila S2 cells were grown under standard conditions (in Schneider's medium (Sigma) supplemented with 10% FBS (Gibco), 100 mg/ml Streptomycin and 100 mg/ml Penicillin at 25°C) and transfected by the calcium phosphate method (74) with pGFP-Ppa. After 48 h of transfection, 0.8 mg/ml G418 was added for selection.
Fly stocks and genetic procedures
Transgenic UAS-CENP-ACID::YFP flies are described in (57). ppaRNAi corresponds to line 9952R-2 from NIG-FLY and is described in (57). Transgenic UAS-Cal1 flies were kindly provided by Dr. Lehner and are described in (24). APC2RNAi, Cdh1RNAi, Cdc20RNAi and Cal1RNAi correspond to lines 106986, 25550, 40500 and 45248 from VDRC, respectively. ey3.5-GAL4, longGMR-GAL4 and elav-GAL4 were obtained from the Bloomington Stock Center. ey3.5-GAL4;UAS-CENP-ACID::YFP and longGMR-GAL4;UAS-CENP-ACID::YFP stocks were obtained by conventional genetic crosses and maintained at 18°C or 25°C.
For RNAi-mediated depletion experiments homozygous ey3.5-GAL4;UAS-CENP-ACID::YFP and longGMR-GAL4;UAS-CENP-ACID::YFP flies were crossed at 29°C to the corresponding homozygous RNAi flies and to white flies as control, except for APC2RNAi where crosses were performed at 25°C. After 3 days adult flies were removed and the crosses were kept at the corresponding temperature until larvae reached the third-instar stage (∼5–6 days at 29°C and 7–8 days at 25°C). When the effects of APC2RNAi and APC10RNAi on endogenous CENP-ACID expression levels were determined in third-instar larvae brains, depletion was induced by elav-GAL4 at 29°C. For Cal1 overexpression in ppaRNAi or Cdh1RNAi flies, homozygous ppaRNAi;UAS-Cal1 and Cdh1RNAi;UAS-Cal1 stocks were obtained by conventional genetic crosses and maintained at 18°C or 25°C. To analyze the effect on CENP-ACID::YFP expression these lines were crossed at 29°C with homozygous ey3.5-GAL4;UAS-CENP-ACID::YFP flies. After three days adult flies were removed and the crosses were kept at 29°C until larvae reached the third-instar stage (∼5–6 days).
Measure of eye size
When eye area was measured, adult flies were collected and kept at –20°C for 24 h. Images were collected using a SZX16 stereomicroscope equipped with an Olympus XC50 camera and CellD software. Eye images were measured and analyzed using Fiji software (75).
Fluorescence microscopy analysis
For direct fluorescence visualization, eye imaginal discs and salivary glands from third-instar larvae were dissected in PBS, fixed in 4% paraformaldehyde for 20 min at room temperature, washed in PBS/0.3% Triton X-100 three times for 10 min and once in PBS for 10 min, incubated for 30 min at room temperature with 0.02ng/μl DAPI in PBS, washed for 5 min in PBS/0.3% Triton X-100 and mounted in Mowiol (Calbiochem-Novabiochem). For immunostaining, eye imaginal discs were dissected and fixed as described above, washed in PBS/0.3% Triton X-100 three times for 10 min and blocked in PBS/0.3% Triton X-100/2% BSA three times for 10 min. Then, discs were incubated overnight at 4°C with the primary antibody diluted in blocking buffer, washed in blocking buffer three times for 10 min, incubated for 2 h at room temperature with secondary antibody in blocking buffer, washed in PBS/0.3% Triton X- 100 three times for 10 min and once in PBS for 10 min, incubated for 30 min at room temperature with 0.02 ng/μl DAPI in PBS, washed for 5 min in PBS/0.3% Triton X-100 and mounted in Mowiol (Calbiochem-Novabiochem). For immunostaining to detect endogenous CENP-ACID, eye imaginal discs were processed as described above, but, after fixation, 0.25 M NaCl was added to the washes, the blocking and the incubation with the primary antibody. For larvae brains, squashes and immunostainings were performed as described in (57). In S2 cells, direct fluorescence microscopy visualization was performed with cells immobilized onto a slide by centrifugation for 10 min at 500 rpm on low acceleration in a TermoShandon Cytospin 4 using a single-chamber Cytospin funnel. Slides were fixed in 4% paraformaldehyde for 10 min, washed with PBS for 15 min and mounted in Mowiol (Calbiochem-Novabiochem). For immunolocalization with specific antibodies, cells were plated on cover slips treated with Concanavalin-A (Sigma) for 2 h at 25°C. Then they were washed with PBS for 10 min, fixed in 4% paraformaldehyde for 15 min, washed with PBS for 15 min, blocked in PBS/0.1% Triton X-100/0.1% BSA for 20 min and incubated with primary antibodies in blocking solution overnight at 4°C. After incubation, cover slips were washed three times for 10 min with blocking solution and incubated for 1 h at room temperature with secondary antibody diluted in blocking solution. Finally, they were washed twice for 10 min in PBS/0.1% Triton X-100, twice in PBS, and mounted in Mowiol (Calbiochem-Novabiochem) containing 0.2 ng/ml DAPI (Sigma). Primary antibodies were αElav (1:100), αPH3 (1:2000), αCENP-ACID (1:300), αProspero (1:10) and αCut (1:100). Secondary antibodies were coupled to Cy3 and Cy5 (Jackson Immunoresearch laboratories) and were used at 1:400 dilutions. Images were collected in a Leica TCS/SPE confocal microscope equipped with LAS/AF software and analyzed with Fiji software (75). To determine nuclei size in salivary glands Fiji software was used to create a mask with DAPI channel and the area of each nucleus was determined. For quantitative analyses of endogenous CENP-ACID levels in eye imaginal discs, fluorescence intensity was determined using the Fiji distribution of ImageJ (75). Integrated density of CENP-ACID spots were calculated using a mask of CENP-ACID channel created from thresholded images on the FeatureJ Laplacian (http://imagescience.org/meijering/software/featurej/) of the regions of interest (anterior or posterior to MF) and running Analyze particles plugin.
EdU incorporation
For EdU incorporation experiments, salivary glands from third-instar larvae were dissected in PBS and incubated with 10μM EdU (ThermoFisher Scientific) for 5 min at room temperature. EdU detection was performed with Click-iT™ Plus EdU Alexa Fluor™ 594 Imaging Kit (ThermoFisher Scientific) following manufacturer's protocol. DAPI staining, mounting and fluorescence visualization were performed as described above.
Western blot (WB) analysis
Total protein extracts were prepared from 35 third-instar larvae salivary glands dissected in PBS. Dissected salivary glands were transferred to 1xPLB/0.05% NP40/2mM PMSF, disrupted by pipetting and boiled 5 min at 95°. Extracts were analyzed by WB with αGFP (1:2000) (to detect the YFP signal), αCal1 (1:2000) and αActin (1:1000) antibodies. Quantitative analyses were carried out with a GS-800 Calibrated Densitometer (Bio-Rad) and Fiji software (75).
FACS sorting
For FACS sorting, cells were fixed in 1% paraformaldehyde for 1 h at 4°C, permeabilized with 70% ethanol and stained with 1 μg/ml DAPI (Sigma). Then, cells were sorted in an Aria SORP flow cytometer (Becton Dickinson) with a UV laser.
RESULTS
Ppa regulates CENP-ACID expression in G1 and S-phase
Previous results showed that the E3-ligase SCFPpa regulates CENP-ACID stability (57). In general, the activity of SCF complexes is cell cycle regulated (58–61). Thus, we analyzed the effect of SCFPpa on CENP-ACID stability across the cell cycle. For this purpose, we took advantage of the cell cycle synchronization that the morphogenetic furrow (MF) induces in the eye imaginal disc of third instar larvae (62,63). MF is a dorso-ventral indentation that moves anterior and induces differentiation. Immediately anterior to MF, asynchronously dividing cells undergo a first synchronized mitosis (FMW). Later, exiting the MF, posterior cells undergo a second synchronized mitosis (SMW), arrest in G1 and differentiate (Figure 1A). In this experimental setting, we induced ectopic expression of a UAS-CENP-ACID::YFP construct in G1-arrested cells using ey3.5-GAL4, which is active posterior to the MF (Supplementary Figure S1A, top). Under these conditions, we detected low CENP-ACID::YFP expression (Figure 1B, top). However, simultaneous depletion of Ppa strongly increased CENP-ACID::YFP levels in G1-arrested posterior cells (Figure 1B, bottom). This increase was not due to a defect on cell cycle progression and proliferation since Ppa depletion did not significantly affect the number of mitotic cells in the posterior region of eye imaginal discs (Figure 1C) or eye size in adult flies (Figure 1D).
Figure 1.
Ppa regulates CENP-ACID expression in G1. (A) Schematic representation of the synchronization induced by the morphogenetic furrow (MF) in the eye imaginal disc. FMW, first mitotic wave. SMW, second mitotic wave. G1, G1-arrested cells. MF moves from posterior to anterior as indicated. (B) The expression of CENP-ACID::YFP in the eye imaginal disc is determined by direct fluorescence (in green) in control ey3.5>CENP-ACID::YFP flies (top) and upon Ppa depletion in ey3.5>CENP-ACID::YFP; ppaRNAi flies (bottom). Immunostainings with αPH3, which marks mitotic cells, are also presented (in red). DNA was stained with DAPI (in white). The position of the MF is indicated. Scale bar corresponds to 25 μm. (C) The number of posterior αPH3-positive cells per disc is presented for control ey3.5>CENP-ACID::YFP flies (N = 10) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 7) (P-value > 0.01, two-tailed t-test; error bars are SEM). D) In the left, the eye phenotypes of control ey3.5>CENP-ACID::YFP flies and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies are presented. Scale bar corresponds to 100 μm. In the right, quantitative analysis of the results showing box plots of the eye area of control ey3.5>CENP-ACID::YFP flies (N = 20) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 20). (P-value > 0.01, two-tailed t-test). (E) In the left, enlarged images of the region indicated by the box in A. Scale bar corresponds to 10 μm. In the right, the percentage of cells showing high levels of mislocalized CENP-ACID::YFP in the posterior region of eye imaginal discs from control ey3.5>CENP-ACID::YFP flies (N = 7) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 5) is presented for total and αPH3-negative and –positive cells (**P-value < 0.001, two-tailed t-test; errors bars are SEM).
We also analyzed the effect of Ppa depletion on CENP-ACID::YFP expression in salivary glands, where ey3.5-GAL4 is also active (Supplementary Figure S1B). Salivary gland cells undergo multiple endoreplication cycles in which, after DNA replication, cells skip G2/M and re-enter G1 (64–66). As in the eye imaginal disc, Ppa depletion increased CENP-ACID::YFP levels, as determined by both immunofluorescence (IF) (Figure 2A) and western blot (WB) analyses (Figure 2B). EdU-incorporation experiments showed that CENP-ACID::YFP levels increased in both EdU-negative G1 cells and replicating EdU-positive cells (Figure 2C). Ppa depletion did not significantly affect the proportion of EdU-positive cells (Figure 2D) or the number of nuclei per gland and their size (Figure 2E), suggesting that it did not significantly affect endocycling of salivary gland cells. These results confirm that Ppa regulates CENP-ACID expression in G1 and that, at least in the specialized salivary glands cells, Ppa is also acting during chromatin replication in S-phase.
Figure 2.
Ppa regulates CENP-ACID expression during DNA replication. (A) The expression of CENP-ACID::YFP in salivary glands is determined by direct fluorescence (in green) in control ey3.5>CENP-ACID::YFP flies (top) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (bottom). DNA was stained with DAPI (in white). Scale bars correspond to 100 μm. (B) Western blot (WB) analysis with αGFP antibodies of the levels of CENP-ACID::YFP expression in salivary glands from control ey3.5>CENP-ACID::YFP flies and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies. Increasing amounts of extract are analyzed (lanes 1–3). αActin antibodies were used as loading control. Quantitative analysis of the results is shown in the bottom (N = 3; ****P-value < 0.00001, two-tailed t-test; errors bars are SEM). (C) As in A, but for salivary glands subjected to EdU-incorporation (in red) to detect replicating cells. Arrows indicate EdU-positive cells. Scale bars correspond to 100 μm. (D) The percentage of EdU-positive salivary gland cells is presented for control ey3.5>CENP-ACID::YFP flies (N = 8) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 12) (P-value > 0.01, two-tailed t-test; error bars are SEM). (E) In the left, the number of nuclei per gland is presented for control ey3.5>CENP-ACID::YFP flies (N = 10) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 24) (P-value > 0.01, two-tailed t-test; error bars are SEM). In the right, nuclei area of salivary glands cells is presented for control ey3.5>CENP-ACID::YFP flies (N = 10) and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies (N = 24) (P-value > 0.01, two-tailed t-test; error bars are SEM).
Next, we performed similar experiments using longGMR-GAL4 to induce UAS-CENP-ACID::YFP expression in the eye imaginal disc cells located most posterior to the MF (Supplementary Figure S1A, bottom). Also in this case, CENP-ACID::YFP levels were very low in control flies and increased upon Ppa depletion (Figure 3A). Concomitantly, a necrotic eye phenotype was observed in ∼60% of adult flies (N = 33) (Figure 3B). We observed that cells showing increased CENP-ACID::YFP levels did not stain with markers of neuronal and cone cell differentiation (Figure 3C, top and Supplementary Figure S2), suggesting that they corresponded to cells that remain undifferentiated in the larval eye imaginal disc. As a matter of fact, secondary and tertiary pigment cells and mechanosensory bristles differentiate later at the pupae stage (62,63,67). These undifferentiated cells can eventually undergo mitosis. In this regard, we observed that cells showing increased CENP-ACID::YFP levels were not stained with αPH3 (Figure 3C, bottom). Similar results were observed in experiments using ey3.5-GAL4. Also in this case, αPH3-positive posterior cells generally showed low CENP-ACID::YFP levels (Figure 1E, left). In fact, upon Ppa depletion, cells showing high levels of mislocalized CENP-ACID::YFP mainly corresponded to αPH3-negative cells (Figure 1E, right). Altogether these results suggest that Ppa is not regulating CENP-ACID::YFP levels in mitosis. In this regard, it is possible that Ppa is not expressed or, alternatively, it is not active during mitosis. Specific αPpa antibodies that could be used to directly address this question are not available. However, using a stable S2 cell line expressing a GFP::Ppa tagged construct under the control of the Ppa promoter, we detected GFP::Ppa expression in αPH3-positive cells (Supplementary Figure S3A), as well as in G2/M FACS-sorted cells (Supplementary Figure S3B). GFP::Ppa expression was also detected in G1- and S-phase sorted cells (Supplementary Figure S3B). Altogether these results suggest that, although Ppa is apparently expressed throughout the cell cycle, its contribution to the regulation of CENP-ACID stability is restricted to G1 and S-phase.
Figure 3.
Ppa does not regulate CENP-ACID expression in differentiated cells in the eye imaginal disc. (A) The expression of CENP-ACID::YFP in the eye imaginal disc is determined by direct fluorescence (in green) in control longGMR>CENP-ACID::YFP flies (left) and Ppa-depleted longGMR>CENP-ACID::YFP; ppaRNAi flies (right). DNA was stained with DAPI (in white). The position of the MF is indicated. Scale bars correspond to 25 μm. (B) The eye phenotypes of control longGMR-GAL4 flies and Ppa-depleted longGMR>ppaRNAi flies, expressing CENP-ACID::YFP (+) or not (–), are presented. Scale bars correspond to 100 μm. In the bottom, the proportions of flies showing strong, mild or no necrotic eye phenotype are presented for the indicated genotypes (N > 56). (C) Immunostaining with αELAV (in blue), which marks neuronal differentiated cells, and αPH3 (in red), which marks mitotic cells, of eye imaginal discs from Ppa-depleted longGMR>CENP-ACID::YFP; ppaRNAi flies. CENP-ACID::YFP expression is determined by direct fluorescence (in green). DNA was stained with DAPI (in white). The position of the MF is indicated. Scale bar corresponds to 25 μm.
APC/C also regulates CENP-ACID expression in G1
Results reported above indicate that the effects of Ppa depletion on CENP-ACID::YFP expression are not uniform during cell cycle progression and differentiation. Yet, in those cell types and cell cycle phases where Ppa depletion caused no effect, CENP-ACID::YFP levels remained low, suggesting that additional factors contribute to the regulation of CENP-ACID stability. In this regard, we tested whether APC/C, a major cell cycle regulated E3-ligase (59–61,68), contributes to CENP-ACID stability. We observed that depletion of APC2, an essential APC/C subunit, increased CENP-ACID::YFP levels in salivary glands (Figure 4A and B), without affecting endocycling since it had no significant effects on the number of nuclei per gland and their size (Figure 4E). These results suggest that APC/C also regulates CENP-ACID expression.
Figure 4.
APC/C regulates CENP-ACID expression. (A) The expression of CENP-ACID::YFP in salivary glands is determined by direct fluorescence (in green) in control ey3.5>CENP-ACID::YFP flies (top) and APC2-depleted ey3.5>CENP-ACID::YFP; APC2RNAi flies (bottom). DNA was stained with DAPI (in white). Scale bars correspond to 100 μm. (B) WB analysis with αGFP antibodies of the levels of CENP-ACID::YFP expression in salivary glands from control ey3.5>CENP-ACID::YFP flies and APC2-depleted ey3.5>CENP-ACID::YFP; APC2RNAi flies. αActin antibodies were used as loading control. Quantitative analysis of the results is shown in the bottom (N = 2; *P-value < 0.01, two-tailed t-test; error bars are SEM). (C) As in A but for control ey3.5>CENP-ACID::YFP flies and Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies. (D) As in B but for control ey3.5>CENP-ACID::YFP flies, Cdc20-depleted ey3.5>CENP-ACID::YFP; Cdc20RNAi flies and Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies Quantitative analysis of the results is shown in the bottom (N ≥ 2; *P < 0.01, ****P < 0.00001, two-tailed t-test; error bars are SEM). (E) In the top, the number of nuclei per gland is presented for control ey3.5>CENP-ACID::YFP flies (N = 10), APC2-depleted ey3.5>CENP-ACID::YFP; APC2RNAi flies (N = 2), Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies (N = 8) and Cdc20-depleted ey3.5>CENP-ACID::YFP; Cdc20RNAi flies (N = 4) (P-value > 0.01, two-tailed t-test; error bars are SEM). In the bottom, nuclei area of salivary glands cells is presented for control ey3.5>CENP-ACID::YFP flies (N = 10), APC2-depleted ey3.5>CENP-ACID::YFP; APC2RNAi flies (N = 2), Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies (N = 8) and Cdc20-depleted ey3.5>CENP-ACID::YFP; Cdc20RNAi flies (N = 4) (P-value > 0.01, two-tailed t-test; error bars are SEM).
APC/C forms two main complexes, APC/CCdc20 and APC/CCdh1, which are active at different cell cycle phases (59–61,68). APC/CCdc20 is active during mitosis and promotes transition from metaphase to anaphase. At the exit from mitosis, Cdh1 replaces Cdc20 and APC/CCdh1 remains active through G1, being degraded at the G1-to-S transition. Later, at the G2-to-M transition, Cdk1 phosphorylation activates APC/CCdc20. Next, we addressed which APC/C form regulates CENP-ACID. We observed that Cdh1 depletion increased CENP-ACID::YFP levels in salivary glands (Figure 4C and D), without significantly affecting endocycling (Figure 4E), which suggest that APC/CCdh1 also regulates CENP-ACID expression in G1. Cdc20 depletion also increased CENP-ACID::YFP expression (Figure 4D), though to a lower extent than Cdh1 depletion. In fact, Cdc20 depletion is not expected to have strong effects in salivary glands since APC/CCdc20 is mainly active in mitosis. In this regard, we observed that Cdc20 depletion in the eye imaginal disc increased CENP-ACID::YFP expression too (Supplementary Figure S4A). Cdc20 depletion in cycling cells induces strong mitotic arrest and affects proliferation (69,70). Accordingly, in the eye imaginal disc, we observed strong proliferation defects and highly increased αPH3-reactivity upon Cdc20 depletion (Supplementary Figure S4A). Therefore, in this case, increased CENP-ACID::YFP expression could be indirect through an effect on cell cycle progression. However, cells showing high levels of mislocalized CENP-ACID::YFP were generally αPH3-negative (Supplementary Figure S4B, top), suggesting that they were not arrested in mitosis. In addition, the increment in the total number of αPH3-positive cells was mainly associated with cells showing no detectable CENP-ACID::YFP mislocalization (Supplementary Figure S4B, bottom), suggesting that cells arrested in mitosis had low CENP-ACID::YFP expression.
Next, we analyzed the contribution of APC/CCdh1 to the regulation of endogenous CENP-ACID expression. We observed that the intensity of immunostaining with αCENP-ACID in the posterior region of the eye imaginal disc is lower than in the anterior region (Figure 5A). However, upon Cdh1 depletion, CENP-ACID expression in the posterior compartment increased and equalized expression in the anterior region (Figure 5A). In addition, specific depletion of APC2 or APC10, another essential APC/C subunit, in larvae brains using an elav-GAL4 driver also increased endogenous CENP-ACID levels (Figure 5B). Altogether these results suggest that APC/CCdh1 regulates expression of endogenous CENP-ACID.
Figure 5.
APC/C regulates endogenous CENP-ACID levels. (A) Endogenous CENP-ACID expression in the eye imaginal disc is determined by immunostaining with αCENP-ACID (in green) in control ey3.5 (top) and Cdh1-depleted ey3.5>Cdh1RNAi flies (bottom). Immunostainings with αPH3, which marks mitotic cells, are also presented (in red). DNA was stained with DAPI (in white). The position of the MF is indicated. Scale bar corresponds to 25 μm. In the bottom, the integrated intensity of fluorescence in posterior versus anterior cells is presented for control ey3.5 flies (N = 17) and Cdh1-depleted ey3.5>Cdh1RNAi flies (N = 13) (****P-value < 0.0001; two-tailed t-test; errors bars are SEM). (B) Immunostaining with αCENP-ACID (in red) of brain squashes from control elav-GAL4 (top), APC2-depleted elav>APC2RNAi (center) and APC10-depleted elav>APC10RNAi larvae (bottom). DNA was stained with DAPI (in blue). Scale bars correspond to 5 μm.
Cal1 protects CENP-ACID from Ppa-mediated degradation, but not from Cdh1-mediated degradation
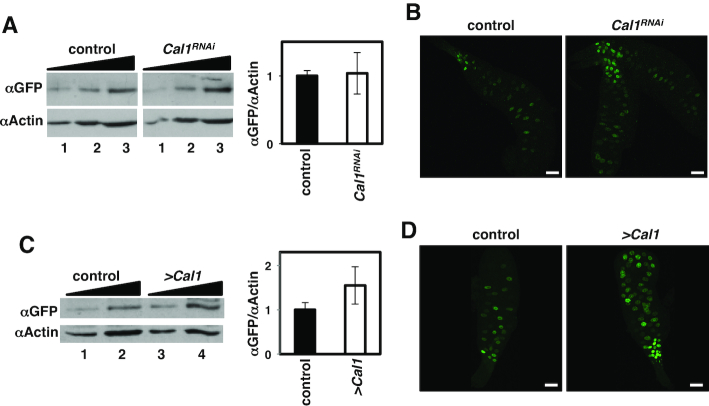
The interaction with Cal1 has been proposed to protect CENP-ACID from proteolytic degradation (25,27). However, we observed that CENP-ACID::YFP levels were not significantly affected upon Cal1 depletion (Figure 6A and B) or overexpression (Figure 6C and D), which is in contrast to previous results in S2 cells showing reduced endogenous CENP-ACID levels upon Cal1 depletion (25,27). To address this apparent contradiction we analyzed the contribution of Cal1 to Ppa- and Cdh1-mediated degradation separately. We observed that Cal1 protected CENP-ACID::YFP from Ppa-mediated degradation since in salivary glands from Cdh1-knockdown larvae, where Ppa principally mediates CENP-ACID::YFP degradation, Cal1 depletion reduced CENP-ACID::YFP levels (Figure 7A and B). Moreover, upon Cal1 overexpression in Cdh1-knockdown glands, IF experiments showed a marked increase of CENP-ACID::YFP levels in ∼30% of the glands (N = 21) (Figure 7D, gland in the right of panel Cdh1RNAi; >Cal1). A similar tendency to increase was detected in WB analyses (Figure 7C). On the contrary, in Ppa-knockdown glands, WB analyses showed that Cal1 depletion did not significantly affect CENP-ACID::YFP levels (Figure 7E and F), suggesting that Cal1 did not protect CENP-ACID::YFP from Cdh1-mediated degradation. Moreover, upon Cal1 overexpression, although global CENP-ACID::YFP levels were not affected (Figure 7G), we observed a marked reduction in the number of YFP-positive cells (Figure 7H). This reduction was accompanied by a change in the pattern of localization of CENP-ACID::YFP, which showed intense fluorescence at the nucleolus region in the center of the nuclei (71,72) and decreased chromosomal signal (Supplementary Figure S5A). Nucleolar CENP-ACID::YFP localization was also observed when Cal1 was overexpressed in Cdh1-depleted and wild type glands (Supplementary Figures S5B and S5C). CENP-ACID::YFP localization at the nucleolus is likely driven by its interaction with Cal1 that is known to localize at the nucleolus in interphase (25,69). Expression of a Cal1::EGFP construct confirmed its nucleolar localization in salivary glands (Supplementary Figure S5D).
Figure 6.
The contribution of Cal1 to CENP-ACID expression. (A) WB analysis with αGFP antibodies of the levels of CENP-ACID::YFP expression in salivary glands from control ey3.5>CENP-ACID::YFP flies and Cal1-depleted ey3.5>CENP-ACID::YFP; Cal1RNAi flies. Increasing amounts of extract are analyzed (lanes 1–3). αActin antibodies were used as loading control. Quantitative analysis of the results is shown in the right (N = 3; P-value > 0.01, two-tailed t-test; errors bars are SEM). (B) The expression of CENP-ACID::YFP in salivary glands is determined by direct fluorescence (in green) in control ey3.5>CENP-ACID::YFP flies (left) and Cal1-depleted ey3.5>CENP-ACID::YFP; Cal1RNAi flies (right). Scale bars correspond to 100 μm. (C) As in A but for control ey3.5>CENP-ACID::YFP flies and ey3.5>CENP-ACID::YFP; UAS-Cal1 flies overexpressing Cal1. (N = 2; P-value>0.01, two-tailed t-test; errors bars are SEM). (D) As in B but for control ey3.5>CENP-ACID::YFP flies and ey3.5>CENP-ACID::YFP; UAS-Cal1 flies overexpressing Cal1. Scale bars correspond to 100 μm.
Figure 7.
Cal 1 protects CENP-ACID from Ppa-mediated degradation. (A) WB analysis with αGFP antibodies of the levels of CENP-ACID::YFP expression in salivary glands from control ey3.5>CENP-ACID::YFP flies, Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies and double Cdh1+Cal1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi; Cal1RNAi flies. Increasing amounts of extract are analyzed (lanes 1–3). Quantitative analysis of the results is shown in the right (N ≥ 2; *P-value <0.01; error bars are SEM). (B) The expression of CENP-ACID::YFP in salivary glands is determined by direct fluorescence (in green) in Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies and double Cdh1+Cal1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi; Cal1RNAi flies. Scale bars correspond to 100 μm. (C) As in A but for control ey3.5>CENP-ACID::YFP flies, Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies and Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi; UAS-Cal1 flies overexpressing Cal1 (N = 3; P-value > 0.01, two-tailed t-test; errors bars are SEM). (D) As in B but for Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi flies and Cdh1-depleted ey3.5>CENP-ACID::YFP; Cdh1RNAi; UAS-Cal1 flies overexpressing Cal1. For the later, two examples are presented with the gland in the right showing increased CENP-ACID::YFP expression. Scale bars correspond to 100 μm. (E) As in A but for control ey3.5>CENP-ACID::YFP flies, Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies and double Ppa+Cal1-depleted ey3.5>CENP-ACID::YFP; ppaRNAi; Cal1RNAi flies (N = 2; P-value > 0.01, two-tailed t-test; errors bars are SEM). (F) As in B but for Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies and double Ppa+Cal1-depleted ey3.5>CENP-ACID::YFP; ppaRNAi; Cal1RNAi flies. Scale bars correspond to 100 μm. (G) As in A but for control ey3.5>CENP-ACID::YFP flies, Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi; UAS-Cal1 flies overexpressing Cal1 (N = 2; P-value > 0.01, two-tailed t-test; errors bars are SEM). (H) As in B but for Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi flies and Ppa-depleted ey3.5>CENP-ACID::YFP; ppaRNAi; UAS-Cal1 flies overexpressing Cal1. Scale bars correspond to 100 μm.
Altogether these results suggest that the Cal1-CENP-ACID::YFP complex is resistant to Ppa-mediated degradation, but not to degradation mediated by Cdh1.
DISCUSSION
Here we have shown that CENP-ACID expression is tightly regulated during cell cycle progression through the combined action of SCFPpa and APC/CCdh1. In G1, APC/CCdh1 and SCFPpa regulate CENP-ACID levels, with SCFPpa acting also in S-phase. The mechanisms regulating CENP-ACID expression in mitosis are less well understood. On one hand, although SCFPpa is likely present in mitosis, it does not regulate CENP-ACID expression levels. It is possible that Ppa is inactivated or, alternatively, that CENP-ACID is resistant to Ppa-mediated degradation in mitosis (see below). On the other hand, CENP-ACID levels increase upon Cdc20 depletion, suggesting that APC/CCdc20, which is active in mitosis, regulates CENP-ACID expression. However, despite Cdc20 depletion in the eye imaginal disc induced a strong mitotic arrest, increased CENP-ACID expression was principally detected in non-mitotic cells. In addition, Cdc20 depletion also increased CENP-ACID expression in the endocycling salivary gland cells that do not undergo mitosis.
The CENP-ACID specific chaperone Cal1 protects CENP-ACID from Ppa-mediated degradation, but not from degradation induced by Cdh1. These observations suggest a model by which SCFPpa can degrade only the pool of CENP-ACID that is not in complex with Cal1, whereas APC/CCdh1 can degrade CENP-ACID in the Cal1-CENP-ACID deposition complex. APC/CCdh1 could mediate degradation of the complex itself or, alternatively, of Cal1, rendering CENP-ACID free for degradation by SCFPpa. Against this second possibility, we observed that endogenous Cal1 levels were not significantly affected upon Cdh1 depletion (Supplementary Figure S6). APC/CCdh1 mediates degradation of CENP-ACID also when it is not in complex with Cal1 since, in Ppa-knockdown conditions, Cal1 depletion did not significantly affect CENP-ACID levels. However, APC/CCdh1 appears to degrade CENP-ACID more efficiently when it is in complex with Cal1 since, upon Cal1 overexpression in Ppa-depleted glands, the levels of CENP-ACID associated with chromosomes were strongly reduced and the remaining CENP-ACID accumulated in the nucleolus, where Cdh1 might not be active. In this regard, Cal1 has been shown to promote CENP-ACID monoubiquitylation by the E3-ligase Cul3/Rdx (73). Thus, it is possible that Cal1 also facilitates CENP-ACID ubiquitylation and degradation by APC/CCdh1.
SCFPpa likely regulates CENP-ACID levels directly since Ppa was shown to physically interact with CENP-ACID (57). Whether APC/CCdh1 is directly responsible for CENP-ACID degradation remains to be determined since co-IP experiments failed to detect an interaction between Cdh1 and CENP-ACID or Cal1. Thus, we cannot exclude the possibility that APC/CCdh1 directly targets an unknown positive regulator of CENP-ACID stability different from Cal1. Further work is required to elucidate the precise molecular mechanism of the contribution of APC/CCdh1 to the regulation of CENP-ACID levels.
In somatic tissues of Drosophila larvae, centromeric CENP-ACID deposition initiates at late telophase and continues during G1 (13), when APC/CCdh1 is active. Similarly, in S2 cells, deposition occurs starting in mitosis and continuing in G1 (11,14). These observations suggest that APC/CCdh1 activity is important to regulate Cal1-CENP-ACID levels during deposition. On the other hand, SCFPpa appears especially important to prevent CENP-ACID misincorporation across chromatin when the bulk of newly synthesized nucleosomes are deposited during DNA replication, as it is active in S-phase when APC/CCdh1 is not. In G1, SCFPpa could also be instrumental in the degradation of CENP-ACID misincorporated at non-centromeric sites.
Our model predicts that the actual contribution of SCFPpa and APC/CCdh1 to the regulation of CENP-ACID levels would depend on the actual proportion of total CENP-ACID that is in complex with Cal1, as well as on the relative activities of both enzymes, thus likely varying between cell types and conditions. This means that the effect of Ppa depletion would depend on the amount of CENP-ACID that is not in complex with Cal1, being less important when the Cal1-CENP-ACID complex is more abundant during mitosis, which might account for the lack of effect observed in mitosis. Along the same lines, the effects of Cal1 depletion would depend on the relative abundance of the Cal1-CENP-ACID complex and the activity of Cdh1. In this regard, endocycling cells, such as those of salivary glands, have very high Cdh1 activity that, together with the overexpression of CENP-ACID, suggest that the proportion of Cal1-CENP-ACID in our experiments must be lower than in other experiments performed in cell types with normal Cdh1 activity and no CENP-ACID overexpression, likely accounting for the different effects of Cal1 depletion observed with respect to experiments performed in S2 cells (25,27). Our model also accounts for the stronger effect of Cdh1 depletion in comparison to Ppa depletion since Cdh1 mediates degradation of CENP-ACID regardless of whether it is in complex with Cal1 or not, whereas Ppa only targets the subset that is not in complex with Cal1. In addition, the extent of Ppa knockdown achieved in our experiments was relatively low, as total ppa mRNA levels were reduced by only ∼1/3 (57).
A necrotic eye phenotype was observed when CENP-ACID expression was driven by longGMR-GAL4 in the most posterior undifferentiated cells of the eye imaginal disc. This phenotype was associated with CENP-ACID overexpression since it was highly enhanced by simultaneous Ppa depletion, which strongly increased CENP-ACID levels and induced its mislocalization across chromatin. In budding yeast, blocking CENP-ACse4 proteolysis leads to its mislocalization and preferential deposition at promoters, resulting in strong changes in gene expression (42). Therefore, preventing CENP-ACID proteolysis could also lead to its preferential deposition at promoters, affect gene expression and, ultimately, interfere with cell differentiation and cause necrosis.
From these studies, proteolysis emerges as a major mechanism regulating CENP-ACID levels. In this regard, regulation at the transcriptional level appears to play a less important role since expression of a CENP-ACID::GFP transgene driven by the endogenous CENP-ACID promoter was detected all across the cell cycle (11). Finally, CENP-A is found overexpressed in various cancers and elevated CENP-A levels correlate with the most aggressive cases (40,41,44,47–52). To what extent, the increased CENP-A content of cancer cells reflects misregulation of the proteolytic pathways that regulate CENP-A stability remains to be determined.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Drs Aaron F. Straight, Sylvia Erhardt, Christian Lehner, Jordan Raff, Yuu Kimata and Maria Lluisa Espinás for materials. We also acknowledge the help of E. Orsi in related experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BFU2015-65082-P]; Generalitat de Catalunya [SGR2014-204, SGR2017-475]; European Community FEDER program; ‘Centre de Referència en Biotecnologia’ of the Generalitat de Catalunya. Funding for open access charge: MINECO [BFU2015-65082-P]; Generalitat de Catalunya [SGR2014-204, SGR2017-475]; European Community FEDER program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Allshire R.C., Karpen G.H.. Epigenetic regulation of centromeric chromatin: old dogs, new tricks. Nat. Rev. Genet. 2008; 9:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Black B.E., Cleveland D.W.. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011; 144:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maddox P.S., Corbett K.D., Desai A.. Structure, assembly and reading of centromeric chromatin. Curr. Opin. Genet. Dev. 2012; 22:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malik H.S., Henikoff S.. Major evolutionay transitions in centromere complexity. Cell. 2009; 138:1067–1082. [DOI] [PubMed] [Google Scholar]
- 5. Torras-Llort M., Moreno-Moreno O., Azorín F.. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 2009; 28:2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKinley K.L., Cheeseman I.M.. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016; 17:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahmad K., Henikoff S.. Centromeres are specilized replication domains in heterochromatin. J. Cell Biol. 2001; 153:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shelby R.D., Monier K., Sullivan K.F.. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000; 151:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S.. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008; 180:1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jansen L.E.T., Black B.E., Foltz D.R., Cleveland D.W.. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007; 176:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lidsky P.V., Sprenger F., Lehner C.F.. Distinct modes of centromere protein dynamics during cell cycle progression in Drosophila S2R+ cells. J. Cell Sci. 2013; 126:4782–4793. [DOI] [PubMed] [Google Scholar]
- 12. Schuh M., Lehner C.F., Heidmann S.. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007; 17:237–243. [DOI] [PubMed] [Google Scholar]
- 13. Dunleavy E.M., Beier N.L., Gorgescu W., Tang J., Costes S.V., Karpen G.H.. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 2012; 10:e1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mellone B.G., Grive K.J., Shteyn V., Bowers S.R., Oderberg I., Karpen G.H.. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011; 7:e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M.. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007; 12:17–30. [DOI] [PubMed] [Google Scholar]
- 16. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M.. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004; 118:715–729. [DOI] [PubMed] [Google Scholar]
- 17. Maddox P.S., Hyndman F., Monen J., Oegama K., Desai A.. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007; 176:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. French B.T., Westhorpe F.G., Limouse C., Straight A.F.. Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev. Cell. 2017; 42:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hori T., Shang W.H., Hara M., Ariyoshi M., Arimura Y., Fujita R., Kurumizaka H., Fukagawa T.. Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev. Cell. 2017; 42:181–189. [DOI] [PubMed] [Google Scholar]
- 20. Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P.. Scm3 is essential to recruit the histone H3 variant CSE4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007; 26:853–865. [DOI] [PubMed] [Google Scholar]
- 21. Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C.. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007; 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- 22. Pidoux A.L., Choi E.S., Abbott J.K.R., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X. et al.. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009; 33:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phansalkar R., Lapierre P., Mellone B.G.. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 2012; 20:493–504. [DOI] [PubMed] [Google Scholar]
- 24. Schittenhelm R.B., Althoff F., Heidmann S., Lehner C.F.. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 2010; 123:3768–3779. [DOI] [PubMed] [Google Scholar]
- 25. Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F.. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008; 183:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N.. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007; 316:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C.C., Dechassa M.L., Bettini E., Ledoux M.B., Belisario C., Heun P., Luger K., Mellone B.G.. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 2014; 204:313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosin L., Mellone B.G.. Co-evolving CENP-A and CAL1 domains mediate centromeric CENP-A deposition across Drosophila species. Dev. Cell. 2016; 37:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G.. HJURP is a cell cycle dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009; 137:485–497. [DOI] [PubMed] [Google Scholar]
- 30. Foltz D.R., Jansen L.E.T., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., Cleveland D.W.. Centromere specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009; 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernad R., Sánchez P., Rivera T., Rodríguez-Corsino M., Boyarchuk E., Vassias I., Ray-Gallet D., Arnaoutov A., Dasso M., Almouzni G. et al.. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 2011; 192:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stankovic A., Guo L.Y., Mata J.F., Bodor D.L., Cao X.J., Bailey A.O., Shabanowitz J., Hunt D.F., Garcia B.A., Black B.E. et al.. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol. Cell. 2017; 65:231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choi E.S., Stralfors A., Castillo A.G., Durand-Dubief M., Ekwall K., Allshire R.C.. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011; 286:23600–23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi E.S., Stralfors A., Catania S., Castillo A.G., Svensson J.P., Pidoux A.L., Ekwall K., Allshire R.C.. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENPA (Cnp1) in fission yeast. PLoS Genet. 2012; 8:e1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collins K.A., Furuyama S., Biggins S.. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004; 14:1968–1972. [DOI] [PubMed] [Google Scholar]
- 36. Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M.. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011; 145:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendiburo M.J., Padeken J., Fülöp S., Schepers A., Heun P.. Drosophila CENH3 is sufficient for centromere formation. Science. 2011; 334:686–690. [DOI] [PubMed] [Google Scholar]
- 38. Moreno-Moreno O., Torras-Llort M., Azorín F.. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006; 34:6247–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R.. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001; 114:3529–3542. [DOI] [PubMed] [Google Scholar]
- 40. Athwal R.K., Walkiewicz M.P., Baek S., Fu S., Bui M., Camps J., Ried T., Sung M.H., Dalal Y.. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet.Chromatin. 2015; 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Filipescu D., Naughtin M., Podsypanina K., Lejour V., Wilson L., Gurard-Levin Z.A., Orsi G.A., Simeonova I., Toufektchan E., Attardi L.D. et al.. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 2017; 31:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hildebrand E.M., Biggins S.. Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 2016; 12:e1005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G.. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014; 53:631–644. [DOI] [PubMed] [Google Scholar]
- 44. Amato A., Schillaci T., Lentini L., Di Leonardo A.. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer. 2009; 8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., Karpen G.H.. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006; 10:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mathew V., Pauleau A.L., Steffen N., Bergner A., Becker P.B., Erhardt S.. The histone-fold protein CHRAC14 influences chromatin composition in response to DNA damage. Cell Rep. 2014; 7:321–330. [DOI] [PubMed] [Google Scholar]
- 47. Hu Z., Huang G., Sadanandam A., Gu S., Lenburg M.E., Pai M., Bayani N., Blakely E.A., Gray J.W., Mao J.H.. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010; 12:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y., Zhu Z., Zhang S., Yu D., Yu H., Liu L., Cao X., Wang L., Gao H., Zhu M.. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE. 2011; 6:e17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma X.J., Salunga R., Tuggle J.T., Gaudet J., Enright E., McQuary P., Payette T., Pistone M., Stecker K., Zhang B.M. et al.. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5974–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qiu J.J., Guo J.J., Lv T.J., Jin H.Y., Ding J.X., Feng W.W., Zhang Y., Hua K.Q.. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumour Biol. 2013; 34:2971–2975. [DOI] [PubMed] [Google Scholar]
- 51. Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F.. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003; 63:3511–3516. [PubMed] [Google Scholar]
- 52. Wu Q., Qian Y.M., Zhao X.L., Wang S.M., Feng X.J., Chen X.F., Zhang S.H.. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012; 77:407–414. [DOI] [PubMed] [Google Scholar]
- 53. Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin- Brown S., Florens L., Workman J.L., Gerton J.L.. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell. 2010; 40:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ranjitkar P., Press M.O., Yi X., Baker R., MacCoss M.J., Biggins S.. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 2010; 40:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheng H., Bao X., Gan X., Luo S., Rao H.. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci. Rep. 2017; 7:8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohkuni K., Takahashi Y., Fulp A., Lawrimore J., Au W.C., Pasupala N., Levy-Myers R., Warren J., Strunnikov A., Baker R.E. et al.. SUMO-Targeted Ubiquitin Ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell. 2016; 27:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moreno-Moreno O., Medina-Giró S., Torras-Llort M., Azorín F.. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr. Biol. 2011; 21:1488–1493. [DOI] [PubMed] [Google Scholar]
- 58. Cardozo T., Pagano M.. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004; 5:739–751. [DOI] [PubMed] [Google Scholar]
- 59. Nakayama K.I., Nakayama K.. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006; 6:369–381. [DOI] [PubMed] [Google Scholar]
- 60. Reed S.I. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 2003; 4:855–864. [DOI] [PubMed] [Google Scholar]
- 61. Vodermaier H.C. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 2004; 14:R787–R796. [DOI] [PubMed] [Google Scholar]
- 62. Charlton-Perkins M., Brown N.L., Cook T.A.. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol. Genet. Genomics. 2011; 286:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Treisman J.E. Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2013; 2:545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Edgar B.A., Orr-Weaver T.L.. Endoreplication cell cycles: more for less. Cell. 2001; 105:297–306. [DOI] [PubMed] [Google Scholar]
- 65. Lee H.O., Davidson J.M., Duronio R.J.. Endoreplication: polyploidy with purpose. Genes Dev. 2009; 23:2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lilly M.A., Duronio R.J.. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005; 24:2765–2775. [DOI] [PubMed] [Google Scholar]
- 67. Baker N.E. Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol. 2001; 12:499–507. [DOI] [PubMed] [Google Scholar]
- 68. van Leuken R., Clijsters L., Wolthuis R.. To cell cycle, swing the APC/C. Biochim. Biophys. Acta. 2008; 1786:49–59. [DOI] [PubMed] [Google Scholar]
- 69. Dawson I.A., Roth S., Akam M., Artavanis-Tsakonas S.. Mutations of the fizzy locus cause metaphase arrest in Drosophila melanogaster embryos. Development. 1993; 117:359–376. [DOI] [PubMed] [Google Scholar]
- 70. Tavormina P.A., Burke D.J.. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998; 148:1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vujatovic O., Zaragoza K., Vaquero A., Reina O., Bernués J., Azorín F.. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012; 40:5402–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marinho J., Casares F., Pereira P.S.. The Drosophila Nol12 homologue viriato is a dMyc target that regulates nucleolar architecture and is required for dMyc-stimulated cell growth. Development. 2011; 138:349–357. [DOI] [PubMed] [Google Scholar]
- 73. Bade D., Pauleau A.L., Wendler A., Erhardt S.. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell. 2014; 28:508–519. [DOI] [PubMed] [Google Scholar]
- 74. Krasnow M.A., Saffman E.E., Kornfeld K., Hogness D.S.. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989; 57:1031–1043. [DOI] [PubMed] [Google Scholar]
- 75. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.