Abstract
The telomeric protein TRF2 is overexpressed in several human malignancies and contributes to tumorigenesis even though the molecular mechanism is not completely understood. By using a high-throughput approach based on the multiplexed Luminex X-MAP technology, we demonstrated that TRF2 dramatically affects VEGF-A level in the secretome of cancer cells, promoting endothelial cell-differentiation and angiogenesis. The pro-angiogenic effect of TRF2 is independent from its role in telomere capping. Instead, TRF2 binding to a distal regulatory element promotes the expression of SULF2, an endoglucosamine-6-sulfatase that impairs the VEGF-A association to the plasma membrane by inducing post-synthetic modification of heparan sulfate proteoglycans (HSPGs). Finally, we addressed the clinical relevance of our findings showing that TRF2/SULF2 expression is a worse prognostic biomarker in colorectal cancer (CRC) patients.
INTRODUCTION
The Telomeric Repeat binding Factor 2 (TRF2) is one of the main regulators of telomere integrity (1). TRF2 suppresses aberrant DNA damage response (DDR) at functional telomeres through the inhibition of the ATM kinase signaling and of the non-homologous end joining (NHEJ) repair pathway (2).
TRF2 has been found overexpressed in various human malignancies and in the vasculature of many cancer types (3–5); it contributes to carcinogenesis in mice (6) and it is regulated by the Wnt/β-catenin pathway (7), WT1 (5) and p53 pathways (8). Consistent with its oncogenic role in human cancers, an increased dosage of TRF2 in a variety of tumor cells enhanced their tumorigenicity, whereas TRF2 depletion reduced tumor growth (9–12). Notably, the role of TRF2 in cancer did not only rely on its effect on telomere protection, but also on its ability to modulate gene expression (13). By combining chromatin immunoprecipitation with high-throughput DNA sequencing (ChIP-Seq), TRF2 has been described to occupy a set of interstitial regions throughout the human genome, referred to as interstitial telomeric sequences (ITSs), as it can act as transcriptional activator (14,15). Another role of TRF2 in transcriptional regulation is to interact with the Repressor Element 1-Silencing Transcription factor (REST) to regulate the expression of neuronal differentiation genes (16–18). The fact that TRF2 may directly control gene expression raises the intriguing possibility that, besides its role in telomere protection it may contribute to several steps in tumor formation, progression and metastasis. Our group showed that an increased dosage of TRF2 can control tumorigenesis, not only via cancer cell-intrinsic mechanisms but also via a cell-extrinsic pathway, through the positive regulation of HS3ST4, a gene encoding for the heparan sulfate (glucosamine) 3-O-sulfotransferase 4, which is involved in post-synthetic modification of Heparan Sulfate Proteoglycan (HSPG) and in the recruitment of NK-cells (10,19).
HSPGs are glycoproteins containing one or more covalently attached heparan sulfate (HS) chains, localized either at the cell surface or in the extracellular matrix, where they act as a hub for a plethora of ligands. Notably, in the last few years, a growing body of literature shed light on the biological significance of these interactions. In particular, it has been evidenced that HSPGs are able to regulate the bio-availability of secreted growth factors and cytokines by binding and storing them in tissues for later use and/or helping to establish molecular gradients in the extracellular behaviour (20–22). Notably, tumors of different histotype including breast, lung, brain, pancreatic, skin, and colorectal cancer are characterized by profound alterations in the fine structure of proteoglycans leading to uncontrolled proliferation, immune-escape, metastasis and differentiation (23). These structural changes are mainly due to alterations in the expression of HS biosynthetic enzymes (3-O-sulfotransferase and 6-O-sulfotransferase), and/or catabolic enzymes such as sulfatases and the heparanase.
Here, we unveiled that TRF2, through the binding to a distal regulatory element, promotes the expression of SULF2, a gene encoding for an endoglucosamine-6-sulfatase known to remove the sulfate group from 6-O position of heparan sulfate (HS) (24–27) with an impact on tumor secretome. Through this mechanism, TRF2 is capable of impairing the capability of HSPGs to bind and sequester signaling molecules containing an heparin-binding domain (28–31), including the angiogenic factor VEGF-A, with a profound impact on tumor vascolarization and, consequently, on tumor growth and metastasis.
MATERIALS AND METHODS
Cell lines, culture conditions, transfection and infection
Colon cancer cell line HCT116 cells were obtained by Dr Vogelstein, Johns Hopkins University. Human cervix carcinoma HeLa cells were purchased by the ATCC. Human breast cancer cell MDA-MB-231 and Human embryonic kidney cells (HEK) were obtained from Dr Eric Gilson. All the cell lines were grown in high glucose Dulbecco modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with L-glutamine, Penicillin/ streptomycin and 10% foetal bovine serum (FBS, Hyclone). HCT116-LUC2 cells were purchased from PerkinElmer (Waltham, MA, USA) and maintained in McCoys medium (EuroClone) supplemented with L-glutamine, Penicillin/streptomycin and 10% heat inactivated FBS (Hyclone). Human umbilical vascular endothelial cells (HUVEC) were purchased from Lonza (Group Ltd, Basel, Switzerland) and maintained in endothelial cell growth medium (EGM-2, Lonza) derived from the endothelial cell basal medium (EBM-2, Lonza) supplemented with serum ad growth factors of EGM-2 BulletKit (Lonza).
For transient RNA interference experiments, siTRF2 (Dharmacon Inc., Chicago, USA) and siCTRL (Santa Cruz Biotechnology; CA, USA) were transfected into HCT116 cells with Interferin (Polyplus) according to the manufacturer's instructions.
Stable TRF2-overexpressing cells (pBabe-puro-mycTRF2), DNA-binding TRF2 mutants (pBabe-puro-mycTRF2ΔM and pBabe-puro-mycTRF2ΔBΔM) and the control counterpart (pBabe-puro-Empty) (32); were obtained by infecting the cells with amphotropic retroviruses generated into Phoenix packaging cells transfected with retroviral vectors, using the JetPEI reagent (Polyplus, New York, NY, USA), according to the manufacturer's instructions.
For stable suppression of TRF2 gene, cells were infected with lentiviral particles produced into HEK293T cells transfected with the packaging pCMVR8.74 and the envelope pMD2.G vectors in combination with the vectors encoding either for a scramble short hairpin sequence (shSCR; N2040 targeting Escherichia coli DNA polymerase) or for one of the two short hairpin sequences directed against TRF2 (shTRF2_N1; N2573 TRCN0000004813 or shTRF2_N2; N2571 TRCN0000004811, which were a gift from Prof Stefan Shoeftner, University of Trieste).
SULF2 overexpressing and silenced cells were prepared by using MISSION lentiviral transduction particles, TRCN0000377275 and SHCLNV-NM_018837 (Sigma), respectively, according to the manifacture's protocol.
Early passages of stably infected cells were used for all experiments.
Drugs and treatments
Where indicated, cells underwent to following treatments: Cobalt Chloride (CoCl2; Sigma Aldrich) 100 μM for 16 h; Heparin (PharmaTex, Milan, Italy) 200 ng/ml for 16 h; Heparinase II from Flavobacterium heparinum (Sigma Aldrich) 15 mU/ml for 2 h; (S)-(+)-camptothecin 0.2 μM for 2 h (CPT, Sigma Aldrich); the ATM-inhibitor KU-55933 (Sigma-Aldrich) was used at 5 μM for 24 h. G-quadruplex ligands Emicoron (33) and RHPS4 (34), were used at 1 μM for 24 and 72 h, respectively. SULF2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007, R&D systems) was dissolved in water and used at 50 mg/kg once a day for 2 weeks.
Real-time PCR
To quantify gene expression by real-time quantitative polymerase chain reaction (qPCR), total RNA was isolated from cell pellets by using TRIzol reagent (Ambion). Quality of the extracted RNA was assessed by 1% agarose gel electrophoresis and from the A260 nm/A280 nm absorbance ratio (Nanodrop 1000, ThermoFisher Scientific). Reverse transcription (RT) from 1 μg of RNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. Real-time rt-PCRs were performed on the obtained cDNAs by using Fast power Syber green master mix (Apply Biosystem) on either QuantStudio 6-Flex (Thermo Fisher Scientific) or 7900HT Fast Real-Time PCR System (Applied Biosystem) thermocycler. For each sample 5 μl of the 1:10 diluted cDNA was mixed with 0.5 μl of each primer (10 μM), 10 μl of the sybr green master mix and water at final volume of 20 μl. Standard qPCR thermal parameters were used: one cycle of 95°C for 10 min then 40 cycles of 95°C for 15 sec and 60°C for 1 min followed by dissociation curve (95°C for 15 s, 60°C for 1 min, 95°C for 15 s).
The primers used for gene analysis were synthesized by Integrated DNA Technologies (BVBA Leuven, Belgium). For each primers’ pair, forward and reverse sequences are specified in the Supplemental Table S1. All experiments were run in triplicate and the gene expression levels were normalized to the β-actin.
Cell proliferation
HUVECs (4 × 104 cells/well in 96 well plates) were seeded in the presence of the indicated CMs and proliferation was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay. At the indicated times, 20 μl MTT (Sigma-Aldrich) were added per each well, and cells were incubated for additional 4 h at 37°C + 5% CO2. The formazan crystals formed were dissolved in 100 μl of isopropanol and the optical density value was recorded at 540 nm on a microplate reader.
Invasion assay
Invasion assay was performed in a 48-microwell modified Boyden chamber using 8-μm-pore-size polycarbonate filters (NeuroProbeInc, MD, USA) coated with Matrigel® (Becton Dickinson). Briefly, 2 × 104 HUVECs resuspended in endothelial cell basal medium (EBM-2, Lonza) were seeded in the upper compartment of each well, while the lower compartments were filled with the indicated CMs. EBM-2 and EGM-2 (Lonza) were used as negative and positive controls, respectively. After 4 h of incubation at 37°C + 5% CO2, cells remaining in the upper side of the filter were scraped off while the cells at the bottom side were fixed with ethanol 70% and stained with crystal violet. The number of invaded cells was counted using phase contrast microscopy (5×). For each well, four randomly selected fields were counted.
In vitro angiogenesis
EC differentiation into tubular structures (TS) was assessed as previously reported (35). The presence of TS was monitored 6 h after plating, and images were captured by phase contrast microscopy. TS formation was evaluated by quantifying the number of branching points in five randomly selected fields.
When indicated, the CMs were incubated for 30 min in presence of 1 μg/ml of the blocking antibody against VEGF-A (clone MAB293; R&D systems) before being assayed.
In vivo angiogenesis
The indicated CMs, mixed with heparin (PharmaTex, Milan, Italy) and 500 μl Matrigel® (BD Biosciences) were injected subcutaneously in C57/BL6 mice or NSG mice, when directly specified. Groups of four mice (two plugs per mouse) were used for each experiment. The CMs were replaced by medium supplemented or not with VEGF-A (100 ng/ml) and Tumor Necrosis Factor α (TNFα; 2 ng/ml) (R&D Systems, Inc. MN, USA) in the positive and negative controls, respectively. Four days after injection, the Matrigel plugs were recovered and processed for hemoglobin quantization using Drabkin solution (Sigma-Aldrich). The optical density of each sample was determined at 540 nm. Results were normalized for 100 mg of Matrigel®.
When specified, CMs were pre-incubated for 30 minutes in presence of 1 μg/ml of the blocking antibody against VEGF-A (clone MAB293; R&D systems) before injection.
For the analysis of cell infiltrate, the recovered Matrigel® plugs were dissociated mechanically in serum-free medium containing DNase I (Roche), collagenase A (Roche) and dispase (BD Biosciences) and then incubated for 30 min at 37 °C under gentle agitation. The obtained cells were analysed by flow cytometry (see ‘Immunofluorescence' section).
Multiplexed immunoassay
The quantitative analysis of secreted molecules in conditioned media (CM) of TRF2-compromised and TRF2 overexpressing HCT116 cells was performed by using the Bio-Plex multiplex system (Bio-Rad, Milan, Italy) based on xMAP technology. that makes use of magnetic beads coated with specific antibodies raised against target analytes. Microspheres are internally labeled with red and infrared fluorophores. Each bead is bound to a specific antibody, thus allowing the simultaneous detection of multiple analytes within one sample. Following reaction of coupled beads with target analytes, a biotinylated antibody is added for the detection, which is then finalized by adding phycoerytrin-conjugated streptavidin. All steps were performed according to manufacturer's instructions for determining the concentration of the indicated molecules. Data were acquired using a Bio-Plex MAGPIX Multiplex Reader system equipped with a Bio-Plex Manager software v. 6.1 (BioRad). All washing steps were performed on the Bio-Plex magnetic wash station (BioRad). Measurements were performed in triplicate on CM samples (50 μl) using the Bio-Plex Pro human cancer biomarker panel 2 (Cat. No. 171AC600M, BioRad), according to the manufacturer's protocol. Standard curves optimization and the calculation of analyte concentrations were performed by using the Bio-Plex Manager software.
Enzyme-linked immunosorbent assay (ELISA)
To evaluate the amount of VEGF-A in the supernatants of infected cells, the cells were incubated in 10 ml of serum-free medium for the indicated times and then the CMs were collected, centrifuged to remove cellular debris, and assayed by ELISA (Quantikine Immunoassay Human VEGF, R&D systems), according to the manufacturer's instructions. Results were normalized to cell number.
Similarly, cell- and membrane-associated VEGF-A was quantified by ELISA on cell lysates and purified membrane, respectively. Results were normalized to protein content.
Cell fractionation
Cell cytoplasm, membrane and nuclei fractions were purified by ProteoExtract Native Membrane Protein Extraction Kit (Calbiochem), according to the manufacturer's instructions.
Western blotting
Western blot analysis was performed as previously reported (36). Expression levels of TRF2 were evaluated by using the mouse mAb anti-TRF2 (Millipore). The DNA damage response was evaluated by using the following antibodies: rabbit mAb anti-Ser1981 p-ATM (Abcam Ltd.); rabbit pAb anti-Thr68 p-CHK2 (Cell Signaling, Beverly, MA, USA); mouse monoclonal anti-γH2AX antibody (Upstate, Lake Placid, NY, USA).
The purity of the subcellular fractions was evaluated by using the following antibodies: mouse mAb anti-EGFR (a kind gift of Dr Oreste Segatto, Regina Elena National Cancer Institute of Rome); mouse mAb anti-Lamin A/C (636, Santa Cruz Biotecnology); mouse mAb anti-GAPDH (clone 6C5, Santa Cruz Biotecnology).
Growth curves
5 × 104 HCT116 cells silenced (shTRF2) or not (shSCR) for TRF2 were seeded in 100 mm Petri dishes. Cell counts and viability (trypan blue dye exclusion) were determined daily, from day 1 to day 7 of culture.
Immunofluorescence analyses
Cell cycle analysis was performed by flow cytometry using a FACScalibur ((BD Biosciences, Heidelberg, Germany). Briefly, adherent cells (2 × 105) were fixed and resuspended in a solution containing propidium iodide at a concentration of 50 μg/ml. Cell percentages in the different phases of the cell cycle (G0/G1, S and G2 + M) were measured using CELLQuest software (BD Biosciences).
Apoptosis was evaluated by annexin V versus PI assay, as described in (37). Briefly, cells were harvested, suspended in annexin-binding buffer (1 × 106 cells/ml), incubated with fluorescein isothiocyanate-annexin V (annexin-FITC) and PI (Molecular Probes) for 15 min at room temperature in the dark, and then immediately analyzed by flow cytometry using a FACScalibur (BD Biosciences). The annexin V positive/PI negative cells were considered apoptotic.
For cell infiltrate analysis, the cells recovered from Matrigel® (see in vivo angiogenesis section) were stained FITC-conjugated anti-CD31 and anti-CD45 antibodies (all from BD Biosciences) and the percentages of endothelial (CD31–/CD45–) and immune (CD31-/CD45+) cells were evaluated by flow cytometry using a CANTO cytometer and DIVA6 software (BD Biosciences).
To evaluate the amount of membrane-associated VEGF-A, viable cells were immunostained with the mouse mAb anti-hVEGF-A antibody (clone C-1; Santa Cruz biotechnology), incubated with a fluorophore conjugated goat anti mouse Alexa Fluor®-488 secondary antibody (Cell Signaling), and analyzed by flow cytometry and fluorescence microscopy, using a FACScalibur (BD Immunocytometry System-BDIS, San Jose, CA, US) and a Zeiss LSM510 inverted confocal microscope (Carl Zeiss, Gottingen, Germany), respectively.
For interphase nuclei telomere-induced foci (TIFs) analysis, cells were fixed in 2% formaldehyde and permeabilized in 0.25% Triton X100 in phosphate buffered saline (PBS) for 5 min at room temperature (RT). For immunolabeling, cells were incubated with primary antibody (RT, 2 h), washed twice in PBS, and finally incubated with the secondary antibodies (RT, 1 h). The following primary antibodies were used: rabbit polyclonal anti-TRF1 antibody (Abcam Ltd., Cambridge, UK); mouse monoclonal anti-γH2AX antibody (Upstate, Lake Placid, NY, USA). The following secondary antibodies were used: goat anti rabbit Alexa Fluor®-555 and goat anti mouse Alexa Fluor®-488 (Cell Signaling). Images were acquired with a Zeiss LSM510 inverted confocal microscope (Carl Zeiss, Gottingen, Germany). Co-localizations were scored in each optical section by scrolling through the z-stack.
For SULF2 expression the cells, fixed and permeabilized as described above, were labelled with a mouse mAb anti-SULF2 (clone 2B4, R&D systems) and then incubated with a fluorophore conjugated goat anti mouse Alexa Fluor®-488 secondary antibody (Cell Signaling). Nuclei were counterstained with DAPI (Sigma Aldrich). Stained cells were analyzed with an Olympus BX53 microscope equipped with epifluorescence and photographs (63X) were taken using a cooled camera devise (ProgRes MF).
Cell senescence
Senescence was evaluated by β-galactosidase staining kit (Cell Signaling Technology) according to the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed as reported in (38). The primers are reported in the Supplemental Table S2. All the real time quantitative PCR (qPCR) were performed using Power SYBR Green Master Mix (Applied Biosystem) in the 7900HT Fast Real-Time PCR System (Applied Biosystem). The specificity of each PCR product was controlled by melting curve. Relative gene expression levels were calculated using the 2-ΔΔCt method.
ENCODE data analysis
The state of chromatin upstream SULF2 gene was investigated using the collection of ENCODE Histone ChIP-seq experiments on HCT-116 cell line (genome assembly hg19) (39). Fold enrichment over input bigwig files were used as inputs for the DeepTools suite version 2.0 (40). Briefly, the computeMatrix tool was used to calculate scores on the genomic region around the TTAGGGTCCACCCTAA sequence (3000 bases upstream and downstream) for H3K27Me3 and H3K4Me1 histone modifications. Profile plots were then created with the plotProfile tool.
Luciferase assay
A 191 bp portion (Supplementary Table S3) of the identified distal regulatory element was cloned from HCT116 cells DNA using KAPA Hifi HotStart Ready Mix PCR kit (KAPA Biosystems) according to the manufacturer's protocol. Primers (fw: 5′- CGCGGATCCCAAGTGTTGGGTTTACAGGCA-3′ and rw: 5′- CCGCTCGAGCCCCAATTCATGCCAATCCA-3′) flanked by specific restriction sites for BamHI and XhoI were used. The PCR product was cloned into the PGL3 promoter vector (Promega) and validated via Sanger sequencing (Microsynth AG, Balgach, Switzerland).
For luciferase assays, 25 × 103 HCT116 cells silenced (shTRF2) or not (shScramble) were transiently transfected in 96-well plates with 10 ng of Renilla vector (Promega) along with 200 ng of pGL3-promoter vector (Promega) or pGL3-promoter vector containing the specified genomic region (Supplementary Table S3) cloned downstream the Luciferase gene. The transfection was performed with the JetPEI reagent (Polyplus) according to the manufacturer's protocol. Forty-eight hours after transfection, cells were washed once with PBS and the luciferase assay (Dual-Luciferase Reporter Assay System, Promega) was performed according to the manufacturer's protocols. Values are expressed as fold increase in luciferase counts over the pGL3-promoter vector and normalized by the Renilla intensities. Luciferase activity was also evaluated in cells transfected with pGL3-promoter vector containing the indicated genomic region, deleted of the binding site for TRF2 (TTAGGGTCCACCCTAA). Deletion mutants were obtained by Q5® Site-Directed Mutagenesis Kit (New England Biolabs) according to manufacturer's protocol.
Xenograft tumor models
CD-1 male nude (nu/nu) mice (5 weeks old and weighing 26–28 g) were purchased from Charles River Laboratories (Calco, Italy).
Nude mice were injected into the hind leg muscles with 5 × 105 shSCR, shTRF2, shSULF2 or shTRF2+SULF2 HCT116 cells/mouse. Tumors were measured in two dimensions using a caliper at the indicated time points and tumor weight was calculated using the formula: a × b2/2, where a and b are the long and short sizes of the tumor, respectively. Each experimental group included five mice.
Tumor dissemination model
Luminescent HCT116 cells (2 × 105) stably transfected with shSCR, shTRF2, shSULF2 or shTRF2+SULF2, were injected in the spleen of 6 weeks old CB17-SCID male mice. After 30 min, the spleen was removed by spleenectomia and the mice were sutured using absorbable surgical sutures. Each experimental group included five mice. Real time tumor dissemination was monitored using the IVIS imaging system 200 series (Caliper Life Sciences, Hopkinton, MA, USA). Briefly, mice were anesthetized with a combination of tiletamine–zolazepam (Telazol, Virbac, Carros, France) and xylazine (xylazine/ Rompun BAYER) given intramuscularly at 2 mg/kg. Mice were injected intraperitoneally with 150 mg/kg D-luciferin (CaliperLife Sciences) and imaged at days 14 and 21 after cells injection. Data were acquired and analyzed using the Living Image Software version 3.0 (Caliper Life Sciences).
Tissue analysis
TRF2 and SULF2 expression was evaluated by immunohistochemistry (IHC) on paraffin embedded sections cut from HCT116 colon cancer derived xenografts or from tissue microarray (TMA), set up as described in (38), from 169 randomly selected human colo-rectal carcinomas (CRCs). Immunostaining was performed using the mouse monoclonal antibodies anti-TRF2 (clone 4A794; Millipore, Billerica, MA, USA), and anti SULF2 (clone G4, Santa Cruz Biotechnology) in an automated stainer (Bond Max III, Leica Biosystem, Milan, Italy) according to the manufacturer's instructions. Microvessel density was evaluated both on tumor xenografts and on human CRCs by staining endothelial cells using two different anti CD31 reagents, a rat mAb (clone SZ31, Dianova GmbH 1:10) in tumor xenografts and a mouse mAb (clone JC70A, Dako, Milan, Italy) in human CRCs. The microvessel density in mouse xenografts was manually performed using ULTRATEK HRP kit (Scy Tek Laboratories, Utah, USA).
The levels of TRF2 and SULF2 were evaluated on tumor cells in terms of intensity and percentage of nuclear (0/1+: low; 2+/3+: high) and nuclear/cytoplasmic (multiplicative score, see statistical methods) staining respectively.
CD31 immunoreactions were evaluated, both in xenograft and in human CRCs, counting the number of vessels in six high-power fields (HPF, 400 × magnification) per section by using a light microscope equipped with a software able to capture images (DM2000 LED, Leica).
Phosphorylated form of H2AX was evaluated by IHC on paraffin embedded sections cut from tumor xenografts using the mouse mAb γH2AX (Ser139, clone JBW301, Millipore).
The immunohistochemical detection of apoptosis was performed by TUNEL assay using a commercial kit (In Situ Cell Death Detection Kit, POD, Roche). The assay was performed according to the manufacturer's instructions.
H2AX nuclear immunoreaction and apoptosis of tumor cells were counted in six high-power fields (400× magnification) per section. Evaluation of the IHC results was performed independently and in blinded manner by two investigators.
For the purposes of the retrospective cross sectional study, 169 colorectal carcinomas (CRC) surgically treated and diagnosed at the Regina Elena Cancer Institute were selected from our files. All CRC included in this study were histopathologically re-evaluated on haematoxylin and eosin stained slides and representative areas were marked prior to tissue microarray (TMA) construction.
TCGA dataset
Standardized TCGA data were obtained from Broad Institute TCGA Genome Data Analysis Center (2016, https://doi.org/10.7908/C11G0KM9).
Statistical analysis
In the IHC experiments, continuous variables were plotted into box-plots, while categorical variables were reported as frequencies and percentage values. The association between variables was tested by the Pearson Chi Square test or Fisher's exact test, when appropriate. The Mann-Whitney U non parametric test was used to compare quantitative variables. Levels of TRF2 expression were scored semiquantitatively based on IHC staining intensity. Low intensity cases displayed a 0/1+ IHC score and were considered negative and high intensity cases presented a 2+/3+ IHC score and were considered positive. SULF2 expression levels were scored semi-quantitatively using the immune-reactive score (IRS, staining intensity x percentage of positive cells). Receiver operative characteristics (ROC) curve analysis was used to estimate the optimal cut-off values able to split patients into group with different level of TRF2 (negative or positive). The cut off for SULF2 IRS was >15 and the cut off for CD31 was >18 number of vessels/HPF. SPSS software (SPSS version 21.0, SPSS Inc., Chicago, Illinois, USA) and MedCalc (14.10.2) programs were used for all the analyses.
Survival analyses were performed by using Kaplan-Meier method. Differences between curves were assessed by the log-rank test; significance was defined at the P ≤ 0.05 level. Patients with high and low signal intensity for a specific gene were defined by considering positive and negative z-score values. A Cox proportional hazard regression model was fitted to include clinical variables.
Study approval
All animal procedures were in compliance with the national and international directives (D.L. 4 March 2014, no. 26; directive 2010/63/EU of the European Parliament and of the council; Guide for the Care and Use of Laboratory Animals, United States National Research Council, 2011) and approved by the Italian Ministry of the health (authorization n. 17/2016-PR issued on date 12 January 2016).
CRC patients surgically treated at the Regina Elena National Cancer Institute received written informed consent and the study was reviewed and approved by the Local Ethic Committee of the same Institute (del. n.180/2014).
RESULTS
TRF2 regulates VEGF-A levels in the secretome of tumor cells and affects angiogenesis
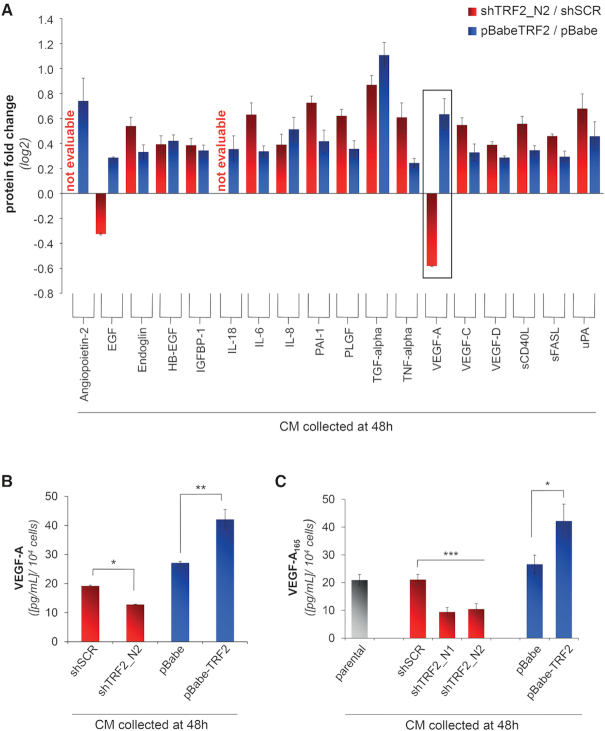
Despite cancer has been long described as a disease consisting of transformed cells with autonomous proliferative, invasive and limitless survival capacities, it is now clear that extrinsic factors (i.e. cytokines/chemokines, growth and angiogenic factors) secreted by cancer cells or by surrounding stromal cells play a fundamental role in tumorigenesis and cancer progression. Here, we questioned if TRF2 is able to affect the secretome of cancer cells. To address this question, HCT116 colon cancer cells were infected with viral particles delivering short hairpin (sh)RNAs against TRF2 (shTRF2_N1 and shTRF2_N2), or the TRF2 cDNA (pBabe-TRF2). The establishment of stable cell lines silenced or overexpressing TRF2 was validated at both mRNA and protein level by real time rt-PCR and western blot analysis (Supplementary Figure S1A–D), respectively. Then, the conditioned media (CMs), obtained by growing the infected cells in serum-free medium for 48 hrs, were assayed by a multiplexed immunoassay based on xMAP technology (Figure 1A). This sensitive screening method allowed the simultaneous detection and quantification of multiple analytes (cytokines, chemokines and growth factors) in a single sample. Notably, on a panel of 18 proteins analysed, we found that Vascular Endothelial Growth Factor-A (VEGF-A) was the only metabolite whose absolute levels were significantly modulated according to TRF2 expression (Figure 1B). These results were confirmed by enzyme-linked immunosorbent assay (ELISA, Figure 1C).
Figure 1.
TRF2 regulates the amount of VEGF-A in the secretome of tumor cells. (A) Luminex/XMAP multiplexed analysis of CMs derived from TRF2-compromised (shTRF2) or -overexpressing (pBabe-TRF2) HCT116 cells and their control counterparts (shScramble/pBabe). CMs were collected 48 h after cell starvation and the expression levels of a panel of secreted chemokines and growth factors involved in angiogenesis were quantified. For each analyte, results are expressed as log2 fold change of protein levels in silenced/overexpressing cells over their controls. (B) Detailed analysis of VEGF-A concentration from A. Histograms show the mean values (±SD) of a single experiment performed in triplicate (*P < 0.1; **P < 0.01; ***P < 0.001; Student's t-test). (C) Concentration of VEGF-A was evaluated by ELISA in the CM of HCT116 silenced (shTRF2_N1 and shTRF2_N2) or overexpressing (pBabe-TRF2) TRF2, collected 48 h after serum-starvation. As control, the amount of VEGF-A was assayed in the CM of HCT116 cells not infected (parental) or infected with viral particles delivering control vectors (shSCR or pBabe). Results were normalized to cell number. Histograms show the mean (±SD) of at least three independent experiments performed in triplicate (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test).
VEGF-A, a member of the PDGF/VEGF growth factor family, is overexpressed in several solid tumors where, promotes the formation of new-vessels favouring tumor growth and dissemination (41). Therefore, the impact of TRF2 on increasing VEGF-A protein in the CMs prompted us to investigate the role of secretome on tumor angiogenesis. HCT116 cells silenced for TRF2 were grown in serum-free medium for 48 h and the obtained CM was assayed for its capability of promoting an angiogenic response in ECs.
Of note, while the medium derived from control cells—parental cell line and cells infected with the scramble shRNA (shSCR)—promoted proliferation and migration of the human umbilical vein endothelial cells (HUVEC), the CM of the cells silenced for TRF2 (shTRF2s) was found to be neither mitogenic nor chemo-attractant (Supplementary Figure S2A and B). Moreover, tubule-formation assays demonstrated that CM derived from the control cells (shSCR and parental cells) was able to promote the differentiation of the HUVEC cells into capillary-like tubular structures (Figure 2A), while the medium collected from TRF2-interfered cells was lacking of any pro-angiogenic activity (Figure 2A). Analogous results were obtained by using amounts of recombinant VEGF-A that recapitulate the concentrations detected in the medium of control and TRF2 silenced cells (Supplementary Figure S2C). Notably, the inhibition of angiogenic response associated with the silencing of TRF2 was neither attributable to changes in proliferation nor to induction of apoptosis or senescence of tumor cells (Supplementary Figure S3). In addition, the angiogenic effect of TRF2 was uncoupled from its role in telomere capping. Indeed, knock-down of TRF2 was not enough to trigger a general activation of DNA damage response (DDR) or telomere deprotection (Supplementary Figures S4A–C) and, consistently with these results, the ATM inhibitor (KU-55933) did not alter the angiogenic response to CM collected from TRF2-proficient or silenced cells (Supplementary Figure S4D).
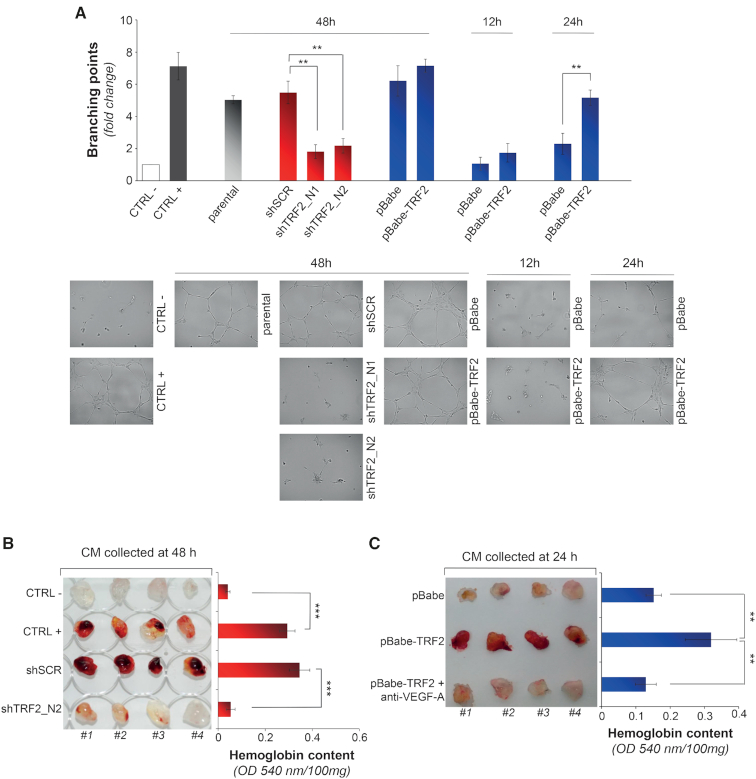
Figure 2.
TRF2 modulation affects the secretome of tumor cells and the angiogenic response of endothelial cells. (A) Tubule formation assay. HUVEC cells were seeded on Matrigel® in the presence of the CMs deriving from HCT116 silenced (shTRF2_N1 and shTRF2_N2) or overexpressing (pBabe-TRF2) TRF2. CMs from cells transduced with control vectors (shScramble or pBabe) or uninfected (parental) were used as controls. EBM-2 supplemented or not with VEGF-A (100 ng/ml) was used as positive and negative control, respectively. The CMs were collected at the indicated times (12, 24 and 48 h). Histograms show the mean number of branching points calculated on five different fields and expressed as fold induction over the negative control. Pictures in the lower panels show representative images of tubular-like structures (5X magnification). Graphs show the mean ±SD of at least three independent experiments performed in triplicate (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test). (B, C) In vivo evaluation of angiogenic response induced by CMs from HCT116 cells silenced (shTRF2_N2) (B) or overexpressing (pBabe-TRF2) TRF2 (C). For each experiment the CMs from control cells (shScramble or pBabe) were also assayed. EBM-2 and EBM-2 supplemented with VEGF-A (100 ng/ml) and TNFα (2 ng/ml) were used as negative and positive controls, respectively. Where indicated, CM was incubated with an anti-VEGF-A165 blocking antibody (1 μg/ml for 30 minutes) and assayed. Left panels: representative pictures show Matrigel® plugs recovered 5 days post-injection. Right panels: histograms represent the average hemoglobin content (±SD) measured in the relative samples and expressed as absorbance (OD540 nm)/mg of Matrigel® (n = 8 plugs per group; *P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test).
Next, to strength the obtained results, we extended our analyses to CM deriving from the cells overexpressing TRF2. Surprisingly, despite the higher levels of secreted VEGF-A (Figures 1B and C), the medium of TRF2-overexpressing cells, collected at 48 h, was unable to promote additional increases in the number of tubules over its control counterpart (Figure 2A). In contrast, when the CMs were collected at shorter times a pro-angiogenic activity attributable to TRF2 overexpression was well appreciable (Figure 2A). These results, corroborated by measurements of VEGF-A performed at the same time points (Figure 1C and Supplementary Figure S5A), suggest that at 48 h the amount of VEGF-A secreted by control cells (pBabe) was already sufficient to promote its maximal functional effect on tube formation.
The role of TRF2 on angiogenesis was also evaluated in vivo by Matrigel assay using CMs of TRF2-interefered and overexpressing cells collected at the nadir of the effect. Analysis of Matrigel plugs injected subcutaneously in the flank of C57BL/6 mice revealed that silencing of TRF2 determines an almost complete inhibition of the angiogenic response, as compared with the not interfered cells (Figure 2B). On the contrary, the plugs containing the CMs derived from TRF2 overexpressing cells appeared more vascularised than their controls cells (Figure 2C). Of note, the effect of TRF2 on tumor angiogenesis was recapitulated on cancer cells of different histotype (Supplementary Figure S6).
Finally, to evaluate if the angiogenic process driven by secretome of TRF2-overexpressing cells was totally attributable to the changes in the VEGF-A levels, the tubule formation assays were performed in presence of a VEGF-A blocking antibody. The results clearly showed that the VEGF-A blockade almost completely inhibited the angiogenic response of TRF2 overexpressing cells both in vitro (Supplementary Figure S5B) and in vivo (Figure 2C).
Since tumor cells have been demonstrated to recruit immune cells that, secreting pro-angiogenic molecules (included VEGF-A), can promote tumor vascularization (42), we questioned whether immune cells participate in the release of VEGF-A mediated by TRF2. To address this point, Matrigel plugs were analysed by FACS analysis for immune cell content (CD31–/CD45+). Interestingly, neither the CM from TRF2 overexpressing cells (pBabe-TRF2) nor that deriving from the compromised cells (shTRF2) produced significant differences in the recruitment of the immune components and the addition of the VEGF-A blocking antibody did not affect the overall immune cell recruitment (Supplementary Figure S7A and B). In line with these data, the CM obtained from cells silenced for TRF2 was found to maintain its angiogenic properties also in immunodeficient NOD SCID γ (NSG) mice (Supplementary Figure S7C). These experiments, even with the limitations due to assaying human secreted factors in a murine system, support the idea that immune system did not participate to TRF2-driven VEGF-A-mediated tumor angiogenesis.
TRF2 induces post-synthetic modification of HSPGs by regulating SULF2 expression
To clarify the mechanism(s) through which TRF2 modulate the VEGF-A levels in the secretome of tumor cells, we tested the possibility that TRF2 activates the transcription of VEGF-A (5,10). However, the analysis of gene expression by quantitative real-time rt-PCR revealed that overall VEGF-A mRNA was not affected by modulation of TRF2 levels (Supplementary Figure S8A). Since VEGF-A transcript undergoes to processes of alternative splicing, the analysis of gene expression was specifically extended to VEGF-A165 (the isoform detected by ELISA), VEGF-A145 and VEGF-A189, the most abundant VEGF-A isoforms. Analysis of these transcripts revealed that none of the mRNAs was significantly affected by TRF2 modulation (Supplementary Figure S8B), permitting to exclude any transcriptional effect of TRF2 on VEGF-A.
Therefore, we evaluated if modulation of TRF2 affected the amount of VEGF-A at protein level. Surprisingly, the ELISA performed on cell lysates showed that, in contrast to what previously observed with the CMs (Figure 1C), the cell-associated VEGF-A was inversely correlated to TRF2 expression (Supplementary Figure S9A), raising the possibility that TRF2 can alter the equilibrium between cell-free and cell-associated amount of VEGF-A.
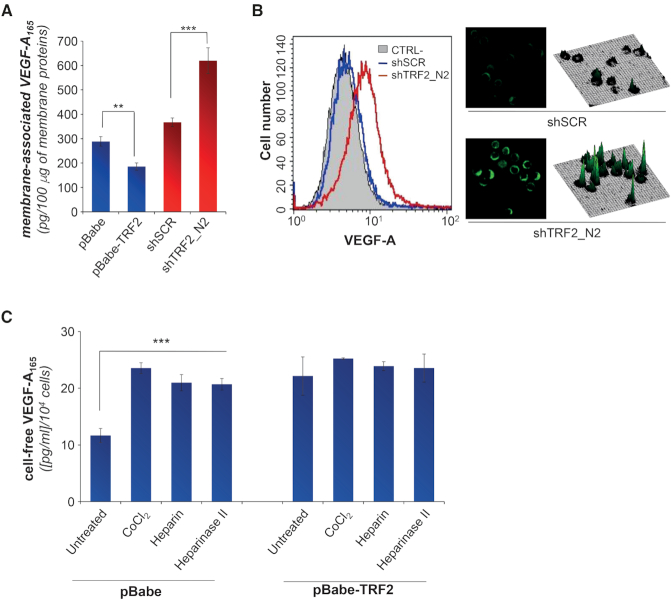
VEGF-A secretion is tightly regulated by the HSPGs (43), a large family of glycoproteins present on both the cell surface and in the extracellular matrix (ECM) (23,44). Indeed, VEGF-A is characterized by the presence of a heparin-binding domain (HBD), a protein-interaction motif that mediates the binding of heparin-like molecules to the HSPGs (43,45,46). Starting from these data we hypothesized that TRF2 might regulate the amount of free VEGF-A by impairing its binding to the cell surface. Notably, analysis of membrane-associated VEGF-A by biochemical (ELISA) and immunofluorescence (FACS and microscopy) techniques showed that the VEGF-A decreased in the TRF2 overexpressing cells and increased in the interfered ones (Figures 3A, B and Supplementary Figure S9B), clearly demonstrating that TRF2 has a crucial role in the regulation of membrane-associated VEGF-A. Consistently with these results, exogenously added heparin (that can compete with the HSPGs for the binding to the VEGF-A (47)), cobalt chloride (CoCl2—known to activate heparanase, an eukaryote HSPG-hydrolysing enzyme (48)) or heparinase II (a prokariotic ortholog of heparanase), increased the amount of free VEGF-A in the control (pBabe) cells (Figure 3C), while TRF2 overexpressing (pBabe-TRF2) cells were quite insensitive to all treatments, clearly demonstrating that TRF2 acts on the binding of VEGF-A to HSPGs.
Figure 3.
TRF2 affects the binding of secreted VEGF-A165 to membrane HSPGs. (A) Amount of membrane-associated VEGF-A165 was quantified by ELISA. The analysis was performed on membrane-enriched lysate fractions obtained from HCT116 cells silenced (shTRF2) or overexpressing (pBabe-TRF2) TRF2 and their controls (shScramble and pBabe). Results were normalized to the total amount of membrane proteins. (B) Immunofluorescence analysis of membrane associated VEGF-A on viable HCT116 cells silenced for TRF2 (shTRF2) and their control (shSCR) counterpart. Left panel: fluorescence-activated cell sorting (FACS) analysis. Histogram shows the fluorescence intensities in the negative control (black line) and in HCT116 silenced (shTRF2, red line) or not (shSCR, blue line) for TRF2. Right panel, results of confocal-microscopy. Representative images acquired at 63x magnification and their respective 3D surface plots. (C) Concentration of VEGF-A evaluated by ELISA in the CM obtained from HCT116 overexpressing (pBabe-TRF2) or not (pBabe) TRF2, untreated or treated with CoCl2 (100 μM for 16 h), heparin (200 ng/ml for 16 h) or heparinase II (15 mU/ml for 2 h). For CM, cells were growth for 24 h in serum-free medium and all the stimuli were directly added in medium. All the histograms show the mean ±SD of at least three independent experiments (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test).
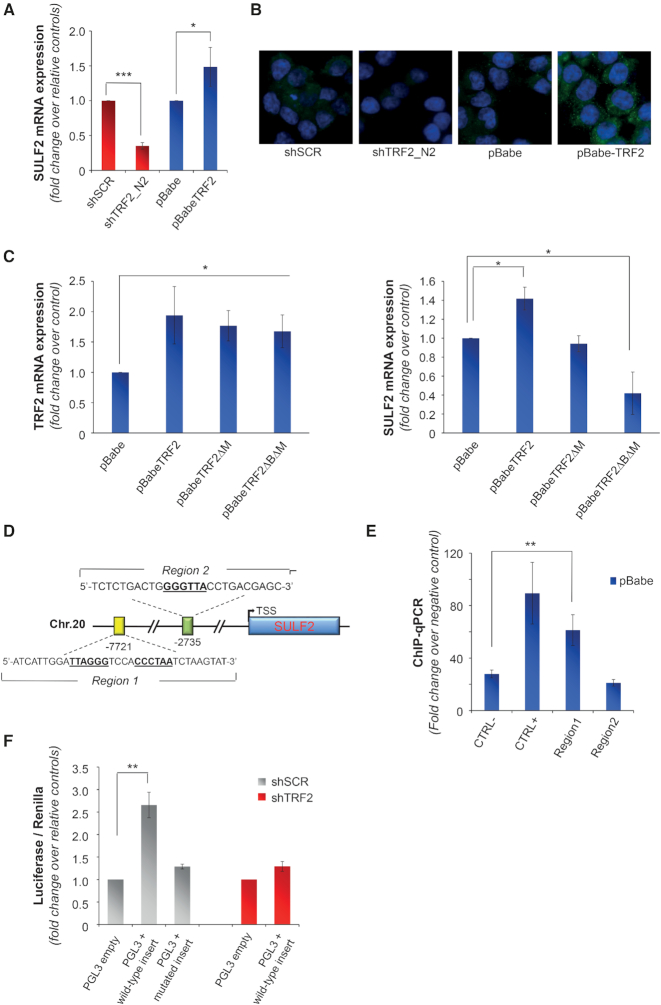
Thus, we examined the possibility that TRF2 directly modulates the expression of HSPGs. Quantitative rt-PCR of genes encoding for different classes of HSPGs revealed that none of the assayed molecules was modulated in response to changes in the levels of TRF2 (Supplementary Figure S10A). Next, as the binding activity of HSPGs to secreted molecules is tightly regulated by post-synthetic modifications (23), the analysis of gene expression in response to TRF2 modulation was also extended to the heparanase (HPSE) and to specific Heparan sulfate sulfotransferases (HSSTs) and sulfatases (SULFs) that, specifically regulating the sulfation of the 6-O position of HS chains, control the binding of HSPGs to VEGF-A (49). Interestingly, modulation of TRF2 determined significant changes in the expression of heparan sulfate 6-O-sulfotransferase-2 (SULF2) at both mRNA and protein levels (Figures 4A and B), without affecting the other enzymes (Supplementary Figure S10B).
Figure 4.
Extracellular release of VEGF-A is regulated by TRF2 through the control of SULF2 expression. (A) Gene expression of SULF2 was evaluated by qPCR in HCT116 cells silenced (shTRF2) or overexpressing (pBabe-TRF2) TRF2 and in their control counterparts (shScramble and pBabe). Results are expressed as fold change of mRNA levels in silenced/overexpressing cells over their controls, after β-actin normalization. (B) IF analysis of SULF2 expression in cells silenced or overexpressing TRF2 and their controls. Representative images acquired at 63x magnification are shown. (C) Expression of TRF2 (left panel) and SULF2 (right panel) was evaluated by qPCR in HCT116 cells overexpressing the wild-type (pBabe TRF2) or the mutated forms of TRF2 (pBabe TRF2ΔM and pBabe TRF2ΔBΔM) and their control counterpart (pBabe). Results are expressed as fold change of mRNA levels in overexpressing cells over their controls, after β-actin normalization. (D) Schematic representation of putative TRF2-binding sites located upstream the transcription starting site (TSS) of SULF2. (E) Real-time qPCR analysis of TRF2-chromatin immunoprecipitates. DNA regions containing (Chr.2 sub-telomeric region) or not (RPLP0) TTAGGG sequences were used as positive and negative control, respectively. (F) pGL3-promoter vector (pGL3 empty) and the vector containing the wild-type or mutant form of the distal regulatory element were co-transfected with a renilla vector (pRL-TK) in control (pBabe) and TRF2 silenced (shTRF2) HCT116 cells and luciferase activity was assayed. Firefly luciferase signal was normalized for the renilla signal to derive the relative luciferase activity. Results are expressed as fold change over the activity measured in the cells transfected with the control vector. The histograms show the mean ± SE of at least three independent experiments performed in triplicate (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test).
Based on the data in the literature showing that TRF2 can regulate gene expression by binding extra-telomeric TTAGGG DNA sequences (5,10,14), we evaluated the effect of TRF2 DNA-binding mutants on SULF2 expression. Notably, the experiments evidenced that overexpression of a TRF2 mutant (TRF2ΔM) lacking of C-terminal DNA-binding domain (Myb domain (50)), was unable to affect SULF2 expression (Figure 4C), indicating that DNA binding by TRF2 is necessary to induce SULF2 expression. On the other side, overexpression of the TRF2 mutant lacking of both the Myb and the Basic domains (TRF2ΔBΔM), known to act as dominant negative (50,51), was found to inhibit, similar to the shTRF2, the expression of SULF2.
Since the expression of certain genes may be affected by either proximal or distal regulatory elements (52,53), we looked for putative TRF2 binding sites within 10 kb upstream the coding sequence of SULF2. As revealed by the quantitative PCR analysis of TRF2-chromatin immunoprecipitates, TRF2 specifically binds an ITS (TTAGGGtccaCCCTAA) located 7721 bp upstream the transcription starting site (TSS) of SULF2 (Figures 4D and E). Notably, Encode data analysis of histone marks enrichment (Supplementary Figures S10C and D) evidenced that the identified TRF2-binding site falls into a genomic region showing distinctive characteristics of the distal regulatory elements (52,53). Starting from this observation, a genome portion of about 200 bp, including the binding site for TRF2, was cloned in the enhancer site of pGL3-promoter vector and its effect on transcription regulation was evaluated by luciferase assay. As reported in the figure 4F, sub-cloning of the indicated sequence induces an increase of ∼2.5-fold in the luciferase expression. In contrast, deletion of the binding site of TRF2, as well as silencing of TRF2 by shRNA, completely abolished the effect observed on luciferase expression (Figure 4F), demonstrating that control of gene expression is due to the direct binding of TRF2 to the identified target region.
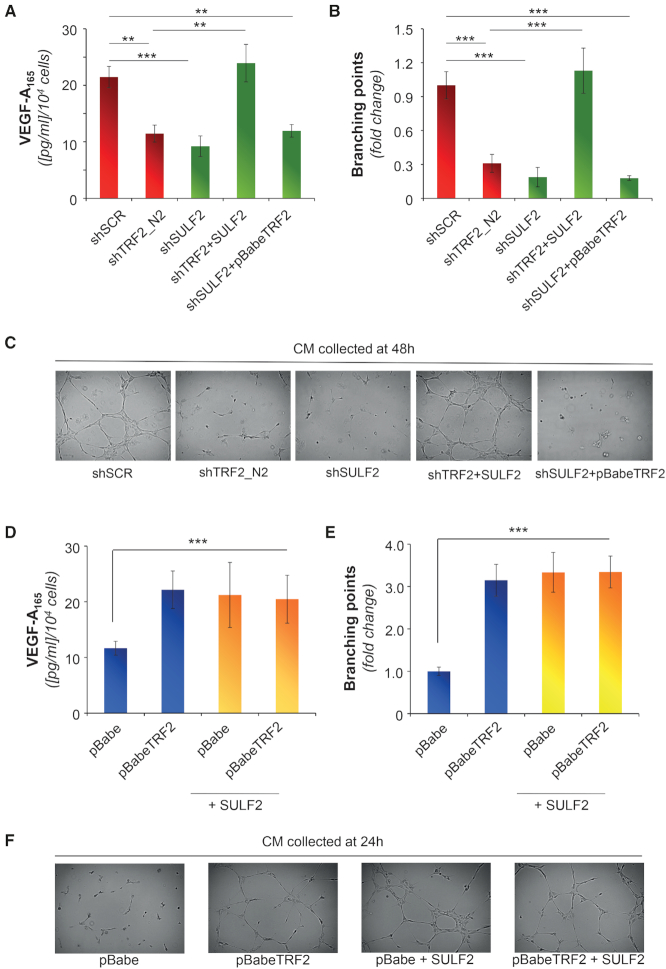
Finally, to define the role of SULF2 on VEGF-A secretion and angiogenesis, cellular levels of SULF2 were modulated (Supplementary Figure S10E), and the effects of protein silencing and overexpression were evaluated by ELISA and tubule formation assay, respectively (Figures 5). Interestingly, the changes of SULF2 expression fully recapitulate those of TRF2, reinforcing the idea that SULF2 could be a downstream effector of TRF2. To validate this hypothesis, TRF2-silenced cells were infected with lentiviral particles delivering a construct encoding for SULF2 or the shRNA against SULF2 (Supplementary Figure S10E) and the effects of the double infections were evaluated in terms of both VEGF-A secretion (Figure 5A) and angiogenesis (Figures 5B and C) Of note, the overexpression of SULF2 counterbalances the absence of TRF2 by completely rescuing the TRF2 activity. In contrast, overexpression of TRF2 was ineffective in cells silenced for SULF2 (Figures 5A–C), confirming the hypothesis that SULF2 is a downstream effector of TRF2 activity. On the other side, the upregulation of SULF2 in TRF2-overexpressing cells (Supplementary Figure S10E) was found to not exacerbate the response mediated by TRF2, clearly indicating that the two molecules act in an epistatic manner (Figures 5D–F and Supplementary Figure S11A).
Figure 5.
SULF2 is a direct target of TRF2 and controls tumor angiogenesis. (A) Concentration of VEGF-A evaluated by ELISA in the CM of empty vector or SULF2 overexpressing HCT116 cells infected with scramble (shSCR), TRF2 (shTRF2) or SULF2 (shSULF2) targeting shRNAs. Results were normalized to cell number. (B, C) CMs described in A were assayed for their capability of inducing capillary structures in HUVEC cells. (B) Mean number of branching points calculated on five different fields and expressed as fold induction over the negative control. (C) Representative images showing tubular-like structures (5X magnification). (D) Concentration of VEGF-A evaluated in the CM obtained from HCT116 cells infected as indicated. CM were obtained from cells growth in serum-free medium for 24 h. Results were normalized to cell number. (E, F) CMs described in D were assayed for their angiogenic potential in vitro. (E) Mean number of branching points calculated as in B. (F) representative tubular-like structure images from E. The histograms show the mean ± SD of at least three independent experiments performed in triplicate (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test).
Direct correlation among TRF2, SULF2 and tumor angiogenesis in colon cancer
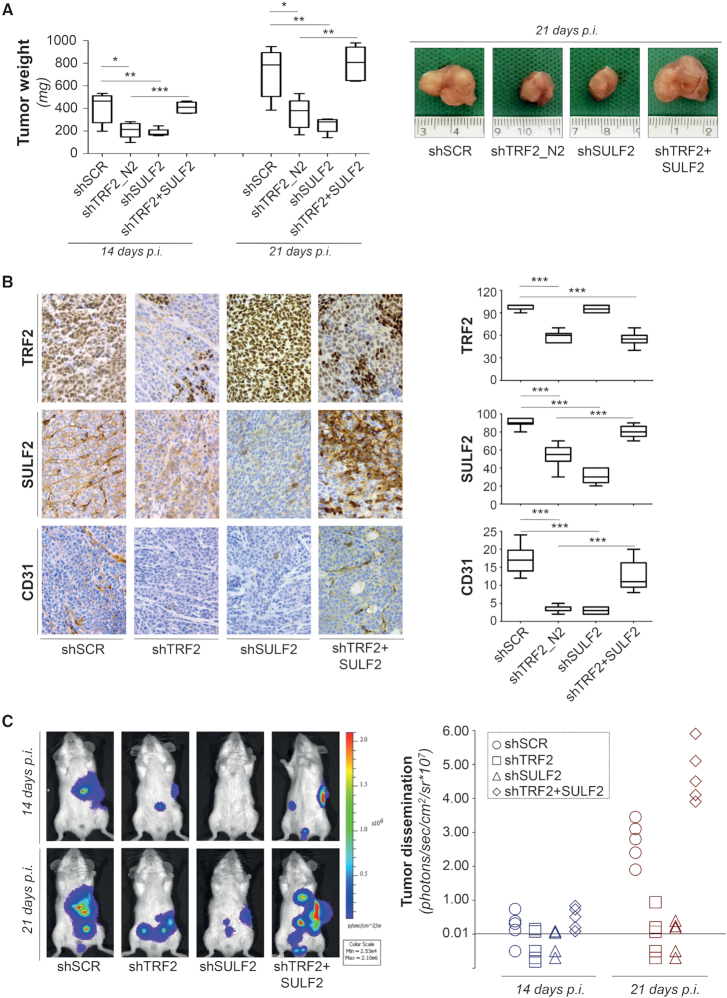
To address the relevance of our findings in vivo, control (shSCR) and TRF2 compromised (shTRF2) HCT116 colon cancer cells were injected in nude mice. As expected, silencing of TRF2 reduced tumor growth of about 50% (Figure 6A and Supplementary Figure S11B), and this difference was maintained during the days following tumor appearance. Interestingly, the analysis of the tumors at day 14 after cell injection revealed that inhibition of tumorigenicity following TRF2 depletion was not associated neither to an increase in apoptosis (Tunel staining) nor to DDR activation (γH2AX staining) (Supplementary Figure S11C). Conversely, a significant reduction of SULF2 expression (P = 0.0002) and a decrease number of microvessels (P = 0.0002) were observed in TRF2 compromised compared to control tumors (Figure 6B), corroborating the key role of TRF2 on tumor angiogenesis. Next, to define the relevance of SULF2 in mediating TRF2 activity, the analyses of tumor growth were also extended to SULF2 depleted cells (shSULF2) and to TRF2 interfered cells overexpressing SULF2 (shTRF2 + SULF2). Of note, while the interference of SULF2 determines an inhibition of tumor growth and angiogenesis comparable with that promoted by TRF2 silencing (Figures 6A and B), the overexpression of SULF2 completely reverts the inhibitory activity of TRF2 knock-down (Figures 6A and B). Finally, treatment with the SULF2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007), determines a reduction of tumor growth in control and, with a higher extent, in TRF2 overexpressing tumors (Supplementary Figure S11D), showing compelling therapeutic potential for cancer with high TRF2 expression.
Figure 6.
TRF2-silencing impairs tumor growth and metastasis by inhibiting SULF2-mediated angiogenesis. (A) Empty vector or SULF2 overexpressing (SULF2) HCT116 cells infected with lentiviral particles carriyng scramble (shSCR), TRF2 (shTRF2) or SULF2 (shSULF2) targeting shRNAs were intramuscularly injected in immunocompromised nude mice and tumor growth was assayed. Left panel: tumor weight was evaluated at the indicated days post-injection. The graph shows the mean ±SD from 5/7 mice per group (*P < 0.1, **P < 0.01, ***P < 0.001; Student's t-test). Right panel: representative images showing the size of tumors excised at day 21 after cell injection. (B) IHC analyses of the tumors from A. Left panel: representative IHC images of tumor stained with antibodies against TRF2, SULF2 and CD31. Right panel: quantitative analysis of the indicated markers. Data are mean (±SD) from two mice per group (*P < 0.1, **P < 0.01, ***P < 0.001; Mann–Whitney test). (C) Luminescent colon cancer cells were injected in the spleen of CB17-SCID mice and after 30 min the spleen was removed by spleenectomia. Real-time tumor dissemination was monitored by the IVIS imaging system 200 series (Caliper Life Sciences, Hopkinton, MA, USA) at day 14 and 21 after tumor cell injection. Left panel: Representative images of tumor dissemination acquired and analyzed using the Living Image Software version 3.0 (Caliper Life Sciences). Right panel: histogram reporting photons of tumor dissemination in each experimental group. The instrumental limit of photons detection for these cell lines was 1 × 105.
Since angiogenesis plays a key role also in tumor metastasization, the same cells used in tumor growth experiments were assayed in a model of spontaneous dissemination in vivo. As evidenced in Figure 6C, interference of either TRF2 or SULF2 determines a reduction of tumor metastases that, compared with the control cells (shSCR), reaches an average inhibition of about the 90% in three out five mice, at day 21 after cell injection. Of relevance, in both shTRF2 and shSULF2, two mice did not develop metastases (Figure 6C, right panel) and, on the other side, SULF2 overexpression completely abolishes the effect of TRF2 silencing on tumor dissemination.
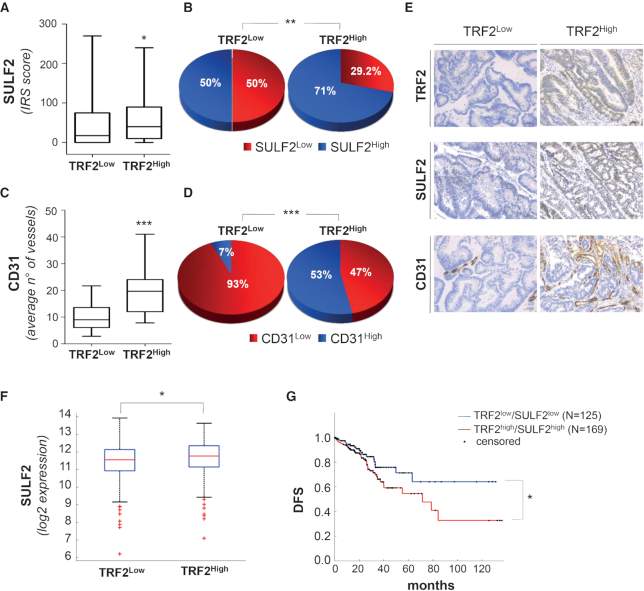
Finally, the clinical relevance of our findings was evaluated on a cohort of 169 CRC patients (123 colon and 46 rectal carcinomas) surgically treated at the Regina Elena National Cancer Institute and the study was reviewed and approved by the Local Ethic Committee of the same Institute (del. n.180/2014). The 169 patients with defined bio-pathological characteristics (Supplemental Table S4) were categorized, depending on the levels of TRF2 expression, into TRF2Low (56/169; 33%) and TRF2High (113/169; 67%). The tumors were stained for SULF2 and the results demonstrated that the median score was significantly (P = 0.046) higher in the sub-set of TRF2High patients, indicating that a positive correlation exists between the two variables (Figure 7A). Moreover, ROC analysis was used to estimate the optimal cut-off values able to split patients into groups with high or low SULF2 levels. Results showed in Figure 7B reports that SULF2 >15 (SULF2High) identify tumors with high level of TRF2 (71% versus 50% P = 0.008). Then, the correlation between TRF2 and vascular density using an anti-CD31 mAb was evaluated in a subset of 30 CRC patients with TRF2Low and 30 with TRF2High. The results demonstrated that the average number of vessels was significantly (P = 0.0001) higher in the TRF2High patients (Figure 7C) and ROC analysis corroborated these results (Figure 7D). Two exemplificative tumors with TRF2Low and TRF2High stained for SULF2 and CD31 were showed in Figure 7E.
Figure 7.
Direct correlation between TRF2 and SULF2 expression in colorectal cancer has prognostic impact on patient survival. (A, B) The correlation between TRF2 and SULF2 expression was evaluated by immunohistochemistry (IHC) using the immune-reactive score (IRS). (A) Box plots show the median values of SULF2 expression in the 56 TRF2Low and in the 113 TRF2High CRCs (*P = 0.046; Student's t-test). (B) Optimal SULF2 cut-off (IRS > 15) was established by ROC analysis and correlation with TRF2 was calculated. Pie charts show the distribution of SULF2 (low and high) in the sub-populations of TRF2Low and TRF2High CRC patients (**P = 0.008; χ2 test). (C, D) Correlation between TRF2 expression and microvessel density (evaluated by CD31). (C) Box plots show average number of vessels evaluated in a sub-set of 30 TRF2Low and 30 TRF2High CRC patients (***P = 0.0001). (D) Optimal CD31 cut-off (vessels/HPF >18) was established by ROC analysis and correlation with TRF2 was evaluated. The pie charts show the distribution of CD31 low and high in the 30 TRF2Low and in the 30 TRF2High CRCs (***P = 0.0001; χ2 test). (E) IHC evaluation of SULF2 and CD31 expression in two representative CRCs showing TRF2Low and TRF2High, respectively. Magnification 40x. Scale bar: 50 μm. (F) Box plot shows the SULF2 mRNA levels evaluated on a cohort of 327 TRF2Low (zscore< 0) and 294 TRF2High (zscore> 0) CRC patients from the TCGA dataset (*P = 0.01; Wilcoxon rank-sum test). (G) Disease-free survival (DFS) evaluated by Kaplan–Meier curves on CRC patients from the TCGA dataset. Patients were stratified on the basis of TRF2 and SULF2 mRNA expression and survival was evaluated in patient subgroups with positive TRF2/SULF2 correlations (TRF2High/SULF2High vs TRF2Low/SULF2Low) (*P = 0.03; log-rank test).
The positive correlation (R = 0.19; P = 0.006) between TRF2 and SULF2 was also found in a larger cohort of CRC patients (N = 621) from The Cancer Genome Atlas (TCGA) dataset (https://doi.org/10.7908/C11G0KM9) stratified on the basis of the TRF2 (TRF2Low 327/621; 53% and TRF2High 294/621; 47%) mRNA (Figure 7F). Moreover, TRF2/SULF2 covariates identified subgroups of patients with a high (P = 0.03) risk of relapse/progression (Figure 7G), suggesting that TRF2/SULF2 overexpression has a prognostic role in CRC patients.
DISCUSSION
We previously described a model in which TRF2 can control tumorigenesis not only via cancer cell-intrinsic mechanisms linked to DNA damage response, apoptosis and/or senescence, but also via non-cell autonomous pathways regulating immunosurveillance of tumor cells (10). Interestingly, this novel function is due to the ability of TRF2 to regulate the expression of HS3ST4, a gene encoding for the heparan sulfate (glucosamine) 3-O-sulfotransferase 4, and to inhibit the recruitment of NK cells (10). These findings reveal an unexpected role of TRF2 as transcriptional regulator, opening the way for a broad impact of TRF2 in cancer formation and progression.
Here, we reveal that TRF2 can alter the secretome of cancer cells and consequently tumor angiogenesis. The high-throughput approach based on multiplexed Luminex X-MAP technology showed that either the down-regulation or the overexpression of TRF2 mostly increased the secretion of growth factors, suggesting that this phenomenon is not controlled by a direct effect of TRF2. This is in agreement with a recent work showing that TRF2 knock-down enhanced the expression of cytokines implicated in inflammation (54). Of note, in this paper the author did not take into consideration cells overexpressing TRF2.
Among the different proteins analysed, only VEGF was found significantly modulated according to TRF2 expression. Consistently with these results, conditioned medium from cells with modulated TRF2 levels was able to interfere with proliferation, migration and angiogenic response of endothelial cells (ECs) both in vitro and in vivo and the effect was strictly dependent on VEGF-A, one of the main regulators of angiogenic response. Interestingly, the role of TRF2 on the secretion of VEGF-A and angiogenesis has been observed in various cell types of different histotype, strongly demonstrating that the role of TRF2 in the formation of new vessels is a general phenomenon. Taken together our data show that TRF2 overexpression in cancer cells can trigger an angiogenic switch, inducing the expression pattern of pro- versus anti-angiogenic molecules (cytokines, chemokines and/or growth factors) of cancer cells, and stimulate the formation of new vessels around the tumor. It was also reported that TRF2 is overexpressed in the vasculature of most human cancer types where it is required for proliferation, migration and tube formation of endothelial cells (5). These two TRF2 pro-angiogenic functions can be considered as the two faces of the same coin in which key players of tumor angiogenesis (endothelial cells from one side and cancer cells from the other) are orchestrated by TRF2. Of note, the role of TRF2 in tumor angiogenesis may also involve the immune system as proposed by us (10) and recently confirmed by an independent research group (54), suggesting that TRF2 can modulate the expression of a panel of cytokines involved in angiogenic balance and inflammatory processes. The fact that, in our experimental models, the role of TRF2 in tumor angiogenesis is independent of the immune system, offered us the opportunity to highlight a novel non cell-autonomous anti-neoplastic effect of TRF2 loss of function not related to telomere deprotection. Indeed, the angiogenic switch induced by TRF2 on several cancer cells is not accompanied by an overt DNA damage response and is not associated with an induction of apoptosis or senescence. Previous studies involving various types of cancer cells suggested that a reduced expression of TRF2 might not be enough to trigger DDR activation (13,55). The uncoupling of TRF2 role in telomere protection from the ability of cancer cells to escape immunosurveillance was attributed to an extratelomeric role of TRF2 in regulating gene expression (10). Our results show that a similar uncoupling between telomere capping and angiogenesis can also occur in tumors cells thanks to the role of TRF2 in increasing the VEGF-A signalling.
Our paper goes one step further questioning about the mechanism(s) through which TRF2 modulates the VEGF-A levels in the secretome of tumor cells. We found that, the expression of VEGF-A was not affected by overexpression/silencing of TRF2, permitting to exclude a direct transcriptional control of VEGF-A by TRF2 protein. Interestingly, by using biochemical and imaging approaches, we found that TRF2 regulates the extracellular release of VEGF-A, altering the equilibrium between cell-free and cell-associated amount of VEGF-A. This property of TRF2 is dependent on the expression of SULF2, a sulfotransferase with a potential oncogenic role that has been found overexpressed in subsets of multiple tumors (56). This enzyme, indeed, inducing post-synthetic modification of HSPGs, is capable of modulating the extracellular release of a number of growth factors containing an heparin-binding domain, including VEGF-A. Mechanistically, the region up-stream the TSS of SULF2 contains a binding site for TRF2 that is localized within a genomic portion showing the properties of a distal regulatory element. Our experiments demonstrated that, binding of TRF2 to its target motif can enhance the promoter activity eventually leading to an increased SULF2 expression. The results reported here clearly demonstrate that SULF2 is a novel direct TRF2-target gene and unveil a novel and so far unreported way through which TRF2 can regulate gene expression. Notably, these data open the question on how the TRF2-binding to a distal regulatory element can favour gene expression. We can envisage at least two mechanisms (TRF2 might) (i) act as a bridge for promoting the binding of transcription factors to DNA or (ii) induce the folding of the DNA required to bring distal-bound transcription factors near to the promoter) that will be further investigated.
These results, together with our previous finding on TRF2-mediated regulation of the sulfotransferase HS3ST4 (10) and, more recently, of two additional genes, GPC6 and versican (VCAN), both involved in the metabolism of HSPGs and GlycoAminoGlycan (GAG) (Cherfils Vicini et al., submitted), strongly indicate that TRF2 can act as a general remodeler of glycocalyx.
We also addressed the functional relevance of our findings demonstrating that the regulation of SULF2 is not only sufficient but also necessary to promote VEGF-A secretion and angiogenesis and, more interesting, genetic and pharmacological targeting of SULF2 impairs tumor growth, opening the way for new therapeutic option for cancer with high levels of TRF2. Notably, SULF2 appears to be a major effector of TRF2′oncogenic function, since overexpression of this target gene restore tumor growth, angiogenesis and metastatic ability of TRF2-dysfunctional cells. This is in line with a large number of high-quality mechanistic studies demonstrating important roles for HS signalling in cancer biology, including proliferation, tumor angiogenesis, metastasis and differentiation (23).
Finally, we translate our results on human patients by showing a direct correlation among TRF2 and SULF2 protein expression on a cohort of CRCs surgically treated in the Regina Elena National Cancer Institute and in the TCGA dataset available online. A direct correlation with tumor angiogenesis was also observed in TRF2High tumors. More interestingly, analysis of patient survival in the TCGA dataset reveled that TRF2/SULF2 overexpression has a prognostic role in CRC patients. Together, our pioneering results may constitute a valuable tool in determining the patient at risk of recurrence and may help to stratify patients who can benefit from anti-tumoral therapies targeting angiogenesis and/or HSPG and their modifying enzymes to enhance chemotherapy efficacy and/or overcome drug resistance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Vogelstein (Johns Hopkins University) for HCT116 cells, Prof Stefan Schoeftner (University of Trieste) for short hairpin constructs for TRF2 silencing, Dr Oreste Segatto (Regina Elena National Cancer Institute) for anti EGFR antibody and Dr Eros Lazzerini Denchi (NIH National Cancer Institute) for pBabe-puro-Empty and pBabe-puro-mycTRF2 constructs (Addgene plasmid #44573). We are also grateful to Prof Grazia Graziani (University of Rome Tor Vergata) for critical reading of the manuscript and Prof Antonella Stoppacciaro (Univestity of Rome La Sapienza) for helpful discussion of tissue analysis data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work in A.B. lab has been financially supported by the Italian Association for Cancer Research (AIRC) [16910]. The E.G. lab was supported by Fondation ARC (labelled program), Ligue Contre le Cancer (Equipe labellisée), Institut National du Cancer (INCa) (TELOCHROM), the LABEX SIGNALIFE (ANR-11-LABX-0028-01) and the UCAJEDI (ANR-15-IDEX-01). The IRCAN CytoMed, PICMI, and Animal core facilities were supported by grants from the Conseil Général 06, the FEDER, GIS IBISA, the Ministère de l’Enseignement Supérieur, the Région Provence Alpes-Côte d’Azur, the Canceropole PACA, the foundation ARC and INSERM. C.L. and E.S. groups were supporeted by AIRC [18637 to C.L., 17121 to E.S.]; R.D. and E.P. are recipients of a fellowship from the Italian Foundation for Cancer Research (FIRC). Funding for open access charge: Italian Ministry of Health - Ricerca corrente 2019.
Conflict of interest statement. None declared.
REFERENCES
- 1. Giraud-Panis M.J., Pisano S., Benarroch-Popivker D., Pei B., Le Du M.H., Gilson E.. One identity or more for telomeres. Front. Oncol. 2013; 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feuerhahn S., Chen L.Y., Luke B., Porro A.. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem. Sci. 2015; 40:275–285. [DOI] [PubMed] [Google Scholar]
- 3. Nakanishi K., Kawai T., Kumaki F., Hiroi S., Mukai M., Ikeda E., Koering C.E., Gilson E.. Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin. Cancer Res. 2003; 9:1105–1111. [PubMed] [Google Scholar]
- 4. Diehl M.C., Idowu M.O., Kimmelshue K.N., York T.P., Jackson-Cook C.K., Turner K.C., Holt S.E., Elmore L.W.. Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res. Treat. 2011; 127:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El Mai M., Wagner K.D., Michiels J.F., Ambrosetti D., Borderie A., Destree S., Renault V., Djerbi N., Giraud-Panis M.J., Gilson E. et al.. The telomeric protein TRF2 regulates angiogenesis by binding and activating the PDGFRbeta promoter. Cell Rep. 2014; 9:1047–1060. [DOI] [PubMed] [Google Scholar]
- 6. Blanco R., Munoz P., Flores J.M., Klatt P., Blasco M.A.. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 2007; 21:206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diala I., Wagner N., Magdinier F., Shkreli M., Sirakov M., Bauwens S., Schluth-Bolard C., Simonet T., Renault V.M., Ye J. et al.. Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Rep. 2013; 14:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujita K., Horikawa I., Mondal A.M., Jenkins L.M., Appella E., Vojtesek B., Bourdon J.C., Lane D.P., Harris C.C.. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat. Cell Biol. 2010; 12:1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biroccio A., Rizzo A., Elli R., Koering C.E., Belleville A., Benassi B., Leonetti C., Stevens M.F., D’Incalci M., Zupi G. et al.. TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur. J. Cancer. 2006; 42:1881–1888. [DOI] [PubMed] [Google Scholar]
- 10. Biroccio A., Cherfils-Vicini J., Augereau A., Pinte S., Bauwens S., Ye J., Simonet T., Horard B., Jamet K., Cervera L. et al.. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013; 15:818–828. [DOI] [PubMed] [Google Scholar]
- 11. Munoz P., Blanco R., Flores J.M., Blasco M.A.. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005; 37:1063–1071. [DOI] [PubMed] [Google Scholar]
- 12. Bai Y., Lathia J.D., Zhang P., Flavahan W., Rich J.N., Mattson M.P.. Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia. 2014; 62:1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye J., Renault V.M., Jamet K., Gilson E.. Transcriptional outcome of telomere signalling. Nat. Rev. Genet. 2014; 15:491–503. [DOI] [PubMed] [Google Scholar]
- 14. Simonet T., Zaragosi L.E., Philippe C., Lebrigand K., Schouteden C., Augereau A., Bauwens S., Ye J., Santagostino M., Giulotto E. et al.. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011; 21:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., Songyang Z.. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011; 21:1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang P., Pazin M.J., Schwartz C.M., Becker K.G., Wersto R.P., Dilley C.M., Mattson M.P.. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr. Biol. 2008; 18:1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang P., Casaday-Potts R., Precht P., Jiang H., Liu Y., Pazin M.J., Mattson M.P.. Nontelomeric splice variant of telomere repeat-binding factor 2 maintains neuronal traits by sequestering repressor element 1-silencing transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16434–16439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ovando-Roche P., Yu J.S., Testori S., Ho C., Cui W.. TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells. 2014; 32:2111–2122. [DOI] [PubMed] [Google Scholar]
- 19. Cherfils-Vicini J., Zizza P., Gilson E., Biroccio A.. A novel pathway links telomeres to NK-cell activity: Implications for immunotherapy. Oncoimmunology. 2014; 3:e27358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitelock J.M., Melrose J., Iozzo R.V.. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008; 47:11174–11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coombe D.R. Biological implications of glycosaminoglycan interactions with haemopoietic cytokines. Immunol. Cell Biol. 2008; 86:598–607. [DOI] [PubMed] [Google Scholar]
- 22. Laguri C., Arenzana-Seisdedos F., Lortat-Jacob H.. Relationships between glycosaminoglycan and receptor binding sites in chemokines-the CXCL12 example. Carbohydr. Res. 2008; 343:2018–2023. [DOI] [PubMed] [Google Scholar]
- 23. Knelson E.H., Nee J.C., Blobe G.C.. Heparan sulfate signaling in cancer. Trends Biochem. Sci. 2014; 39:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ai X., Do A.T., Kusche-Gullberg M., Lindahl U., Lu K., Emerson C.P. Jr. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 2006; 281:4969–4976. [DOI] [PubMed] [Google Scholar]
- 25. Viviano B.L., Paine-Saunders S., Gasiunas N., Gallagher J., Saunders S.. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 2004; 279:5604–5611. [DOI] [PubMed] [Google Scholar]
- 26. Lamanna W.C., Frese M.A., Balleininger M., Dierks T.. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J. Biol. Chem. 2008; 283:27724–27735. [DOI] [PubMed] [Google Scholar]
- 27. Rosen S.D., Lemjabbar-Alaoui H.. Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin. Ther. Targets. 2010; 14:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morimoto-Tomita M., Uchimura K., Bistrup A., Lum D.H., Egeblad M., Boudreau N., Werb Z., Rosen S.D.. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005; 7:1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uchimura K., Morimoto-Tomita M., Bistrup A., Li J., Lyon M., Gallagher J., Werb Z., Rosen S.D.. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006; 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uchimura K., Morimoto-Tomita M., Rosen S.D.. Measuring the activities of the Sulfs: two novel heparin/heparan sulfate endosulfatases. Methods Enzymol. 2006; 416:243–253. [DOI] [PubMed] [Google Scholar]
- 31. Hammond E., Khurana A., Shridhar V., Dredge K.. The role of heparanase and sulfatases in the modification of heparan sulfate proteoglycans within the tumor microenvironment and opportunities for novel cancer therapeutics. Front. Oncol. 2014; 4:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okamoto K., Bartocci C., Ouzounov I., Diedrich J.K., Yates J.R. 3rd, Denchi E.L.. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013; 494:502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Franceschin M., Rizzo A., Casagrande V., Salvati E., Alvino A., Altieri A., Ciammaichella A., Iachettini S., Leonetti C., Ortaggi G. et al.. Aromatic core extension in the series of N-cyclic bay-substituted perylene G-quadruplex ligands: increased telomere damage, antitumor activity, and strong selectivity for neoplastic over healthy cells. ChemMedChem. 2012; 7:2144–2154. [DOI] [PubMed] [Google Scholar]
- 34. Gowan S.M., Heald R., Stevens M.F., Kelland L.R.. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001; 60:981–988. [DOI] [PubMed] [Google Scholar]
- 35. Salvati E., Zizza P., Rizzo A., Iachettini S., Cingolani C., D’Angelo C., Porru M., Randazzo A., Pagano B., Novellino E. et al.. Evidence for G-quadruplex in the promoter of vegfr-2 and its targeting to inhibit tumor angiogenesis. Nucleic Acids Res. 2014; 42:2945–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biroccio A., Amodei S., Antonelli A., Benassi B., Zupi G.. Inhibition of c-Myc oncoprotein limits the growth of human melanoma cells by inducing cellular crisis. J. Biol. Chem. 2003; 278:35693–35701. [DOI] [PubMed] [Google Scholar]
- 37. Biroccio A., Benassi B., Filomeni G., Amodei S., Marchini S., Chiorino G., Rotilio G., Zupi G., Ciriolo M.R.. Glutathione influences c-Myc-induced apoptosis in M14 human melanoma cells. J. Biol. Chem. 2002; 277:43763–43770. [DOI] [PubMed] [Google Scholar]
- 38. Rizzo A., Iachettini S., Salvati E., Zizza P., Maresca C., D’Angelo C., Benarroch-Popivker D., Capolupo A., Del Gaudio F., Cosconati S. et al.. SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res. 2017; 45:1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Claesson-Welsh L., Welsh M.. VEGFA and tumour angiogenesis. J. Intern. Med. 2013; 273:114–127. [DOI] [PubMed] [Google Scholar]
- 42. Bruno A., Pagani A., Pulze L., Albini A., Dallaglio K., Noonan D.M., Mortara L.. Orchestration of angiogenesis by immune cells. Front. Oncol. 2014; 4:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vempati P., Popel A.S., Mac Gabhann F.. Extracellular regulation of VEGF: isoforms, proteolysis, and vascular patterning. Cytokine Growth Factor Rev. 2014; 25:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hacker U., Nybakken K., Perrimon N.. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 2005; 6:530–541. [DOI] [PubMed] [Google Scholar]
- 45. Keyt B.A., Berleau L.T., Nguyen H.V., Chen H., Heinsohn H., Vandlen R., Ferrara N.. The carboxyl-terminal domain (111-165) of vascular endothelial growth factor is critical for its mitogenic potency. J. Biol. Chem. 1996; 271:7788–7795. [DOI] [PubMed] [Google Scholar]
- 46. Krilleke D., Ng Y.S., Shima D.T.. The heparin-binding domain confers diverse functions of VEGF-A in development and disease: a structure-function study. Biochem. Soc. Trans. 2009; 37:1201–1206. [DOI] [PubMed] [Google Scholar]
- 47. Ferrara N. Binding to the extracellular matrix and proteolytic processing: two key mechanisms regulating vascular endothelial growth factor action. Mol. Biol. Cell. 2010; 21:687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu J., Song X., He Y.Q., Freeman C., Parish C.R., Yuan L., Yu H., Tang S.. Heparanase and vascular endothelial growth factor expression is increased in hypoxia-induced retinal neovascularization. Invest. Ophthalmol. Vis. Sci. 2012; 53:6810–6817. [DOI] [PubMed] [Google Scholar]
- 49. Vives R.R., Seffouh A., Lortat-Jacob H.. Post-Synthetic regulation of HS Structure: The yin and yang of the sulfs in cancer. Front. Oncol. 2014; 3:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P. et al.. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007; 14:147–154. [DOI] [PubMed] [Google Scholar]
- 51. van Steensel B., Smogorzewska A., de Lange T.. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998; 92:401–413. [DOI] [PubMed] [Google Scholar]
- 52. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meng H., Bartholomew B.. Emerging roles of transcriptional enhancers in chromatin looping and promoter-proximal pausing of RNA polymerase II. J. Biol. Chem. 2018; 293:13786–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benhamou Y., Picco V., Raybaud H., Sudaka A., Chamorey E., Brolih S., Monteverde M., Merlano M., Lo Nigro C., Ambrosetti D. et al.. Telomeric repeat-binding factor 2: a marker for survival and anti-EGFR efficacy in oral carcinoma. Oncotarget. 2016; 7:44236–44251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takai K.K., Hooper S., Blackwood S., Gandhi R., de Lange T.. In vivo stoichiometry of shelterin components. J. Biol. Chem. 2010; 285:1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lui N.S., Yang Y.W., van Zante A., Buchanan P., Jablons D.M., Lemjabbar-Alaoui H.. SULF2 expression is a potential diagnostic and prognostic marker in lung cancer. PLoS One. 2016; 11:e0148911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.