Abstract
Activation of the checkpoint protein Tel1 requires the Mre11–Rad50–Xrs2 (MRX) complex, which recruits Tel1 at DNA double-strand breaks (DSBs) through direct interaction between Tel1 and Xrs2. However, in vitro Tel1 activation by MRX requires ATP binding to Rad50, suggesting a role also for the MR subcomplex in Tel1 activation. Here we describe two separation-of-functions alleles, mre11-S499P and rad50-A78T, which we show to specifically affect Tel1 activation without impairing MRX functions in DSB repair. Both Mre11-S499P and Rad50-A78T reduce Tel1–MRX interaction leading to poor Tel1 association at DSBs and consequent loss of Tel1 activation. The Mre11-S499P variant reduces Mre11–Rad50 interaction, suggesting an important role for MR complex formation in Tel1 activation. Molecular dynamics simulations show that the wild type MR subcomplex bound to ATP lingers in a tightly ‘closed’ conformation, while ADP presence leads to the destabilization of Rad50 dimer and of Mre11–Rad50 association, both events being required for MR conformational transition to an open state. By contrast, MRA78T undertakes complex opening even if Rad50 is bound to ATP, indicating that defective Tel1 activation caused by MRA78T results from destabilization of the ATP-bound conformational state.
INTRODUCTION
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that must be accurately repaired to ensure genomic stability and avoid cell death. Cells can repair DSBs by either non-homologous end joining (NHEJ) or homologous recombination (HR) (1). HR is initiated by nucleolytically processing of the 5′ strands of each DSB end in a process referred to as resection (2). The resulting 3′-ended single-stranded DNA (ssDNA) tails are covered by Rad51 to form helical filaments that catalyze invasion of the homologous double-stranded DNA (dsDNA) molecules to initiate HR (3).
The highly conserved MRX/MRN complex (Mre11–Rad50–Xrs2 in budding yeast; MRE11–RAD50–NBS1 in mammals) is one of the first protein complexes to be recruited to DSBs, where it plays key roles both in promoting DNA end resection and in maintaining the DSB ends tethered to each other for their repair (4,5). Mre11 is a dimer that possesses both 3′-to-5′ exonuclease and ssDNA endonuclease activities in vitro (6,7). MRX initiates resection of DNA DSB ends by catalyzing an endonucleolytic cleavage of the 5′-terminated DSB end, with the support of the Sae2 protein (CtIP in mammals) (8–10). This MRX-Sae2-mediated DNA cleavage generates an entry site for the nucleases Exo1 and Dna2, the latter working together with the helicase Sgs1 (9–16).
The core MR complex exists as a heterotetrameric assembly, where an Mre11 dimer interacts with two Rad50 molecules. Each Rad50 polypeptide possesses N- and C-terminal domains that fold back onto each other to form two complete ATPase sites on a Rad50 dimer (17). The DNA binding MR head is formed by the two Rad50-ATPase domains and the Mre11 nuclease domain that bind to the base of the Rad50 coiled-coils (5). At the apex of the Rad50 coiled-coils, it lies a hook domain that can dimerize with the hook domain of another Rad50 molecule via tetrahedral coordination of a zinc ion, thereby tethering broken DNA ends together (18–20).
Upon ATP binding, Rad50 closes into a rigid conformation, in which the head domains interact with each other to form a central groove that can accommodate dsDNA (21–26). This Rad50 (ATP-bound) closed state renders dsDNA inaccessible to the Mre11 nuclease active site and stimulates both DNA binding and tethering activities of the complex (21–27). By contrast, in the ATP-free or hydrolyzed state, the Rad50 subunits are flexible and relatively open, suggesting that ATP hydrolysis causes a conformational change in the Rad50 dimer, which opens the globular domain and makes DNA accessible to the Mre11 nuclease active sites (21–27).
MRX/MRN is also necessary for activation of the protein kinase Tel1 (ATM in mammals) (28–31), which responds to DNA damage by phosphorylating downstream effector kinases to coordinate cell cycle progression with DSB repair (32). The exact molecular mechanism of Tel1/ATM activation by MRX/MRN remains to be elucidated. Indeed, MRX/MRN is required to recruit Tel1/ATM to sites of DNA DSBs through direct interaction between Tel1/ATM and the C-terminal domain of the Xrs2/NBS1 subunit (33–37). Accordingly, we have previously identified the mre11-H98Y, mre11-K292E and mre11-R389C mutations, which impaired Tel1 recruitment to DSBs by directly affecting MRX–DNA interaction and therefore MRX recruitment to DSBs (38). While the role of K292 was difficult to envisage, H98 is directly implicated in stabilization of the Mre11 dimerization domain, and its substitution with Y impaired Mre11 dimer formation that is required for MRX–DNA association (25,38). By contrast, the R389 residue directly contacts the phosphodiesteric bridge of the 3′ DNA terminus and its substitution with C likely reduced Mre11-DNA association by eliminating the positive charge (38).
However, Mre11 and Rad50 appear to participate in Tel1/ATM activation independently of the Xrs2/NBS1 subunit. In fact, in vitro Tel1/ATM also interacts with the MR subcomplex, which is sufficient to stimulate the activity of Tel1/ATM, as well as its binding to DNA in both Schizosaccharomyces pombe and mammals (30,31,37). Moreover, in vitro activation of human ATM by MRN requires ATP binding but not ATP hydrolysis (28), suggesting that MRX/MRN activates Tel1/ATM when it is in the ATP-bound state. Consistent with this hypothesis, the L802W aminoacid substitution in Pyrococcus furiosus Rad50 (equivalent to the I1192W, I1214W and L1211W substitutions in S. pombe, Saccharomyces cerevisiae and humans, respectively) destabilized the ATP-bound state and impaired Tel1/ATM-mediated checkpoint signalling (27). However, the R805E aminoacid substitution in P. furiosus Rad50 (equivalent to the R1195E, R1217E and R1214E substitutions in S. pombe, S. cerevisiae and humans, respectively), which reduces ATP hydrolysis and therefore should stabilize the ATP-bound state, also impaired Tel1 activation (27). Thus, other experiments are required to understand the role of Rad50 and Mre11 in Tel1/ATM activation.
To better understand how the MR subcomplex participates in Tel1/ATM activation, we searched for separation of functions mre11 and rad50 alleles that impaired Tel1 activation but still retained MRX functions in DSB repair. As the lack of Tel1 causes telomere shortening (39) and hypersensitivity to camptothecin (CPT) but not to other genotoxic treatments (40), these mre11 and rad50 mutations have been searched among clones showing both decreased viability in the presence of CPT and short telomeres. Here we report the identification and characterization of the separation-of-function mre11-S499P and rad50-A78T mutant alleles, which we show to specifically abolish Tel1 activation without impairing MRX functions in DSB repair. Both the mre11-S499P and rad50-A78T mutations reduced MRX–Tel1 interaction leading to poor Tel1 association to DNA DSBs. The Rad50-A78T variant did not affect MRX complex formation, while the S499P aminoacid substitution, which is located at the Mre11–Rad50 interface, reduced the interaction between Mre11 and Rad50, suggesting that Mre11–Rad50 complex formation is important for Tel1/ATM activation. Molecular dynamics simulations revealed that wild type MR assumes a tightly closed conformation in the presence of ATP, whereas the Rad50-A78T variant under the same conditions destabilizes both the interaction between the two Rad50 subunits and their binding to Mre11, thus facilitating the conversion of the complex to an open conformation. Altogether, our data indicate that the ATP-bound conformation of the Mre11–Rad50 subcomplex has a key role in binding and activating Tel1 in response to DSBs.
MATERIALS AND METHODS
Yeast strains and growth conditions
Strain genotypes are listed in Supplementary Table S1. Strain JKM139, used to detect DSB resection, was kindly provided by J. Haber (Brandeis University, Waltham, USA). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone) supplemented with 2% glucose (YEPD), 2% raffinose (YEPR) or 2% raffinose and 3% galactose (YEPRG). Gene disruptions were generated by one-step PCR disruption method. All the experiments have been performed at 27°C.
Search for mre11 and rad50 mutants
The screen for mre11 and rad50 mutations has been carried out as previously described (38). Briefly, genomic DNA from strains carrying either the HPHMX gene located 250 bp downstream of the MRE11 stop codon or the KANMX gene located 570 bp upstream of the RAD50 ORF was used as template to amplify by low-fidelity PCR the MRE11 and the RAD50 coding region, respectively. Thirty independent PCR reaction mixtures were prepared, each containing 5U EuroTaq DNA polymerase (Euroclone), 10 ng genomic DNA, 500 ng each primer, 0.5 mM each dNTP (dATP, dTTP, dCTP), 0.1 mM dGTP, 0.5 mM MnCl2, 10 mM ß-mercaptoethanol, 10 mM Tris–HCl (pH9), 50 mM KCl and 1.5 mM MgCl2. The resulting PCR amplification products, containing the MRE11 or RAD50 coding sequence and the HPHMX or KANMX resistance gene, respectively, were used to transform a wild type strain. Three thousand transformants were selected and then assayed by drop tests for hypersensitivity to high doses of camptothecin but not to phleomycin or methyl methanesulfonate (MMS). The selected clones were then analyzed by Southern blot for telomere shortening similar to tel1Δ cells.
DSB resection
DSB end resection at the MAT locus in JKM139 derivative strains was detected on alkaline agarose gels, by using a single-stranded probe complementary to the unresected DSB strand, as previously described (41). Quantitative analysis of DSB resection was performed by calculating the ratio of band intensities for ssDNA and total amount of DSB products.
ChIP and qPCR
ChIP analysis was performed with anti-HA (12CA5) or anti-Myc antibodies (Ab32 from Abcam) antibodies as previously described (41). Quantification of immunoprecipitated DNA was achieved by quantitative real-time PCR (qPCR) on a Bio-Rad MiniOpticon apparatus. Triplicate samples in 20 μl reaction mixture containing 10 ng of template DNA, 300 nM for each primer, 2x SsoFast™ EvaGreen® supermix (Bio-Rad #1725201) (2x reaction buffer with dNTPs, Sso7d-fusion polymerase, MgCl2, EvaGreen dye, and stabilizers) were run in white 48-well PCR plates Multiplate™ (Bio-Rad #MLL4851). The qPCR program was as follows: step 1, 98°C for 2 min; step 2, 98°C for 5 s; step 3, 60°C for 10 s; step 4, return to step 2 and repeat 30 times. At the end of the cycling program, a melting program (from 65°C to 95°C with a 0.5°C increment every 5 s) was run to test the specificity of each qPCR. Data are expressed as fold enrichment at the HO-induced DSB over that at the non-cleaved ARO1 locus, after normalization of each ChIP signals to the corresponding input for each time point. Fold enrichment was then normalized to the efficiency of DSB induction.
Plasmid religation assay
The centromeric pRS316 plasmid was digested with the BamHI restriction enzyme before being transformed into the cells. Parallel transformation with undigested pRS316 DNA was used to determine the transformation efficiency. Efficiency of re-ligation was determined by counting the number of colonies that were able to grow on medium selective for the plasmid marker and was normalized to the transformation efficiency for each sample. The re-ligation efficiency in mutant cells was compared to that of wild type cells that was set up to 100%.
Western blotting and immunoprecipitation
Trichloroacetic acid protein extracts were separated on 10% polyacrylamide gels and Rad53 detection was carried out by using anti-Rad53 polyclonal antibodies (ab104232) from Abcam. Immunoprecipitations were performed as previously described (42), with the following modifications: protein extracts were prepared in a lysis buffer containing 50 mM HEPES (pH 7.5), 140 mM NaCl, 1 mM EDTA (pH 7.5), 10% glycerol, 0.5% NP40, 1 mM phenylmethylsulfonyl fluoride, 60 mM β-glycerophosphate, 1 mM sodium orthovanadate and a protease inhibitor cocktail (Roche Diagnostics).
Structural models
A starting structural model of Mre11–Rad50 (ATP-bound) complex was built by homology modelling (HM) technique, as previously described (38). Briefly, Mre11 and Rad50 subunits were built using as templates the homologs of S. pombe (PDB ID: 4FCX) and Chaetomium thermophilum (PDB ID: 5DA9), respectively. Then, the Mre11–Rad50 complex was assembled using the tetrameric structure of the complex of Methanocaldococcus jannaschii (PDB ID: 5F3W). Maestro Mutagenesis Wizard (https://www.schrodinger.com/maestro) was used to mutate alanine 78 to threonine and to change ATP into ADP to obtain ATP-bound Mre11–Rad50 (MR-ATP), ADP-bound Mre11–Rad50 (MR-ADP), ATP-bound Mre11–Rad50A78T (MRA78T-ATP) and ADP-bound Mre11–Rad50A78T (MRA78T-ADP) systems. Proper protonation states of amino acids were assigned using Maestro Protein Preparation Wizard at pH 7.0 (43). All models were refined through Molecular Dynamics (MD) simulations. MD simulations were performed using Gromacs 5.1.2 (https://www.gromacs.org) and CHARMM36 force field (44). All bonds involving hydrogen were constrained by linear constrained solver algorithm (45). Non-bonded interaction pair list was calculated with Verlet algoritm (46) every 20 fs using a cutoff of 1.2 nm. Long range electrostatic interactions were treated using PME method (47) setting a cut-off of 1.2 nm. Each complex was solvated in a dodecahedral box with TIP3P water model (48) at a minimum distance of 1.2 nm from solute to box edges. K+ ions were added as counter ions to obtain electroneutral models.
Refinement of Homology Modelling (HM) structures
Structures obtained by HM were subsequently optimized by steepest descent energy minimization with a limit of 50000 steps and a convergence criterion on the maximum force (<10 kJ mol−1 nm−1). Temperature and pressure were equilibrated at 300 K with NVT simulation of 4 ns and 1 atm with NPT simulation of 400 ps, respectively. MD simulations were carried out for 500 ns in NPT ensemble at 300 K and 1 atm with a time step of 2 fs. All trajectories of MR-ATP, MR-ADP, MRA78T-ATP, MRA78T-ADP systems were analysed using Gromos algorithm (49) for cluster analysis with a cutoff of 0.30, 0.35, 0.35 and 0.32 respectively. Centroids of the first five clusters of each model were used as starting points for further simulations.
MD production
Productive MD simulations were performed within a NPT thermodynamic ensemble at 300 K and 1 atm. More in detail, five simulations (200 ns) were carried out using centroid structures of the first five clusters obtained from previous simulations, as starting points of each system. To improve conformational sampling, two independent replicas were carried out using different initial atomic velocities for each structure for a total of ten simulations on each system. As a result, 2000 ns of simulation time have been collected for each model.
Post-production trajectory analysis
Analysis tools of GROMACS package were used to perform the analyses on the concatenated trajectories. Backbone root-mean-square deviation (RMSD) was used to evaluate trajectory stability of each replica, computed using the initial structure of each replica as reference. Principal Component Analysis (PCA) was performed on ATP/ADP and backbone atoms in order to identify globally correlated motions from MD trajectories. This method is based on dimensionality reduction of trajectory through the construction and diagonalization of the covariance matrix (50). Free Energy Landscapes (FELs) were constructed using the probability distribution along two reaction coordinates (the first two eigenvectors, PC1 and PC2) in order to identify the dominant conformations with lower energy for each system. The joint-probability distribution is estimated by ΔG(X) = –kBTlnP(X), where kB is the Boltzmann constant, T is the absolute temperature and P(X) is the probability distribution along reaction coordinates X.
RESULTS
Identification of mre11 and rad50 mutations that specifically affect Tel1 activation
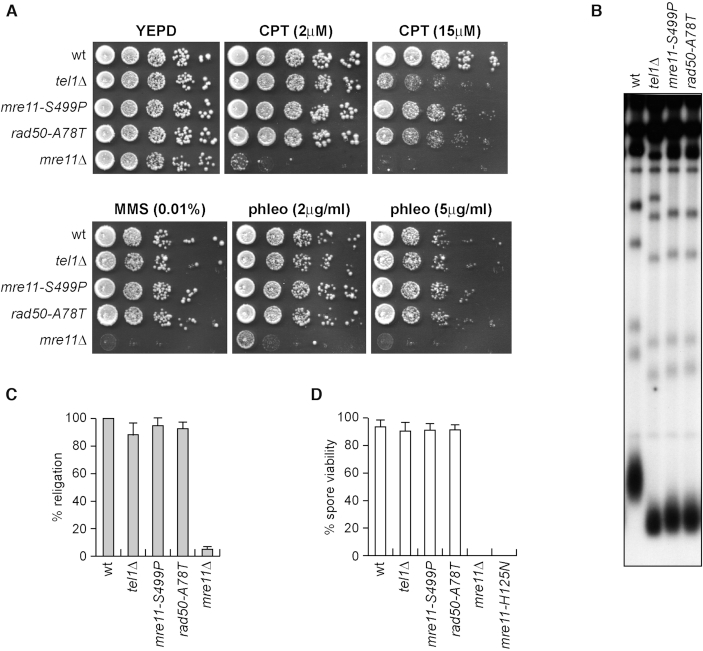
To identify separation of function mre11 and rad50 mutations that specifically affect Tel1 activation without impairing MRX functions in DSB repair, we took advantage of the finding that Tel1-deficient cells are hypersensitive to high doses of the type I topoisomerase inhibitor camptothecin (CPT), but not to other genotoxic agents like methyl methanesulfonate (MMS) and phleomycin (40) (Figure 1A). We used low-fidelity PCR to random mutagenize the MRE11 and RAD50 genes, followed by transformation with the linear MRE11 or RAD50 PCR products into wild type cells in order to replace the corresponding MRE11 or RAD50 wild type sequence with the mutagenized DNA fragments (38). Transformants were then screened for low viability in the presence of high doses of CPT, while retaining wild type survival to phleomycin and MMS treatment. As the lack of Tel1/ATM causes telomere shortening (39), transformant clones specifically showing CPT hypersensitivity were subsequently analyzed for telomere length by Southern blot. This analysis allowed us to identify the mre11-S499P mutation, causing the replacement of the Mre11 Ser499 residue with Pro, and the rad50-A78T mutation, causing the replacement of the Rad50 Ala78 residue with Thr. Both mre11-S499P and rad50-A78T cells lost viability at high doses of CPT, although their CPT hypersensitivity was less severe than that of tel1Δ cells (Figure 1A). Furthermore, they did not lose viability in the presence of MMS or phleomycin, differently from mre11Δ cells (Figure 1A), indicating that they did not affect MRX functions in DNA damage resistance. Finally, they showed telomere shortening similar to tel1Δ cells (Figure 1B).
Figure 1.
The mre11-S499P and rad50-A78T mutations phenocopy TEL1 deletion with respect to CPT hypersensitivity and telomere length. (A) Exponentially growing cells were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without CPT, MMS or phleomycin. (B) Telomere length. XhoI-cut genomic DNA from exponentially growing cells was subjected to Southern blot analysis using a poly(GT) telomere-specific probe. (C) Plasmid re-ligation assay. Cells were transformed with the same amounts of BamHI-linearized pRS316 plasmid DNA. Data are expressed as percentage of re-ligation relative to wild type that was set up at 100% after normalization to the corresponding transformation efficiency of the uncut plasmid. (D) Spore viability. Diploid cells homozygous for the indicated mutations were induced to enter in meiosis followed by tetrad dissection on YEPD plates. At least 40 tetrads for each strain have been analyzed. Mean values are represented with error bars denoting S.D. (n = 3).
MRX is essential to repair DSBs by both NHEJ and HR, whereas Tel1 has a very minor role in both processes (41,51). We asked whether the mre11-S499P and rad50-A78T mutations affect MRX function in NHEJ by measuring the ability of mre11-S499P and rad50-A78T cells to re-ligate a plasmid that was linearized before being transformed into the cells. The efficiency of plasmid re-ligation was similar in wild type, tel1Δ, mre11-S499P and rad50-A78T cells (Figure 1C), while it was dramatically decreased in mre11Δ cells, indicating that the mre11-S499P and rad50-A78T mutations do no affect MRX functions in DSB repair by NHEJ.
The MRX complex is essential to generate and resect meiotic DSBs, which are created by the Spo11 transesterase that forms a covalent linkage between a conserved tyrosine residue and the 5′ end of the cleaved strand (52). Spore viability is impaired by the lack of either the Mre11 nuclease activity or any MRX subunit (53). We therefore induced meiosis in wild type, mre11-S499P/mre11-S499P and rad50-A78T/rad50-A78T diploid cells, followed by tetrad dissection and spore viability analysis. Spore viability was not reduced by the tel1Δ, mre11-S499P and rad50-A78T mutations, while it was dramatically decreased in diploid cells either lacking Mre11 (mre11Δ/mre11Δ) or expressing a nuclease defective mre11 allele (mre11-H125N/mre11-H125N) (Figure 1D), indicating that Mre11-S499P and Rad50-A78T variants maintain Mre11 functions in meiosis.
The mre11-S499P and rad50-A78T mutations phenocopy TEL1 deletion with respect to DSB resection and checkpoint signalling
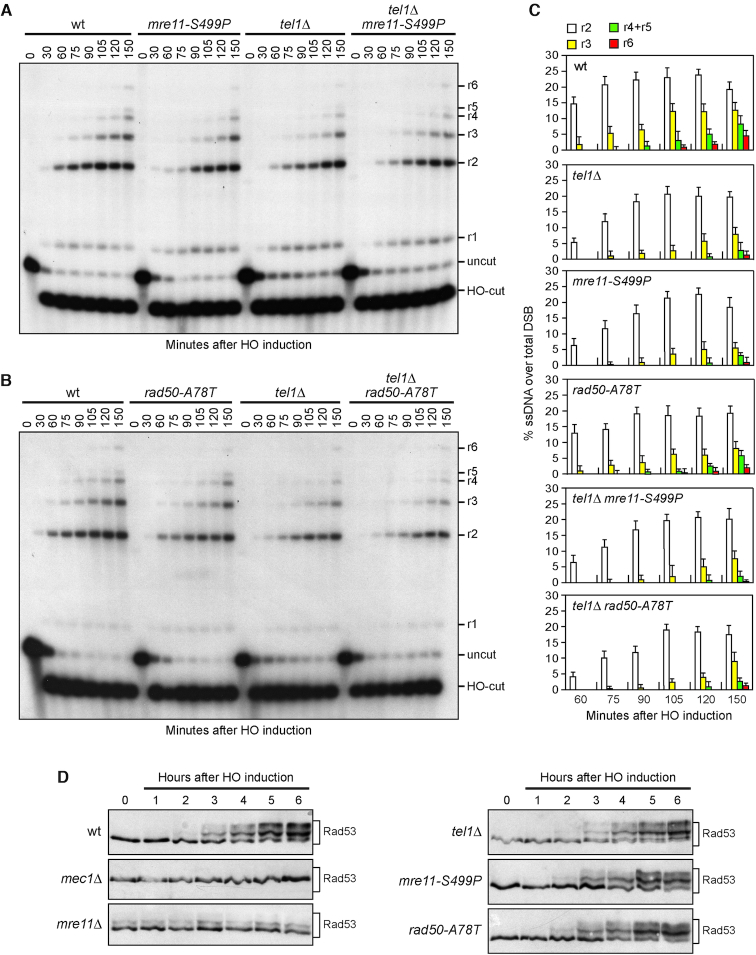
During HR, MRX is required to generate 3′-ended RPA-coated ssDNA at the DSB ends that catalyses strand invasion. This ssDNA also induces activation of a Mec1-dependent checkpoint response (54,55). The lack of any MRX subunit severely reduces both ssDNA generation at the DSB ends and Mec1 activation (56,57). By contrast, Tel1 has a minor role in DSB signalling compared to Mec1, as tel1Δ cells caused only a very slight reduction both in ssDNA generation at the DSB ends and in checkpoint activation in response to a single DSB (51). To investigate the effect of the mre11-S499P and rad50-A78T alleles on both processes, we used JKM139 derivative strains, where a single DSB at the MAT locus can be generated by expressing the HO endonuclease gene under the control of a galactose-inducible promoter. The HML and HMR loci were deleted in these strains to prevent DSB repair by gene conversion (57). Resection of DNA regions flanking the HO-induced DSB renders the DNA sequence resistant to cleavage by restriction enzymes, resulting in the appearance of resection intermediates that can be detected by Southern blot analysis with a probe that anneals to the 3′ end at one side of the break. Both mre11-S499P and rad50-A78T cells showed only a slight reduction in the efficiency of ssDNA generation at the HO-induced DSB and the extent of this defect was similar to that of tel1Δ cells (Figure 2A–C). This resection defect was not exacerbated in mre11-S499P tel1Δ and rad50-A78T tel1Δ double mutants (Figure 2A–C), indicating that the mre11-S499P and rad50-A78T mutations reduce DSB resection by altering the same pathway affected by TEL1 deletion.
Figure 2.
The mre11-S499P and rad50-A78T mutations phenocopy TEL1 deletion with respect to DSB resection and checkpoint activation. (A, B) DSB resection. YEPR exponentially growing cell cultures were arrested in nocodazole and then transferred to YEPRG at time zero to induce HO expression. SspI-digested genomic DNA was separated and hybridized with a single-stranded MAT probe that anneals to the unresected 3′ end at one side of the break. 5′-3′ resection progressively eliminates SspI sites, producing larger SspI fragments (r1 through r6) that can be detected by the probe. (C) Densitometric analysis. The experiment as in (A, B) was independently repeated and the mean values are represented with error bars denoting S.D. (n = 3). (D) Rad53 phosphorylation. Exponentially growing YEPR cultures were transferred to YEPRG (time zero) to induce HO, followed by western blot analysis with anti-Rad53 antibodies.
To monitor checkpoint activation, we followed Rad53 phosphorylation, which is required for Rad53 activation and is detectable as a decrease of Rad53 electrophoretic mobility. As previously reported (51), phosphorylated Rad53 was not detectable in galactose-induced JKM139 mec1Δ cells. Furthermore, HO induction did not cause Rad53 phosphorylation in mre11Δ cells, where a basal level of Rad53 phosphorylation was detectable even in the absence of DSB formation possibly due to DNA replication defects (Figure 2D). By contrast, mre11-S499P, rad50-A78T and tel1Δ cells phosphorylated Rad53 with kinetics similar to that of wild type cells (Figure 2D). Altogether, these data indicate that both Mre11-S499P and Rad50-A78T mutant variants behave like TEL1 deletion with respect to DSB resection and checkpoint activation.
The mre11-S499P and rad50-A78T mutations specifically abolish Tel1-mediated checkpoint activation
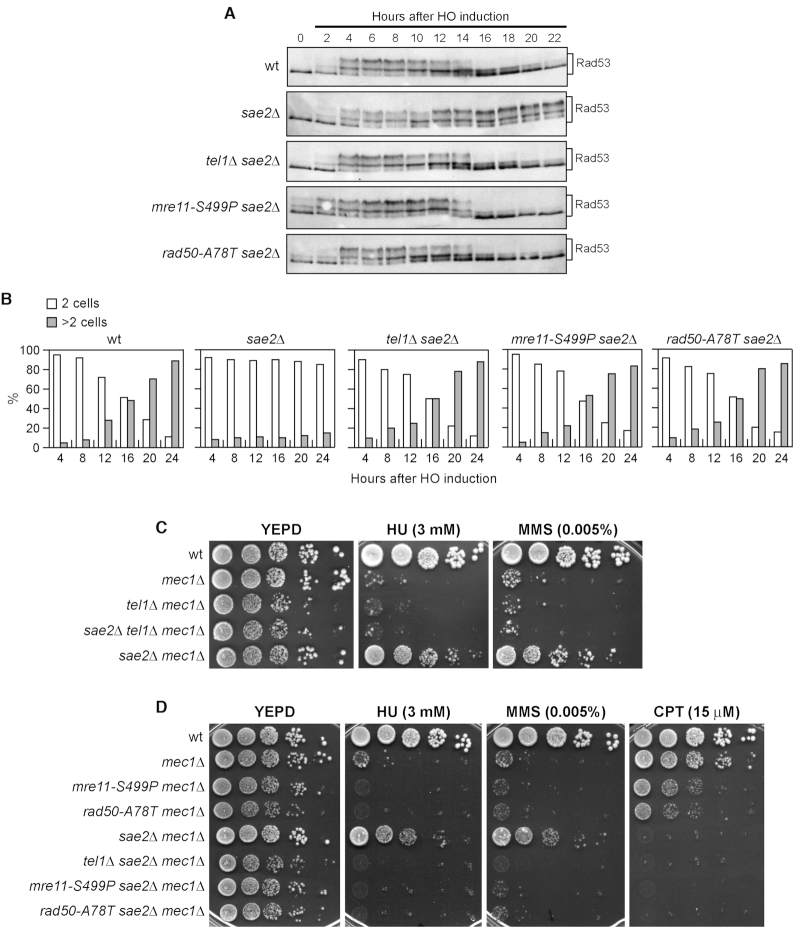
The checkpoint response triggered by a single unrepairable DSB can be eventually turned off, allowing cells to resume cell cycle progression through a process that is called adaptation (54,58). In the absence of Sae2, cells display heightened Tel1 activation, which is associated with persistent Rad53 phosphorylation that prevents cells from adapting to an unrepaired DSB (59–61). If the mre11-S499P and rad50-A78T mutations affect Tel1 activation, they would be expected to suppress the adaptation defect of sae2Δ cells by reducing Tel1-dependent Rad53 phosphorylation. As shown in Figure 3A, Rad53 phosphorylated forms were detectable with similar kinetics ∼2–4 h after galactose addition in wild type, sae2Δ, tel1Δ sae2Δ, mre11-S499P sae2Δ and rad50-A78T sae2Δ cells. Consistent with DSB-induced checkpoint activation, when cells arrested in G1 with α-factor were spotted on galactose-containing plates to induce HO, all the above cell cultures accumulated large budded cells within 4 h (Figure 3B). However, sae2Δ cells showed persistent Rad53 phosphorylation (Figure 3A) and most of them remained arrested as large budded cells for at least 24 h (Figure 3B). By contrast, the amounts of phosphorylated Rad53 decreased in sae2Δ mre11-S499P and sae2Δ rad50-A78T cells (Figure 3A), which formed microcolonies with more than two cells ∼12–16 h after HO induction with kinetics similar to those observed in sae2Δ tel1Δ cells (Figure 3B). Therefore, both Mre11-S499P and Rad50-A78T rescue the Tel1-mediated sae2Δ adaptation defect, indicating that they impair Tel1 signalling.
Figure 3.
The mre11-S499P and rad50-A78T mutations suppress the adaptation defect of sae2Δ cells. (A) Rad53 phosphorylation during adaptation. Exponentially growing YEPR cultures were transferred to YEPRG (time zero), followed by western blot analysis with anti-Rad53 antibodies. (B) Adaptation assay. YEPR G1-arrested cell cultures of the strains in (A) were plated on galactose-containing plates (time zero). At the indicated time points, 200 cells for each strain were analyzed to determine the frequency of large budded cells (two cells) and of cells forming microcolonies of more than two cells. (C, D) Drop test. Exponentially growing cells were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without HU, MMS and CPT at the indicated concentrations. All strains carried SML1 deletion to keep mec1Δ cells viable.
The unscheduled Tel1 activation in sae2Δ cells was shown to suppress the hypersensitivity to hydroxyurea (HU) and MMS caused by the lack of Mec1 (59) (Figure 3C). In fact, SAE2 deletion suppressed the MMS and HU hypersensitivity of mec1Δ cells, but not that of mec1Δ tel1Δ cells (Figure 3C). Consistent with the inability of Mre11-S499P and Rad50-A78T to activate Tel1, the lack of Sae2 failed to restore resistance to HU and MMS of mre11-S499P mec1Δ and rad50-A78T mec1Δ cells. In fact, mre11-S499P sae2Δ mec1Δ and rad50-A78T sae2Δ mec1Δ cells lost viability in the presence of HU or MMS to the same extent as mre11-S499P mec1Δ and rad50-A78T mec1Δ cells (Figure 3D). By contrast, due to the requirement of Sae2 in repairing CPT-induced DNA lesions (2,4), SAE2 deletion did not restore CPT resistance not only in mec1Δ tel1Δ, mre11-S499P mec1Δ and rad50-A78T mec1Δ cells, but also in mec1Δ cells (Figure 3D). Finally, consistent with an increased CPT sensitivity caused by the mre11-S499P and rad50-A78T mutations, mre11-S499P mec1Δ and rad50-A78T mec1Δ cells were more sensitive to CPT compared to mec1Δ cells (Figure 3D).
Mre11-S499P and Rad50-A78T reduce Tel1–MRX interaction and Tel1 association to DNA DSBs
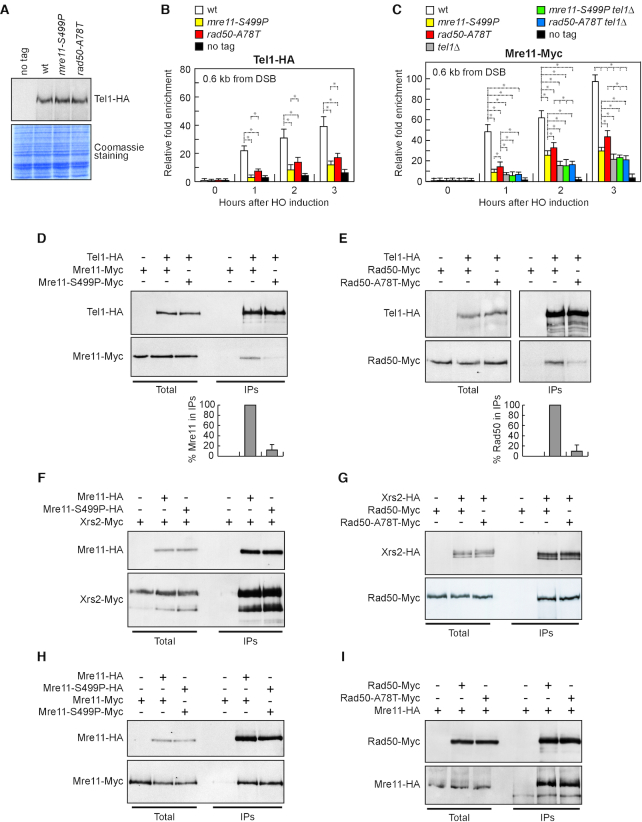
To investigate whether the defective Tel1 signalling activity in mre11-S499P and rad50-A78T cells was due to decreased Tel1 persistence to DSBs, we measured Tel1 association at DSBs by chromatin immunoprecipitation (ChIP) and quantitative real time PCR (qPCR). Although similar amount of Tel1 were detected in protein extracts from wild type, mre11-S499P and rad50-A78T cells (Figure 4A), the amount of Tel1 bound at the HO-induced DSB was dramatically lower in both mre11-S499P and rad50-A78T cells than in wild type cells (Figure 4B). This finding indicates that Mre11-S499P and Rad50-A78T reduce Tel1 association to DNA DSBs.
Figure 4.
Tel1 association to DSBs and Tel1–MRX interactions. (A) Western blot analysis with anti-HA antibodies of protein extracts prepared from exponentially growing cells. The same amounts of extracts were separated by SDS-PAGE and stained with Coomassie Blue as loading control. (B, C) ChIP analysis. Exponentially growing YEPR cell cultures were transferred to YEPRG at time zero. Relative fold enrichment of Tel1-HA (B) and Mre11-Myc (C) proteins at the indicated distances from the HO cleavage site was determined after ChIP and qPCR analysis. Plotted values are the mean values with error bars denoting S.D. (n = 3). *P < 0.05 (Student's t-test). (D, E) Protein extracts from exponentially growing cells that were treated with phleomycin (15 μg/ml) for 1 hour were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (total) or after immunoprecipitation (IPs) with anti-HA antibody. Graphs represent the amount of Mre11-S499P-Myc (D) and Rad50-A78T-Myc (E) in IPs relative to Mre11-Myc and Rad50-Myc, respectively, which were set up to 100%. Plotted values are the mean values with error bars denoting S.D. (n = 3). An immunoblot from one of these experiments is shown. (F–H) Protein extracts prepared from exponentially growing cells were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (Total) or after immunoprecipitation (IPs) with anti-HA antibody. (I) As in F-H, but after immunoprecipitation with anti-Myc antibody.
Indeed, when Tel1 is recruited by MRX at DSBs (33–35), it promotes MRX association/persistence in a positive feedback loop (41). Thus, we measured Mre11 association at DSBs in both mre11-S499P and rad50-A78T cells either in the presence or in the absence of Tel1. We found that the tel1Δ, mre11-S499P and rad50-A78T alleles decreased the amount of Mre11 bound at DSBs, with tel1Δ cells showing the strongest effect (Figure 4C). If the decrease in Mre11 association to DSBs caused by the mre11-S499P and rad50-A78T mutations were due to defective MRX–DNA interaction rather than to failure of Tel1 in promoting MRX persistence at DSBs, tel1Δ mre11-S499P and/or tel1Δ rad50-A78T cells would be expected to show a further decrease of the amount of MRX bound at DSBs compared to tel1Δ cells. However, the amount of Mre11 bound at DSBs in both mre11-S499P tel1Δ and rad50-A78T tel1Δ double mutants was similar to that of tel1Δ single mutant (Figure 4C), suggesting that the poor Mre11 persistence to DSBs in both mre11-S499P and rad50-A78T cells is a consequence of the decreased Tel1 association at DSBs.
Next, we analyzed the ability of MS499PRX and MRA78TX to interact with Tel1 by coimmunoprecipitation. When Tel1 was immunoprecipitated with anti-HA antibodies, a reduced amount of Mre11-S499P-Myc (Figure 4D) or Rad50-A78T-Myc (Figure 4E) could be detected in HA-tagged Tel1 immunoprecipitates compared to wild type Mre11-Myc and Rad50-Myc, respectively. Altogether, these data indicate that the mre11-S499P and rad50-A78T mutations reduce Tel1 association to DSBs by impairing MRX–Tel1 interaction.
The mre11-S499P mutation reduces Mre11–Rad50 interaction
MRX was shown to recruit Tel1 to DSBs through a direct interaction between Tel1 and Xrs2 (33), prompting us to test whether the defective MRX–Tel1 interaction in mre11-S499P and/or rad50-A78T cells was due to reduced ability of Xrs2 to interact with MS499PR or MRA78T. However, similar amounts of Xrs2-Myc could be detected in immunoprecipitates of HA-tagged Mre11 and Mre11-S499P (Figure 4F). Furthermore, similar amounts of Rad50-Myc and Rad50-A78T-Myc could be detected in immunoprecipitates of Xrs2-HA (Figure 4G). Thus, the reduced MRX–Tel1 interaction in both mre11-S499P and rad50-A78T cells is not due to a poor Xrs2 ability to interact with MS499PR or MRA78T.
Mre11-S499P turned out to be proficient also in Mre11 dimer formation. In fact, when Mre11 was immunoprecipitated with anti-HA antibodies from protein extracts of MRE11-MYC/MRE11-HA and mre11-S499P-MYC/mre11-S499P-HA diploid cells, the amount of Mre11-S499P-Myc detected by anti-Myc antibodies in immunoprecipitates of Mre11-S499P-HA was similar to that of wild type Mre11-Myc detected in immunoprecipitates of Mre11-HA (Figure 4H). Furthermore, the rad50-A78T mutation does not impair the ability of Rad50 to interact with Mre11, as similar amount of Mre11-HA could be detected in immunoprecipitates of Rad50-Myc and Rad50-A78T-Myc (Figure 4I).
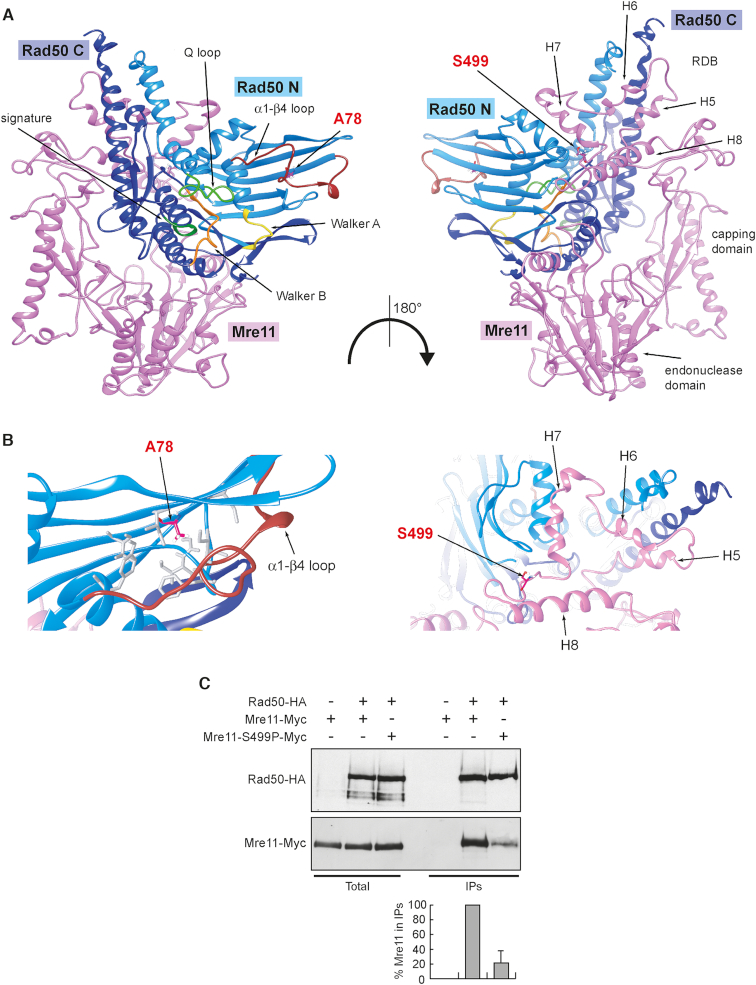
The structure of the Rad50 binding domain (RDB) is still unknown in eukaryotic MR complexes, although the only structure available from C. thermophilum (21) suggests the presence of a helix-loop-helix motif spanning three hydrophobic helices clanging to Rad50 coiled coil region and stabilized by a fourth helix. We have previously proposed a computational model for ScMre11 Rad50 RDB (38), where the helix-loop-helix motif (H5 and H6 in Figure 5A and B) is flanked by an additional helix (H7 in Figure 5A and B) that contacts the globular domain of Rad50. The S499 residue is localized on the flexible loop leading to the fourth helix (H8 in Figure 5A and B), which proved to be relevant for the stability of the RBD (38). The Ser499 substitution with the more rigid proline residue would impinge on this connector flexibility that is required to accommodate the necessary mobility of the RBD-Rad50 interface during the large-scale conformational changes occurring upon ATP hydrolysis by Rad50. This analysis raises the possibility that the mre11-S499P mutation could affect Mre11–Rad50 interaction. We directly tested this prediction by immunoprecipitating Rad50 with anti-HA antibodies from protein extracts of MRE11-MYC RAD50-HA and mre11-S499P-MYC RAD50-HA cells. A reduced amount of Mre11-S499P-Myc compared to Mre11-Myc could be detected in immunoprecipitates of HA-tagged Rad50 (Figure 5C), indicating that the S499P amino acid substitution affects Mre11–Rad50 interaction. In any case, the amount of Rad50 bound to Mre11 appears to be sufficient to support MRX function in DSB repair, as mre11-S499P cells did not show major defects in DSB resection and NHEJ.
Figure 5.
The mre11-S499P mutation is localized at the Mre11–Rad50 interface and reduces Mre11–Rad50 interaction. (A) Structural prediction of S. cerevisiae Mre11–Rad50 heterodimer, obtained as previously described (38), showing the localization of S499 residue in the Rad50-binding domain (RBD) of Mre11 and of A78 residue in the N-terminal of Rad50. Pink, Mre11. Light blue, N-terminal lobe of Rad50 (Rad50 N). Dark blue, C-terminal lobe of Rad50 (Rad50 C). (B) Detailed view of the molecular surroundings of the residues affected by the mutations. The hydrophobic residues surrounding A78 in Rad50 are shown as gray sticks. (C) Mre11–Rad50 interaction. Protein extracts prepared from exponentially growing cells were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (Total) or after immunoprecipitation (IPs) with anti-HA antibody. Graph represents the amount of Mre11-S499P-Myc relative to Mre11-Myc that was set up to 100%. Plotted values are the mean values with error bars denoting S.D. (n = 3). An immunoblot from one of these experiments is shown.
Structural insights of ATP- and ADP-bound MR subcomplex by molecular dynamics simulations
In vitro activation of human ATM by MRN requires ATP but not Mre11 nuclease activity (28), suggesting that MRX/MRN activates Tel1/ATM when it is present in the ATP-bound state. To investigate further this hypothesis, we analysed the conformations of the MR subcomplex bound to either ATP (MR-ATP) or ADP (MR-ADP) by molecular dynamics (MD). Ten replicas of 200 ns were carried out on the two aforementioned systems in order to obtain a total 2 μs trajectory for each set of simulations. All replicas immediately reached a stable value of backbone root-mean-square deviation (RMSD), thus ensuring that the calculated parameters reflect the real behaviour of the investigated systems (Supplementary Figure S1A and B). Principal Component Analysis (PCA) was used to identify globally correlated motions from MD trajectories. PCA analysis was performed on the protein backbone and ATP/ADP atoms trajectories to inspect the functional motions collected during the simulations. This method calculates eigenvectors (PC) and eigenvalues, which describe the direction and the magnitude of concerted motions, respectively. The first two eigenvectors (PC1 and PC2) calculated from the MD trajectories account for most of the total variance (Supplementary Figure S2, top). Interestingly, while they reveal only minor motions for the MR-ATP system (Supplementary Figure S3A), they show the separation of two Rad50 subunits from each other and the disengagement of Mre11 endonuclease site from Rad50 in the MR-ADP system (Supplementary Figure S3B). These two movements combined together describe the initial steps of MR complex opening, an event that occurs after ATP hydrolysis by Rad50 at the resection onset.
Movements along PC1 and PC2 were used as reaction coordinates to build 3D histograms of the percentage of existence in each molecular conformation, from which free energy landscape (FEL) graphs were calculated (Figure 6A and B). All FELs show a good overlap among replicas, which highlights that the transitions among all basins are predicted to occur at the simulation temperature (300 K) (Supplementary Figure S4, top). FELs allow the identification of free energy basins, which correspond to the energetically favoured protein conformations. Since the projection of a multidimensional trajectory in a two-dimensional space (PC1 and PC2) involves a large reduction in dimensionality, FEL plots might show only a partial picture of the protein conformational scenario. For this reason, we confirmed FEL results with cluster analysis derived from the concatenated trajectory, in order to retrieve all conformational variance stored in the entire simulation that allows estimating which conformations are more favourable for each system with respect to energy content. The results of the cluster analysis were in agreement with the FELs (data not shown).
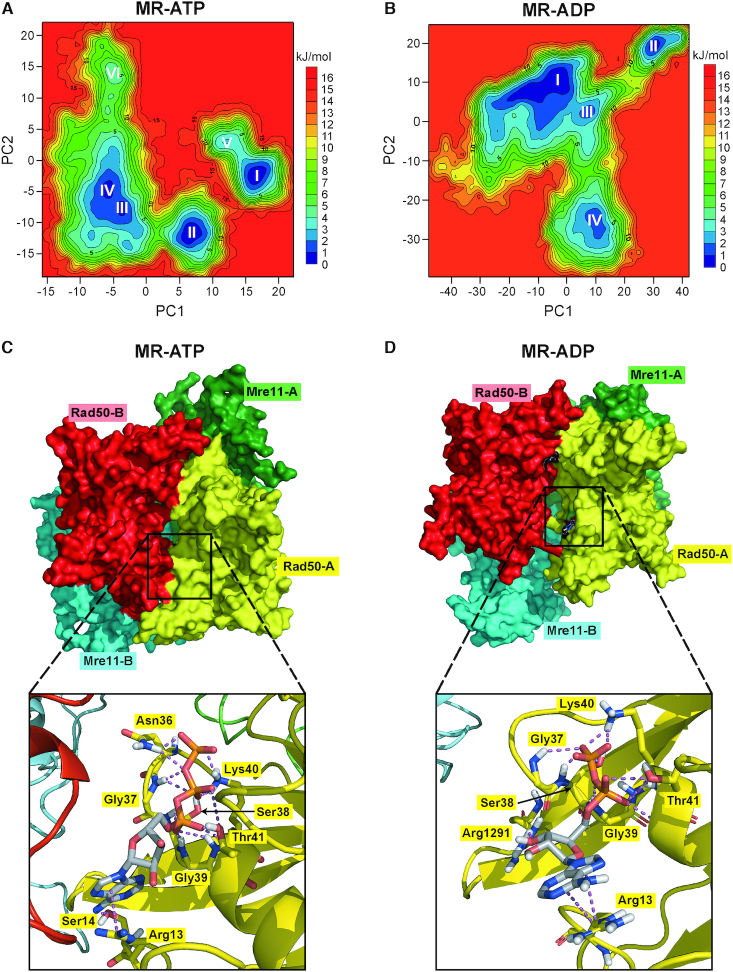
Figure 6.
MD simulations to identify stable MR-ATP and MR-ADP conformations. (A, B) FELs were evaluated for MR-ATP (A) and MR-ADP (B). Analyses have been carried out using the projection of concatenated trajectories along the first and the second principal components (PC1 and PC2) as reaction coordinates. Basins are progressively numbered according to their energetic stability. Energy values are reported in kJ/mol. (C, D) The most energetically favoured conformations shown are derived from the FELs and cluster analysis for MR-ATP (C) (corresponding to basin I in panel A) and MR-ADP (D) (corresponding to basin I in panel B). Rad50 subunits are in red and yellow; Mre11 subunits are in cyan and green. Close-up views of one of the nucleotide binding sites are shown at the bottom. Nucleotides and residues interacting with ATP/ADP are shown as sticks; each residue is colored according to the chain it belongs to.
All basins, progressively numbered with roman numbers from the lowest to the highest energy, in MR-ATP FELs (Figure 6A) correspond to MR conformations in which the two Rad50 subunits are tightly bound to each other and bury the ATP molecules in their interface (Figure 6C) (Supplementary Dataset S1). This is consistent with ATP requirement for the MR complex to assemble in a ‘closed’ conformation (22–25). By contrast, basins in the MR-ADP FELs (Figure 6B) correspond to different MR conformations, suggesting a higher flexibility of the complex. In particular, the first basin (I) represents MR complexes in which the two Rad50 subunits have drifted apart (Figure 6D) (Supplementary Dataset S2), whereas the second basin (II) represents a conformation where, although the two Rad50 subunits are closer than in basin I, they are not tightly bound as in MR-ATP (Supplementary Figure S5A), suggesting that the Rad50 dimer interface in MR-ADP is different from that in MR-ATP complex. The third and fourth basins (III and IV) describe a transition state between the previous conformations (Supplementary Figure S5B for basin III).
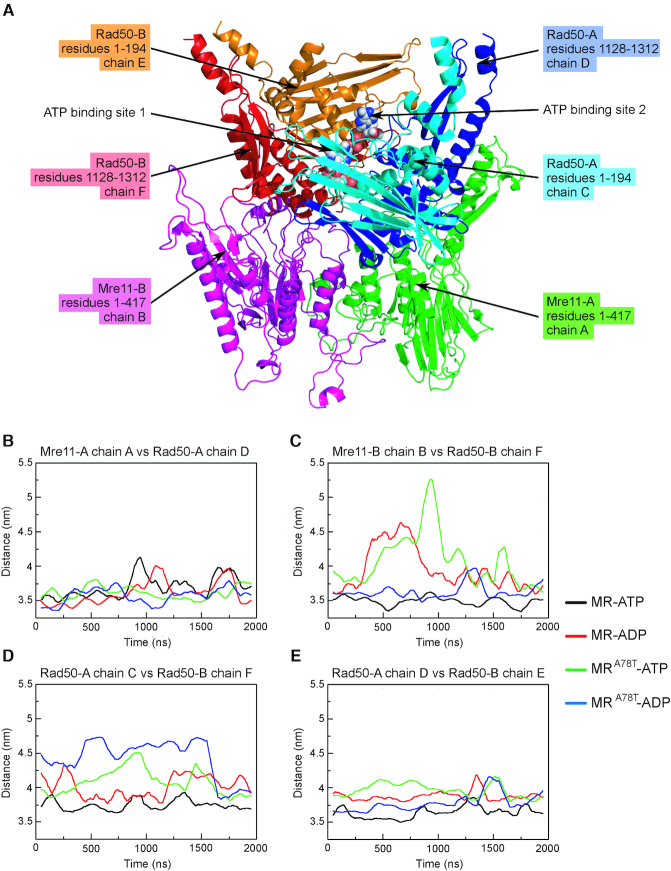
These motions can be monitored by measuring the distance between the centers of mass (COM) of the different MR complex subunits (Figure 7A) (Mre11 and Rad50 monomers are indicated as A and B). In particular, we monitored the distances over the interfaces between each Mre11–Rad50 dimer (Mre11-A chain A versus Rad50-A chain D in Figure 7B; Mre11-B chain B versus Rad50-B chain F in Figure 7C) and those between Rad50-Rad50 dimers, corresponding to the two nucleotide binding sites (ATP binding site 1, between Rad50-A chain C and Rad50-B chain F, in Figure 7D; ATP binding site 2, between Rad50-A chain D and Rad50-B chain E, in Figure 7E). MR bound to ADP undergoes dissociation of Mre11-B chain B from Rad50-B chain F (Figure 7C) and opening of the ATP binding site 1 (Figure 7D). By contrast, the ATP binding site 2 is permanently less tight in the MR-ADP compared to the MR-ATP system (Figure 7E), further suggesting that the two Rad50 subunits have lower affinity for each other in presence of ADP than in the presence of ATP. The observation of an asymmetric opening of Rad50 dimer was expected, due to the ATP-driven cooperativity and allosteric control typical of ABC-ATPases superfamily to which Rad50 belongs with (17). The cooperativity implies that the opening of the two ATP binding sites does not occur simultaneously, but the opening of the first site induces the opening of the second one. However, changes in the quaternary structure and/or allosteric transitions are very slow and occur at ms/s time scale, still unachievable in MD simulations for large molecular systems (62,63).
Figure 7.
Distance between the centers of mass (COM) of the MR complex subunits. (A) Labeling of the different chains in the model used for simulations. Mre11 and Rad50 monomers are indicated as A and B. Mre11-A and Mre11-B monomers are in green and purple, respectively; N-terminal and C-terminal parts of Rad50-A monomer are in light blue and dark blue, respectively; N-terminal and C-terminal parts of Rad50-B monomer are in orange and red, respectively. (B–E) Graphs show distances among centres of mass of the indicated chains as monitored along the concatenated trajectory of the MD simulations.
The observed variations of COM distances represent a genuine interchain movement and it is not due to an internal rearrangement of the centre of gravity of subunits. In fact, the overall folding of each subunit is not changing significantly over the simulation, as demonstrated by the lack of variation in the radius of gyration (Rg) (Supplementary Figure S6).
Altogether, these analyses indicate that the MR subcomplex, in the presence of ATP, lingers in a tightly closed conformation with the two ATP molecules deeply buried at the interface between the Rad50 subunits (Supplementary Movie S1). By contrast, the presence of ADP leads to the loosening of the Rad50 homodimer, which exposes the ADP molecules, and to a destabilization of Mre11–Rad50 association (Supplementary Movie S2).
Structural studies describing the conformational transitions occurring on thermophilic archaean Rad50 upon binding to ATP analogues showed that the main difference is a rotation of the C-terminal lobe with respect to the N-terminal lobe (Figure 8A and B) (23,25), which reduces the affinity of Rad50 subunits for each other (25). The Q loop release from Mg2+ coordination upon ATP hydrolysis allows the switch in the hinge helix and the release of the region spanning from residue 50 to 65, named α1-β4 loop; interestingly, the same movements have been observed by NMR also in the P. furiosus Rad50-R805E mutant protein (64). Indeed, we found that the α1-β4 loop shows higher mobility in MR-ADP than in MR-ATP simulations (Supplementary Figure S7A). All of these events can be visualized when the most stable conformations of Rad50 subunit along the MD simulations for MR-ATP and MR-ADP systems are superimposed by structural alignment on the N-terminal lobe (Figure 8C), which shows that the Rad50 subunit undergoes the typical conformation changes observed in MR crystals in the presence or not of ATP analogues (compare Figure 8B and C). Moreover, our results suggest that ADP presence loosens not only the association of Rad50 monomers, allowing the initial opening of Rad50 dimer, but also the Rad50–Mre11 association, both events being required to open the MR complex for its engagement in DSB resection.
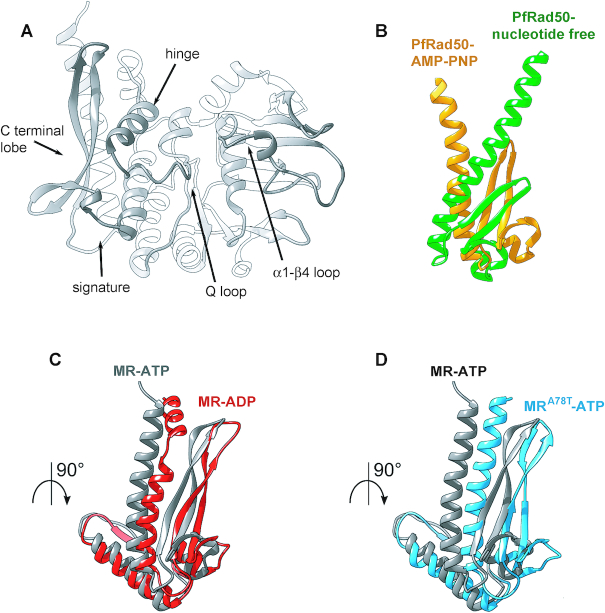
Figure 8.
The rad50-A78T mutation induces similar conformational changes in Rad50 as ATP hydrolysis. (A) Cartoon of the centroid structures of one Rad50 subunit (chains C and D) from the absolute minimum corresponding clusters from MR-ATP showing the main features involved in the conformational change upon ADP versus ATP binding. (B) View of the C-terminal lobes of the structure of P. furiosus Rad50 bound to AMP-PNP (PDB:3qku; orange) superimposed to nucleotide-free Rad50 (PDB:3qks; green). (C, D) View of the C-terminal lobes of centroid structures of the energetic absolute minimum of Rad50 subunit from MR-ATP MD simulations (gray) superimposed by structural alignment to the N-terminal region of MR-ADP (C) and MRA78T-ATP (D) energetically favourite Rad50 structures.
The A78T mutation destabilizes the MR-ATP conformation
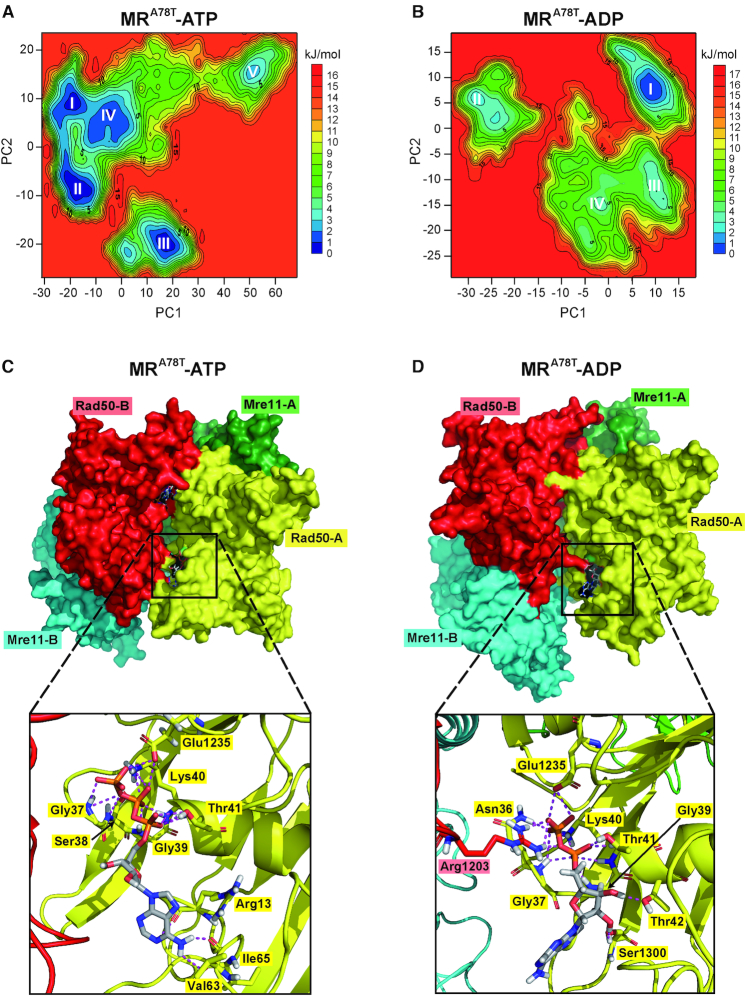
The A78 residue is located within a hydrophobic region of Rad50, facing the α1-β4 loop residues (Figure 5B). To investigate the structural impact of the A78T mutation, ATP- and ADP-bound MRA78T complexes have been subjected to MD analyses. Again, ten replicas of 200 ns immediately reached a stable value of backbone root-mean-square deviation (RMSD) (Supplementary Figure S1C and D). As for MR simulations, the first two eigenvectors (PC1 and PC2) calculated from the MD trajectories account for most of the total variance (Supplementary Figure S2, bottom) and describe the initial steps of MRA78T complex opening (Supplementary Figure S3C and D). Movements along PC1 and PC2 were used as reaction coordinates to build 3D histograms of the percentage of existence in each molecular conformation, from which FELs graphs were calculated (Figure 9A and B). Again, all FELs show a good overlap among replicas (Supplementary Figure S4, bottom) and the energy basins, identified in the FELs and progressively numbered from the lowest to the highest, were consistent with the results of cluster analysis (data not shown).
Figure 9.
MD simulations to identify stable MRA78T-ATP and MRA78T-ADP conformations. (A, B) FELs were evaluated for MRA78T-ATP (A) and MRA78T-ADP (B). Analyses have been carried out using the projection of concatenated trajectories along the first and the second principal components (PC1 and PC2) as reaction coordinates. Basins are progressively numbered according to their energetic stability. Energy values are reported in kJ/mol. (C, D) The most energetically favoured conformations shown are derived from the FELs and cluster analysis for MRA78T-ATP (C) (corresponding to basin II in panel A), and MRA78T-ADP (D) (corresponding to basin I in panel B). Rad50 subunits are in red and yellow; Mre11 subunits are in cyan and green. Close-up views of the nucleotide binding site are shown at the bottom. Nucleotides and residues interacting with ATP/ADP are shown as sticks; each residue is colored according to the chain it belongs to.
The A78T substitution in Rad50 completely changes the behaviour of the MR subcomplex in the presence of ATP (Supplementary Movie S3). In fact, the most stable structures for MRA78T-ATP show a less tight conformation compared to MR-ATP, i.e. one of the nucleotide binding pockets is accessible to the solvent in the most stable conformation (basin I in Supplementary Figure S5C), while both are exposed in all the other energetically similar conformations (basins II and III) (Figure 9C for basin II) (Supplementary Dataset S3). Moreover, the structure associated with basin V also shows the disengagement of Rad50 from the endonucleolytic site of Mre11 (Supplementary Figure S5D). The open conformation is stabilized further in MRA78T-ADP, as shown by the higher existence probability of basin I (Figure 9B and D) (Supplementary Movie S4) (Supplementary Dataset S4). The other structures are less stable and have low probability of being sampled, suggesting they are not relevant.
We found that the increment in COM distances in MRA78T is accentuated compared to that of wild type MR (Figure 7). In fact, the variations that were characteristic of the MR-ADP system (see in particular Figure 7C and D) can be detected with enhanced intensity in the MRA78T system, albeit in the presence of ATP. Furthermore, MRA78T-ADP shows the maximum distance between the two Rad50 subunits (Figure 7D). These findings indicate that the A78T substitution in Rad50 causes the MR complex to undergo conformational motions similar to those observed for wild type MR-ADP, even when MRA78T is bound to ATP.
All of these events can be visualized when the most stable conformations of Rad50 subunit along the MD simulations for MR-ATP and MRA78T-ATP systems are superimposed (Figure 8D). The conformational changes occurring in the P. furiosus Rad50 protein upon binding to ATP (25), described as a rotation of the C-terminal lobe with respect to the N-terminal lobe (Figure 8B), are also observed when the most energetically stable conformation of wild type Rad50 bound to ATP is superimposed to Rad50-A78T, notwithstanding the presence of ATP (Figure 8D), with the mutant protein showing a more dramatic effect on the hinge helix switch than the Rad50-ADP itself.
As aforementioned, comparison between Rad50 Root-Mean-Square Fluctuation (RMSF) profiles of MR-ATP and MR-ADP complexes shows an increased mobility within the region spanning from residue 50 to 65 (α1–β4 loop) (Supplementary Figure S7A). The same is observed for Rad50-A78T RMSF profiles in MRA78T-ATP MD simulations (Supplementary Figure S7B). Interestingly, as already described, the A78 residue is opposite to the α1–β4 loop residues and is located within a hydrophobic region of Rad50 (Figure 5B). Hence, its substitution with the polar threonine residue can easily destabilize this hydrophobic region, thus increasing its mobility. This event would be sufficient to induce conformational rearrangements in MRA78T bound to ATP similar to those observed when wild type MR is bound to ADP. Consistent with this hypothesis, the high mobility of the α1-β4 loop has been previously described as one of the events in the conformational transition occurring in Rad50 upon ATP hydrolysis (23,25,64).
DISCUSSION
Activation of Tel1/ATM checkpoint kinase by MRX/MRN is mechanistically poorly understood. Indeed, MRX/MRN is required to activate Tel1/ATM by recruiting it to DSBs through direct interaction between Tel1/ATM and Xrs2/NBS1 (33–37). Accordingly, the Mre11 H98Y and R389C aminoacid substitutions reduce Tel1 recruitment to DSBs by directly affecting MRX–DNA interaction (38). However, also the MR subcomplex by itself appears to contribute to Tel1/ATM activation, as it was shown to interact with Tel1/ATM and to recruit it to DNA independently of Xrs2/Nbs1 in both mammals and S. pombe (30,31,37). Furthermore, both the zinc hook and the coiled-coil domains of Rad50 are important for Tel1/ATM activation (28,65,66). This function of the zinc hook can be bypassed by the addition of a heterologous homodimerization domain that links the Rad50 monomers together (28), suggesting a role for Rad50 in maintaining an MRX/MRN conformation capable to sustain Tel1/ATM activation.
Here we show that the separation-of-function mre11-S499P and rad50-A78T mutations specifically impair activation of Tel1/ATM. In particular, both mre11-S499P and rad50-A78T cells phenocopy tel1Δ cells with respect to DNA damage resistance, telomere metabolism, DSB resection and checkpoint signalling. The defective Tel1 activation by both Mre11-S499P and Rad50-A78T is due to a poor Tel1 association to DSBs that is caused by reduced MRX–Tel1 interaction. This decreased MRX–Tel1 association does not depend on impaired Xrs2-MR interaction, as Xrs2 interacts equally well with wild type MR, MS499PR and MRA78T subcomplexes. Interestingly, the mre11-S499P mutation is localized at the Mre11–Rad50 interface and reduces Mre11–Rad50 association, suggesting that Rad50 plays an important function in Tel1/ATM activation that is specifically impaired by the rad50-A78T mutation.
In vitro activation of human ATM by MRN was shown to require ATP binding but not ATP hydrolysis (28), raising the possibility that MRX/MRN activates Tel1/ATM when it is present in the ATP-bound state. By molecular dynamics, we showed that, in the presence of ATP, wild type MR lingers in a tightly closed conformation, with the two ATP molecules deeply buried at the interface between the Rad50 subunits. In the presence of ADP, the Rad50 homodimer loosens up due to the conformational change occurring in Rad50 when bound to ADP compared to ATP. ADP-bound Rad50 exposes the nucleotide binding sites, and its association with Mre11 is also destabilized. These events are the earlier stages of the large-scale conformational changes occurring upon ATP hydrolysis and required for MRX engagement in resection (22–25).
The rad50-A78T mutation induces conformational rearrangements in MRA78T bound to ATP similar to those observed when wild type MR is bound to ADP, thus impinging on the ability of MRA78T-ATP complex to maintain the closed conformation. In particular, substitution of A78 with T already induces a higher mobility of the α1-β4 loop in the ATP-bound state, triggering a series of motions that lead to the C-terminal lobe rotation and interfering with the dimer interface affinity. Interestingly, similar motions have been revealed by NMR also in the P. furiosus Rad50-R805E mutant (64), suggesting that the inability of this mutant to elicit Tel1 activation is caused by a spontaneous conformational change that occurs independently of ATP hydrolysis (which is actually impaired in this mutant) (27). In any case, these changes in the stability of the ATP-bound MRA78T conformational state greatly reduce Tel1 activation, but do not impair the ability of the mutant complex to support both DNA damage resistance and ligation of broken DNA ends, suggesting different requirements for the ATP-bound conformational state in promoting either Tel1 activation or efficient DSB repair.
In summary, we provide evidence that the ATP-bound Mre11–Rad50 conformation is important to sustain Tel1/ATM binding and activation. The finding that the in vitro binding of Xrs2/NBS1 with Mre11–Rad50 is increased in the presence of ATP (29) supports the view that these Mre11 and Rad50 structural features reflect an important function of MRX in Tel1/ATM activation. Given the evolutionarily conservation of both the MRX complex and Tel1, these structure–function relationships might apply to human cells to improve our knowledge of the consequences of MRN and ATM dysfunctions in human diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Lucchini for critical reading of the manuscript and J. Haber for yeast strains. We acknowledge Consorzio Interuniversitario del Nord-Est per il Calcolo Automatico (CINECA) for granting us the access to the high-performance computing resource MARCONI, on which all molecular dynamics simulations have been run.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AIRC under IG 2017 – ID. 19783 project – P.I. Longhese Maria Pia; Progetti di Ricerca di Interesse Nazionale (PRIN) 2015 (to M.P.L.); Fondazione Italiana per la Ricerca sul Cancro (FIRC) (to C.C.). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Villa M., Cassani C., Gobbini E., Bonetti D., Longhese M.P.. Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell Mol. Life Sci. 2016; 73:3655–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daley J.M., Gaines W.A., Kwon Y., Sung P.. Regulation of DNA pairing in homologous recombination. Cold Spring Harb. Perspect. Biol. 2014; 6:a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gobbini E., Cassani C., Villa M., Bonetti D., Longhese M.P.. Functions and regulation of the MRX complex at DNA double-strand breaks. Microb. Cell. 2016; 3:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Syed A., Tainer J.A.. The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018; 87:263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paull T.T., Gellert M.. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998; 1:969–979. [DOI] [PubMed] [Google Scholar]
- 7. Trujillo K.M., Yuan S.S., Lee E.Y., Sung P.. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998; 273:21447–21450. [DOI] [PubMed] [Google Scholar]
- 8. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 9. Mimitou E.P., Symington L.S.. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008; 455:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11–Rad50–Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L. et al.. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010; 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C.. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011; 25:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F. et al.. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014; 53:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reginato G., Cannavo E., Cejka P.. Physiological protein blocks direct the Mre11–Rad50–Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 2017; 31:2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang W., Daley J.M., Kwon Y., Krasner D.S., Sung P.. Plasticity of the Mre11–Rad50–Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 2017; 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hopfner K.P., Karcher A., Shin D.S., Craig L., Arthur L.M., Carney J.P., Tainer J.A.. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000; 101:789–800. [DOI] [PubMed] [Google Scholar]
- 18. Hopfner K.P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A., Karcher A., Henderson B., Bodmer J.L., McMurray C.T. et al.. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002; 418:562–566. [DOI] [PubMed] [Google Scholar]
- 19. Wiltzius J.J., Hohl M., Fleming J.C., Petrini J.H.. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 2005; 12:403–407. [DOI] [PubMed] [Google Scholar]
- 20. Hohl M., Kwon Y., Galván S.M., Xue X., Tous C., Aguilera A., Sung P., Petrini J.H.. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 2011; 18:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seifert F.U., Lammens K., Stoehr G., Kessler B., Hopfner K.P.. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016; 35:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lammens K., Bemeleit D.J., Möckel C., Clausing E., Schele A., Hartung S., Schiller C.B., Lucas M., Angermüller C., Söding J. et al.. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011; 145:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim H.S., Kim J.S., Park Y.B., Gwon G.H., Cho Y.. Crystal structure of the Mre11–Rad50-ATPγS complex: understanding the interplay between Mre11and Rad50. Genes Dev. 2011; 25:1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y., Sung S., Kim Y., Li F., Gwon G., Jo A., Kim A.K., Kim T., Song O.K., Lee S.E. et al.. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016; 35:743–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams G.J., Williams R.S., Williams J.S., Moncalian G., Arvai A.S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P. et al.. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011; 18:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Möckel C., Lammens K., Schele A., Hopfner K.P.. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012; 40:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deshpande R.A., Williams G.J., Limbo O., Williams R.S., Kuhnlein J., Lee J.H., Classen S., Guenther G., Russell P., Tainer J.A. et al.. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014; 33:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J.H., Mand M.R., Deshpande R.A., Kinoshita E., Yang S.H., Wyman C., Paull T.T.. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J. Biol. Chem. 2013; 288:12840–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y.. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003; 22:5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J.H., Paull T.T.. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004; 304:93–96. [DOI] [PubMed] [Google Scholar]
- 31. Lee J.H., Paull T.T.. ATM activation by DNA double-strand breaks through the Mre11–Rad50-Nbs1 complex. Science. 2005; 308:551–554. [DOI] [PubMed] [Google Scholar]
- 32. Gobbini E., Cesena D., Galbiati A., Lockhart A., Longhese M.P.. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair (Amst.). 2013; 12:791–799. [DOI] [PubMed] [Google Scholar]
- 33. Nakada D., Matsumoto K., Sugimoto K.. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003; 17:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Falck J., Coates J., Jackson S.P.. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434:605–611. [DOI] [PubMed] [Google Scholar]
- 35. You Z., Chahwan C., Bailis J., Hunter T., Russell P.. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005; 25:5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berkovich E., Monnat R.J., Kastan M.B.. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007; 9:683–690. [DOI] [PubMed] [Google Scholar]
- 37. Limbo O., Yamada Y., Russell P.. Mre11–Rad50-dependent activity of ATM/Tel1 at DNA breaks and telomeres in the absence of Nbs1. Mol. Biol. Cell. 2018; 29:1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cassani C., Gobbini E., Vertemara J., Wang W., Marsella A., Sung P., Tisi R., Zampella G., Longhese M.P.. Structurally distinct Mre11 domains mediate MRX functions in resection, end-tethering and DNA damage resistance. Nucleic Acids Res. 2018; 46:2990–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ritchie K.B., Petes T.D.. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000; 155:475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menin L., Ursich S., Trovesi C., Zellweger R., Lopes M., Longhese M.P., Clerici M.. Tel1/ATM prevents degradation of replication forks that reverse after topoisomerase poisoning. EMBO Rep. 2018; 19:e45535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cassani C., Gobbini E., Wang W., Niu H., Clerici M., Sung P., Longhese M.P.. Tel1 and Rif2 regulate MRX function in end-tethering and repair of DNA double-strand breaks. PLoS Biol. 2016; 14:e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baldo V., Testoni V., Lucchini G., Longhese M.P.. Dominant TEL1-hy mutations compensate for Mec1 lack of functions in the DNA damage response. Mol. Cell Biol. 2008; 28:358–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W.. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013; 27:221–234. [DOI] [PubMed] [Google Scholar]
- 44. Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D. Jr.. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017; 14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M.. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997; 18:1463–1472. [Google Scholar]
- 46. Grubmüller H., Heller H., Windemuth A., Schulten K.. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991; 6:121–142. [Google Scholar]
- 47. Darden T., York D., Pedersen L.. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993; 98:10089–10092. [Google Scholar]
- 48. Mahoney M.W., Jorgensen W.L.. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000; 112:8910–8922. [Google Scholar]
- 49. Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W.F., Mark A.E.. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 1999; 38:236–240. [Google Scholar]
- 50. Maisuradze G.G., Liwo A., Scherega H.A.. Relation between free energy landscapes of proteins dynamics. J. Chem. Theory Comput. 2010; 6:583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mantiero D., Clerici M., Lucchini G., Longhese M.P.. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007; 8:380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. [DOI] [PubMed] [Google Scholar]
- 53. Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., Ogawa T.. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998; 95:705–716. [DOI] [PubMed] [Google Scholar]
- 54. Pellicioli A., Lee S.E., Lucca C., Foiani M., Haber J.E.. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell. 2001; 7:293–300. [DOI] [PubMed] [Google Scholar]
- 55. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 56. Ivanov E.L., Sugawara N., White C.I., Fabre F., Haber J.E.. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell Biol. 1994; 14:3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E.. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998; 94:399–409. [DOI] [PubMed] [Google Scholar]
- 58. Toczyski D.P., Galgoczy D.J., Hartwell L.H.. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997; 90:1097–1106. [DOI] [PubMed] [Google Scholar]
- 59. Usui T., Ogawa H., Petrini J.H.. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001; 7:1255–1266. [DOI] [PubMed] [Google Scholar]
- 60. Clerici M., Mantiero D., Lucchini G., Longhese M.P.. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006; 7:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clerici M., Trovasi C., Galbiati A., Lucchini G., Longhese M.P.. Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 2014; 33:198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zwier M.C., Chong L.T.. Reaching biological timescales with all-atom molecular dynamics simulations. Curr. Opin. Pharmacol. 2010; 10:745–752. [DOI] [PubMed] [Google Scholar]
- 63. Bhabha G., Biel J.T., Fraser J.S.. Keep on moving: discovering and perturbing the conformational dynamics of enzymes. Acc. Chem. Res. 2015; 48:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boswell Z.K., Rahman S., Canny M.D., Latham M.P.. A dynamic allosteric pathway underlies Rad50 ABC ATPase function in DNA repair. Sci. Rep. 2018; 8:1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hohl M., Kochańczyk T., Tous C., Aguilera A., Krężel A., Petrini J.H.. Interdependence of the Rad50 hook and globular domain functions. Mol. Cell. 2015; 57:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park Y.B., Hohl M., Padjasek M., Jeong E., Jin K.S., Krężel A., Petrini J.H., Cho Y.. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 2017; 24:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.