Abstract
RNA secondary structures have been increasingly recognized to play an important regulatory role in post-transcriptional gene regulation. We recently showed that RNA G-quadruplexes, which serve as cis-elements to recruit splicing factors, play a critical role in regulating alternative splicing during the epithelial-mesenchymal transition. In this study, we performed a high-throughput screen using a dual-color splicing reporter to identify chemical compounds capable of regulating G-quadruplex-dependent alternative splicing. We identify emetine and its analog cephaeline as small molecules that disrupt RNA G-quadruplexes, resulting in inhibition of G-quadruplex-dependent alternative splicing. Transcriptome analysis reveals that emetine globally regulates alternative splicing, including splicing of variable exons that contain splice site-proximal G-quadruplexes. Our data suggest the use of emetine and cephaeline for investigating mechanisms of G-quadruplex-associated alternative splicing.
INTRODUCTION
RNA secondary structures have been shown to play key roles in gene expression through regulating alternative splicing (1–4), translational efficiency (5,6), and transcriptional termination (7). One type of RNA secondary structures is the RNA G-quadruplex, which is formed within guanine-rich sequences. These structures can be formed within a single strand or between multiple strands of RNA, where four G-tracts of two or more guanines separated by short stretches of other nucleotides are assembled in layered loops bound together through Hoogsteen hydrogen bonding.
In recent years, emerging evidence has indicated the importance of RNA G-quadruplexes in regulating key cellular processes including translational regulation (8–10), 3′ end processing (11–13), alternative splicing (3,14) and mRNA localization (15). Many studies have also indicated that RNA G-quadruplex structures are associated with human diseases, including neurological disorders (16) and cancer (17,18). Although increasing effort has been directed toward understanding the importance of RNA G-quadruplexes in biology, our knowledge on RNA G-quadruplex structures is still in its infancy. Additional approaches are needed to provide a more complete picture of G-quadruplexes genome wide while illustrating their specific functions in living cells. Studies have identified several small molecules that are capable of targeting RNA G-quadruplex structures to modulate biological functions (19–23). However, these compounds were limited to bind to a specific G-quadruplex structure in one particular gene instead of detecting RNA G-quadruplexes genome-wide. Furthermore, studies on identification of small molecules that affect G-quadruplex-mediated alternative splicing have been limited.
Alternative splicing of pre-messenger RNA occurs in up to 95% human genes during development or in response to extracellular stimuli (24,25). During these processes, a single gene can produce protein isoforms with different, even opposing functions. Disruption of splice site recognition and misregulation of alternative splicing causes many human diseases, including autoimmune disorders (26), neurodegenerative diseases (27) and cancer progression (28,29). Therefore, alternative splicing is an important and potential target for the treatment of various types of human disorders. Alternative splicing is principally regulated by recruitment of RNA binding proteins to conserved cis-regulatory elements on the pre-mRNA. Some progress has been made in the discovery of small molecules designed to selectively target splicing factors and alternative splicing events (30–32). The identification of small molecules targeting RNA secondary structures that are associated with disease-relevant alternative splicing, such as G-quadruplexes, has been very limited.
In this study, we utilized a dual-color splicing reporter containing a G-quadruplex secondary structure proximal to the splice sites that allowed us to measure changes in cassette exon inclusion and skipping dependent on the adjacent G-quadruplex. Using this splicing reporter, we performed a high-throughput screen to identify small molecules that affect G-quadruplex-dependent alternative splicing. We found that the analogous small molecules emetine and cephaeline regulate alternative splicing by interfering with G-quadruplex structures. We further demonstrate that these small molecules modulate splicing when G-quadruplexes are located either upstream or downstream of the cassette exon. In addition, we show that emetine-regulated alternative splicing affects G-quadruplex associated alternative splicing across the transcriptome. Therefore, our study identifies new small molecule compounds with the potential to regulate alternative splicing by targeting G-quadruplexes.
MATERIALS AND METHODS
Plasmids and primers
The bichromatic fluorescent reporters, CRQ, CRQ-G4m2, CRQ-G4m and scrambled sequences, were cloned into EcoRI and XhoI sites of the RG6 vector, a kind gift of Dr. Cooper TA, Baylor College of Medicine (33). The cloned sequences are listed in Supplementary Table S1. The EcoRI and XhoI sites are located in the intron 20 nucleotides downstream of the variable exon 5′ splice site (3). To determine whether the location of the G-quadruplex affects alternative splicing, these sequences were also inserted into BsiW1 and Not1 sites of the RG6 vector, which are located 63 nucleotides upstream of the variable exon. The full sequence of the minigene is included in the Supplementary Materials.
Cell lines and generation of stable cell lines
To generate the stable cell lines expressing the bichromatic fluorescent reporters, CRQ and CRQ-G4m reporters were transfected into HEK 293A cells, termed HEK 293A CRQ and HEK 293A CRQ-G4m. Cells stably expressing the reporters were selected using G418 and inspected visually under a fluorescent microscope for EGFP and dsRed. After two weeks of selection, HEK 293A CRQ were sorted for EGFP-positive cells and HEK 293A CRQ-G4m were sorted for dsRed-positive cells by flow cytometry for downstream screening.
Cell culture
HEK 293FT, HEK 293A CRQ and HEK 293A CRQ-G4m cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and streptomycin, 2 mM l-glutamine in 5% CO2 at 37°C. Maintenance of HMLE-Twist and MCF10A were described previously (28,34).
High-throughput screening
The screen was performed on the ImageXpress Micro Confocal High-Content Imaging System. HEK 293A CRQ cells (3000 per well) were seeded into 384-well plates overnight prior to treatment with the MicroSource Custom Collection library. This library contains 2700 compounds: 50% of the compounds are approved drugs, 35% are natural products and 15% are compounds with established and diverse bioactivity. HEK 293A CRQ-G4m cells, which solely express dsRed, were used as a positive control for dsRed in images. HEK 293A CRQ cells that were treated with DMSO were used as a positive control for EGFP in images. Images of each well were recorded under a 10× microscope immediately after addition of the compounds (10 μM) as a day 0 timepoint. After 48 h of treatment, images were taken and analyzed to obtain the numbers of dsRed cells in each well. The library was screened in duplicate. Percentage of dsRed cells were calculated by the number of red colored cells in the fluorescence images divided by the total number of cells in the corresponding images. A cut-off with total cell number ≥ 600 and percent of red cells ≥7% was set to select for positive hits. After the screen, a dose response assay and semi-quantitative RT-PCR analysis were performed for each positive compound to filter out false positives.
Reporter transfection assay
For reporter transfection assays, HEK 293FT (1 × 105) cells were seeded in a 24-well plate prior to transfection. The bichromatic fluorescent reporters (800 ng) were transfected using Lipofectamine 2000 (Life Technologies). After 24 h transfection, small molecule compounds or DMSO were added into each well. Fluorescence images were taken under 10× microscopy after 48 h of drug treatment. RNA was extracted after 48 h of drug treatment to perform RT-PCR analysis of splicing.
Circular dichroism spectroscopy
To conduct the circular dichroism spectroscopy measurement, RNA and DNA oligonucleotides were purchased from Integrated DNA Technologies after purification by HPLC and dissolved in Nuclease-free water at a concentration of 100 μM. The sequences were as follows:
CRQ RNA oligo: 5′-GGGCGGCGGGCGGGCUGGGG-3′
I-8 RNA oligo: 5′-GCUUUGGUGGUGGAAUGGUGCUAUGUGG-3′
I-8 DNA oligo: 5′-GCTTTGGTGGTGGAATGGTGCTATGTGG-3′
TBA DNA oligo: 5′-GGTTGGTGTGGTTGG-3′
cMyc DNA oligo: 5′-TGGGGAGGGTGGGGAGGGTGGGGAAGG-3′
RNA and DNA oligonucleotides (5 μM) were prepared in annealing buffer (10 mM Tris–HCl, pH 7.4 and 100 mM K+) and annealed by heating to 95°C and then cooling down slowly to room temperature. After annealing, different amounts of emetine were added and the circular dichroism of RNA oligonucleotides was measured at 20°C by a Jasco J-815 spectropolarimeter. An average of five CD spectra ranging from 220 to 320 nm were collected at a bandwidth of 1 nm in a 0.1-mm path length cuvette, with a response time of 2 s/nm and continuous scan mode.
RNA extraction, semi-quantitative and real-time RT-PCR
Total RNA was isolated from cultured cells with total RNA kit (Omega Bio-tek). Isolated total RNA (125 ng) was used for reverse transcription in a 10 μl volume with the GoScript Reverse Transfection System to produce cDNA (Promega). For semi-quantitative RT-PCR, 20 μl reactions were set up consisting of 16.9 μl of the pre-mixed PCR Mix (PCR buffer, dNTP, MgCl2 and water), 1 μl of template cDNA, 2 μl of primers and 0.1 μl of Hot StarTaq DNA polymerase (QIAGEN). The PCR reaction was run in a linear range and was no more than 30 cycles. The primer sequences are included in Supplemental Table S1. The PCR products were separated on 2% agarose gels containing ethidium bromide. The relative signal intensity of each band was obtained and PSI values were calculated using the intensities of the inclusion band divided by the sum of the intensities of inclusion and skipping bands. For real-time quantitative PCR, Bio-Rad iQ SYBR Green Supermix was used on a BioRad CFX instrument. 0.25 μl cDNA from the reverse transcription reaction was included in a 20 μl reaction using 1.6 μl primers and 2× GoTaq qPCR Master Mix (Promega). Primers were pre-mixed to include the forward and reverse oligo for each primer pair, at a concentration of 10 pMol/ μl for each primer. The reaction was performed with initial denaturation at 95°C for 2 min, 40 cycles of amplification and quantification (10 s at 95°C and 30 s at 60°C). The primer sequences are summarized in Supplemental Table S1. Primer specificity was checked using melt-curve analysis after PCR. Relative gene expression measurements were performed using the 2−ΔCt method using the PCR product of TBP as an internal control (35)
Native PAGE assay
RNA oligonucleotides were incubated with or without the presence of emetine for 1 hour and then resolved in a 10% Native polyacrylamide gel. For hnRNPF binding to RNA oligonucleotides, 4 μM of RNA was incubated with recombinant hnRNPF protein in G-quadruplex formation buffer (10 mM Tris, 100 mM KCl, pH 7.5) on ice for 2 h with or without the presence of emetine. After incubation, RNA-protein complexes were resolved in an 8% Native polyacrylamide gel. RNA species were visualized using SYBR-Gold.
RNA pull-down
The 5′ biontinylated RNA oligonucleotides were synthesized by Integrated DNA technologies. The RNA sequences were as follows:
Biotin-I-8: 5′-biotin-GACUUGGUGGUGGAAUGGUCA-3′
Biotin-I-8-G4m: 5′-biotin-GACUUGAUGAUGAAAUGAUCA-3′
Biotin-CRQ: 5′-biotin-GACUGGGCGGCGGGCGGGCUGGGGCA-3′
Biotin-CRQ-G4m: 5′-biotin-GACUGAGCGCCGAGCGCGCUGAGGCA-3′
Biontinylated RNA oligonucleotides (400 pmol) were immobilized to streptavidin beads at 4°C for 2 h before incubation with 100 μg HEK293FT nuclear extraction in 300 μl binding buffer (20 mM Tris, 200 mM NaCl, 6 mM EDTA, 5 mM potassium fluoride, 5 mM β-glycerophosphate, 2 mg/ml aprotinin at pH 7.5) at 4°C for 2 h. Conjugated beads were washed with binding buffer for three times and boiled at 95°C for 5 min in 60 μl sodium dodecyl sulfate buffer. The samples were centrifuged and analyzed by western blot.
RNA sequencing, G-quadruplex prediction and gene set enrichment analysis
RNA was extracted using Trizol from HMLE-Twist and MCF10A cells that were treated with DMSO or emetine (10 μM) for 24 h. 10 μM of emetine gave similar results to 20 μM on splicing changes of endogenous genes, and this lower dose of emetine was used for RNAseq to avoid potential secondary effects caused by the higher dose. Two biological replicates were performed per experimental condition. Poly-A selected RNA-seq libraries were generated using TruSeq Stranded mRNA Library Prep Kits (Illumina) and subjected to 100 bp PE stranded RNA sequencing on an Illumina HiSeq 4000. RNA-seq reads were aligned to the human genome (GRCh37, primary assembly) and transcriptome (Gencode v24 backmap 37 comprehensive gene annotation) using STAR v2.5.3a (36) with alignEndsType set to ‘EndToEnd’. Differential alternative splicing was quantified using rMATS v3.2.5 (37) using the same transcriptome. Significant differentially spliced cassette exons after emetine treatment were called using only unique junction reads and the following cutoffs: FDR < 0.05, delta PSI ≥0.1, average junction reads per cassette event ≥20. Predicted G-quadruplexes were curated from a strand-specific list of all sequences in the GRCh37 version of the human genome matching the regular expression GxNy, where N equals any series of nucleotides other than G, x ≥ 2 or 3 and 1 ≤ y ≤ 7 to identify all sequences with G-tracts of 2–3 or more separated by 1–7 of any other base. Bedtools v2.25.0 and custom bash and python scripts were used to intersect emetine-dependent cassette exons, the flanking variable exons, and the 250 nucleotides flanking each splice site with these G-quadruplexes. RNA sequencing data has been deposited under accession number GSE113505.
Gene set enrichment analysis (GSEA) was conducted by first computing differentially expressed genes between emetine-treated and untreated cells using cuffdiff v2.2.1 (38). Genes were filtered to drop lowly expressed genes with FPKM <10 in both untreated and emetine-treated cells. Genes were then ranked in descending order by a custom ranking statistic computed by multiplying the -log10P-value reported by cuffdiff times the log2 Fold Change in gene expression comparing emetine treated cells to untreated cells. The GSEA pre-rank algorithm was run with 100 permutations using a curated set of gene sets associated with EMT to determine if emetine-regulated genes were significantly associated with EMT transcriptional signatures (39,40).
RESULTS
The small molecule emetine inhibits G-quadruplex-dependent alternative splicing
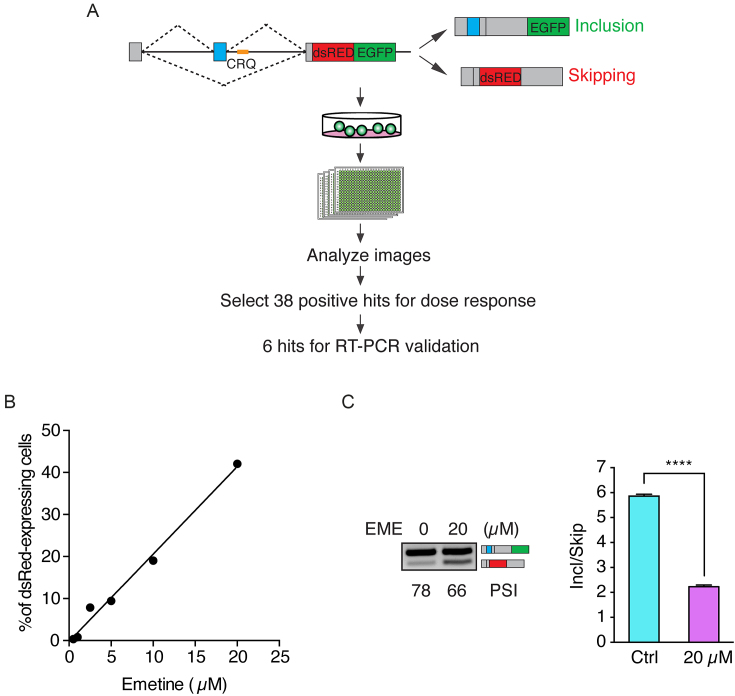
We previously constructed a bichromatic fluorescent splicing reporter, which contains a cassette exon and a known G-quadruplex containing sequence, CRQ, inserted just downstream of the 5′ splice site (3). The splicing reporter expresses EGFP as a readout of alternative exon inclusion and dsRed when exon skipping occurs (Figure 1A). CRQ is a 21-nucleotide (nt) RNA sequence that forms a bona-fide G-quadruplex secondary structure (Supplementary Figure S1A, and (3)). In accordance with our previous report that the G-quadruplex sequence of CRQ promotes exon inclusion, cells expressing this CRQ reporter primarily produce the exon inclusion isoform and show EGFP fluorescence (Supplementary Figure S1B and (3)). Disrupting the G-quadruplexes in CRQ results in the production of the exon skipping isoform and shifts the fluorescence from EGFP to dsRed (Supplementary Figure S1B and (3)). To identify compounds that regulate alternative splicing by disrupting G-quadruplexes, we developed an imaging assay to quantify changes in the ratio of EGFP and dsRed signals in an HEK 293A cell line stably expressing the CRQ-splicing reporter. The predominant green fluorescence and minute level of red fluorescence in these cells provided an optimal system to identify compounds that inhibit CRQ-mediated exon inclusion, which would result in a shift of splicing from the green inclusion isoform to the dsRed skipping isoform. We employed the MicroSource Custom Collection library that contained 2700 small molecule compounds and treated the CRQ-reporter expressing HEK 293A cells in 384-well plates. After 48 hours of treatment, cells in individual wells were imaged by ImageXpress microscopy and monitored for changes in total cell number as well as an increase in the dsRed signal, which represents exon skipping events. After initial identification of 38 hits that showed increased percentage of red-colored cells (Supplementary Table S2), we conducted a secondary screen by dose-response assay and found six out of the 38 compounds that exhibited a dose-dependent increase in red signals (Supplementary Table S3). Next, we performed semi-quantitative RT-PCR analysis to measure changes of alternative splicing upon the compound treatment. The efficiency of alternative splicing is represented by the Percent Spliced In (PSI) index, which measures the percentage of transcripts with the variable exon spliced in. Among the six compounds, only emetine, obtusaquinone, and sanguinarine sulfate showed effects on inhibiting alternative splicing. The three other compounds did not show splicing changes indicating that their effects on increasing the red signals are secondary and are not related to CRQ splicing alterations. Among the three positive compounds, sanguinarine sulfate is most toxic to cells, the effect of obtusaquinone on the splicing is much weaker compared to that of emetine, and emetine showed the highest potency on inhibiting CRQ-dependent splicing (Figure 1B and C, Supplementary Figure S1C and D) and less toxicity. Thus, we chose to focus on emetine, a natural alkaloid produced from ipecas root (41) in this study. Quantitative real-time PCR (qPCR) also confirmed that emetine treatment caused a shift of alternative splicing to the skipped isoform (Figure 1C).
Figure 1.
A high-throughput screen to identify modulators of G-quadruplex-dependent alternative splicing. (A) Flowchart of the high-throughput screening assay. The minigene splicing reporter is depicted on the top panel. The CRQ G-quadruplex sequence is colored in yellow. Inclusion of the variable exon (in blue) results in the production of EGFP. Skipping of the variable exon generates dsRed. (B) Dose response relationship between emetine and the percentage of dsRed-expressing cells. (C) Semi-quantitative RT-PCR (left) and qRT-PCR analysis (right) of emetine (20 μM) treatment on HEK 293A CRQ for 48 h. Error bars represent S.E.M. ****P < 0.0001; Student's t-test.
Emetine regulates alternative splicing by interfering with G-quadruplex secondary structures
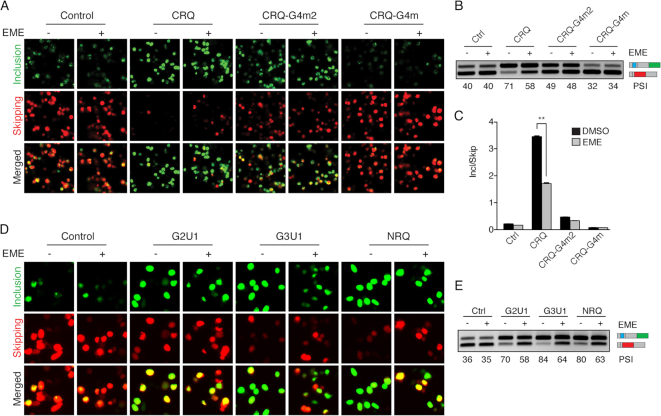
To assess whether emetine inhibits exon inclusion by interfering with G-quadruplexes, we compared the effect of emetine in G-quadruplex containing CRQ minigene to the control non-CRQ minigene. We found that the control non-G-quadruplex containing minigene did not show any changes in splicing in response to emetine treatment (Figure 2A–C). Insertion of the G-quadruplex containing CRQ resulted in an increase in exon inclusion, but this increase was impaired when cells were treated with emetine, suggesting that emetine inhibits the G-quadruplex-dependent increase in exon inclusion (Figure 2A–C). Furthermore, we introduced mutations in the CRQ guanine tetrads to abolish the G-quadruplex structure (Supplementary Figure S1A). We substituted the middle two guanine nucleotides with two adenosines in CRQ, which prevented the sequence from forming a G-quadruplex without dramatically altering the guanine composition of the sequence (Supplementary Figure S1A). This G-quadruplex mutant, termed CRQ-G4m2, partially abrogated CRQ-mediated exon inclusion, resulting in a decrease in the ratio of exon inclusion to skipping (Figure 2A–C). Emetine treatment with this minigene showed minimal changes in alternative splicing, suggesting that emetine loses its inhibitory effect in exon inclusion in the absence of an intact G-quadruplex (Figure 2A–C). Disrupting the G-quadruplex by substituting all four GGG repeats (CRQ-G4m), which completely diminished G-quadruplex-mediated exon inclusion (Figure 2A–C), also abolished the effect of emetine-mediated inhibition on exon inclusion (Figure 2A–C), once again indicating that the G-quadruplex is required for emetine-mediated inhibition of exon inclusion.
Figure 2.
Emetine inhibits exon inclusion in a G-quadruplex-dependent manner. (A) Fluorescence images of HEK 293FT cells transfected with CRQ and its mutants with or without the presence of emetine (20 μM). The fluorescent protein EGFP is a readout for variable exon inclusion and dsRed for variable exon skipping. (B, C) Semi-quantitative RT-PCR (B) and qRT-PCR (C) showing that emetine inhibits exon inclusion only of the CRQ minigene but not control or CRQ mutant constructs whose G-quadruplex structures were destroyed. (D) Fluorescence images of HEK 293FT cells transfected with different G-quadruplex-containing splicing constructs with or without the presence of emetine. (E) Semi-quantitative RT-PCR results showing that emetine inhibits exon inclusion in different G-quadruplex-containing splicing constructs. Error bars represent S.E.M. **P < 0.01; Student's t-test.
To determine whether emetine affects G-quadruplexes in addition to CRQ, we examined the effect of emetine on an RNA sequence, termed I-8, that was shown to form a G-quadruplex structure (Supplementary Figure S2A) (3). Insertion of the I-8 sequence into the minigene resulted in an enhanced exon inclusion (Supplementary Figure S2B–D). Similar to the results observed with CRQ, emetine treatment resulted in a significant decrease in exon inclusion from 56% to 48% in the I-8 splicing minigene, but showed little effect on the I-8 mutants where the G-quadruplexes were destroyed (Supplementary Figure S2B–D).
To thoroughly determine whether the effect of emetine on G-quadruplexes is dependent on the numbers of guanine tetrads, we analyzed three additional G-quadruplex sequences including two G-quadruplex sequences containing GU repeats, G2U1 (two guanine tetrads), G3U1 (three guanine tetrads), and one previously reported G-quadruplex sequence named NRQ (Supplementary Figure S3A) (42). In all cases, emetine inhibited exon inclusion of the G-quadruplex-containing splicing reporter minigene. Emetine showed a slightly stronger effect on three guanine tetrads than on two guanine tetrads (Figure 2D and E). These results further indicate that emetine inhibits alternative splicing in a G-quadruplex-dependent manner.
Emetine directly perturbs G-quadruplex secondary structures
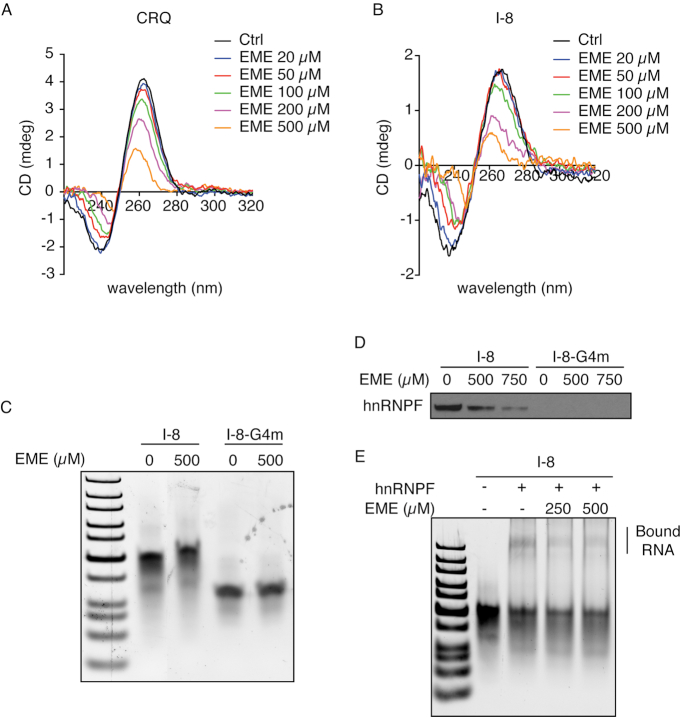
To directly examine whether emetine disrupts quadruplex structural integrity, we conducted circular dichroism (CD) spectroscopy measurements using RNA oligonucleotides that contained the 21-nt CRQ sequences. The CD spectrum of CRQ showed a positive peak at 264 nm and a negative peak at 240 nm (Figure 3A), which is a characteristic feature of a parallel RNA G-quadruplex structure (43). Increasing the concentrations of emetine resulted in a decrease in the CD spectra peak, suggesting that emetine interferes with the G-quadruplex structure (Figure 3A). Similarly, emetine perturbed the characteristic G-quadruplex CD spectrum of the I-8 RNA in a dose-dependent manner (Figure 3B).
Figure 3.
The interaction between emetine and G-quadruplex secondary structure. (A, B) CD spectra analysis of CRQ (A) and I-8 (B) RNA oligonucleotides in the presence of different concentrations of emetine. (C) Native gel images showing locations of I-8 Wild Type (WT) and its G4m mutant RNA oligonucleotides with or without the presence of emetine. (D) RNA pulldown followed by western blot analysis showing that the addition of emetine disrupts the interaction between hnRNPF and I-8. (E) RNA electrophoretic mobility shift assay using I-8 RNA and recombinant hnRNPF protein on native polyacrylamide gels.
To examine whether emetine directly interacts with G-quadruplex containing RNA oligonucleotides, we incubated RNA I-8 and its mutant I-8-G4m with emetine and performed gel electrophoresis. The I-8 RNA ran slower than the same length G4m RNA oligo, presumably due to formation of the I-8 G-quadruplex (3). Notably, the I-8 RNA incubated with emetine migrated more slowly than I-8 alone, whereas its mutated G4m with emetine migrated at similar speed as G4m alone (Figure 3C). These results suggest that emetine interacts with the G-quadruplex containing I-8 RNA and formed a stable complex.
Our previous work identified that the splicing factor hnRNPF is a G-quadruplex binding protein (3). We next determined whether emetine disrupts the interaction between hnRNPF and the G-quadruplex containing RNA. Biotin-labeled I-8 or CRQ RNA oligonucleotides were immobilized onto the streptavidin beads and incubated with HEK293FT nuclear extracts. The hnRNPF protein that interacted with the RNA was then pulled down by the streptavidin beads and quantitated by immunoblot analysis. Notably, addition of emetine abolished the interaction between hnRNPF and the I-8 or CRQ RNAs in a dose-dependent manner (Figure 3D, Supplementary Figure S3B). Moreover, native gel-shift assay using the recombinant hnRNPF and the I-8 RNA showed that the super-shifted I-8 RNA-hnRNPF higher-order complexes were disrupted with the addition of emetine (Figure 3E), indicating that emetine abolished the interaction of hnRNPF with the I-8 RNA. In aggregate, these results reveal that emetine directly interferes with the G-quadruplex structure and protein binding to G-quadruplexes.
Cephaeline modulates alternative splicing in a G-quadruplex dependent manner
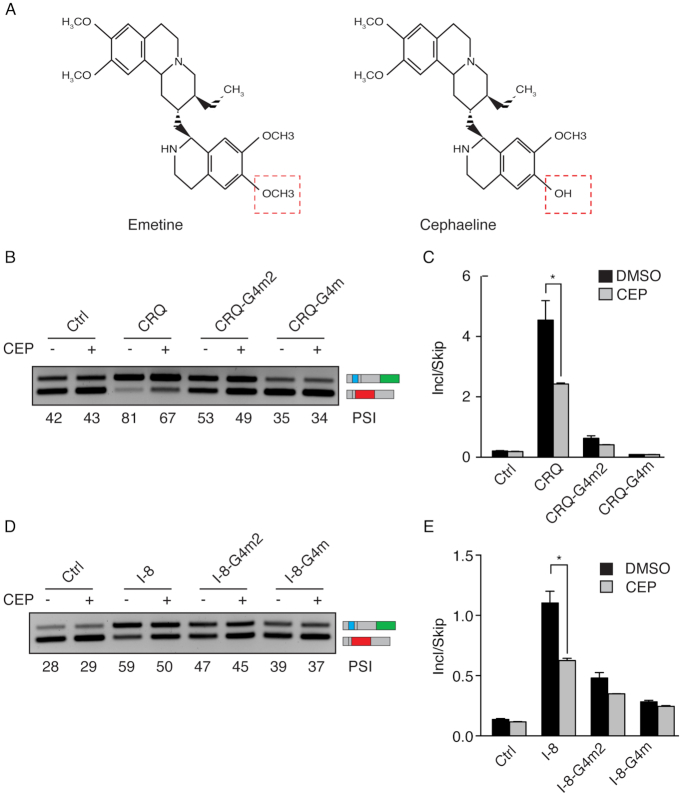
Our results suggested that the small molecule emetine inhibits G-quadruplex-dependent exon inclusion. We speculated that small molecules with a similar structure to emetine would exhibit similar properties in disrupting G-quadruplexes and regulating alternative splicing. Cephaeline is a closely related analog to emetine (Figure 4A). Cephaeline and Emetine differ only by the presence of one methoxy group in emetine that is present as a hydroxyl group in cephaeline. To investigate whether cephaeline regulates alternative splicing in a G-quadruplex dependent manner, we transfected HEK 293FT cells with the CRQ and CRQ-mutated splicing minigenes and observed that cephaeline treatment caused a decrease in CRQ-dependent exon inclusion from PSI 81–67%. By contrast, cephaeline showed minor effects on G4m2 and no effect on G4m splicing minigenes (Figure 4B and C). In addition, cephaeline treatment also showed a decrease in the G-quadruplex containing I-8 minigene, but not the G-quadruplex mutated I-8 minigene. (Figure 4D and E). These results indicate that cephaeline modulates G-quadruplex-dependent alternative splicing in the same manner as of its analog emetine.
Figure 4.
Cephaeline modulates alternative splicing in a G-quadruplex-dependent manner. (A) Comparison of the chemical structures of emetine and cephaeline. The structures differ only by the moieties labeled in red boxes. (B, C) Semi-quantitative RT-PCR images (B) and qRT-PCR results (C) of HEK 293FT cells transfected with CRQ and its mutant splicing minigenes showing that cephaeline (CEP) inhibits exon inclusion of the CRQ splicing minigene and not the mutated splicing minigenes. (D, E) Effects of CEP on the I-8 G-quadruplex and its mutated splicing minigenes in HEK 293FT cells. Semi-quantitative RT-PCR images (D) and qRT-PCR results (E) are shown. Error bars represent S.E.M. *P < 0.05; Student's t-test.
RNA G-quadruplexes are transcribed from DNA, and their DNA sequence may also form G-quadruplexes. Because transcription and splicing are coupled processes and because DNA G-quadruplexes have been shown to slow down the rate of transcription (44), we further examined whether the observed RNA G-quadruplex-dependent alternative splicing was due to changes in transcription elongation. We transfected HEK 293FT cells with the splicing minigenes CRQ and its mutant CRQ-G4m and treated these cells with two transcription inhibitors, camptothecin (CPT) and 5,6-Dichlorobenzimidazole 1-β-d-ribofuranoside (DRB). Notably, CPT and DRB had little effects on the splicing of CRQ and its mutant CRQ-G4m (Supplementary Figure S4A), excluding the contribution of transcription on G-quadruplex-mediated splicing changes. Moreover, emetine has been used as a translation inhibitor (45). To exclude the possibility that splicing changes caused by emetine were due to translation inhibition, we examined the effect of the translation inhibitor cyclohexamide (CHX) on CRQ-mediated exon inclusion. CHX exhibited little changes in alternative splicing of the CRQ minigene (Supplementary Figure S4B), illustrating that the effect of emetine on modulating alternative splicing is not due to translation inhibition.
The effect of G-quadruplex structure on alternative splicing is not location-dependent
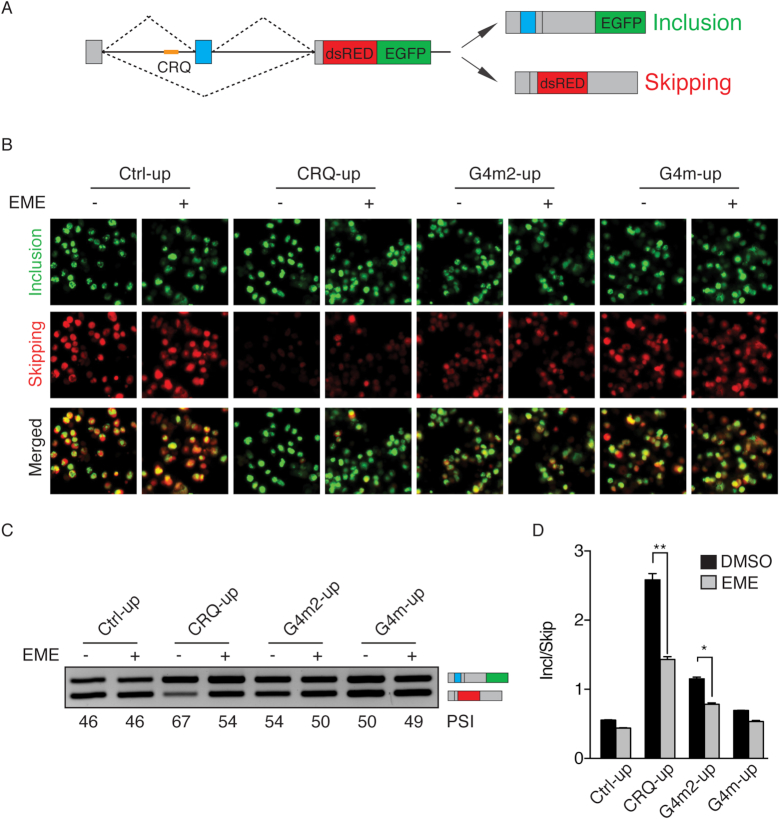
The above data examining the effect of disruption of G-quadruplexes on alternative splicing were conducted on constructs with the G-quadruplexes located downstream of the variable exon. To test whether the location of the G-quadruplex affects alternative splicing, we introduced the same sequence of CRQ in the intron upstream of the cassette exon (CRQ-up) (Figure 5A). Transfection of the CRQ-up splicing minigene in HEK 293FT cells showed an increase in exon inclusion and decrease in exon skipping, as represented by the switch of dsRed to EGFP (Figure 5B) and by semi-quantitative RT-PCR analysis (Figure 5C) and qRT-PCR (Figure 5D). Furthermore, the CRQ-up mutants, G4m2-up and G4m-up, were also less effective in promoting exon inclusion compared to the CRQ-up construct (Figure 5B–D). These results are consistent with the effect of CRQ inserted in the intron downstream of the cassette exon (Figure 2A–C), indicating that the effect of G-quadruplexes on exon inclusion is not location-dependent.
Figure 5.
Emetine regulates alternative splicing through disrupting G-quadruplexes in a location-independent manner. (A) Diagram of the fluorescent splicing reporter construct that contains the CRQ G-quadruplex in the intron upstream of the variable exon. (B) Fluorescent images of HEK 293FT cells after the transfection with CRQ-up or its mutant minigene reporters that contain sequences inserted in the intron upstream of the variable exon with or without the presence of emetine. (C, D) Semi-quantitative RT-PCR images (C) and qRT-PCR results (D) showing that emetine promotes exon skipping of the CRQ-up minigene with no or much less effects on the control or mutant minigene constructs. Error bars represent S.E.M. *P < 0.05, **P < 0.01; Student's t-test.
We next sought to determine whether emetine inhibits exon inclusion of the CRQ-up construct. We transfected CRQ-up and its mutated minigenes in HEK 293FT cells and found that emetine inhibited exon inclusion only of the CRQ-up minigene but not of the mutants (Figure 5B–D), indicating that emetine also inhibits G-quadruplex-mediated splicing when the G-quadruplexes are located in the intron upstream of the variable exon.
G-quadruplexes are enriched near alternative exons regulated by emetine
After observing that emetine regulates alternative splicing in a G-quadruplex-dependent manner, we aimed to identify the effect of emetine on alternative splicing globally across the transcriptome. We performed RNA sequencing (RNAseq) using two human cell lines, MCF10A epithelial cells and HMLE-Twist mesenchymal cells, with or without emetine treatment. We chose these two cell lines representing different cell states aiming to identify the generality of the effects of emetine regardless of specific cell state.
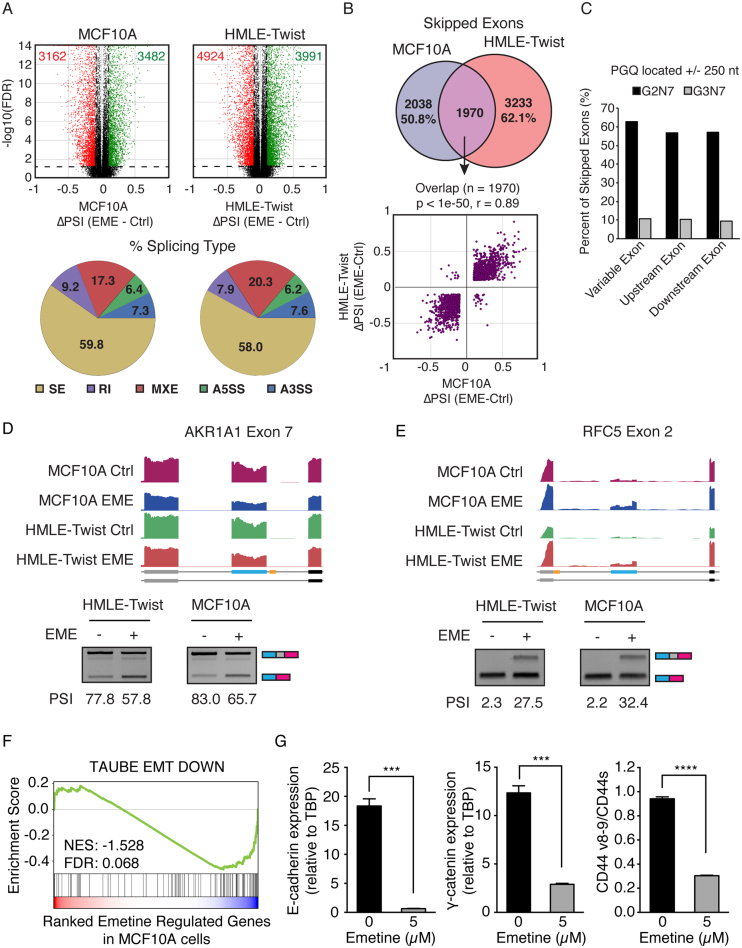
RNAseq analysis showed that emetine treatment caused significant differential splicing of thousands of splicing events in both cell lines, of which nearly 60% represented skipped exon (SE) events (Figure 6A). We focused on SE events as they were the most frequently regulated by emetine and the most common form of alternative splicing (25). To identify a stringent set of SE regulated by emetine, we overlapped the SE events in MCF10A and HMLE-Twist and identified 1970 events shared between both cell lines that were strongly positively correlated (Pearson r = 0.89, Figure 6B). To test whether emetine mediates alternative splicing of cassette exons containing proximal G-quadruplexes, we identified all SE containing potential G-quadruplexes (PGQ) inside the cassette exon, flanking exons, or within 250 nucleotides of a splice site. We found that ∼60% of emetine regulated SE contained a PGQ with 2-nt guanine tracts and ∼10% contained a more stringent PGQ with 3-nt guanine tracts (Figure 6C). As examples, two of the emetine-regulated cassette exons identified as containing an exon-proximal G-quadruplex, AKR1A1 exon 7 and RFC5 exon 2, were previously identified to contain G-quadruplexes capable of regulating alternative splicing (46). Experimental validation of these two SEs showed that emetine treatment promoted AKR1A1 exon 7 skipping and RFC5 exon 2 inclusion in both HMLE-Twist and MCF10A cells (Figure 6D and E). Bcl-x undergoes alternative splicing through the use of alternative 5′ splice sites (A5′SS) in its exon 2 to generate Bcl-xS and Bcl-xL. Emetine was reported to promote the production of Bcl-xS (47). In agreement with previous results, our RNAseq data and RT-PCR validation also showed increased production of Bcl-xS when cells were treated with emetine (Supplementary Figure S5A and B). Interestingly, two G-quadruplexes were recently identified in the Bcl-x exon 2 near the alternative 5′ splice sites (48). The location of these two G-quadruplexes were shown in yellow boxes in Supplementary Figure S5A. Taken together, these results illustrate that emetine regulates alternative splicing events that are enriched for G-quadruplexes globally across the transcriptome.
Figure 6.
Emetine globally affects G-quadruplex associated alternative splicing. (A) Top Panel: Differential alternative splicing events identified after emetine treatment in MCF10A (left) and HMLE-Twist (right) cells. Alternative splicing events that show FDR ≤0.05 and ΔPSI ≥0.2 or ≤–0.2 were colored as green and red dots, respectively. Bottom panel: Pie charts showing percentage of each splicing type represented in emetine differential splicing events. The majority of events are skipped exon (SE) events. (B) Top panel: Venn diagram showing common skipped exons regulated by emetine in both MCF10A and HMLE-Twist cells. Bottom panel: The common set of cassette exons show highly correlated regulation in both cells types. (C) Bar charts showing the percentage of skipped exon splicing events containing a G2N7 or G3N7 predicted G-quadruplex (PGQ) proximal to splice sites. (D, E) Genome browser tracts of RNA sequencing data and semi-quantitative RT-PCR validation showing emetine treatment inhibits exon inclusion of AKR1A1 Exon 7 (D) and promotes inclusion of RFC5 Exon 2 (E). The G-quadruplexes are depicted in yellow in the schematics of the exon annotation. (F) GSEA analysis of emetine regulated genes in MCF10A cells. Genes downregulated by emetine are also downregulated during EMT. (G) qRT-PCR of MCF10A cells showing that emetine treatment resulted in a decrease in expression of epithelial markers E-cadherin and γ-catenin, and a decrease in the ratio between CD44v and CD44s. Error bars represent S.E.M. ***P < 0.001, ****P < 0.0001; Student's t-test.
Since CD44 splice isoform switching to the variable exon skipped CD44s isoform is essential for EMT (34), and because our results using a CD44 minigene reporter showed that emetine promotes CD44 variable exon skipping through the I-8 G-quadruplex (Supplementary Figure S2), we also examined whether cells treated with emetine showed an EMT signature. We performed GSEA analysis using differentially expressed genes from RNAseq data of the emetine treated epithelial MCF10A cells. Noticeably, GSEA analysis showed significant negative enrichment of an EMT-downregulated gene set (49) in response to emetine treatment (Figure 6F), suggesting that emetine favors the loss of an epithelial phenotype. Experimental validation indeed supports this observation. We found that emetine treatment in MCF10A cells resulted in a decrease in expression of epithelial markers E-cadherin and γ-catenin (Figure 6G). In accordance with our previous findings that CD44 isoform switching from CD44v to CD44s is essential for cells to undergo EMT (34), analysis on endogenous CD44 splice isoforms also showed a decrease in the ratio between CD44v and CD44s (Figure 6G). Consistent results are observed in other additional epithelial cells, HMLE and MCF7, in response to emetine treatment (Supplementary Figure S5C and D). These experimental results support the notion that emetine promotes an EMT cell state.
DISCUSSION
In recent years, emerging evidence has indicated the importance of RNA G-quadruplex structures in regulating key cellular functions. Compared to DNA G-quadruplexes which can form both antiparallel and parallel structures, RNA G-quadruplexes are less diverse in terms of strand orientation under physiological conditions and prefer to adopt a parallel-stranded structure due to the contribution of the 2′-hydroxyl group. The formation of RNA G-quadruplexes has been shown to play critical roles in gene regulation. It is therefore important to identify small molecular compounds capable of targeting these structures to denature or stabilize them.
In this study, we used a dual-output splicing reporter that contains a bona-fide G-quadruplex element and performed a cell-based small-molecule high-throughput screen to identify small molecules that alter G-quadruplex-dependent alternative splicing. High-throughput screening approaches based on alternative splicing have been used in many studies (32,50–52). Compared to these methods, our screening system takes into account the role of RNA secondary structures on alternative splicing regulation. In addition to the dual fluorescence signals as our screen readout, we applied RT-PCR validation for promising compounds, thus increasing the specificity in targeting alternative splicing and eliminating false-positive hits.
We identified emetine as a compound capable of regulating alternative splicing by interfering with the G-quadruplexes. Through mutational analysis, CD-spectra, and gel shift assays, we showed that emetine disrupts RNA G-quadruplex secondary structure to modulate alternative splicing. These results were bolstered by similar results from our complementary analysis using cephaeline, an analog of emetine. We observed that increasing the dosage of emetine appeared with concomitant increase in cell death. Because emetine exhibited no effects on the splicing of G-quadruplex mutants, we concluded that the splicing changes of the G-quadruplex reporters are not due to the cell death caused by emetine. In support of our view, emetine was previously identified to regulate Bcl-x alternative splicing and promote the production the Bcl-xS splice isoform (47,48), and this result was also validated in our experimental system. Since Bcl-xS antagonizes the anti-apoptotic activity of Bcl-xL, we speculate that the decreased viability in emetine-treated cells may reflect the increased production of Bcl-xS in these cells. As a future direction, it will be of great interest to investigate the contribution of these G-quadruplexes on Bcl-x alternative splicing, which is closely connected to cancer cell survival.
We wish to note that emetine seemed to also bind to DNA G-quadruplex structures. We examined two known DNA G-quadruplexes, the antiparallel DNA G-quadruplexes from Thrombin binding aptamer (TBA) and the parallel DNA G-quadruplex in the cMyc promoter. We found that emetine had detectable effects on both TBA and cMyc. While the effect of emetine on TBA was weaker, the effect on cMyc was similar to that on CRQ RNA and I-8 RNA (Supplementary Figure S6A). The I-8 DNA oligonucleotides, however, were not able to form a typical G-quadruplex (Supplementary Figure S6B), ruling out the possibility that I-8 DNA G-quarduplex may contribute to splicing changes upon emetine treatment. Our minigene splicing assays on using transcription inhibitors also support the idea that emetine does not affect splicing through potential DNA G-quadruplex-mediated transcription changes.
From our RNAseq analysis, we found that many emetine-regulated exon skipping events contain putative G-quadruplex forming sequences proximal to the splice sites, suggesting that emetine may have a global role in regulating G-quadruplex-dependent alternative splicing. Our experiments have shown that emetine directly interacts with G-quadruplexes to regulate alternative splicing. However, it is important to point out that emetine was previously shown to inhibit protein translation by binding to the 40S subunit of the ribosome (41). In addition to its direct effect on alternative splicing, we cannot fully exclude the possibilities that the alternative splicing changes caused by emetine may be due to inhibiting the expression of splicing regulators (47). Interestingly, using our RNAseq data, we analyzed the differential gene expression of RNA binding protein (RBPs) in response to emetine treatment to investigate emetine's indirect effects through RBPs. Significant differentially expressed genes were identified using a cutoff as 2-fold. None of the differentially expressed RBPs was previously reported to bind to RNA G-quadruplexes. In accordance with this, the expression level of hnRNPF, which affects G-quadruplex-mediated alternative splicing, did not change in the presence of emetine. However, whether emetine also affects other RNA biosynthetic pathways to further regulate G-quadruplex will be an interesting subject for future investigation.
An exciting finding was that emetine promotes an EMT cell state, supported by evidence from our RNAseq data analysis and experimental validation. These results are congruent with the fact that emetine CD44 variable exon skipping is essential for EMT and our findings that emetine promotes CD44 variable exon skipping through a G-quadruplex I-8 sequence. Because EMT is hijacked by cancer cells during tumor metastasis, we postulate that small molecules may alter cell states that are associated with cancer progression.
To date, few studies have evaluated the effect of small molecules on regulating G-quadruplex-mediated alternative splicing. TMPYP4 has been shown to disrupt G-quadruplex structures when binding to RNA (53,54), but it has additional effects beyond potential disruption of RNA G-quadruplexes (22,55). Our study identifies emetine as a splicing modulatory compound. By interfering with RNA G-quadruplex structures, emetine may affect cellular functions by impacting G-quadruplex-mediated alternative splicing. Emetine can serve as a reagent to help understand the mechanisms underlying how RNA binding proteins regulate alternative splicing through interactions with G-quadruplexes. It may also be used for the identification of splicing targets that are regulated by G-quadruplex structures. Thus, our studies may inspire researchers to consider the role of RNA secondary structures in addition to the linear sequences in regulating alternative splicing.
Taken together, the presented study is the first to screen compounds that regulate alternative splicing by perturbing RNA G-quadruplex structures rather than RNA linear sequences. Our findings open new horizons for the identification of small molecules capable of regulating alternative splicing by targeting RNA secondary structures.
DATA AVAILABILITY
RNA sequencing data has been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE113505.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Chi-Hao Luan and Sara Fernandez Dunne at the High Throughput Analysis Laboratory, Northwestern University for assisting the high throughput screening and analysis. We thank Dr Jin Wang at Baylor College of Medicine for suggestions on cephaeline.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [R01GM110146 and R01CA182467 to C.C., F30CA196118 to S.E.H.]; C.C. is a CPRIT Scholar in Cancer Research [RR160009]. Funding for open access charge: NIH R01GM110146.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rubtsov P.M. Role of pre-mRNA secondary structures in the regulation of alternative splicing. Mol. Biol. 2016; 50:823–830. [DOI] [PubMed] [Google Scholar]
- 2. Shepard P.J., Hertel K.J.. Conserved RNA secondary structures promote alternative splicing. RNA. 2008; 14:1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang H., Zhang J., Harvey S.E., Hu X., Cheng C.. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017; 31:2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L.. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016; 5:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tellam J., Smith C., Rist M., Webb N., Cooper L., Vuocolo T., Connolly G., Tscharke D.C., Devoy M.P., Khanna R.. Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M.. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014; 505:696. [DOI] [PubMed] [Google Scholar]
- 7. Kriner M.A., Groisman E.A.. RNA secondary structures regulate three steps of Rho-dependent transcription termination within a bacterial mRNA leader. Nucleic Acids Res. 2017; 45:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J.. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008; 14:1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beaudoin J.D., Perreault J.P.. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013; 41:5898–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arora A., Suess B.. An RNA G-quadruplex in the 3′ UTR of the proto-oncogene PIM1 represses translation. RNA Biol. 2011; 8:802–805. [DOI] [PubMed] [Google Scholar]
- 13. Decorsiere A., Cayrel A., Vagner S., Millevoi S.. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011; 25:220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.P., Hall J., Mergny J.L., Hainaut P., Van Dyck E.. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011; 32:271–278. [DOI] [PubMed] [Google Scholar]
- 15. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.-L., Moine H.. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 17. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeraati M., Moye A.L., Wong J.W., Perera D., Cowley M.J., Christ D.U., Bryan T.M., Dinger M.E.. Cancer-associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression. Sci. Rep. 2017; 7:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsuda Y., Sato S.-i., Asano L., Morimura Y., Furuta T., Sugiyama H., Hagihara M., Uesugi M.. A small molecule that represses translation of G-Quadruplex-Containing mRNA. J. Am. Chem. Soc. 2016; 138:9037–9040. [DOI] [PubMed] [Google Scholar]
- 20. Miglietta G., Cogoi S., Marinello J., Capranico G., Tikhomirov A.S., Shchekotikhin A., Xodo L.E.. RNA G-Quadruplexes in Kirsten Ras (KRAS) oncogene as targets for small molecules inhibiting translation. J. Med. Chem. 2017; 60:9448–9461. [DOI] [PubMed] [Google Scholar]
- 21. Weldon C., Dacanay J.G., Gokhale V., Boddupally Peda Venkat L., Behm-Ansmant I., Burley G.A., Branlant C., Hurley L.H., Dominguez C., Eperon I.C.. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018; 46:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bugaut A., Rodriguez R., Kumari S., Hsu S.-T.D., Balasubramanian S.. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010; 8:2771–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Su Z., Zhang Y., Gendron Tania F., Bauer Peter O., Chew J., Yang W.-Y., Fostvedt E., Jansen-West K., Belzil Veronique V., Desaro P. et al.. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-Associated defects in c9FTD/ALS. Neuron. 2014; 83:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413. [DOI] [PubMed] [Google Scholar]
- 25. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evsyukova I., Somarelli J.A., Gregory S.G., Garcia-Blanco M.A.. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biology. 2010; 7:462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mills J.D., Janitz M.. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol. Aging. 2012; 33:1012–1024. [DOI] [PubMed] [Google Scholar]
- 28. Xu Y., Gao X.D., Lee J.-H., Huang H., Tan H., Ahn J., Reinke L.M., Peter M.E., Feng Y., Gius D.. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014; 28:1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu S., Cheng C.. Alternative RNA splicing and cancer. Wiley Interdiscip. Rev. RNA. 2013; 4:547–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soret J., Bakkour N., Maire S., Durand S., Zekri L., Gabut M., Fic W., Divita G., Rivalle C., Dauzonne D. et al.. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. PNAS. 2005; 102:8764–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arslan A.D., He X., Wang M., Rumschlag-Booms E., Rong L., Beck W.T.. A high throughput assay to identify small molecule modulators of alternative pre-mRNA splicing. J. Biomol. Screen. 2013; 18:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoilov P., Lin C.H., Damoiseaux R., Nikolic J., Black D.L.. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orengo J.P., Bundman D., Cooper T.A.. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res. 2006; 34:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown R.L., Reinke L.M., Damerow M.S., Perez D., Chodosh L.A., Yang J., Cheng C.. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 2011; 121:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 36. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. PNAS. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. et al.. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 40. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akinboye E.S., Bakare O.. Biological activities of emetine. Open Nat. Prod. J. 2011; 4:8–15. [Google Scholar]
- 42. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vorlickova M., Kejnovska I., Sagi J., Renciuk D., Bednarova K., Motlova J., Kypr J.. Circular dichroism and guanine quadruplexes. Methods. 2012; 57:64–75. [DOI] [PubMed] [Google Scholar]
- 44. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noensie E.N., Dietz H.C.. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 2001; 19:434–439. [DOI] [PubMed] [Google Scholar]
- 46. Dardenne E., Polay Espinoza M., Fattet L., Germann S., Lambert M.P., Neil H., Zonta E., Mortada H., Gratadou L., Deygas M. et al.. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014; 7:1900–1913. [DOI] [PubMed] [Google Scholar]
- 47. Boon-Unge K., Yu Q., Zou T., Zhou A., Govitrapong P., Zhou J.. Emetine regulates the alternative splicing of Bcl-x through a protein phosphatase 1-dependent mechanism. Chem. Biol. 2007; 14:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weldon C., Behm-Ansmant I., Hurley L.H., Burley G.A., Branlant C., Eperon I.C., Dominguez C.. Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat. Chem. Biol. 2017; 13:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taube J.H., Herschkowitz J.I., Komurov K., Zhou A.Y., Gupta S., Yang J., Hartwell K., Onder T.T., Gupta P.B., Evans K.W. et al.. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng S., Damoiseaux R., Chen L., Black D.L.. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res. 2013; 23:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han H., Braunschweig U., Gonatopoulos-Pournatzis T., Weatheritt R.J., Hirsch C.L., Ha K.C.H., Radovani E., Nabeel-Shah S., Sterne-Weiler T., Wang J. et al.. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell. 2017; 65:539–553. [DOI] [PubMed] [Google Scholar]
- 52. Rousseaux M.W., de Haro M., Lasagna-Reeves C.A., De Maio A., Park J., Jafar-Nejad P., Al-Ramahi I., Sharma A., See L., Lu N.. TRIM28 regulates the nuclear accumulation and toxicity of both alpha-synuclein and tau. Elife. 2016; 5:e19809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zamiri B., Reddy K., Macgregor R.B. Jr, Pearson C.E.. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014; 289:4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morris M.J., Wingate K.L., Silwal J., Leeper T.C., Basu S.. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic. Acids Res. 2012; 40:4137–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cammas A., Millevoi S.. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017; 45:1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data has been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE113505.