Abstract
UV light induces cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone (6-4) photoproducts (6-4PPs), which can result in carcinogenesis and aging, if not properly repaired by nucleotide excision repair (NER). Assays to determine DNA damage load and repair rates are invaluable tools for fundamental and clinical NER research. However, most current assays to quantify DNA damage and repair cannot be performed in real time. To overcome this limitation, we made use of the damage recognition characteristics of CPD and 6-4PP photolyases (PLs). Fluorescently-tagged PLs efficiently recognize UV-induced DNA damage without blocking NER activity, and therefore can be used as sensitive live-cell damage sensors. Importantly, FRAP-based assays showed that PLs bind to damaged DNA in a highly sensitive and dose-dependent manner, and can be used to quantify DNA damage load and to determine repair kinetics in real time. Additionally, PLs can instantly reverse DNA damage by 405 nm laser-assisted photo-reactivation during live-cell imaging, opening new possibilities to study lesion-specific NER dynamics and cellular responses to damage removal. Our results show that fluorescently-tagged PLs can be used as a versatile tool to sense, quantify and repair DNA damage, and to study NER kinetics and UV-induced DNA damage response in living cells.
INTRODUCTION
Our genome is continuously exposed to various types of DNA damage. If not repaired correctly, DNA lesions may result in mutations, cellular senescence or cell death, which can eventually lead to various pathological conditions including carcinogenesis and aging (1). To counteract these deleterious effects of DNA damage, cells have evolved a variety of mechanisms, including several DNA repair pathways (2). Nucleotide excision repair (NER) is one of the most versatile DNA repair pathways, as it removes a wide variety of DNA helix-destabilizing lesions. Prominent examples of NER substrates are the UV-induced cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone (6-4) photoproducts (6-4PPs). The biological importance of NER is illustrated by the severe clinical symptoms of human disorders caused by inherited NER defects, including the cancer-prone xeroderma pigmentosum (XP) syndrome or the premature aging disorder Cockayne's syndrome (CS) (3).
NER is initiated by two sub-pathways that differ in their mode of damage recognition. Global genome NER (GG-NER) detects lesions in the entire genome, by the main DNA damage binding protein XPC (4). XPC recognizes DNA-helix distortions such as induced by 6-4PP lesions, but needs the activity of the UV-DDB complex, composed of DDB1 and DDB2, to detect mildly helix-destabilizing CPD lesions (5,6). Transcription-coupled NER (TC-NER) is initiated when DNA damage located in the actively transcribed strand blocks elongating RNA polymerase II, which results in the recruitment of the TC-NER factors CSA, CSB and UVSSA (7,8). Once the DNA lesion is recognized, general transcription factor II H (TFIIH) is recruited (9,10) to unwind the DNA surrounding the damage (11) and to verify the lesion together with XPA (12,13). The endonucleases XPG and ERCC1/XPF subsequently remove a ∼30 nucleotide long fragment of DNA around the lesion (14). Finally, the DNA is restored back to its original state by DNA synthesis and ligation steps (15,16).
Recent studies have shown that NER is a tightly regulated, multistep pathway that requires many proteins and post-translational modifications for the efficient and accurate transition between the successive reaction steps (3,17–19). Additionally, as NER takes place in the complex chromatin and nuclear environment, many factors involved in chromatin remodeling (3,20,21), transcription (22), or replication (23) influence NER activity, and most likely many other involved factors are awaiting their discovery. Therefore, assays to quantify DNA damage and repair rates are invaluable tools to investigate the roles of such factors and to obtain new fundamental insights into the molecular mechanism of NER. Moreover, assays to detect impairments or deficiencies in NER activity have been crucial for the diagnosis of NER-deficient patients and can be used as indicators for predispositions to mutations, the onset of cancer, or DNA damage-induced aging (24–27).
Over the years, several assays were developed to quantitatively monitor UV-induced DNA damage and NER-mediated repair. Traditionally, NER activity is measured by determining the rate of UV-induced DNA repair synthesis, the last step of the NER reaction (28–30), or by determining the levels of CPDs in the DNA in time using T4 endonuclease V (31). Over the years, several other assays have been developed to monitor upstream NER activity, including UV-damage removal (32), NER-induced incisions (33) or quantification of excision products (34). TC-NER is often determined indirectly by quantifying the recovery of RNA synthesis (RRS) (35,36), or by using host cell reactivation assays (37). Alternatively, TC-NER can be measured in a direct manner by strand-specific repair assays (38), or by more recently developed single-cell assays, such as the modified COMET-FISH procedure (39), or the TC-NER specific UDS assay (40). Direct detection and quantification of UV-induced DNA damage and its removal in time can be accomplished using antibodies specifically recognizing CPD or 6-4PP lesions in combination with immunofluorescence or ELISA procedures (32). Although proven to be useful in studying UV-induced DNA repair, these assays depend highly on the quality of the antibodies and have specific limitations. For instance, antibody-based detection of CPD or 6-4PP lesions requires DNA denaturation, to allow DNA damage recognition by these antibodies. For example in immunofluorescence experiments, this denaturation may interfere with co-staining of other proteins of interest. Importantly, most of these assays require cell fixation, which makes them incompatible with live-cell applications, and therefore can only provide endpoint measurements. To overcome these issues, measurements of the DNA damage binding kinetics of fluorescently-tagged NER factors can be used to evaluate repair activity in living cells (41–43). However, these binding kinetics do not provide a direct measurement of DNA damage quantities, as the DNA damage-induced binding of the NER proteins is not influenced exclusively by the DNA damage load, but can also be regulated by post-translational modifications or chromatin remodelers (42,44–49).
While NER is the only mechanism to repair UV-induced DNA damage in placental mammals, an alternative damage removal mechanism known as photo-reactivation (PR) remained preserved through evolution in other branches of life, ranging from bacteria to non-placental mammals (50,51). In contrast to NER-mediated repair, which is a complex mechanism that requires the activity of at least 30 proteins (3), PR is the direct reversal of CPD or 6-4PP lesions by one single damage specific photolyase (PL). PLs recognize the helix distortions created by CPD and 6-4PPs and bind to them through moderately strong ionic interactions. These interactions further destabilize the distorted DNA helix and lead to a flipping out of the DNA lesion into the active site of the PL, forming a highly stable complex (52,53). In contrast to the binding of PLs to DNA lesions, which is independent of light, the catalytic reversal of pyrimidine dimers to the original bases requires the absorption of a photon. Catalysis by PLs is achieved by light-initiated cycloconversion of the cyclobutane ring joining the two pyrimidines, which encompasses first the adsorption of a 333–500 nm photon by the chromophore MTHF, second the energy transfer from the blue light photon to the Flavin cofactor (FADH-), and third the electron transfer from FADH- to the cyclobutane ring, which splits the pyrimidine dimer and forms a flavin radical (FADH·). The catalytic cycle is completed when the electron is transferred back to the cofactor, restoring catalytically active, fully reduced FADH- (53–55). The entire reaction takes ∼1 ns for both types of PLs (53). The repair-independent binding of PLs to CPDs or 6-4PPs and their very fast damage removal makes PLs an attractive tool to study UV-induced damage and its repair. However thus far, PLs have mainly been used to test the specific cellular responses to either CPD or 6-4PP after removing the other type of lesion by PR (56–59).
Here we show that fluorescently labelled PLs provide a versatile and sensitive tool to locate, quantify and repair UV-induced DNA damage in real time in living cells. Fluorescence recovery after photobleaching (FRAP)-based mobility studies of PLs allow to quantitatively determine DNA damage load, as well as repair kinetics. Furthermore, we show that PLs can be activated by the 405 nm laser light during live cell imaging experiments to photo-reactivate DNA damage, which facilitates studying the behavior of NER factors and the DNA damage response upon DNA repair in living cells.
MATERIALS AND METHODS
Cell lines and constructs
VH10 hTERT immortalized human fibroblasts, XP4PA SV40 immortalized XP-C fibroblasts and HCT116 human colon cancer cells were cultured in DMEM/F10 and RPMI/F10 media, respectively, containing 10% FCS and 1% penicillin–streptomycin in a humidified incubator at 37°C and 5% CO2. To generate a lentiviral 6-4PP-PL-mCherry-3xNLS-HA expression vector, Arabidopsis thaliana 6-4PP-PL cDNA (60), missing the first 57 nucleotides corresponding to a mitochondrial localization signal, was first cloned into pENTR/D-TOPO vector (Invitrogen). mCherry-3xNLS-HA was ordered as gBlocks Gene Fragment (Integrated DNA technologies) and ligated to the C-terminal end of the 6-4PP-PL cDNA in the pENTR/D-TOPO vector, using AscI and EcoRI. Then 6-4PP-PL-mCherry-3xNLS-HA was cloned into pLenti CMV Puro DEST using Gateway cloning (Invitrogen). CPD-PL-mCherry (48), or 6-4PP-PL-mCherry-3xNLS-HA expressing lentiviral vectors were used to make the corresponding lentiviruses using the third generation system (61). GFP-DDB2 expressing VH10 cells (44), or GFP-XPC expressing HCT116 cells were transduced with the generated lentiviruses and cells stably expressing CPD-PL-mCherry or 6-4PP-PL-mCherry-3xNLS-HA were selected with puromycin.
GFP-XPC expressing HCT116 cells were generated by a CRISPR/Cas9 mediated knock-in strategy where HCT116 cells were co-transfected with a lentiCRISPR v2 vector containing an XPC guide RNA (5′-GCTCGGAAACGCGCGGCCGG-3′) targeting right after the XPC start codon and a linearized homology-directed repair (HDR) template. GFP-XPC DDB2-/- HCT116 cells were generated by transfection of GFP-XPC HCT116 cells with a lentiCRISPR v2 vector containing a DDB2 guide RNA (5′-TATTACGCCCCAGGAACAAG-3′). The HDR template to generate a GFP-XPC knock-in was generated in a single PCR step using 200 bp primers. The primers were designed in a way that 30 bp of each primer anneals to the FLAG-GFP construct and the remaining 170 bp anneals to the human genomic XPC sequence. Furthermore, the PAM sequence was mutated by 5 silent mutations which were introduced in the region targeted by the XPC gRNA to prevent Cas9 cutting the integrated HDR template. The following primers were used to generate the HDR template: forward primer (5′CCGCAGTTTTTTAGTGGCCACGGGTATGGGGTGGAGCTTCCTTTAGGGGCGTGACTAGGCCTCCAACGAAGGGGCGTGGCCAAGCGCACCGCCTCGGGGCGGGGCCGGCGTTCTAGCGCATCGCGGCCGGGTGCGTCACTCGCGAAGTGGAATTTGCCCAGACAAGCAACATGGACTACAAGGACGACGATGACAAGGTG-3′), reverse primer (5′-GCCTCTGGGCCTCCTCCGCCCACCGGCGGCGTCTCCCGCGAAGCCCGCTGGGCCTCGCTCTCACCCTCCTCCTCCTCCTCACGCCGGGCCTTGCTCTTGGCCTTGGATTTCTGGCTGCGCAGTTCGCGTCCCCGCGGCTCCCCGCCTGCGGCTCTCTTCCGAGCGAGATGCTTGTACAGCTCGTCCATGCCGAGAGTGAT-3′). The PCR generated template was cloned into pCR-Blunt II-TOPO vector and then the vector was digested with EcoRI to generate the linearized GFP-XPC construct.
Transfected HCT116 cells were selected by puromycin for 2 days and stable GFP-XPC expressing cells were FACS sorted. Then single cell clones were picked and clones were selected using genotyping, and western blotting to check for expression of the full-length GFP-XPC protein and the concomitant loss of wild type XPC expression.
RNA interference
Cells were transfected with the indicated siRNAs (150 pmol) using RNAiMax (Thermo Fisher Scientific) 48–72 h prior to the experiment, according to manufacturer's protocol. The siRNAs were purchased from Dharmacon: control (siGENOME Non_Targeting siRNA#5, D-001210-05) and XPF (siGENOME ERCC4 siRNA, M-019946-00).
Inflication of UV-induced DNA damage
Cells were washed with PBS, and after PBS removal the cells were exposed to UV-C light from a 254 nm germicidal lamp (Philips). Local UV-C damage was inflicted through an isopore membrane filter (Millipore) with a pore size of 5 μm (62).
Photo-reactivation
After PBS wash, cells were covered with a thin layer of HBSS (Thermofisher) and then placed at a distance of 10 cm under white-light tubes (General Electric Lightning Polylux LX F36W/840) for 10 min at 37°C. Mock-treated samples were covered with aluminum foil during photo-reactivation (PR).
Western blotting
Cells were lysed in 2× sample buffer and boiled for 10 min at 95°C. The proteins were subsequently separated by SDS-PAGE and transferred to PVDF membranes (0.45 μm). Membranes were blocked with 5% BSA in PBS-T (PBS containing 0.05% Tween 20) for 1 h at room temperature (RT) and blotted with the following primary antibodies: CPD-PL and 6-4PP-PL (rabbit polyclonal, 1:500) (58,59), RFP mCherry (rat monoclonal, 1:1000, 5F8, Chromotek), DDB2 (rabbit monoclonal, 1:1000, EPR981, abcam), Ku70 (goat polyclonal, 1:1000, M-19, sc-1487, Santa Cruz), tubulin (mouse monoclonal, 1:3000, B-5-1-2, sc-23948, Santa Cruz), XPC (rabbit polyclonal, 1:1000, A301-122A, Bethyl) or XPF (mouse monoclonal, 1:500, 3F2/3, sc-136153, Santa Cruz). After five times washing with PBS-T, the membranes were blotted with the following corresponding secondary antibodies from Sigma Aldrich: CF™ 680 Goat anti-Rabbit IgG (1:5000) and CF™ 770 Goat anti-Mouse IgG (1:5000). The blots were imaged with the Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence
Cells were grown on 24 mm coverslips and fixed in 2% paraformaldehyde containing PBS Triton X-100 (0.1%). After five times washing with PBS Triton X-100, the coverslips were blocked in PBS+ (PBS containing 0.15% glycine and 0.5% BSA). A denaturation step of 5 min using freshly diluted NaOH (0.07 M) in PBS was performed to make DNA lesions accessible for the CPD (mouse monoclonal, 1:1000, TDM-2, Cosmo Bio) or 6-4PP (mouse monoclonal, 1:300, 64M2, Cosmo Bio) primary antibodies. Following an incubation of 1–2 h at RT with primary antibodies diluted in PBS+, the coverslips were washed with PBS Triton X-100 five times and PBS+ once. Then the coverslips were incubated with 488, 555 or 639 Alexa Fluor secondary antibody conjugates (Invitrogen) diluted in PBS+ for 1 h at RT. After the coverslips were washed again as described above, they were embedded in Vectashield Mounting Medium with DAPI (Vector Laboratories). The coverslips were imaged using a LSM 700 microscope equipped with a Plan-Apochromat 40×/1.3 NA oil immersion lens (Carl Zeiss MicroImaging Inc.). The ImageJ software (63) was used to quantify the CPD and 6-4PP signals in the generated images. The DAPI signal was used to determine the nuclei and the mean fluorescence intensities measured in the nuclei were used to plot the graphs. For CPD and 6-4PP removal assay, cells were globally UV irradiated with 10 J/m2 and 16 J/m2 respectively, and fixed after the indicated time points. CPD and 6-4PP staining was performed as described above. Fluorescence levels were quantified in at least 70 cells per sample by measuring the background-corrected overall nuclear fluorescence, which was set at 100% for 0 h after UV irradiation.
Colony survival assay
Cells were seeded in triplicate in six-well plates (300 cells/well) and treated with the indicated UV-C doses the next day. After one week, the colonies were fixed and stained with 0.1% Brilliant Blue R (Sigma), and counted using GelCount (Oxford Optronix Ltd.).
Live cell confocal laser-scanning microscopy
All live cell imaging experiments were performed at 37°C and 5% CO2 using a Leica SP5 laser-scanning confocal microscope with a 63×/1.4 NA HCX PL APO CS oil immersion objective. Fluorescence recovery after photobleaching (FRAP) experiments were performed as described previously (42), in short; a narrow strip (512 × 16 pixels at zoom 9) along the nucleus was bleached 94 ms with 100% power of 488 nm laser for the GFP and 42 ms with 100% power of 561 nm laser for the mCherry signal. The signal in this strip was measured pre-bleach for 3.6 s and post-bleach for 20 s every 400 ms with 0.2% power of the 488 nm laser for GFP-XPC. FRAP of the mCherry-tagged PLs was performed by measuring pre-bleach for 2.5 s and post-bleach for 20 s every 100 ms with 3% power of the 561 nm laser for the mCherry signal of the PLs. To analyze fluorescence recovery, measured fluorescence intensities were first background corrected, then normalized to the average pre-bleach fluorescence signal which was set at 1. Immobile fractions were calculated using the following formula: Immobile fraction (%) = 1 − ((average fluorescence intensity of UV-C irradiated cells − the first post-bleach data point)/(average fluorescence intensity of mock-treated cells − the first post-bleach data point)). The average fluorescence intensities are calculated over the measurements of the last 10 s. For local repair during live cell imaging experiments, the fluorescence intensity of PL-mCherry was monitored every 2.585 s, both inside and outside the local damage within the nucleus (at zoom 10). PR was performed by exposure of the DNA damage to five frames of 5% 405 nm laser light. The power output of the 405 nm laser was measured to be 0.063 mW at 10% laser power. Data were corrected for background fluorescence signal outside the cell and normalized to average fluorescence signal at the local damage before PR, which was set at 1.
RESULTS
Generation and characterization of CPD-PL and 6-4PP-PL-expressing cells
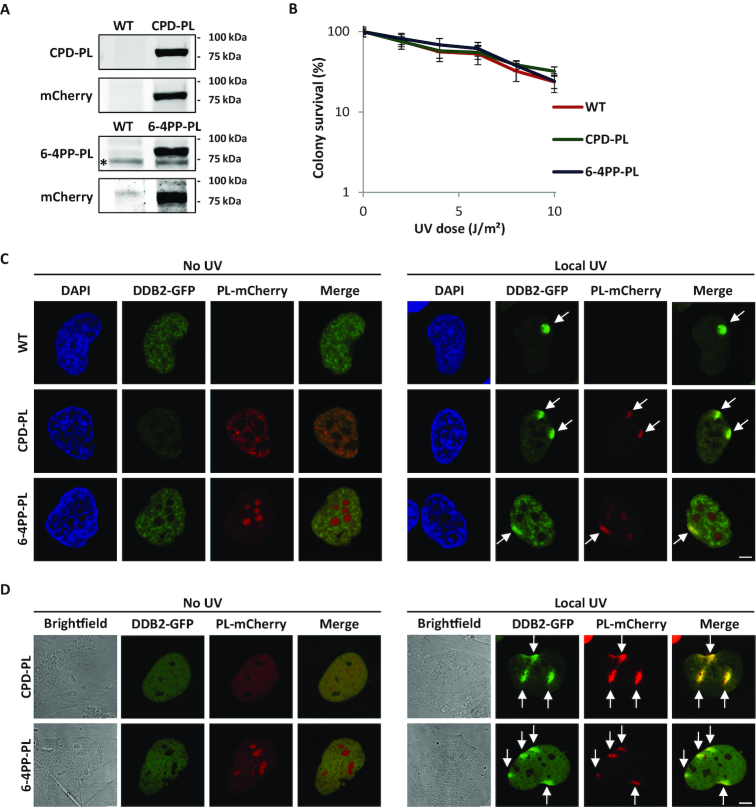
To develop a method to quantify UV-induced DNA damage and its repair kinetics directly in living cells, we first tested whether the ability of PLs to specifically detect UV-induced lesions could be exploited to generate live cell damage markers by fluorescently labeling them. For this purpose, we generated lentiviral vectors (64) expressing Potorous tridactylis CPD-PL (58) or Arabidopsis thaliana 6-4PP-PL (65) tagged with mCherry fluorescent protein at their C-terminus. In addition, three NLS sequences were added after the mCherry-tag of the 6-4PP PL to ensure nuclear expression. These lentiviruses were used to transduce GFP-DDB2 expressing VH10 (hTERT immortalized human fibroblast) cells (44) to stably express either CPD PL-mCherry or 6-4PP PL-mCherry (referred as CPD-PL or 6-4PP-PL, respectively). Western blot analysis showed that the generated VH10 cell lines express full-length PL-mCherry fusion proteins (Figure 1A). To be able to use these PL-mCherry proteins as UV damage markers, it is important that PL expression does not interfere with NER-mediated repair of UV-induced lesions. As shown by UV colony survival experiments, both CPD-PL and 6-4PP-PL-expressing cells showed a similar UV sensitivity as wild type (WT) VH10 cells (Figure 1B), indicating that the expression of these fusion proteins does not affect endogenous DNA repair activity. To corroborate this, we compared the kinetics of 6-4PP and CPD removal in PL-expressing cells and WT VH10 cells (Supplementary Figure S1A and B). This revealed that the endogenous repair of CPDs and 6-4PPs by NER was similar in PL-expressing cells and in wild type cells, but was strongly diminished in NER deficient XP-C cells.
Figure 1.
Characterization of mCherry-tagged photolyase-expressing cells. (A) Western blot of lysates of VH10 cells stably expressing GFP-DDB2 (WT), or co-expressing GFP-DDB2 and either CPD-PL-mCherry (CPD-PL, upper panel, expected size 75kDa) or 6-4PP-PL-mCherry (6-4PP-PL, lower panel, expected size 85kDa). Blots were stained with the indicated antibodies. Relevant marker sizes are indicated and * indicates an unspecific band. (B) UV-C sensitivity of WT or PL-expressing VH10 cells, determined by colony-forming ability (mean ± SEM). Percentage of surviving cells is plotted against the applied UV-C dose, colony number at 0 J/m2 is set at 100%. (C and D) Representative images of WT and PL-expressing VH10 cells. Cells were either non-irradiated (no UV, left panel) or locally irradiated with 60 J/m2 UV-C (Local UV, right panel). Cells were either fixed directly after DNA damage induction (C) or monitored directly by live cell imaging (D). Arrows indicate local UV damage. Scale bar: 5 μm.
The fusion to a mCherry tag allowed direct visualization of PLs and showed that both CPD-PL and 6-4PP-PL are expressed mainly in the nucleus (left panels of Figure 1C and D). While the CPD-PL is excluded from the nucleoli, the 6-4PP-PL was enriched in the nucleoli (bottom panels of Figure 1C and D), however, the mechanism behind this different nucleolar localization is unknown. As PLs bind CPDs and 6-4PPs light independently, but need white light to initiate catalysis, we subsequently tested whether the PLs were capable of binding to UV-induced DNA damage, while cells were kept in the dark. Both CPD-PL and 6-4PP-PL accumulated at local UV damage induced through micropore filters (62), as shown by a co-localization with the DNA damage recognizing protein DDB2 (Figure 1C, right panel). Of note, the exogenous expression of the PLs did not block DDB2 recruitment to sites of DNA damage. Furthermore, both CPD-PL and 6-4PP-PL co-localized with the respective lesion-specific antibodies (Supplementary Figure S1C). Importantly, CPD-PL and 6-4PP-PL were efficiently recruited to locally induced DNA damage in living cells (Figure 1D, right panel), demonstrating that PLs can be used to directly detect UV-induced CPD and 6-4PP lesions in living cells, which is not possible with photo lesion-specific antibodies.
CPD-PL and 6-4PP-PL as quantitative, real-time, damage and repair markers in living cells
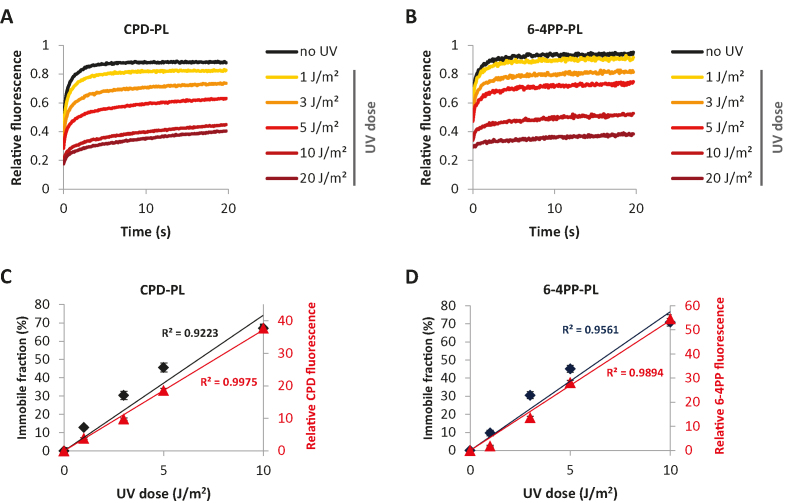
Both CPD-PL and 6-4PP-PL were able to precisely detect the UV-induced DNA damage without interfering with NER activity (Figure 1B-D, Supplementary Figures S1A and B). Binding of repair proteins to DNA damage generally immobilizes them on chromatin, which can be quantified by fluorescence recovery after photo-bleaching (FRAP) (43,66). Therefore, we performed FRAP experiments to quantitatively assess differences in the chromatin-bound fraction of PLs in response to different UV doses. FRAP of PLs showed that both CPD-PL and 6-4PP-PL are highly mobile in unperturbed cells (no UV) (Figure 2A and B), indicating that PLs are not stably bound to chromatin in the absence of DNA damage. Interestingly, both CPD-PL and 6-4PP-PL were immobilized in a dose-dependent manner after UV irradiation (Figure 2A and B). From these FRAP curves, we determined the immobile fractions of the PLs (Supplementary Figure S2A and B), which revealed a linear increase for both CPD-PL and 6-4PP-PL with increasing UV doses up to 10 J/m2. To assess whether the PL immobilization correlates with the actual quantity of CPDs and 6-4PPs, we quantified the relative amount of CPDs and 6-4PPs induced at these UV doses by immunofluorescence using photo lesion-specific antibodies (32) (Supplementary Figure S2C–E). Importantly, this revealed that the PL immobilization determined by PL FRAP (Figure 2C and D, primary Y-axis) at the indicated UV doses correlates very well with antibody-detected CPD and 6-4PP damage loads (Figure 2C and D, secondary Y-axis). This shows that FRAP of PLs allows a direct, relative quantification of UV-induced DNA damage in living cells. Interestingly, above 10 J/m2, hardly any increase in PL immobilization was observed. This is most likely caused by limiting amounts of non-bound PLs at higher UV doses, in line with the almost complete immobilization of PLs at 10 J/m2 (Figure 2A and B). This may indicate that PL expression levels influence UV-induced PL immobilization. To test this, we compared the UV-induced PL immobilization in cells with low and high PL expression levels. This revealed that PL expression levels determine the dynamic range of PL mobility (Supplementary Figure S2F and G). Cells with low PL expression levels showed an increased immobilization at lower UV doses (e.g. 1 and 3 J/m2). However, this dose-dependent increase in immobilization levelled off around 5 J/m2. (Supplementary Figure S2F and G, left panels). In contrast, cells with high PL expression showed a reduced immobilization at low UV doses, but PL immobilization continued to increase at high damage loads (e.g. 10–20 J/m2) (Supplementary Figure S2F and G, right panels). These experiments demonstrate the importance of using cells with similar PL expression levels to avoid variation due to differences in the dynamic range of PL immobilization.
Figure 2.
mCherry-tagged PLs as quantitative and real-time damage markers in living cells. (A and B) UV dose-dependent immobilization of CPD-PL (A) and 6-4PP-PL (B). PL-expressing VH10 cells were non-irradiated or global UV-irradiated with the indicated UV doses and were directly analyzed by FRAP. The plotted values were normalized over the average pre-bleach signal (n = 20 cells from two independent experiments). (C and D) Immobile fractions of CPD-PL (C) and 6-4PP-PL (D) at the indicated UV-C doses were plotted together with the relative quantity of CPD and 6-4PP lesions at the same UV-C doses as determined by the mean fluorescence intensities in immunofluorescence assays using lesion-specific antibodies.
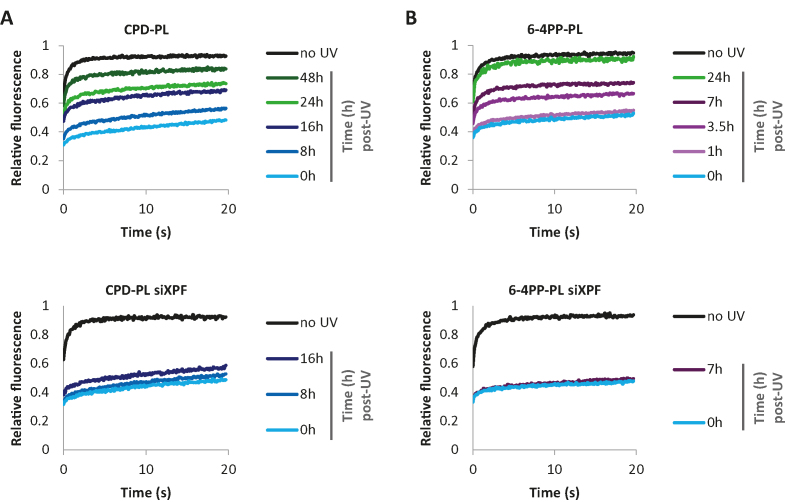
To test whether FRAP of PLs can also be used to study live-cell repair kinetics of CPDs and 6-4PPs, we UV irradiated cells and determined PL immobilization in time (Figure 3, Supplementary Figure S3A and B). In line with the previous experiments, both CPD-PL and 6-4PP-PL were strongly immobilized immediately upon UV exposure (10 J/m2). As expected, this immobilization decreased over time, reflecting the repair of CPD and 6-4PP lesions. While 6-4PP-PL was quickly mobilized, with a 50% reduction at 7h post UV and an almost complete mobilization at 24h post UV, the reduction in binding to damaged DNA by CPD-PL was much slower, in line with previously shown differences in repair rates of CPD and 6-4PP lesions (42,44,67). The mobilization of PLs over time was almost completely blocked by siRNA-mediated depletion of the NER factor XPF (Figure 3 lower panel, Supplementary Figure S3A and B, lower panel and Supplementary Figure S3C), indicating that the mobilization of PLs in time represented repair of CPD and 6-4PP by NER. These results demonstrate that FRAP of PLs enables the real-time monitoring of DNA damage load in living cells and thus provides a sensitive method to detect perturbations of the NER reaction in living cells.
Figure 3.
mCherry-tagged PLs to monitor DNA repair in living cells (A) CPD-PL and (B) 6-4PP-PL expressing VH10 cells were transfected with the control (upper panel) or XPF siRNAs (siXPF) (lower panel). The plotted PL mobilities in non-irradiated or globally UV-C irradiated (10 J/m2) cells were determined by FRAP at the indicated time points post UV irradiation (n ≥ 25 cells from 2 independent experiments for control siRNA experiment, n ≥ 15 for siXPF).
Lesion-specific repair of UV-induced DNA damage in living cells
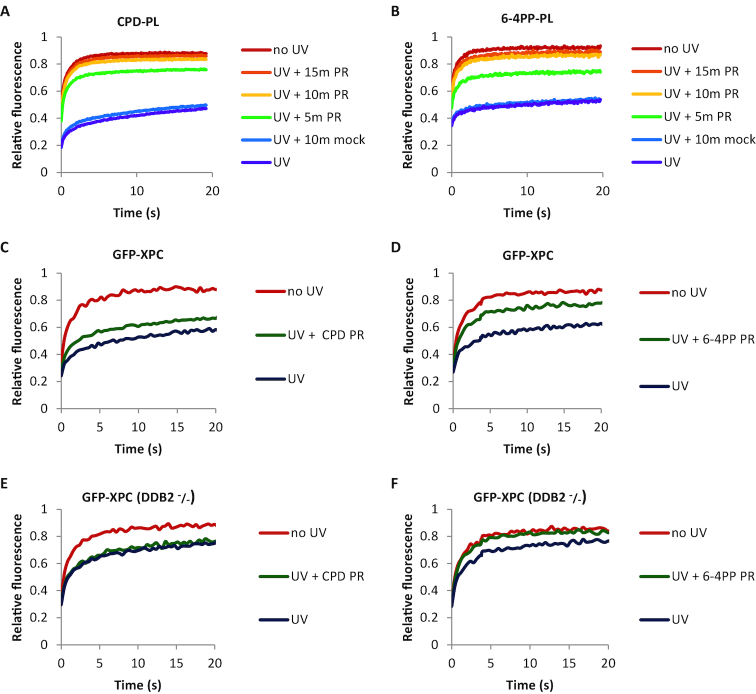
In addition to their use as quantitative live-cell damage markers, PLs can be used to specifically remove either CPD or 6-4PP lesions by direct reversal of the DNA damage using energy from near UV light (300–500 nm) (68,69). First, we determined optimal PR times for the repair of CPD and 6-4PP lesions (Figure 4A and B). PL-expressing cells were UV-irradiated and DNA damage was photo-reactivated with white light for the indicated times. While 5 min of PR was not enough for complete removal of DNA damages, 10 min PR resulted in a PL mobility similar to that of non-irradiated cells, indicative of an almost complete removal of DNA lesions (Figure 4A and B, Supplementary Figure S4A and B). Of note, the PL mobility was not affected in cells that were shielded from the white light during PR (UV + 10 min mock).
Figure 4.
Lesion-specific repair of UV-induced DNA damage in living cells. (A and B) Mobility of (A) CPD-PL and (B) 6-4PP-PL as determined by FRAP analysis. Cells were non-irradiated (no UV), globally UV-C irradiated with 10 J/m2 (UV), or globally UV-irradiated with 10 J/m2 and photo-reactivated (UV + PR) for the indicated times by exposure to white light at 37°C. Cells were directly analyzed by FRAP after each treatment. “UV + 10 min mock” cells were UV-irradiated and mock photo-reactivated by shielding from white light during PR. (n ≥ 20 cells from two independent experiments) (C and D) GFP-XPC mobility was determined by FRAP in non-irradiated, globally UV-C irradiated (10 J/m2), or globally UV-C irradiated (10 J/m2) and photo-reactivated (10 min PR) in CPD-PL (C) or 6-4PP-PL (D) expressing cells. (E and F) Similarly, GFP-XPC mobility analysis was performed in DDB2 deficient cells (GFP-XPC DDB2–/–) (n ≥ 20 cells from 2 independent experiments).
Having determined the optimal PR conditions, we assessed the previously described PR specificity of each PL. For this purpose, UV-induced DNA damage was photo-reactivated and the CPD or 6-4PP lesions were detected using immunofluorescence with specific antibodies. As expected, we observed an almost complete loss of CPDs following PR in CPD-PL-expressing cells, while the quantity of 6-4PP lesions was not affected (Supplementary Figure S4C). In 6-4PP-PL-expressing cells, removal of only 6-4PP lesions, but not of CPD, was observed upon PR (Supplementary Figure S4C).
After confirming that PL-expressing cells can specifically repair CPD or 6-4PP lesions, we made use of this feature to study live-cell DNA binding kinetics of XPC, the main damage sensor in GG-NER (4). For this purpose, we co-expressed GFP-XPC and CPD-PL or 6-4PP-PL in HCT116 cells (Supplementary Figure S4D, left panel), and performed FRAP experiments to simultaneously asses the mobility of mCherry-tagged PLs and GFP-tagged XPC (Figure 4C and D). As shown in the FRAP curves (Figure 4C and D) and the respective immobile fractions (Supplementary Figure S4E and F), UV irradiation led to the binding of GFP-XPC to damaged DNA resulting in its immobilization. Upon PR of each type of photo lesion, GFP-XPC immobilization was reduced, however not to the same extent as in non-irradiated cells. This is most likely due to the fact that XPC has affinity for both CPD and 6-4PP lesions (4,70,71). PR of 6-4PPs and CPDs was successful as shown by the mobilization of both PLs upon PR (Supplementary Figure S4G and H). We observed more increase in the GFP-XPC mobilization upon PR of 6-4PPs compared to CPDs. Although XPC is able to directly recognize 6-4PP lesions, DDB2 facilitates this recognition and is crucial for XPC to detect CPD lesions (5,6,72). This suggests that the residual damage binding of XPC, following 6-4PP removal, represents DDB2-mediated binding to CPDs. To test this, we performed the same FRAP experiments in DDB2-deficient cells (Supplementary Figure S4D, right panel). In line with a stimulatory effect of DDB2 on XPC damage recognition, the UV-induced XPC immobilization was reduced by approximately 50% in DDB2 deficient cells (Figure 4E and F, Supplementary Figure S4I and J). Furthermore, in the absence of DDB2, GFP-XPC immobilization fully recovered after PR of 6-4PPs (Figure 4F and Supplementary Figure S4J), indicating that the UV-induced immobilization of XPC in DDB2-deficient cells is caused solely by 6-4PPs. In contrast, PR of CPDs (Supplementary Figure S4I) did not affect the XPC immobilization (Figure 4E), which confirms that XPC does not bind CPDs in DDB2-deficient cells. These experiments illustrate firstly, that the PL-mediated removal of specific UV-induced lesions can provide important quantitative insights into the behavior of NER factors on specific types of DNA lesions. Secondly, the direct comparison of PL and XPC mobility by FRAP in the same cell following the same UV exposure illustrated that fluorescently-labelled PLs can quantify DNA damage with a bigger dynamic range than XPC, as shown by the bigger immobile fraction of PLs (Supplementary Figure S4K). Thirdly, the mobility of PLs was not affected by the presence or absence of DDB2, which like PLs directly binds to DNA lesions (3) (compare Supplementary Figure S4G and I, and Supplementary Figure S4H and J). This shows that the dynamic range of PLs as live cell damage markers is not influenced by competitive substrate binding of PLs and DDB2.
Local repair of UV-induced DNA damage in living cells
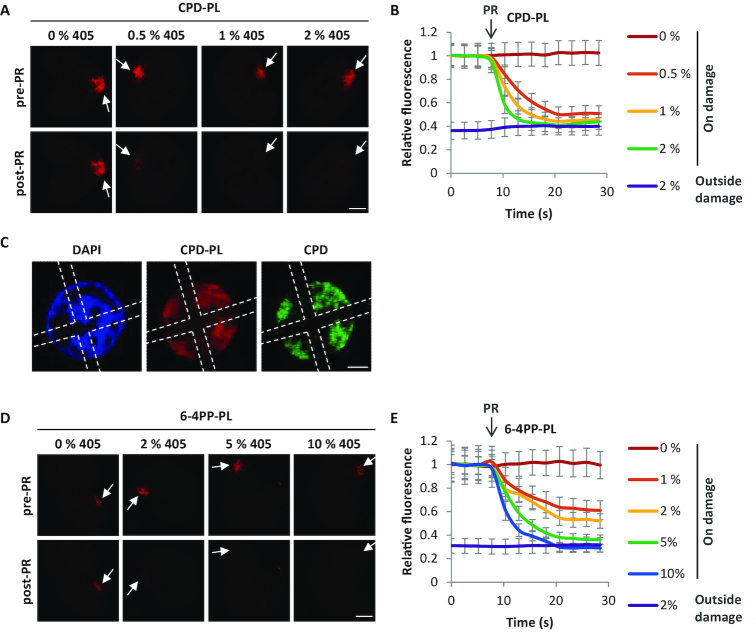
A limitation of the PR-based DNA damage removal described above is that full PR takes ∼10 minutes and needs to be performed before live-cell imaging. This interferes with the real-time measurement of the effects of DNA damage removal on proteins of interest in the cells. To improve our system, we set out to perform PR during live-cell imaging. As Potorous tridactylis CPD-PL and Arabidopsis thaliana 6-4PP-PL have absorption spectra that peak between 360 and 450 nm (65,73), we tested whether it was possible to remove UV-induced damage by activating the PLs using a 405 nm laser during live cell imaging. CPD-PL-expressing cells were locally UV-irradiated resulting in accumulation of CPD-PL at sites of DNA damage (Figure 5A, upper panel). These locally accumulated CPD-PLs were subsequently exposed to different intensities of the 405 nm laser, which almost instantaneously released the damage-accumulated PLs already at 0.5% 405 nm laser power (Figure 5A and B), reaching complete PR at 1% laser power. To exclude that the loss of fluorescence at the damage site was caused by photo bleaching of mCherry, the 405 nm laser was also activated at a region outside the damage within the nucleus, which did not result in any reduction in signal intensity (Supplementary Figure S5A). To further confirm that PL exposure to the 405 nm laser induced CPD removal by PR, we first photo-reactivated damaged DNA in a specific region (marked with the cross) in the nucleus and then stained the cells with a CPD specific antibody (Figure 5C). CPD lesions within the marked area were completely removed. In line with this, the mCherry signal of CPD-PL was reduced in the 405 nm laser-exposed region. This can be explained by its release and its subsequent binding to the areas in the nucleus where the damage is not removed. Additionally, 6-4PP lesions could also be removed upon PR by 6-4PP-PL, however, this required slightly higher 405 nm laser intensities (>5%) (Figure 5D and E, and Supplementary Figure S5B).
Figure 5.
Local repair of UV-induced DNA damage in living cells. (A) Representative images of CPD-PL-expressing VH10 cells before and 13 s after PR (pre-PR and post-PR, respectively) using 405 nm laser at the indicated intensities. Arrows indicate local UV damage. Cells were locally UV-irradiated (60 J/m2), the local DNA damage spot and a region of the exact same size outside the damage within the nucleus were exposed after 7.8 s (indicated by arrow and PR) to the indicated intensities of 405 nm laser for 13 s. Scale bar: 5 μm.(B) Relative mCherry fluorescence signal of CPD-PL was quantified inside and outside the DNA damage within the nucleus and normalized to pre-PR intensities at the local damage (n = 8 cells, mean ± SEM). (C) Representative immunofluorescence images of CPD-PL-expressing VH10 cells after PR. Global UV-irradiation (10 J/m2) of the cells was followed by photo-reactivation of the damaged DNA by 5% 405 nm laser for 13 s in a specific region (region marked with dotted line) in the nucleus and the cells were subsequently fixed and stained with CPD antibody using immunofluorescence. (D) Representative images of locally UV-irradiated (60 J/m2) 6-4PP-PL-expressing VH10 cells before and 13 s after PR (pre-PR and post-PR, respectively) using 405 nm laser at the indicated intensities as described above in Figure 4A. Arrows are indicating the local UV-C damage spots. Scale bar: 5 μm. (E) Relative mCherry fluorescence signal of 6-4PP-PL was quantified inside and outside the DNA damage within the nucleus and normalized to pre-PR intensities at the local damage (n = 8 cells, mean ± SEM).
Importantly, this live-cell PR is compatible with GFP imaging, as the PR-based repair is hardly triggered by the 488 nm laser at intensities that are commonly used for imaging GFP-tagged factors (Supplementary Figure S5C-H). Altogether, these results show that PLs can be used to photo-reactivate UV-induced DNA damage in real-time in living cells, using the 405 nm laser. In conclusion, while the induction of DNA damage in living cells has been an available tool for many years (74) and resulted in many important mechanistic insights in the repair reaction, in this study, we introduce the repair of specific UV-induced DNA damage in living cells as a unique tool to study the dissociation of DNA repair factors and behavior of other cellular processes upon damage removal.
DISCUSSION
The currently available assays to investigate UV-induced DNA damage and repair have proven to be invaluable tools to study NER factors in both fundamental and clinical research. However, these assays cannot be performed in living cells, and are therefore confined to endpoint measurements instead of monitoring the DNA damage quantities in real time. Therefore, in this study, we developed a novel method using fluorescently-tagged PLs to directly recognize and quantify UV-induced DNA damage in a highly sensitive manner in living cells.
For this purpose, we made use of the high affinity of PLs for UV-induced DNA damage, which was confirmed by their accumulation at locally induced UV damage (Figure 1C and D), and their immobilization on damaged DNA during FRAP. Both CPD-PL and 6-4PP-PL showed a strong and reproducible UV dose-dependent immobilization. This approach allowed to quantitatively monitor the relative DNA damage loads (Figure 2A-D) and NER-mediated repair kinetics in a highly sensitive manner (Figure 3). FRAP of PLs proved to be highly sensitive and enabled the detection of physiological relevant damage loads as low as 1 J/m2, which are difficult to quantify with other techniques. Furthermore, we observed a linear and relatively large dynamic range of PL immobilization between 0 and 10 J/m2 UV, enabling precise quantification of the DNA damage loads. Of note, at higher UV doses (20 J/m2) the dose-dependent immobilization was not linear anymore, which might be caused by limiting amounts of non-chromatin bound PLs, in line with the almost complete immobilization of PLs at 10 J/m2 (Figure 2A and B).
In line with this, in cells with higher PL expression levels this levelling off of PL immobilization at higher UV doses was reduced, indicating that cells with higher PL levels are more suitable to quantify high damage loads (>5 J/m2) (Supplementary Figure S2F and G, right graphs). On the other hand, our experiments show that cells with low PL expression levels allow a more sensitive detection of low damage loads (<5 J/m2) (Supplementary Figure S2F and G, left graphs). Together these data show that the dynamic range of FRAP-based UV damage detection using fluorescently-tagged PLs can be adjusted to experimental needs by choosing cells with distinct PL expression levels. PL expression levels can easily be fine-tuned in the used lentiviral transduction system by the choice of promoter (64). These data furthermore show the importance of using cells with similar PL expression levels when studying PL kinetics in different conditions. To achieve a very homogenous expression of fluorescently-tagged PLs, thereby potentially even increasing the precision of PL-mediated damage quantification, CRISPR/Cas9-mediated genomic targeting of PL expression cassettes to safe harbor loci like ROSA26 or AAVS1 (75) could be used.
Furthermore, mutated PLs that are still capable of binding the UV-induced DNA damage, but are incapable of PR, might be developed, as these catalytically dead PLs will be insensitive to unintentional day light exposure during experimental handling. Of note, the FRAP-based PL assay is already very sensitive, as shown by the direct comparison of the PL immobilization to that of XPC (Supplementary Figure S4K), the main damage sensor of GG-NER (4). XPC is one of NER factors that shows the highest immobilization on UV-damaged DNA (42,76,77), however our FRAP data show that the fluorescently labeled PLs detect damage with even higher sensitivity than XPC.
The precise correlation between PL immobilization and DNA damage load is most likely explained by the fact that exogenously expressed PLs, which function as single proteins, are most likely not regulated by the activity of other proteins, post-translational modifications or other forms of regulation in mammalian cells. Especially, these types of regulation have been shown to influence the direct correlation of the NER factor immobilization with the DNA damage quantity (17–19,45,47–49,78). The direct recognition of DNA damage with high affinity makes PLs ideally suited to visualize DNA damage in both living and fixed cells. Fluorescently-tagged PLs can therefore be used as sensitive, lesion-specific quantitative damage markers while studying the accumulation of other proteins at sites of local UV-induced damage (62,74).
Of note, although the PLs have high affinity for DNA damage and are highly immobilized on damaged DNA, they did not interfere with NER. No differences were observed in the UV survival of parental and PL-expressing cells (Figure 1B). In line with this, PL expression did not block the accumulation of DDB2 on local UV damages (Figure 1C) or inhibit the NER-mediated repair, as shown by PL immobilization in time (Figure 3). The absence of interference with the NER reaction might be explained by a transient binding of the PLs to DNA damage, thereby allowing NER factors to access DNA lesions. In line with this, even though a large fraction of the PLs were immobilized following UV exposure, these PLs were not long-term immobilized on chromatin but were rather continuously released and rebound as evidenced by the continuous increase in the FRAP curves over time (Figure 2A and B). Overall, our data showed that the use of fluorescently-labeled PLs is a robust and sensitive new method for the direct detection and quantification of UV-induced DNA damage in living cells.
In addition to their use as sensitive DNA damage markers, the fluorescently-tagged PLs can also be used to revert the UV-induced damage by PR. In this case, the fluorescent tag could be used to monitor directly the DNA damage reversion by assessing PL immobilization. This feature allowed us to determine the minimally required PR times for CPD-PL and 6-4PP-PL (Figure 4A and B). Our results showed that 10–15 min of PR by exposure to white light was enough for both PLs to repair almost all of the lesions (Figure 4A and B). This minimal PR duration is much shorter than the PR times of 1–4 h used in most previous studies performed in mammalian cells (6,79–83). The lesion-specific repair by CPD-PL and 6-4PP-PLs can be used to investigate the lesion-specific behavior of NER factors (6,84). These fluorescently-tagged PLs can be also used in combination with NER factors with different fluorescent tags, to simultaneously study the dynamic behavior of NER factor of interest while confirming the successful PR of CPD or 6-4PP lesions by PLs in the same FRAP experiment. As a proof of principle, we compared the GFP-XPC kinetics before and after PR, by simultaneously monitoring PR-mediated repair by PL mobility (Figure 4E and F). This approach could be applied to investigate the behavior of any fluorescently-tagged repair factor while monitoring the progress of either PR-based or endogenous repair over time in living cells.
Despite the short required PR times, white light mediated-PR is technically incompatible with live cell imaging. To study cellular processes directly upon, or even during PR we developed PR using a 405 nm laser as a new method to repair DNA lesions during live-cell imaging. As the CPD-PL and 6-4PP-PL absorption spectra peak between 360 and 450 nm, we photo-reactivated PLs using the 405 nm laser, which is commonly available in live cell imaging setups. Using laser pulses as short as 12.5 s, with relatively low laser power, allowed us to efficiently photo-reactivate DNA damage in live cell imaging experiments (Figure 5). Of note, while the 405 nm laser light can also be used to generate DNA damage, (85,86), the laser intensity used for efficient PR is more than 10-fold lower than the intensities required to induce DNA damage (74). Our results indicated that PR is rather specific for 405 nm laser, as PLs were not efficiently activated by the 488 nm laser at settings normally used for imaging GFP-tagged factors (Supplementary Figure S5C-H).
Thus far, mainly association kinetics (Kon) of NER factors were studied using local DNA damage infliction during live cell imaging (74). This has been a powerful tool to study the accumulation kinetics and recruitment order of fluorescently-tagged NER factors and has revealed crucial information about molecular mechanism and interdependencies of NER factors (41,87). Thus far the dissociation kinetics (Koff) of NER factors following DNA repair were more difficult to address, as the endogenous NER-mediated repair is expected to happen in a stochastic manner over time. Interestingly, our 405 nm laser-assisted live-cell repair method enables to almost instantaneously remove DNA damage. This approach can be used to gain important insights into the release of NER factors, repair times and stability of NER intermediates. In addition, as the PLs are lesion-specific, these parameters could be specifically attributed to CPD or 6-4PP lesions. PR activity of the CPD and 6-4PP-PLs can also be used simultaneously to repair the vast majority of UV-induced lesions directly. This allows testing whether specific cellular effects are caused by the DNA damage itself, or by other types of damages generated by UV exposure, such as membrane, protein, or RNA damage (88–90). Additionally, 405 nm laser-mediated PR can be used to instantly repair sub-nuclear regions, which could be used to determine the contribution of DNA damage (in cis) or signaling pathways (in trans) to transcription inhibition, replication stress, or other cellular effects following UV damage (91–93).
In conclusion, here we describe how fluorescently-labeled PLs can be used as highly sensitive UV-induced DNA damage markers to quantitatively determine damage load and repair in real-time, in living cells. Moreover, the instant repair of DNA damage by activating PLs during live cell imaging opens new possibilities to assess the cellular effects following damage removal. In addition, lentiviral expression is highly efficient to stably express PLs in a broad range of cell lines. Overall, the methods described here are a valuable extension of the current toolbox to study factors involved in the UV-induced DNA damage response, and will contribute to a better understanding of the molecular mechanism of NER in living cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Optical Imaging Centre (OIC) at the Erasmus MC for their support with microscopes and image analysis and Dr A. Theil for FACS sorting. Dr H. Lans generously provided us with the lentiviral CPD-PL-mcherry construct. We thank Prof. B. van der Horst and Dr I. Chaves for 6-4PP Photolyase cDNA and antibodies against 6-4PP and CPD PLs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Oncode Institute which is partly financed by the Dutch Cancer Society; Dutch Organization for Scientific Research ZonMW TOP Grant [912.12.132]; TOP ALW grant [854.11.002]; Horizon Zenith [935.11.042]; VIDI ALW [864.13.004]; NWO Graduate Programme Erasmus MC – Medical Genetics Grant [022.004.002]; Erasmus MC fellowship. Funding for open access charge: Dutch Science Organization (NWO).
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. [DOI] [PubMed] [Google Scholar]
- 2. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014; 15:465–481. [DOI] [PubMed] [Google Scholar]
- 4. Sugasawa K., Ng J.M., Masutani C., Iwai S., van der Spek P.J., Eker A.P., Hanaoka F., Bootsma D., Hoeijmakers J.H.. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998; 2:223–232. [DOI] [PubMed] [Google Scholar]
- 5. Wakasugi M., Kawashima A., Morioka H., Linn S., Sancar A., Mori T., Nikaido O., Matsunaga T.. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002; 277:1637–1640. [DOI] [PubMed] [Google Scholar]
- 6. Fitch M.E., Nakajima S., Yasui A., Ford J.M.. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003; 278:46906–46910. [DOI] [PubMed] [Google Scholar]
- 7. Vermeulen W., Fousteri M.. Mammalian transcription-coupled excision repair. Cold Spring Harbor Perspect. Biol. 2013; 5:a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 9. Yokoi M., Masutani C., Maekawa T., Sugasawa K., Ohkuma Y., Hanaoka F.. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000; 275:9870–9875. [DOI] [PubMed] [Google Scholar]
- 10. Volker M., Mone M.J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., van Zeeland A.A., Mullenders L.H.. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001; 8:213–224. [DOI] [PubMed] [Google Scholar]
- 11. Compe E., Egly J.M.. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012; 13:343–354. [DOI] [PubMed] [Google Scholar]
- 12. Sugasawa K., Akagi J., Nishi R., Iwai S., Hanaoka F.. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 2009; 36:642–653. [DOI] [PubMed] [Google Scholar]
- 13. Camenisch U., Dip R., Schumacher S.B., Schuler B., Naegeli H.. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat. Struct. Mol. Biol. 2006; 13:278–284. [DOI] [PubMed] [Google Scholar]
- 14. Staresincic L., Fagbemi A.F., Enzlin J.H., Gourdin A.M., Wijgers N., Dunand-Sauthier I., Giglia-Mari G., Clarkson S.G., Vermeulen W., Scharer O.D.. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009; 28:1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G. et al.. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010; 37:714–727. [DOI] [PubMed] [Google Scholar]
- 16. Moser J., Kool H., Giakzidis I., Caldecott K., Mullenders L.H., Fousteri M.I.. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell. 2007; 27:311–323. [DOI] [PubMed] [Google Scholar]
- 17. van Cuijk L., Vermeulen W., Marteijn J.A.. Ubiquitin at work: the ubiquitous regulation of the damage recognition step of NER. Exp. Cell Res. 2014; 329:101–109. [DOI] [PubMed] [Google Scholar]
- 18. Dijk M., Typas D., Mullenders L., Pines A.. Insight in the multilevel regulation of NER. Exp. Cell Res. 2014; 329:116–123. [DOI] [PubMed] [Google Scholar]
- 19. Dantuma N.P., van Attikum H.. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016; 35:6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mandemaker I.K., Vermeulen W., Marteijn J.A.. Gearing up chromatin: A role for chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus. 2014; 5:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steurer B., Marteijn J.A.. Traveling rocky Roads: The consequences of Transcription-Blocking DNA lesions on RNA polymerase II. J. Mol. Biol. 2017; 429:3146–3155. [DOI] [PubMed] [Google Scholar]
- 23. Novarina D., Amara F., Lazzaro F., Plevani P., Muzi-Falconi M.. Mind the gap: keeping UV lesions in check. DNA Repair. 2011; 10:751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Boer J., Hoeijmakers J.H.. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000; 21:453–460. [DOI] [PubMed] [Google Scholar]
- 25. Lehmann A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003; 85:1101–1111. [DOI] [PubMed] [Google Scholar]
- 26. DiGiovanna J.J., Kraemer K.H.. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 2012; 132:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Decordier I., Loock K.V., Kirsch-Volders M.. Phenotyping for DNA repair capacity. Mut. Res. 2010; 705:107–129. [DOI] [PubMed] [Google Scholar]
- 28. Latimer J.J., Kelly C.M.. Unscheduled DNA synthesis: the clinical and functional assay for global genomic DNA nucleotide excision repair. Methods Mol. Biol. 2014; 1105:511–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cleaver J.E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968; 218:652–656. [DOI] [PubMed] [Google Scholar]
- 30. Friedberg E.C. The discovery that xeroderma pigmentosum (XP) results from defective nucleotide excision repair. DNA Repair. 2004; 3:183. [PubMed] [Google Scholar]
- 31. Mellon I., Bohr V.A., Smith C.A., Hanawalt P.C.. Preferential DNA repair of an active gene in human cells. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:8878–8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi N., Katsumi S., Imoto K., Nakagawa A., Miyagawa S., Furumura M., Mori T.. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001; 14:94–102. [DOI] [PubMed] [Google Scholar]
- 33. Langie S.A., Knaapen A.M., Brauers K.J., van Berlo D., van Schooten F.J., Godschalk R.W.. Development and validation of a modified comet assay to phenotypically assess nucleotide excision repair. Mutagenesis. 2006; 21:153–158. [DOI] [PubMed] [Google Scholar]
- 34. Hu J., Adar S., Selby C.P., Lieb J.D., Sancar A.. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015; 29:948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mayne L.V., Lehmann A.R.. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982; 42:1473–1478. [PubMed] [Google Scholar]
- 36. Nakazawa Y., Yamashita S., Lehmann A.R., Ogi T.. A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA repair. 2010; 9:506–516. [DOI] [PubMed] [Google Scholar]
- 37. Johnson J.M., Latimer J.J.. Analysis of DNA repair using transfection-based host cell reactivation. Methods Mol. Biol. 2005; 291:321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bohr V.A., Smith C.A., Okumoto D.S., Hanawalt P.C.. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985; 40:359–369. [DOI] [PubMed] [Google Scholar]
- 39. Guo J., Hanawalt P.C., Spivak G.. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013; 41:7700–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wienholz F., Vermeulen W., Marteijn J.A.. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2017; 45:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., Vermeulen W.. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009; 186:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Cuijk L., van Belle G.J., Turkyilmaz Y., Poulsen S.L., Janssens R.C., Theil A.F., Sabatella M., Lans H., Mailand N., Houtsmuller A.B. et al.. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 2015; 6:7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vermeulen W. Dynamics of mammalian NER proteins. DNA Repair. 2011; 10:760–771. [DOI] [PubMed] [Google Scholar]
- 44. Pines A., Vrouwe M.G., Marteijn J.A., Typas D., Luijsterburg M.S., Cansoy M., Hensbergen P., Deelder A., de Groot A., Matsumoto S. et al.. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012; 199:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robu M., Shah R.G., Purohit N.K., Zhou P., Naegeli H., Shah G.M.. Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E6847–E6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niida H., Matsunuma R., Horiguchi R., Uchida C., Nakazawa Y., Motegi A., Nishimoto K., Sakai S., Ohhata T., Kitagawa K. et al.. Phosphorylated HBO1 at UV irradiated sites is essential for nucleotide excision repair. Nat. Commun. 2017; 8:16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang L., Zhang Q., Jones K., Patel M., Gong F.. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle. 2009; 8:3953–3959. [DOI] [PubMed] [Google Scholar]
- 48. Aydin O.Z., Marteijn J.A., Ribeiro-Silva C., Rodriguez Lopez A., Wijgers N., Smeenk G., van Attikum H., Poot R.A., Vermeulen W., Lans H.. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res. 2014; 42:8473–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang Y., Wang X., Bao S., Guo R., Johnson D.G., Shen X., Li L.. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17274–17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dulbecco R. Reactivation of ultra-violet-inactivated bacteriophage by visible light. Nature. 1949; 163:949. [DOI] [PubMed] [Google Scholar]
- 51. Thompson C.L., Sancar A.. Photolyase/cryptochrome blue-light photoreceptors use photon energy to repair DNA and reset the circadian clock. Oncogene. 2002; 21:9043–9056. [DOI] [PubMed] [Google Scholar]
- 52. Husain I., Sancar G.B., Holbrook S.R., Sancar A.. Mechanism of damage recognition by Escherichia coli DNA photolyase. J. Biol. Chem. 1987; 262:13188–13197. [PubMed] [Google Scholar]
- 53. Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J. Biol. Chem. 2008; 283:32153–32157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang Y., Baxter R., Smith B.S., Partch C.L., Colbert C.L., Deisenhofer J.. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17701–17706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang H., Saxena C., Quan D., Sancar A., Zhong D.. Femtosecond dynamics of flavin cofactor in DNA photolyase: radical reduction, local solvation, and charge recombination. J. Phys. Chem. B. 2005; 109:1329–1333. [DOI] [PubMed] [Google Scholar]
- 56. Sancar G.B. Enzymatic photoreactivation: 50 years and counting. Mut. Res. 2000; 451:25–37. [DOI] [PubMed] [Google Scholar]
- 57. Eker A.P., Quayle C., Chaves I., van der Horst G.T.. DNA repair in mammalian cells: Direct DNA damage reversal: elegant solutions for nasty problems. Cell. Mol. Life Sci.: CMLS. 2009; 66:968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schul W., Jans J., Rijksen Y.M., Klemann K.H., Eker A.P., de Wit J., Nikaido O., Nakajima S., Yasui A., Hoeijmakers J.H. et al.. Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J. 2002; 21:4719–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jans J., Schul W., Sert Y.G., Rijksen Y., Rebel H., Eker A.P., Nakajima S., van Steeg H., de Gruijl F.R., Yasui A. et al.. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol.: CB. 2005; 15:105–115. [DOI] [PubMed] [Google Scholar]
- 60. Jans J., Garinis G.A., Schul W., van Oudenaren A., Moorhouse M., Smid M., Sert Y.G., van der Velde A., Rijksen Y., de Gruijl F.R. et al.. Differential role of basal keratinocytes in UV-induced immunosuppression and skin cancer. Mol. Cell. Biol. 2006; 26:8515–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L.. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998; 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mone M.J., Volker M., Nikaido O., Mullenders L.H., van Zeeland A.A., Verschure P.J., Manders E.M., van Driel R.. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001; 2:1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., Kaufman P.D.. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009; 4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakajima S., Sugiyama M., Iwai S., Hitomi K., Otoshi E., Kim S.T., Jiang C.Z., Todo T., Britt A.B., Yamamoto K.. Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998; 26:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Houtsmuller A.B., Vermeulen W.. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 2001; 115:13–21. [DOI] [PubMed] [Google Scholar]
- 67. Mitchell D.L. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 1988; 48:51–57. [DOI] [PubMed] [Google Scholar]
- 68. Sancar G.B. DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mut. Res. 1990; 236:147–160. [DOI] [PubMed] [Google Scholar]
- 69. Zhao X., Liu J., Hsu D.S., Zhao S., Taylor J.S., Sancar A.. Reaction mechanism of (6-4) photolyase. J. Biol. Chem. 1997; 272:32580–32590. [DOI] [PubMed] [Google Scholar]
- 70. Batty D., Rapic'-Otrin V., Levine A.S., Wood R.D.. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000; 300:275–290. [DOI] [PubMed] [Google Scholar]
- 71. Hey T., Lipps G., Sugasawa K., Iwai S., Hanaoka F., Krauss G.. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002; 41:6583–6587. [DOI] [PubMed] [Google Scholar]
- 72. Moser J., Volker M., Kool H., Alekseev S., Vrieling H., Yasui A., van Zeeland A.A., Mullenders L.H.. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair. 2005; 4:571–582. [DOI] [PubMed] [Google Scholar]
- 73. Yasui A., Eker A.P., Yasuhira S., Yajima H., Kobayashi T., Takao M., Oikawa A.. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994; 13:6143–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W.. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 2007; 120:2731–2740. [DOI] [PubMed] [Google Scholar]
- 75. Sadelain M., Papapetrou E.P., Bushman F.D.. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011; 12:51–58. [DOI] [PubMed] [Google Scholar]
- 76. Nishi R., Alekseev S., Dinant C., Hoogstraten D., Houtsmuller A.B., Hoeijmakers J.H., Vermeulen W., Hanaoka F., Sugasawa K.. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair. 2009; 8:767–776. [DOI] [PubMed] [Google Scholar]
- 77. Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W. et al.. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 2008; 121:2850–2859. [DOI] [PubMed] [Google Scholar]
- 78. Puumalainen M.R., Lessel D., Ruthemann P., Kaczmarek N., Bachmann K., Ramadan K., Naegeli H.. Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity. Nat. Commun. 2014; 5:3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chigancas V., Sarasin A., Menck C.F.. CPD-photolyase adenovirus-mediated gene transfer in normal and DNA-repair-deficient human cells. J. Cell Sci. 2004; 117:3579–3592. [DOI] [PubMed] [Google Scholar]
- 80. Asahina H., Han Z., Kawanishi M., Kato T. Jr, Ayaki H., Todo T., Yagi T., Takebe H., Ikenaga M., Kimura S.H.. Expression of a mammalian DNA photolyase confers light-dependent repair activity and reduces mutations of UV-irradiated shuttle vectors in xeroderma pigmentosum cells. Mut. Res. 1999; 435:255–262. [DOI] [PubMed] [Google Scholar]
- 81. de Lima-Bessa K.M., Armelini M.G., Chigancas V., Jacysyn J.F., Amarante-Mendes G.P., Sarasin A., Menck C.F.. CPDs and 6-4PPs play different roles in UV-induced cell death in normal and NER-deficient human cells. DNA Repair. 2008; 7:303–312. [DOI] [PubMed] [Google Scholar]
- 82. You Y.H., Lee D.H., Yoon J.H., Nakajima S., Yasui A., Pfeifer G.P.. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001; 276:44688–44694. [DOI] [PubMed] [Google Scholar]
- 83. Chigancas V., Miyaji E.N., Muotri A.R., de Fatima Jacysyn J., Amarante-Mendes G.P., Yasui A., Menck C.F.. Photorepair prevents ultraviolet-induced apoptosis in human cells expressing the marsupial photolyase gene. Cancer Res. 2000; 60:2458–2463. [PubMed] [Google Scholar]
- 84. Chigancas V., Lima-Bessa K.M., Stary A., Menck C.F., Sarasin A.. Defective transcription/repair factor IIH recruitment to specific UV lesions in trichothiodystrophy syndrome. Cancer Res. 2008; 68:6074–6083. [DOI] [PubMed] [Google Scholar]
- 85. Martin R.F., Kelly D.P., Roberts M., Nel P., Tursi J., Denison L., Rose M., Reum M., Pardee M.. Comparative studies of UV-induced DNA cleavage by analogues of iodoHoechst 33258. Int. J. Radiat. Biol. 1994; 66:517–521. [DOI] [PubMed] [Google Scholar]
- 86. Martin R.F., Murray V., D’Cunha G., Pardee M., Kampouris E., Haigh A., Kelly D.P., Hodgson G.S.. Radiation sensitization by an iodine-labelled DNA ligand. Int. J. Radiat. Biol. 1990; 57:939–946. [DOI] [PubMed] [Google Scholar]
- 87. Luijsterburg M.S., Dinant C., Lans H., Stap J., Wiernasz E., Lagerwerf S., Warmerdam D.O., Lindh M., Brink M.C., Dobrucki J.W. et al.. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009; 185:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schwarz T. UV light affects cell membrane and cytoplasmic targets. J. Photochem. Photobiol.. B, Biol. 1998; 44:91–96. [DOI] [PubMed] [Google Scholar]
- 89. Pattison D.I., Davies M.J.. Actions of ultraviolet light on cellular structures. Exs. 2006; 131–157. [DOI] [PubMed] [Google Scholar]
- 90. Wurtmann E.J., Wolin S.L.. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009; 44:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sirbu B.M., Cortez D.. DNA damage response: three levels of DNA repair regulation. Cold Spring Harbor Perspect. Biol. 2013; 5:a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tresini M., Marteijn J.A., Vermeulen W.. Bidirectional coupling of splicing and ATM signaling in response to transcription-blocking DNA damage. RNA Biol. 2016; 13:272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.