Abstract
While the number of human miRNA candidates continuously increases, only a few of them are completely characterized and experimentally validated. Toward determining the total number of true miRNAs, we employed a combined in silico high- and experimental low-throughput validation strategy. We collected 28 866 human small RNA sequencing data sets containing 363.7 billion sequencing reads and excluded falsely annotated and low quality data. Our high-throughput analysis identified 65% of 24 127 mature miRNA candidates as likely false-positives. Using northern blotting, we experimentally validated miRBase entries and novel miRNA candidates. By exogenous overexpression of 108 precursors that encode 205 mature miRNAs, we confirmed 68.5% of the miRBase entries with the confirmation rate going up to 94.4% for the high-confidence entries and 18.3% of the novel miRNA candidates. Analyzing endogenous miRNAs, we verified the expression of 8 miRNAs in 12 different human cell lines. In total, we extrapolated 2300 true human mature miRNAs, 1115 of which are currently annotated in miRBase V22. The experimentally validated miRNAs will contribute to revising targetomes hypothesized by utilizing falsely annotated miRNAs.
INTRODUCTION
MicroRNAs have a major regulatory impact on gene expression by facilitating sequence-specific RNA interference. Mediated by Argonaute proteins and other components of RISC (RNA-induced Silencing Complex), miRNAs bind complementary sequences within mRNA transcripts, which results in decreased expression levels of target proteins (1–3). Variations in miRNA levels have been reported for patients’ solid tissues, blood and other body fluids, making miRNAs promising candidates for markers in a manifold of diseases (4–15). The still increasing information density on miRNAs necessitates collecting sequences and annotations in public databases. The reference repository miRBase, currently holds information about 1917 human precursors and 2656 mature miRNAs (release 22) (16).
Since the start of the miRNA registry that later developed into miRBase (17), the number of deposited miRNAs has constantly increased mostly due to high-throughput sequencing of small RNAs. The challenge of the exploding number of miRNAs is an increase of false-positive entries in miRBase and other databases. Since the best possible evidence for each miRNA is among the primary aims of miRBase (18), explicit action was taken since release 5 to reduce the number of false-positives. In 2014, miRBase defined criteria for high-confidence miRNAs, which represented only 16% of the human miRNAs annotated in release 21 (19). In 2018, a new version of miRBase was released, which incorporated additional sequencing data that have been considered to annotate all miRNAs, leading to 26% high-confidence human miRNA annotations (16). Notably, there is a striking drop of high-confidence miRNAs in later versions of miRBase. The increasing number of questionable miRNAs in late miRBase releases apparently results from the aforementioned increasing use of NGS-based approaches (20). This challenge was also reported for mouse miRNAs. Here, nearly a third of the annotations in miRBase version 14.0 was discounted as non-authentic miRNAs (21).
Certainly, there is a need for universal definition of criteria to define true miRNAs (22–25). Consistent naming system and precise in silico prediction models can contribute to the quality of miRNA databases. Both highly specific databases (such as miRGeneDB containing a few but high-likely miRNAs (26)) and sensitive databases (such as miRCarta that aims at providing high-likely miRNAs but also a broad collection of published miRNA candidates (27)) are required. However, the quality of respective miRNA databases finally always depends on the availability of highly reliable positive and negative training sets, i.e. miRNAs that have been verified by suitable experimental methods. Like NGS-based approaches, polymerase chain reaction (PCR) also entails an elevated risk of identifying false-positive miRNAs due to its nature as an amplification based approach (28,29). Northern blotting (NB) represents a rather solid technique for the detection of single miRNAs. However, there is an obvious decline of miRNAs detected by NB due to its time-consuming design and its consequently low-throughput character. Only for 3.6% of all human miRNAs listed in miRBase V22, there is evidence of miRNA expression by NB, according to experiments listed in miRBase. Frequently, only endogenous expression of a RNA fragment matching the miRNA sequence is provided (17,20,26,30).
To estimate the total number of human miRNAs, we used a high-throughput in silico and a low-throughput experimental approach. First, 28 866 human small RNA sequencing data sets containing 363.7 billion sequencing reads were mapped to the human genome and to 24 127 mature miRNA candidates. After excluding likely false-positives, we selected 108 miRNA precursors giving rise to 205 mature forms, i.e. 84 known and 119 novel candidates to be tested for processing in HEK 293T cells. Endogenous expression of 11 selected miRNAs (5 of which were not tested in HEK 293T) was analyzed in 11 additional human cell lines derived from different tissues including, liver, lung, prostate, bone marrow, cervix, placenta, mammary gland, testis, B- and T-lymphocytes, and keratinocytes. For NB, we used radiolabeled probes designed to detect both the precursors and the according 5p and 3p mature forms. A sketch of the study set-up is presented in Figure 1.
Figure 1.
Workflow of the analysis to estimate the number of true human miRNAs. Samples containing NGS data were collected from SRA, TCGA and data uploaded to miRMaster, and the obtained samples and reads were filtered. Three sets of miRNAs were created: miRBase high-confidence (HC), miRBase low-confidence set (LC) and other (Other). Using in silico high-throughput validation and experimental low-throughput validation steps, the probabilities that a miRNA will pass the validation procedure have been calculated for each miRNA set respectively. Finally, the number of true miRNAs was estimated using the original miRNA counts and the computed probabilities.
MATERIALS AND METHODS
miRBase-data analysis
All data from miRBase used in this study are available at the miRBase download section (http://www.mirbase.org/ftp.shtml). Changes between two subsequent miRBase releases are collected in miRNA.diff files. miRNA.dead files contain entries that have been removed from the database. Data accumulation and comparisons were carried out both manually and bioinformatically. If not mentioned explicitly, all analyses done in this manuscript are performed on mature miRNAs.
miRCarta/miRMaster analysis
To obtain a collection of as many as possible NGS data sets, we integrated data from miRCarta (27), as well as additional data from the sequence read archive that was added between February and November 2017. A detailed description of the data collection and filtering can be found in our recently published study (31). In brief, the sample collection stems from three different sources, i.e. the sequence read archive (32), the cancer genome atlas (TCGA) and data sets analyzed with miRMaster (33). miRMaster contains data sets from users that applied it for data analysis and volunteered to make their data available in aggregated form for secondary analysis. Altogether, 28 866 human small RNA sequencing data sets containing 363.7 billion reads were integrated. A stringent quality filtering was applied to verify the data integrity. The majority of reads had to match to the human genome (to avoid contamination by other species that erroneously have been annotated as human samples) and of those the majority was not allowed to match to mRNAs (to avoid transcriptome sequencing erroneously annotated as small RNA sequencing and low quality data sets that contain many fragmented mRNAs). From the remaining 20 488 data sets, valid mature miRNAs have been described by applying three criteria on the read profile of their precursors. To this end, we mapped the reads of all data sets against all precursors and allowed no mismatches. We normalized the expression of each read to reads per million mapped to miRNAs to account for different sequencing depth and library composition. First, we determined the 5′ homogeneity of the dominant miRNA and required that over 50% of the normalized reads that mapped to the miRNA start at the same 5′ position. Thereby, we accounted for the fact that miRNAs have a very low 5′ end variability. Second, we considered the reads that did not map in accordance with Dicer processing. Therefore, we determined which fraction of the reads did not map with a variability of two bases at the 5′ end and five bases at the 3′ end to the annotated miRNAs. If this fraction accounted for more than 25% of the normalized reads, we discarded the precursor. Third, we determined the number of valid stacks that could be found by evaluating the coverage profile of the precursors. We defined a stack as the longest stretch of bases for which the most covered base differed from the lowest covered one by at most 20%. A stack was considered valid if it spanned between 16 and 29 bases. We required at least one valid stack per precursor. This criteria accounts for coverage profiles that exhibit clear read stacks, as expected from miRNA precursors. Finally, we kept all miRNAs that resulted from the precursors that fulfilled all criteria.
IsomiR analysis
IsomiR variants for miRNAs of miRBase V22 were determined using miRMaster (33), based on the 20 488 NGS data sets described above. Briefly, reads were mapped to all annotated miRNA precursors while allowing up to two non-template additions at both ends and one mismatch in between. IsomiRs were then determined relative to the coordinates of the annotated miRNAs in miRBase. Reads were counted when their mapping position differed at most two nucleotides at the 5′ end and five nucleotides at the 3′ end. Finally, an isomiR was determined to be present when totaling at least 2% of the total reads per million mapped to miRNA normalized counts.
Construction of miRNA expression vectors
Inserts for miRNA expression vectors with pSG5 backbone (Stratagene, now Agilent Technologies, Santa Clara, California) were synthesized and cloned into pEX-A2 vectors by Eurofins Genomics (Ebersberg, Germany). Therefore, hsa-miRNA precursor sequences were pasted into the UCSC genome browser BLAT tool (GRCh38/hg38 assembly) and correspondent DNA sequences with 100 additional bases up- and downstream and flanking EcoRI/BamHI/BglII restriction sites were used for custom gene synthesis by Eurofins Genomics (Ebersberg, Germany). Lyophilisates were reconstituted at 100 ng/μl with H2O and 1 μg of DNA was digested using appropriate EcoRI/BamHI/BglII restriction enzymes. Pre-mir inserts were subcloned into pSG5 vector. Positive clones were determined by colony-PCR, restriction digestion and sanger sequencing (Seq-It, Kaiserslautern, Germany and Eurofins Genomics, Ebersberg, Germany). pSG5-miRNA expression vectors that were not synthesized by Eurofins Genomics have been cloned by PCR amplification before.
Cell culture and transfection
A549, HaCaT, HeLa and HUH-7 cells were cultivated at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin, 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany). SHSY-5Y cells were cultivated at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (Life Technologies, Darmstadt, Germany) supplemented with 20% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany). MCF-7, PC-3, DG-75 and Jurkat cells were cultivated at 37°C and 5% CO2 in RPMI 1640 Medium (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany). JEG-3 cells were cultivated at 37°C and 5% CO2 in Ham’s F12 Nutrient Mixture Medium (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany). Tera-1 cells were cultivated at 37°C and 5% CO2 in McCoy’s 5A Medium (Life Technologies, Darmstadt, Germany) supplemented with 15% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany).
HEK 293T cells for transfections were purchased from Leibnitz Institute DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) and cultivated at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and antibiotics (100 U/ml Penicillin and 100 μg/ml Streptomycin) (Life Technologies, Darmstadt, Germany). A total of 2.4 × 106 cells were seeded in 100-mm dishes and transiently transfected using PolyFect Transfection Reagent (Qiagen, Hilden Germany) according to the manufacturer’s recommendations. In brief, 24 h after seeding, 8 μg of pSG5-miRNA expression plasmid DNA diluted in 300 μl of DMEM without supplements were used for the transfection. Cells and transfection complexes were incubated for 48 h at 37°C and 5% CO2 to allow for miRNA overexpression.
RNA extraction
Total RNA including miRNA from cell lines was purified manually using miRNeasy Mini Kit (Qiagen, Hilden Germany) according to the manufacturer’s protocol. Therefore, cell-culture DMEM was completely removed and the monolayer was carefully washed with 1 ml of phosphate buffered saline. About 700 μl of QIAzol Lysis Reagent was used to disrupt the cells by using a cell-scraper and vortexing. After adding 140 μl of chloroform, lysates were mixed thoroughly and centrifuged for 15 min at 12 000 g at 4°C to allow for phase separation. RNA was precipitated with a 1.5 vol. of 100% ethanol, washed and eluted in 2 × 40 μl H2O RNase-free. Quality and quantity of isolated total RNA including miRNA were determined using NanoDrop 2000 UV-Vis Spectrophotometre (ThermoFisher Scientific, Waltham, Massachusetts, USA) with A260/280 ∼2 and A260/230 ∼ 1.8 and Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) with RIN > 7.5.
Microarray analysis
miRNA abundance analysis of 12 samples was performed using Agilent microarrays for the Human miRBase V21 that contain probes for 2549 mature human miRNAs (Agilent Technologies). The procedures were performed as described previously according to the manufacturer’s recommendations (34). A total of 100 ng total RNA from 12 cell lines (HEK 293T, PC-3, Tera-1, SHSY-5Y, HUH-7, DG-75, Jurkat, HeLa, JEG-3, MCF7, HaCaT and A549) (see also section ‘Cell culture and transfection’ for details) was processed using the miRNA Complete Labeling and Hyb Kit (Agilent Technologies) to generate fluorescently labeled miRNA. The microarrays were loaded and incubated at 55°C for 20 h with rotation. After washing, microarrays were scanned with the Agilent G2565CA Microarray Scanner System at 3 μm in double path mode. Raw data were acquired using Agilent AGW Feature Extraction software version 10.10.11 (Agilent Technologies). Background subtraction and quantile normalization of raw data were performed using R scripts (version 3.0.2) (35).
Northern blotting
For NB, 20 μg of total RNA including miRNA extracted by using miRNAeasy Mini Kit (Qiagen, Hilden Germany) was separated in 12% denaturing urea-polyacrylamide gels using SequaGel UreaGel System (National Diagnostics, Nottingham, UK) and 1x TBE running buffer. All buffers and solutions for NB were prepared using DEPC-treated H2O to eliminate nuclease activity. A ssRNA marker was used to estimate the sizes of bands independently of the influence of external factors (temperature etc.) (RiboReady™ Color Micro RNA ladder, VWR, Radnor, PA, USA or Low range ssRNA Ladder and microRNA Marker, New England Biolabs, Frankfurt am Main, Germany). To check for loading control, the gel was stained with ethidiumbromide (10 mg/ml EtBr in 1× TBE) or 1× SYBR™ Gold in 1× TBE (Invitrogen/ThermoFisher Scientific, Waltham, Massachusetts, USA) and documented with a ChemiDoc Touch Imaging System (Bio-Rad, Munich, Germany). For semi-dry electroblotting, RNA was transferred to a Hybond N nylon membrane (GE Healthcare Life Sciences, Freiburg, Germany) for 30 min at 15 V. RNA was cross-linked chemically to the membrane using N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (Sigma-Aldrich, Munich, Germany) for 2 h at 55°C. After cross-linking of endogenous RNA, the membranes were cut in half to prevent overlapping during hybridization. The generation of radiolabeled RNA probes was performed using miRVana miRNA Probe Construction Kit (Ambion/ThermoFisher Scientific, Waltham, Massachusetts, USA) following the manufacturer’s instructions. Therefore, ssDNA templates composed of the full-length miRNA-of-interest sequence and an 8 nt T7 promoter sequence (5′-CCTGTCTC-3′) added to the 3′ end were hybridized to the T7 promoter primer. The remaining nucleotides corresponding to the template were added using Klenow DNA polymerase resulting in the desired dsDNA template. Next, the dsDNA template was in vitro transcribed using T7 RNA polymerase and radiolabeled UTP or GTP, if the probe was only containing two UTPs or less (Hartmann Analytic, Braunschweig, Germany). Template DNA was removed by DNase I digestion.
Pre-hybridization for 30 min was performed at 55°C in 5× SSC, 7% SDS, 1× blocking solution (Roche Diagnostics, Rotkreuz, Suisse), 20 mM Na2HPO4, 1× Denhardt’s solution (Invitrogen/ThermoFisher Scientific, Waltham, Massachusetts, USA). Hybridization of radiolabeled probes to the cross-linked RNA membranes was performed at 55°C overnight. Blots were washed twice 15 min in 5× SSC, 1% SDS and twice 15 min in 1× SSC, 1% SDS at 55°C and exposed to a storage phosphor screen overnight. Screens were documented using a Typhoon scanner (GE Healthcare Life Sciences, Freiburg, Germany). Here, contrast and brightness were automatically adjusted by Typhoon Scanner Control Software (GE Healthcare Life Sciences, Freiburg, Germany) during scanning according to the darkest spot on the area. Whole NB images in this manuscript were further manually adjusted in terms of contrast and brightness.
Estimation of the validation rate
To provide an estimate of the total number of miRNAs, we considered the three groups, A: p1 = 897 high-confidence miRNAs from the miRBase; B: p2 = 1759 low-confidence miRNAs from the miRBase; C: p3 = 21 471 miRNAs not contained in the miRBase but predicted in various other studies separately from each other. For all three groups, we computed a high-throughput based exclusion rate based on the NGS data sets mentioned above (denoted as h1, h2, h3, respectively) as well as a low throughput validation rate (l1, l2, l3, respectively). We further assumed that high-throughput exclusion and low throughput validation are independent of each other. This makes the total estimated number of miRNAs to be: 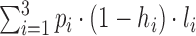 .
.
RESULTS
Exclusion of likely false-positives by high-throughput data analysis
We collected 28 866 human small RNA sequencing samples containing 363.7 billion reads. Following stringent quality filtering, 8418 samples were excluded because of wrong annotations or low data quality. The remaining samples were mapped to 14 738 human miRNA precursor candidates totaling 24 094 mature miRNA candidates. These can be split in three basic sets: A: p1 = 897 high-confidence miRNAs from miRBase V22; B: p2 = 1759 low-confidence miRNAs from miRBase V22; C: p3 = 21 471 miRNAs not contained in miRBase but predicted in various other studies. For mapping of those billions of small RNA sequencing reads, only 4.9% of high-confident miRNAs did not fulfill the quality criteria (h1 = 0.049). For low-confidence miRNAs from miRBase, this rate already increased to 38.6% (h2 = 0.386). For the set of miRNA candidates, not yet annotated in miRBase, this rate increased to 70.1% (h3 = 0.701). In total, 853 of 897 high-confidence miRBase miRNAs, 1080 of 1759 low-confidence miRBase miRNAs and 6412 of 21 471 novel candidates fulfilled the high-throughput criteria (details are provided in Supplementary Table S1).
Detection of IsomiR variants
For 2873 miRNAs from miRBase V22, we found at least one isomiR variant with additional nucleotides either at their 5p or at their 3p end (Supplementary Table S3). From our training set, miR-6829–5p was found to have the highest number (16) of isomiRs detected. miR-34a-3p showed the most isomiRs (9) from our positively validated miRNA set. The sequences and corresponding reads per million mapped (RPMMM) are listed in Table 2. The standard sequence for miR-34a-3p, designated as ‘0F_0T’ (zero changes at the five prime end and zero changes at the three prime end) was represented by 34.75% RPMMM. The next highest number of RPMMM (16.52%) was found for variant ‘0F_1T’ with one additional nucleotide at the 3p end. The third highest number of RPMMM (13.79%) was found for isomiR variant ‘1F_1T’ with one additional nucleotide at the 5p and one additional nucleotide at the 3p end. MiRNA isomiR variants were not further validated by northern blotting due to the lack of specificity of RNA probes for the nucleotide changes at 5p or 3p ends.
Table 2.
Relative isomiR read counts (> 2%) detected for hsa-miR-34a-3p
| sequence | iso_type | Relative RPMMM (%) | Total RPMMM | Total reads |
|---|---|---|---|---|
| CAAUCAGCAAGUAUACUGCCC | 0F_-1T | 4.00 | 2079.53 | 13 287 |
| CAAUCAGCAAGUAUACUGCC | 0F_-2T | 2.10 | 1089.15 | 6082 |
| CAAUCAGCAAGUAUACUGCCCU | 0F_0T | 34.75 | 18 060.38 | 93 848 |
| CAAUCAGCAAGUAUACUGCCCUA | 0F_1T | 16.52 | 8585.07 | 46 920 |
| CAAUCAGCAAGUAUACUGCCCUAG | 0F_2T | 6.09 | 3164.45 | 19 520 |
| AAUCAGCAAGUAUACUGCCCU | 1F_0T | 8.90 | 4623.40 | 22 909 |
| AAUCAGCAAGUAUACUGCCCUA | 1F_1T | 13.79 | 7167.57 | 40 377 |
| AAUCAGCAAGUAUACUGCCCUAG | 1F_2T | 5.87 | 3053.31 | 21 018 |
| AUCAGCAAGUAUACUGCCCUAG | 2F_2T | 3.20 | 1664.71 | 9701 |
Iso-types are defined as additional or absent nucleotides on 5′ (F) or 3′ (T) ends, respectively. RPMMM = Reads Per Million Mapped to MiRNAs. Iso-type 0F_0T represents the commonly known sequence from miRBase V22.
Validation of exogenous miRNAs by northern blotting
To select miRNAs for experimental validation, we employed a recently developed algorithm that acknowledges key criteria characteristic for miRNAs. The complete listing of criteria to define high-confident miRNAs is given in Backes et al. (20) (see also our implemented web-server for ranking potential miRNA candidates at www.ccb.uni-saarland.de/novomirank). After excluding precursors that did not meet the criteria of high-confident miRNAs, we employed an experimental validation step to further identify false-positive miRNAs. From each of the three abovementioned groups, we selected a representative number for low-throughput validation as described in the ‘Materials and Methods’ section. Altogether, 203 mature miRNAs (originating from 108 precursor molecules) were tested in this validation step. To this end, miRNA precursor sequences were cloned into pSG5-miRNA expression plasmids and recombinantly expressed in HEK 293T cells. Since the likelihood for true positives was highest in set A and lowest in set C, the test set sizes were selected accordingly, with respect to miRBase V21, which was the most recent version at the time of the study setup: we analyzed 51 (40 in V21) high-confidence miRNAs from miRBase V22 (set A), 33 (41 in V21) low-confidence miRNAs from miRBase V22 (set B) and 119 (122 in V21) novel miRNA candidates from set C. Out of the three miRNA candidates that were not present in V21 yet, one (hsa-miR-9903) has been added to the high-confidence set and was successfully validated by us, while the other two (hsa-miR-12129, hsa-miR-10527–5p) were added to the low-confidence set and did not pass our validation. Notably, the three miRNA precursor candidates predicted by us to encode both 5p and 3p miRNA have been added to miRBase V22 as giving rise to only one mature form. In addition to miR-9903–3p, miR-9903–5p was also positively validated by us and should be included in miRBase.
Validation of endogenous miRNAs by northern blotting
To search for endogenous miRNAs with expression levels high enough to be identifiable by northern blotting, we analyzed 12 human cell lines by microarray. In detail, miRNA abundance analysis was performed using Agilent microarrays based on miRBase V21 that contains probes for 2549 mature human miRNAs. The 12 cell lines were derived from prostate (PC-3), testis (Tera-1) bone marrow (SHSY-5Y), liver (HUH-7), lung (A549), B (DG-75)- and T-lymphocytes (Jurkat), cervix (HeLa), placenta (JEG-3), mammary gland (MCF7), keratinocytes (HaCaT) and HEK 293T (kidney) as a reference. Based on the array results, we selected 11 miRNAs with high expression levels to be detectable by northern blotting (see Table 3 for array data of selected miRNAs and Supplementary Table S3 for array data of V22 miRNAs). Notably, the miRNAs, miR-4284 and miR-1260a that were upon the most recently deposited miRNAs, i.e. with a very high ID, did not yield signals of the expected size in any of the analyzed cell lines. miR-7975 that, according to the microarray, showed the strongest expression in all 12 cell lines exhibited several potential signals but no precise assignment to mature or precursor forms was possible. All other miRNAs including 137–3p, 148a-3p, 155–5p, 16–5p, 19b-3p, 20a-5p, 23a-3p and 23b-3p yielded signals in the expected size range for the mature form as shown in Figure 2. While the miRNAs 16–5p, 19b-3p, 20a-5p, 23a-3p and 23b-3p were detected in all 12 cell lines, miR-137–3p was found in 5 cell lines only, miR-148a-3p in 10 cell lines and miR-155–5p in 5 cell lines (Figure 2B). Out of the 11 miRNAs, the miRNAs 137–3p, 148a-3p, 155–5p, 23a-3p and 23b-3p were also identified by the exogenous expression analysis.
Table 3.
Quantile normalized microarray data for selected miRNAs in 12 cell lines
| miRNA | HEK 293T | A549 | HUH7 | SHSY- 5Y | HaCaT | HeLa | MCF7 | JEG3 | Tera1 | PC3 | DG75 | Jurkat |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-16–5p | 9.733 | 9.428 | 9.733 | 9.946 | 9.501 | 9.428 | 9.946 | 7.474 | 8.832 | 9.213 | 10.403 | 11.161 |
| hsa-miR-19b-3p | 11.404 | 9.036 | 10.220 | 10.756 | 9.946 | 10.220 | 8.729 | 9.733 | 10.220 | 9.309 | 11.161 | 11.404 |
| hsa-miR-20a-5p | 10.936 | 8.832 | 10.062 | 10.550 | 9.863 | 9.501 | 8.609 | 9.428 | 9.863 | 9.036 | 10.936 | 10.936 |
| hsa-miR-23a-3p | 4.952 | 9.733 | 7.036 | 8.454 | 9.733 | 10.062 | 8.522 | 5.197 | 7.182 | 9.428 | 7.595 | 5.986 |
| hsa-miR-23b-3p | 5.578 | 9.213 | 6.527 | 7.893 | 7.437 | 8.177 | 7.437 | 4.101 | 4.481 | 7.256 | 5.777 | 5.219 |
| hsa-miR-137–3p | 0.583 | 4.467 | 2.095 | 7.474 | 1.728 | 4.345 | -0.094 | 0.148 | 0.444 | 2.142 | 0.208 | 0.118 |
| hsa-miR-148a-3p | 7.523 | 1.577 | 8.123 | 5.049 | 4.710 | 1.636 | 6.445 | 3.961 | 7.394 | 6.384 | 6.527 | 8.307 |
| hsa-miR-155–5p | -0.300 | 0.246 | -0.062 | -0.143 | 1.118 | 3.984 | -0.235 | 0.256 | 3.346 | 0.263 | 5.435 | 3.612 |
| hsa-miR-1260a | 11.891 | 11.161 | 11.891 | 11.891 | 11.404 | 11.404 | 11.891 | 11.891 | 10.936 | 11.404 | 11.404 | 10.756 |
| hsa-miR-4284 | 11.161 | 10.936 | 11.404 | 11.161 | 10.062 | 11.161 | 11.161 | 9.636 | 10.062 | 10.550 | 9.863 | 10.403 |
| hsa-miR-7975 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 | 16.004 |
Figure 2.

Northern blots of endogenous miRNAs. (A) The 11 analyzed cell lines that are indicated on top of the figure were derived from lung (A549), liver (HUH-7), bone marrow (SHSY-5Y), keratinocytes (HaCaT), cervix (HeLa), mammary gland (MCF7), placenta (JEG-3), Testis (Tera-1), prostate (PC-3), B-lymphocytes (DG-75) and T-lymphocytes (Jurkat). HEK-293T RNA was used as a reference to compare signals to exogenously expressed miRNAs. The endogenous mature forms are shown for the miRNAs indicated on the left side of the figure. (B) The number of mature and premature forms of the endogenous miRNA expressed in the 12 cells lines as indicated in Figure 2A.
Taken together, from our exogenous and endogenous experiments, we successfully validated 51 of 54 high-confidence miRNAs (94.4%; l1 = 0.944), 10 of 35 low-confidence miRNAs (28.6%, l2 = 0.286) and 22 of 119 miRNAs that have not yet been annotated in miRBase V22 (18.5%, l3 = 0.185). Details are provided in Supplementary Table S2. Our simplified model described in the ‘Materials and Methods’ section lets us estimate that 806 of the 897 miRBase high-confidence miRNAs are true positives (89.8%), 309 of the 1759 low-confidence miRNAs (17.5%) and 1185 of the 21 471 potential candidates (5.5%).
Specific analysis of mature and precursor miRBase miRNAs
Out of the 89 miRNAs from miRBase V22 (54 high- and 35 low-confidence), we detected northern blot signals for 61 mature miRNAs (51 high- and 10 low-confidence). All of these signals for mature miRNAs approximately matched the expected size range. Most of them were not discovered in the endogenous controls. mir-1260a and mir-23c showed two signals in the size range of their mature miRNAs, signal intensities virtually corresponded to those found in control cells. We defined miRNAs as positive when signals for both a precursor and a mature form were detected and are stronger for the overexpression compared to control RNA lysates. However, in the majority of cases, the size of the precursor did not correspond to the size indicated for the respective stem-loop forms by miRBase. NB examples for positive, negative and doubtful miRNAs from miRBase V22 are shown in Figure 3.
Figure 3.
Representative NB results for a positive, questionable and negative miRNA in HEK 293T cells. (A) NB for hsa-miR-155–5p from high-confidence set A showing distinct bands for its precursor (p) and mature (m) form. (B) Hybridization against hsa-miR-1260a (low-confidence set B) detects two small RNA fragments with similar signal intensities for the control. (C) Probing for hsa-miR-6776–5p (low-confidence set B) did not result in any specific bands. Ethidium bromide staining of RNA gels was used as a loading control.
Considering all cases, we found six recombinants and two endogenous miRNAs, as already described above, with signals neither in the size range of the precursor nor in the range of the mature forms. The only high-confident miRNAs that were not confirmed by our northern blot analysis were miR-6511a-5p and miR-6511a-3p, both of which showed signals for potential precursor forms, albeit in different sizes, but not for any of both mature forms and miR-26b-3p where only a premature form could be detected. These results suggest that even in the high-confidence set a certain number of false-positive miRNAs exist.
For the low-confident miRNAs, exogenous and endogenous expression analysis failed to confirm 22 and 3 miRNAs, respectively, that did not show signals for the mature and precursor form. About 19 of these had at least some signals designated as potential precursor form or unclear signals (details provided in Table 1). The remaining 10 miRNAs were confirmed by the identification of both the mature and the precursor forms. Although our analysis is largely consistent with the miRBase qualification of many low-confident miRNAs, our data also indicate a considerable number of false-negative miRNAs among the low-confident set in miRBase.
Table 1.
Comparison between NB signal intensities of 5p versus 3p mature miRNA and corresponding precursor forms from set A (high-confidence), set B (low-confidence) and set C (miRNA candidates)
| set | miRNA (candidate) | pre | mat | pre versus mat | pre | mat | pre versus mat |
|---|---|---|---|---|---|---|---|
| A | 10a | moderate | strong | weaker | weak | strong | weaker |
| A | (endo) 16 | moderate | strong | weaker | – | – | – |
| A | (endo) 19b | – | – | – | weak | strong | weaker |
| A | (endo) 20a | – | – | – | weak | strong | weaker |
| A | 23a | moderate | weak | stronger | weak | strong | weaker |
| A | 26b | moderate | moderate | weaker | moderate | n. d. | pre only |
| A | 27b | weak | weak | equal | strong | weak | stronger |
| A | 34a | moderate | strong | weaker | moderate | weak | stronger |
| A | 101 | weak | near bg | stronger | moderate | weak | stronger |
| A | 122 | weak | strong | weaker | moderate | moderate | stronger |
| A | 125a | weak | strong | weaker | moderate | strong | weaker |
| A | 137 | – | – | – | moderate | strong | weaker |
| A | 140 | weak | weak | weaker | weak | weak | weaker |
| A | 142 | moderate | moderate | stronger | near bg | moderate | weaker |
| A | 143 | moderate | moderate | equal | strong | weak | stronger |
| A | 145 | weak | strong | weaker | weak | weak | weaker |
| A | 148a | strong | weak | stronger | weak | strong | weaker |
| A | (endo) 148a | – | – | – | weak | moderate | weaker |
| A | 155 | moderate | strong | weaker | moderate | weak | stronger |
| A | 181a | moderate | strong | weaker | weak | weak | stronger |
| A | 191 | weak | near bg | stronger | strong | near bg | stronger |
| A | 193a | moderate | moderate | stronger | weak | strong | weaker |
| A | 195 | moderate | strong | weaker | weak | weak | stronger |
| A | 205 | strong | moderate | stronger | strong | strong | equal |
| A | 301a | weak | weak | stronger | moderate | strong | weaker |
| A | 361 | moderate | strong | weaker | moderate | weak | stronger |
| A | 375 | – | – | – | moderate | strong | weaker |
| A | 483 | moderate | moderate | equal | strong | strong | stronger |
| A | 497 | weak | strong | weaker | weak | near bg | stronger |
| A | 874 | weak | weak | stronger | near bg | near bg | equal |
| A | 6511a | moderate | n. d. | pre only | weak | n. d. | pre only |
| A | 9903 | strong | strong | weaker | weak | strong | weaker |
| B | 23b | moderate | weak | stronger | weak | strong | weaker |
| B | #23c | – | – | – | strong | weak | stronger |
| B | #133b | – | – | – | moderate | strong | weaker |
| B | #630 | – | – | – | near bg | n. d. | pre only |
| B | #665 | – | – | – | near bg | n. d. | pre only |
| B | 939 | n. d. | n. d. | – | n. d. | n. d. | – |
| B | #1202 | weak | near bg | stronger | – | – | – |
| B | 1228 | weak | n. d. | pre only | weak | n. d. | pre only |
| B | 1229 | moderate | n. d. | pre only | strong | weak | stronger |
| B | 1238 | weak | weak | equal | weak | n. d. | pre only |
| B | #1246 | weak | near bg | stronger | – | – | – |
| B | #1260a | n. d. | near bg | mat only | – | – | – |
| B | #3137 | weak | weak | equal | – | – | – |
| B | #3148 | moderate | strong | weaker | – | – | – |
| B | 3162 | weak | moderate | weaker | n. d. | n. d. | – |
| B | #4534 | – | – | – | weak | n. d. | pre only |
| B | #4721 | – | – | – | moderate | n. d. | pre only |
| B | 6776 | n. d. | n. d. | – | n. d. | near bg | mat only |
| B | 6829 | near bg | n. d. | pre only | weak | n. d. | pre only |
| B | 6865 | n. d. | moderate | mat only | weak | n. d. | pre only |
| B | 10527 | weak | moderate | weaker | n. d. | moderate | mat only |
| B | 12129 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | novel-241 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | novel-1002 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | novel-1037 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | novel-1043 | near bg | n. d. | pre only | weak | near bg | weaker |
| C | novel-1236 | weak | moderate | stronger | weak | n. d. | pre only |
| C | novel-1521 | n. d. | n. d. | – | near bg | n. d. | pre only |
| C | novel-1564 | n. d. | moderate | mat only | n. d. | near bg | mat only |
| C | novel-1790 | weak | near bg | stronger | n. d. | n. d. | – |
| C | novel-1887 | near bg | weak | weaker | weak | n. d. | pre only |
| C | novel-2295 | moderate | weak | stronger | weak | weak | weaker |
| C | pnm-18 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-339 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-501 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-1089 | near bg | strong | weaker | near bg | strong | weaker |
| C | pnm-1523 | n. d. | weak | mat only | n. d. | near bg | mat only |
| C | pnm-1609 | weak | n. d. | pre only | n. d. | n. d. | – |
| C | pnm-1728 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-2012 | moderate | near bg | stronger | weak | moderate | weaker |
| C | pnm-2523 | near bg | near bg | stronger | near bg | near bg | stronger |
| C | pnm-2908 | n. d. | near bg | mat only | n. d. | n. d. | – |
| C | pnm-3375 | n. d. | weak | mat only | n. d. | n. d. | – |
| C | pnm-4607 | moderate | strong | weaker | n. d. | near bg | mat only |
| C | pnm-4828 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-4927 | weak | near bg | stronger | strong | moderate | stronger |
| C | pnm-6141 | n. d. | n. d. | – | n. d. | near bg | mat only |
| C | pnm-6147 | n. d. | n. d. | – | weak | near bg | stronger |
| C | pnm-6785 | n. d. | n. d. | – | weak | near bg | stronger |
| C | pnm-7379 | weak | moderate | weaker | moderate | moderate | equal |
| C | pnm-7519 | moderate | weak | stronger | n. d. | near bg | mat only |
| C | pnm-8500 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-8679 | strong | moderate | stronger | weak | weak | equal |
| C | pnm-8692 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-8893 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-9262 | weak | near bg | stronger | n. d. | n. d. | – |
| C | pnm-10387 | n. d. | n. d. | – | near bg | near bg | equal |
| C | pnm-10468/-71/-72 | n. d. | n. d. | – | n. d. | near bg | mat only |
| C | pnm-10470 | n. d. | n. d. | – | n. d. | near bg | mat only |
| C | pnm-10565 | strong | near bg | stronger | n. d. | near bg | mat only |
| C | pnm-10945 | n. d. | n. d. | – | near bg | near bg | stronger |
| C | pnm-11436 | weak | near bg | stronger | weak | near bg | stronger |
| C | pnm-11712 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-12346 | n. d. | near bg | mat only | n. d. | n. d. | – |
| C | pnm-12352 | weak | near bg | stronger | weak | near bg | stronger |
| C | pnm-12395 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-13945 | weak | weak | equal | moderate | moderate | equal |
| C | pnm-14137 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-14397 | n. d. | n. d. | – | n. d. | near bg | mat only |
| C | pnm-15272 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-15546 | n. d. | weak | mat only | n. d. | weak | mat only |
| C | pnm-16556 | strong | near bg | stronger | moderate | near bg | pre only |
| C | pnm-17724 | weak | moderate | weaker | strong | weak | stronger |
| C | pnm-20077 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-20714 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-21981 | near bg | n. d. | pre only | n. d. | near bg | mat only |
| C | pnm-22155 | n. d. | n. d. | – | n. d. | n. d. | – |
| C | pnm-22472 | weak | n. d. | pre only | weak | n. d. | pre only |
| C | pnm-23093 | strong | n. d. | pre only | strong | near bg | stronger |
| C | pnm-23453 | weak | near bg | stronger | strong | n. d. | pre only |
#miRNAs are predicted to only give rise to one mature form. n. d.: not detectable, near bg: near background. miRCarta IDs of novel and miRA candidates were shown in Supplementary Table S5.
Ratio of precursor and mature -5p/-3p miRNA forms
As for variances in signal intensities, 49 miRNAs showed stronger signals for the precursor than for the mature form indicating a reduced processing efficiency (Table 1). As abovementioned, miR-6511a-5p and 24 other miRNAs showed only signals for the precursor but not for the mature form. Overall, the 5p-forms appear to be more efficiently processed into mature miRNAs than the 3p-forms. Out of the 14 miRNAs, which gave rise to one mature form only according to miRBase V22, we confirmed 4 miRNAs, 2 of which, including mir-133b and mir-3148, showing stronger signals for their mature form than for the precursor (Table 1).
Failed confirmation of miRNAs in recent miRBase releases
Grouping miRNAs to the according miRBase version of which the miRNA has first been listed highlights that mature miRNAs confirmed by our NB based approach are enriched in early miRBase versions. In detail, all 33 miRNAs that were taken from miRBase versions 1 through 7 have been experimentally validated. Version 8 of miRBase was the first version to contain a miRNA that showed no clear signals. Here, for mir-630, we only detected a very faint signal in precursor size. Out of 26 miRNAs taken from version 10, 20 have been verified by our NB analysis. Only 3 out of 20 miRNAs taken form version 17 onwards have been verified. Consistent with the doubts raised in previous studies, these results further question the nature of miRNAs with high ID numbers recently deposited in miRBase that are mostly identified by NGS studies only.
Stability of estimated human miRNome size between miRBase V21 and V22
At the time of the study setup miRBase V22 was not yet released. Therefore, we selected miRNAs for analysis in accordance to the proportions of the high- and low-confidence miRNA annotations of miRBase V21, containing 544 and 2044 miRNAs respectively. Extrapolating the miRNome size based on the data of miRBase V21, 509 miRNAs of the high-confidence set were considered true positives (93.6%), 627 of the low-confidence miRNAs (30.7%) and 1213 of the candidate miRNAs (5.6%), resulting in an estimated miRNome size of 2349 miRNAs. The release of miRBase V22 had a large impact on the proportions of the high- and low-confidence sets, increasing the size of the high-confidence set by 65% to 897 miRNAs. While the proportions changed substantially, our estimate remained stable and decreased only by 49 miRNAs (2.1%).
Likely false-positive miRNAs and their targets
There were no miRNA targets for miRNAs that were predicted in various studies but not deposited in miRBase (set C). By contrast, target genes have been annotated for almost all miRNAs taken from miRBase V21. At the time of writing, no annotations were available for miRNAs from miRBase V22. Here, we compared the targetomes of miRNAs verified by our above applied criteria versus the targetomes of miRNAs not confirmed in our analysis. To this end, we considered only miRNA targets that have been classified as verified targets by miRTarBase (strong evidence). In the set of validated miRNAs, each miRNA had a median of 144 validated target genes. In the set of the not-validated miRNAs, each had a decreased number of 65 (median) target genes. While the difference was statistically significant (P=0.009) as determined by Wilcoxon Rank-sum test, these results demonstrate substantial numbers of targets for not-validated miRNAs. For example, both mature forms of hsa-mir-939 that we could not validate in this study have been associated with complex target gene sets of 199 for the 5p and 439 targets for the 3p form. Altogether, 16 miRNAs not-validated by our approach have recently been associated with 2844 experimentally validated targets.
DISCUSSION
Messenger RNA transcripts have been frequently confirmed as endogenous molecules by NB. Due to their rather low endogenous expression few endogenous miRNAs are identifiable by NB. Although there are no studies that systematically analyzed endogenous miRNAs by NB, there are data sets listing endogenous miRNAs according to their signal intensities (e.g. GSM1513689 deposited in GEO). The overall low endogenous expression of miRNAs necessitates an exogenous expression system, which also allows monitoring the processing of the precursor into the mature form. We chose HEK 293T cell culture as expression system that stems from a kidney of a healthy aborted fetus and that allows high transfection efficiency and high expression rates for the pSG5 vector (36,37). The identification of both the precursor and the mature form by NB indicates processing of a miRNA and strongly argues in favor of a true miRNA.
The use of an exogenous expression system also allowed to systematically compare the processing of the 5p and the 3p form for each of the analyzed miRNAs. Depending on the tissue, the cell, and the applied condition, both mature forms have been reported as functional (38). In case of miRNAs for which no mature but a precursor form can be detected by our NB procedure, it is conceivable that the amount of miRNA was too low to yield distinct signals. Overall, our data indicated that the 5p-forms are more frequently processed into mature miRNAs than the 3p-forms. This observation is consistent with previous publications that analyzed miRNA strand selection with regards to thermodynamic stability (39,40). Although our validation pipeline was not optimized for the detection of splicing-derived miRNAs, we obtained positive NB signals for mirtron precursor mir-1229 and its mature 3p form but not for mir-1228 and mir-1238. To the best of our knowledge, the only studies that also identified mirtrons by NB were by Schamberger et al. (41), who reported mirtron mir-1226 as one out of three candidates tested and Hubé et al. (42) who detected 6 out of 56 short intron derived miRNA candidates. Agotrons, another potential exception for our validation system, do not appear as miRNA like signals or even not as a premature like form on northern blots as they differ in size up to 100 nt and irrespective of their association with Ago proteins. For the specific validation of exogenous agotrons via northern blotting they should be coexpressed along with Argonaute proteins for stabilizing effects (43). In contrast to the detection of single-nucleotide polymorphisms, isomiR detection is not readily possible by quantitative Real Time-PCR or northern blotting. These methods do not allow to discriminate between miRNAs with length variants of up to 5 nt at their 5p or 3p ends. Almost all of the studies describing isomiR variants use enzyme-based methods making the bias-free validation of miRNA isoforms that were detected by NGS a major challenge (44–46).
For a substantial number of miRNAs, we failed to establish confirmation by NB. This should raise the awareness that a presumed miRNA may not be a true miRNA, even if other methods, i.e. microarray or qRT-PCR, suggest high expression values or other studies already provided evidence for its functionality. For example, we failed to validate mir-939 exogenously and miR-4284 and miR-7975 endogenously, although others have already provided functional evidence for its derived miRNAs (47–53 and many others). Failed confirmation by NB does however not necessarily disqualify a miRNA as true miRNA. As addressed above, one has to differentiate between miRNAs that show a signal for the precursor miRNA only but no processing, a signal only for the processed form, and cases without any or with only faint signals. While the latter cases likely do not represent true miRNAs, even the lack of identification of the two forms does not necessarily disprove that a tested sequence represents a true miRNA. Tissue specific factors that are required for processing in a given cell may not be present in the HEK cells used (21). Finally, signal intensities also depend on the amount of miRNA expressed and processed and the number of radiolabeled nucleotides in probes. To minimize the influence of biased labeling, we used radiolabeled GTP for all oligonucleotide probes that contained only two or less UTPs.
The abovementioned limitations are also found in other systems like knockout systems for Drosha/Dicer that have been used to confirm true canonical miRNAs. In this system, a failure of processing a miRNA precursor in the Drosha/Dicer mutants may be due to the specific physiology of these mutants. A failure of processing a miRNA precursor in the according wild-type cells can likewise be linked to a specific cell type. In addition, most of the studies that use knockouts for Drosha/Dicer characterize the analyzed miRNA candidates by NGS and PCR entailing the problems related to these techniques. In sum, knockout experiments of proteins processing miRNAs do not rule out erroneous confirmation of false-positive miRNAs or erroneous disproval of real miRNAs (54).
Complementary to experimental settings that manipulate miRNA expression of a specific cell type, endogenous miRNA expression can be analyzed. To this end, we tested 11 human cell lines in addition to HEK 293T cells by northern blotting and found signals for several miRNAs in the size range of the mature sequences. For endogenously detected precursors, the sizes varied to a degree comparable to the range detected for the exogenous analyses. Notably, and as shown in Figure 4, the signal sizes of the endogenous miRNAs (mature and premature forms) correspond to those of the exogenous miRNAs further supporting the validity of the data obtained for the induced miRNAs. The relatively weak intensity of the signals of the endogenous miRNAs show, however, that only a minor portion of the miRNAs can be identified without induced overexpression.
Figure 4.
Comparison of exogenous and endogenous miRNA expression by northern blotting. The mature form is indicated by ‘m’ and the premature form by ‘p’. The left part of the figure shows exogenous expression of miR-148a-3p in HEK 293T cells. HEK 293T cells that were transfected with an empty vector are shown as a control (ctrl). The right part of the figure shows endogenous expression of miR-148a-3p in 12 cells lines as specified in Figure 2A. To show endogenous miRNAs, the signal intensity of the endogenous miRNAs apparently is enhanced as compared to the signal intensity of the exogenous miRNAs due to the very strong signal of overexpressed miR-148a-3p (compare backgrounds of exo- and endogenous northern blots and see the comparison between HEK 293T cells that were used as a control for the transfection analysis (ctrl) shown in the left part of the figure and the HEK 293T cells that were compared with other cells shown in the right part of the figure). As described in the ‘Materials and Methods’ section, contrast and brightness were adjusted by the software during scanning according to the darkest spot on the blot.
Most miRNAs that have failed the northern blot validation step were detected by less NGS reads and in less NGS samples compared to miRNAs that passed this step. However, there are also examples of miRNAs that were highly expressed in many NGS samples or microarray data sets but still failed validation by northern blotting. For example, miR-26b-3p, miR-6511a-3p and miR-1246 were all detected in over 10 000 samples, with in total more than 230 000 reads per miRNA. In addition, three miRNAs, i.e. miR-7975, miR-1260a and miR-4284, which were upon the top 16 miRNAs expressed in all 12 cell lines analyzed, failed the northern blot filtering. Furthermore, the opposite was also found. Among the validated miRNAs, miR-1202, miR-3148, miR-3137 and miR-3162–5p were all detected in <100 samples, when requiring at least 10 reads per sample. Accordingly, a read number based filtering would surely not be sufficient to eliminate false-positive miRNAs.
In the light of the different possibilities to identify and confirm miRNAs, it is certainly not justified to prematurely limit a quality control scheme to few methods. NB is certainly only one quality criterion for a true miRNA but nevertheless an essential one. Arguments for NB as a method to confirm true miRNAs are the possibility of a high degree of standardization, the visual demonstration of the product sizes, the omission of amplification and ligations steps, and the possibility to show processing from the precursor into the mature form by using an exogenous expression system. We strongly suggest including NB in future database formats. To provide highest data integrity, data repositories with NB data should provide (i) complete documentation of the NB by scanned images with brightness/contrast settings without image adjustments, (ii) the method of RNA extraction, (iii) the size of the primary transcript that is (over-) expressed in the cell type analyzed, (iv) the minimal sequence surrounding a miRNA hairpin that assures correct processing of mature miRNAs, (v) the respective cells used for validation analyses and (vi) if applicable endogenous NB signal intensities and their correlation with qRT-PCR data and/or microarray and/or NGS data.
In summary, we found the highest number of confirmed miRNAs for the high-confidence set of miRNAs of miRBase V22, particularly among the miRNAs deposited in miRBase releases 1 through 10. We also present a set of miRNAs with a precursor and the mature forms confirmed by the NB pipeline, which have not yet been annotated in miRBase. In total, our data indicate 2300 human mature miRNAs ∼50% (1115) of which are annotated in miRBase V22. Since the high- and low-throughput validation presented in this study are not independent of each other, our model represents certainly only an estimate of the total number of miRNAs. Based on the presented data, our estimation, however, likely indicates the upper number of what we can expect as the final extent of the true human miRNome.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: J.A., U.F. and M.M. performed NB experiments. J.A. performed miRNA microarrays and edited raw NB images. M.H. and M.A.-H. prepared expression plasmids for 22 and 6 miRNAs, respectively. F.A.G. provided expression plasmids for 26 miRNAs and 4 cell lines. T.F., C.B. and V.G. performed the bioinformatics analyses. V.G. prepared the graphical abstract and Figure 1. J.A., T.F., H.-P.L., A.K. and E.M. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Michael J. Fox Foundation (US), Grant ID: 14446. Funding for open access charge: Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hart M., Nolte E., Wach S., Szczyrba J., Taubert H., Rau T.T., Hartmann A., Grasser F.A., Wullich B.. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014; 12:250–263. [DOI] [PubMed] [Google Scholar]
- 5. Petriella D., De Summa S., Lacalamita R., Galetta D., Catino A., Logroscino A.F., Palumbo O., Carella M., Zito F.A., Simone G. et al.. miRNA profiling in serum and tissue samples to assess noninvasive biomarkers for NSCLC clinical outcome. Tumour Biol. 2016; 37:5503–5513. [DOI] [PubMed] [Google Scholar]
- 6. Drusco A., Nuovo G.J., Zanesi N., Di Leva G., Pichiorri F., Volinia S., Antenucci A., Costinean S., Bottoni A. et al.. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014; 9:e96670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alles J., Menegatti J., Motsch N., Hart M., Eichner N., Reinhardt R., Meister G., Grasser F.A.. miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing. FEBS Open Bio. 2016; 6:251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wen Y., Han J., Chen J., Dong J., Xia Y., Liu J., Jiang Y., Dai J., Lu J., Jin G. et al.. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int. J. Cancer. 2015; 137:1679–1690. [DOI] [PubMed] [Google Scholar]
- 9. Akers J.C., Hua W., Li H., Ramakrishnan V., Yang Z., Quan K., Zhu W., Li J., Figueroa J., Hirshman B.R. et al.. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017; 8:68769–68779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abu-Halima M., Meese E., Keller A., Abdul-Khaliq H., Radle-Hurst T.. Analysis of circulating microRNAs in patients with repaired Tetralogy of Fallot with and without heart failure. J. Transl. Med. 2017; 15:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller A., Meese E.. Can circulating miRNAs live up to the promise of being minimal invasive biomarkers in clinical settings. Wiley Interdiscip. Rev. RNA. 2016; 7:148–156. [DOI] [PubMed] [Google Scholar]
- 12. Ludwig N., Nourkami-Tutdibi N., Backes C., Lenhof H.P., Graf N., Keller A., Meese E.. Circulating serum miRNAs as potential biomarkers for nephroblastoma. Pediatr. Blood Cancer. 2015; 62:1360–1367. [DOI] [PubMed] [Google Scholar]
- 13. Abu-Halima M., Hammadeh M., Backes C., Fischer U., Leidinger P., Lubbad A.M., Keller A., Meese E.. Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil. Steril. 2014; 102:989–997. [DOI] [PubMed] [Google Scholar]
- 14. Leidinger P., Backes C., Deutscher S., Schmitt K., Mueller S.C., Frese K., Haas J., Ruprecht K., Paul F., Stahler C. et al.. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013; 14:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keller A., Leidinger P., Bauer A., Elsharawy A., Haas J., Backes C., Wendschlag A., Giese N., Tjaden C., Ott K. et al.. Toward the blood-borne miRNome of human diseases. Nat. Methods. 2011; 8:841–843. [DOI] [PubMed] [Google Scholar]
- 16. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2018; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004; 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozomara A., Griffiths-Jones S.. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Backes C., Meder B., Hart M., Ludwig N., Leidinger P., Vogel B., Galata V., Roth P., Menegatti J., Grasser F. et al.. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 2016; 44:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. et al.. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010; 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meng Y., Shao C., Wang H., Chen M.. Are all the miRBase-registered microRNAs true? A structure- and expression-based re-examination in plants. RNA Biol. 2012; 9:249–253. [DOI] [PubMed] [Google Scholar]
- 23. Wang X., Liu X.S.. Systematic curation of miRBase annotation using integrated small RNA High-Throughput sequencing data for C. elegans and drosophila. Front. Genet. 2011; 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen T.B., Kjems J., Bramsen J.B.. Enhancing miRNA annotation confidence in miRBase by continuous cross dataset analysis. RNA Biol. 2011; 8:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown M., Suryawanshi H., Hafner M., Farazi T.A., Tuschl T.. Mammalian miRNA curation through next-generation sequencing. Front. Genet. 2013; 4:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fromm B., Billipp T., Peck L.E., Johansen M., Tarver J.E., King B.L., Newcomb J.M., Sempere L.F., Flatmark K., Hovig E. et al.. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu. Rev. Genet. 2015; 49:213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Backes C., Fehlmann T., Kern F., Kehl T., Lenhof H.P., Meese E., Keller A.. miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Res. 2017; 46:D160–D167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krawczak M., Reiss J., Schmidtke J., Rosler U.. Polymerase chain reaction: replication errors and reliability of gene diagnosis. Nucleic Acids Res. 1989; 17:2197–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyerhans A., Vartanian J.P., Wain-Hobson S.. DNA recombination during PCR. Nucleic Acids Res. 1990; 18:1687–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N.. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fehlmann T., Backes C., Alles J., Fischer U., Hart M., Kern F., Langseth H., Rounge T., Umu S.U., Kahraman M. et al.. A high-resolution map of the human small non-coding transcriptome. Bioinformatics. 2017; 34:1621–1628. [DOI] [PubMed] [Google Scholar]
- 32. Kodama Y., Shumway M., Leinonen R. International Nucleotide Sequence Database, C. . The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fehlmann T., Backes C., Kahraman M., Haas J., Ludwig N., Posch A.E., Wurstle M.L., Hubenthal M., Franke A., Meder B. et al.. Web-based NGS data analysis using miRMaster: a large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017; 45:8731–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stahler C., Meese E. et al.. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016; 44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keller A., Backes C., Haas J., Leidinger P., Maetzler W., Deuschle C., Berg D., Ruschil C., Galata V., Ruprecht K. et al.. Validating Alzheimer's disease micro RNAs using next-generation sequencing. Alzheimers Dement. 2016; 12:565–576. [DOI] [PubMed] [Google Scholar]
- 36. Graham F.L., Smiley J., Russell W.C., Nairn R.. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977; 36:59–74. [DOI] [PubMed] [Google Scholar]
- 37. Thomas P., Smart T.G.. HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods. 2005; 51:187–200. [DOI] [PubMed] [Google Scholar]
- 38. Biasiolo M., Sales G., Lionetti M., Agnelli L., Todoerti K., Bisognin A., Coppe A., Romualdi C., Neri A., Bortoluzzi S.. Impact of host genes and strand selection on miRNA and miRNA* expression. PLoS One. 2011; 6:e23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Zhou Y.H., Chen W., Khaitovich P.. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009; 10:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 41. Schamberger A., Sarkadi B., Orban T.I.. Human mirtrons can express functional microRNAs simultaneously from both arms in a flanking exon-independent manner. RNA Biol. 2012; 9:1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hube F., Ulveling D., Sureau A., Forveille S., Francastel C.. Short intron-derived ncRNAs. Nucleic Acids Res. 2017; 45:4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen T.B., Veno M.T., Jensen T.I., Schaefer A., Damgaard C.K., Kjems J.. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016; 7:11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Magee R., Telonis A.G., Cherlin T., Rigoutsos I., Londin E.. Assessment of isomiR discrimination using commercial qPCR methods. Noncoding RNA. 2017; 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pillman K.A., Goodall G.J., Bracken C.P., Gantier M.P.. miRNA length variation during macrophage stimulation confounds the interpretation of results: implications for miRNA quantification by RT-qPCR. RNA. 2019; 25:232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nejad C., Pepin G., Behlke M.A., Gantier M.P.. Modified polyadenylation-based RT-qPCR increases selectivity of amplification of 3′-MicroRNA isoforms. Front. Genet. 2018; 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hou S., Fang M., Zhu Q., Liu Y., Liu L., Li X.. MicroRNA-939 governs vascular integrity and angiogenesis through targeting gamma-catenin in endothelial cells. Biochem. Biophys. Res. Commun. 2017; 484:27–33. [DOI] [PubMed] [Google Scholar]
- 48. Aghdaei F.H., Soltani B.M., Dokanehiifard S., Mowla S.J., Soleimani M.. Overexpression of hsa-miR-939 follows by NGFR down-regulation and apoptosis reduction. J. Biosci. 2017; 42:23–30. [DOI] [PubMed] [Google Scholar]
- 49. Zhang J.X., Xu Y., Gao Y., Chen C., Zheng Z.S., Yun M., Weng H.W., Xie D., Ye S.. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol. Cancer. 2017; 16:18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Chen C., Wu M., Zhang W., Lu W., Zhang M., Zhang Z., Zhang X., Yuan Z.. MicroRNA-939 restricts Hepatitis B virus by targeting Jmjd3-mediated and C/EBPalpha-coordinated chromatin remodeling. Sci. Rep. 2016; 6:35974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo Z., Shao L., Zheng L., Du Q., Li P., John B., Geller D.A.. miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5826–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang F., Nam S., Brown C.E., Zhao R., Starr R., Ma Y., Xie J., Horne D.A., Malkas L.H., Jove R. et al.. A novel berbamine derivative inhibits cell viability and induces apoptosis in cancer stem-like cells of human glioblastoma, via up-regulation of miRNA-4284 and JNK/AP-1 signaling. PLoS One. 2014; 9:e94443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y., Shen Z., Jiang H., Lai Z., Wang Z., Jiang K., Ye Y., Wang S.. MicroRNA4284 promotes gastric cancer tumorigenicity by targeting ten-eleven translocation 1. Mol. Med. Rep. 2018; 17:6569–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim Y.K., Kim B., Kim V.N.. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





