Abstract
Members of the pentatricopeptide repeat (PPR) protein family are sequence-specific RNA-binding proteins that play crucial roles in organelle RNA metabolism. Each PPR protein consists of a tandem array of PPR motifs, each of which aligns to one nucleotide of the RNA target. The di-residues in the PPR motif, which are referred to as the PPR codes, determine nucleotide specificity. Numerous PPR codes are distributed among the vast number of PPR motifs, but the correlation between PPR codes and RNA bases is poorly understood, which hinders target RNA prediction and functional investigation of PPR proteins. To address this issue, we developed a modular assembly method for high-throughput construction of designer PPRs, and by using this method, 62 designer PPR proteins containing various PPR codes were assembled. Then, the correlation between these PPR codes and RNA bases was systematically explored and delineated. Based on this correlation, the web server PPRCODE (http://yinlab.hzau.edu.cn/pprcode) was developed. Our study will not only serve as a platform for facilitating target RNA prediction and functional investigation of the large number of PPR family proteins but also provide an alternative strategy for the assembly of custom PPRs that can potentially be used for plant organelle RNA manipulation.
INTRODUCTION
Pentatricopeptide repeat (PPR) proteins constitute a large protein family, the members of which serve as single-stranded RNA (ssRNA)-binding proteins. These proteins are particularly abundant in terrestrial plants. More than 400 members of this family have been identified in Arabidopsis and rice (1). Increasing evidence have indicated that nuclear-encoded PPR proteins are targeted exclusively to mitochondria and plastids (2), where these proteins post-transcriptionally modulate gene expression by affecting RNA cleavage (3), splicing (4), editing (5), translation (6) and stability (7). Some PPRs, functioning as restorers of fertility, process aberrant mitochondrial RNA transcripts to overcome cytoplasmic male sterility (CMS) in staple crops (8–13). PPR mutants always exhibit metabolic disorders of specific organellar RNAs, leading to a myriad of plant developmental defects, such as aberrant organelle biogenesis (14,15), retarded leaf emergence, restricted root growth, delayed flowering (16), aborted embryo and endosperm development (15,17–19), defects in kernel growth (15,20) and abnormal stress response (21,22). Nevertheless, characterization of the RNA targets of PPRs to elucidate the molecular mechanism by which mutation results in unfavorable developmental phenotypes remains challenging.
PPR proteins are typically characterized by tandem degenerate repeats consisting of 35-amino-acid motifs (2). In recent years, we and other research groups have determined the crystal structures of various PPR proteins (23–27). All these structures exhibit an overall right-handed superhelical α-solenoid structure that consists of successive tandem repeats, each of which contains a pair of antiparallel α-helices. The structures of PPR proteins in complex with RNA reveal that PPRs recognize ssRNA in a modular and sequence-specific manner (23,27). Within a certain repeat, the combinatorial di-residues at the 5th and 35th positions (corresponding to residues 6 and 1′ in Barkan et al. (28) and residues 4 and ii in Yagi et al. (29)) are responsible for specific RNA base recognition. These di-residues are referred to as the PPR code. The nature of the modular repeat structure of PPR proteins provides a framework for deciphering PPR codes.
To date, several PPR codes have been biochemically validated using artificially designed PPRs (25,30). For example, threonine and asparagine (TN), threonine and aspartic acid (TD), asparagine and serine (NS), and asparagine and aspartic acid (ND) at the 5th and 35th positions correspond to the nucleotides adenine (A), guanine (G), cytosine (C) and uracil (U), respectively (2). These four types of di-residue combinations recognize the corresponding bases with Watson–Crick edges via distinct hydrogen bond networks (27). The fifth position is the major determinant of RNA base specificity. The presence of asparagine at this position results in a preference for pyrimidines, whereas the presence of serine or threonine at the same position is strongly correlated with a preference for purines. The amino acid at the 35th position is the second major determinant. The presence of asparagine at this position is correlated with base A or C, whereas aspartate exhibits a preference for base G or U (2). Although great progress has been made in elucidating the RNA base-recognition mechanisms of several PPR codes, our knowledge of PPR codes remain limited. Considering the large number of PPR codes distributed in natural PPR proteins, additional PPR codes and the correlation of these codes with RNA bases need to be further investigated.
In this study, we launched a systematic exploration to decipher PPR codes. We first analyzed the distribution frequency of PPR codes from P-type PPR proteins derived from 65 land plants and 62 codes were selected for further studies, accounting for 85% of the total code distribution frequency. We then developed a hierarchical assembly method for high-throughput construction of designer PPR (dPPR) proteins. Using this method, 62 dPPR proteins harboring the most frequent PPR codes were constructed. The RNA-binding specificity of these 62 dPPRs was examined by electrophoretic mobility shift assay (EMSA) and isothermal titration calorimetry (ITC). We characterized the correlation between RNA bases and PPR codes, which enriched the pool of PPR codes. Based on the results, we developed the online PPRCODE web server to facilitate target RNA prediction for PPR proteins (http://yinlab.hzau.edu.cn/pprcode/). The results of this study will not only serve as a platform for functional investigation of a large number of PPR proteins, but also provide an effective strategy for custom assembly of dPPRs that can potentially be used for plant organelle RNA manipulation.
MATERIALS AND METHODS
PPR protein sequence analysis
The P-type PPR protein sequences from land plants (65 species), algae (16 species) and protists (11 species) were derived from a previous report and the PPR motifs were extracted from these sequences (http://ppr.plantenergy.uwa.edu.au/) (31). PPR protein sequences from fungi (161 species) and metazoans (41 species) were obtained via the Ensembl Genomes Archive website (http://ensemblgenomes.org/). The PPR motifs of these PPR proteins were obtained via PPRCODE server analysis (please see more additional details in the section of titled ‘PPRCODE server construction’). Amino acids at the 5th and 35th positions of each PPR motif were extracted as the PPR code. These codes were used to analyze the distribution frequency.
Designer PPR (dPPR) motif construction
Individual monomers with different codes were initially optimized and amplified by overlapping polymerase chain reaction (PCR) using long primers. These monomers were cloned into a modified pPR18 vector and the clones were verified by DNA sequencing. To assemble the individual monomers in a specific order, we altered the DNA sequence based on codon degeneracy at the glycine–leucine (GL) junction between each pair of monomers to obtain unique 4-bp sticky-end ligation adapters. We performed a digestion/ligation reaction (10 μl) with 100 ng of each monomer, 0.5 μl of BsaI/BsmAI (10 U/μl), 0.5 μl of T4 DNA ligase (400 U/μl), 1 μl of 10× T4 DNA ligase buffer and 1 μl of pPR18 vector in a thermocycler with the following program: 37°C for 30 min, followed by 80°C for 5 min. Another 0.5 μl of T4 DNA ligase was then added to the reaction mixture. Subsequently, the mixture was incubated at 25°C for 10 min. The reaction mixture was then transformed into Escherichia coli DH5α competent cells. Positive clones were examined by PCR screening and verified by DNA sequencing. The resultant ligated 9-mer repeats were cloned into a modified pET21b backbone vector that already harbored a complete PPR repeat with a GL junction that was divided to obtain a full 10-PPR repeat assembly. Details regarding the construction steps can be found in the Supplementary Methods. The primers used are listed in Supplementary Table S1.
Protein expression and purification
The plasmids were transformed into E. coli BL21 (DE3). One liter of lysogeny broth supplemented with 100 mg ml−1 of ampicillin was inoculated with a transformed bacterial preculture and shaken at 37°C until the cell density reached an OD600 of ∼1.0–1.2. Protein expression was subsequently induced with 0.2 mM isopropyl-β-d-thiogalactoside at 16°C for 12 h. The cells were then collected by centrifugation, homogenized in buffer A (25 mM Tris–HCl, pH 8.0, 150 mM NaCl), and lysed using a high-pressure cell disrupter (JNBIO, China). The cell debris was removed by centrifugation at 20 000 × g at 4°C for 1 h. Then, the supernatant was loaded onto a column equipped with Ni2+ affinity resin (Ni-NTA, Qiagen), washed with buffer B (25 mM Tris–HCl, pH 8.0, 150 mM NaCl, 15 mM imidazole) and then eluted with buffer C (25 mM Tris–HCl, pH 8.0, 250 mM imidazole). The protein was then separated by anion-exchange chromatography (Source 15Q, GE Healthcare) via a linear NaCl gradient in buffer A. The purified protein was then concentrated and subjected to gel filtration chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare) in a buffer containing 25 mM Tris–HCl, pH 8.0, 150 mM NaCl and 5 mM dithiothreitol. Protein purity was examined by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and visualized by Coomassie blue staining. The peak fractions were collected and then stored at −80°C.
Electrophoretic mobility shift assay (EMSA)
5′ FAM-labeled ssRNA oligonucleotides (5′-gUUUUNNUUUUc-3′, where NN represents AA, GG, CC or UU) were synthesized (TaKaRa, Japan). The proteins were incubated with 100 nM RNA probes in final binding reactions that contained 25 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 40 mM NaCl, 250 ng ml−1 heparin and 10% glycerol at 25°C for 30 min. The reactants were then resolved on 8% native acrylamide gels (37.5:1 acrylamide:bis-acrylamide) in 0.5× Tris-glycine buffer under a 15 V cm−1 electric field for 2 h. The gels were subsequently screened using a Typhoon Trio Imager (Amersham Biosciences). Three technical replicates were performed for EMSA.
Isothermal titration calorimetry (ITC)
To quantitatively measure the RNA-binding affinity of the dPPRs, ITC experiments were performed at 25°C using Auto-iTC200 isothermal titration calorimeter (MicroCal). The RNA probes (5′-gUUUUNNUUUUc-3′, where NN represents AA, GG, CC or UU) were synthesized using an ABI-3400 synthesizer as previously described (32). RNA (120 μM) was dissolved in reaction buffer containing 20 mM HEPES, pH 7.5 and 150 mM NaCl (100 μl) and titrated against 10 μM protein in the same buffer (400 μl). The first injection (0.5 μl) was followed by 19 2-μl injections. The heat of dilution was measured by injecting RNA into buffer alone. The values were subtracted from the experimental curves before data analysis. The stirring rate was 750 rpm. The MicroCal ORIGIN software, supplied with the instrument, was used to determine the site-binding model that produced a good fit (low × 2 value) for the resulting data. Two technical replicates were performed for ITC.
PPRCODE server construction
To facilitate the application of the identified codes to target RNA prediction, the PPRCODE web server was constructed. This server utilizes the ScanProsite program (33) to detect and annotate PPR motifs using the PPR signature PS51375. The di-residues of the PPR motifs obtained from ScanProsite analysis were located at the sixth position of one motif and the first position of the next motif, which was inconvenient for PPR code analysis. Thus, we modified each PPR motif with a single one amino acid shift backwards to ensure that the PPR code within the entire motif conformed to the 5′ and 35′ scheme. PPR codes were extracted from the modified PPR motifs. The correlated RNA bases were provided for each PPR code.
RESULTS
Distribution frequency of PPR codes in land plants
PPR proteins usually contain a tandem array of 2–30 PPR repeats (2). With respect to the canonical P-type motif, previous studies have revealed that the amino acids at the 5th and 35th positions within each motif play crucial roles in RNA base recognition (23,27); the di-residues are referred to as the PPR code (23,28). Theoretically, there are 20 possible residues at both the 5th and 35th positions; thus, 400 di-residue combinations of PPR codes could be distributed within the PPR motifs. However, to date, the association between PPR codes and RNA bases is poorly understood, which limits target RNA prediction for PPR proteins and functional investigation of these proteins. To wrestle with this problem, a systematic investigation of the correlation between PPR codes and RNA bases was conducted.
We first assessed the PPR code distribution within the natural PPR motifs. The P-type PPR protein sequences from 65 land plants were collected, and canonical 35-amino-acid PPR repeats were identified (31) (Supplementary Table S2). Collectively, a total of 186456 PPR motifs were identified (Supplementary Table S2). Then, 391 PPR codes were derived from these motifs, which is very close to the theoretical value (Supplementary Table S3). The code frequency exhibited a biased distribution. Among these codes, ‘ND’ was the PPR code with the highest distribution frequency, accounting for more than 19% of the codes, followed by ‘NN’, ‘TD’, ‘SD’, ‘TN’, ‘SN’ and ‘NS’ (Supplementary Figure S1). Notably, the correlated RNA bases recognized by these seven PPR codes have already been predicted (28,29) and biochemically and structurally validated (27). However, base recognition by the remaining PPR codes stays largely elusive. In this study, the cutoff value of the coverage of the code distribution frequency was designated as 85%. A total of 62 PPR codes that met this criterion were selected for further analysis (Supplementary Figure S1).
Development of modular assembly method for dPPR construction
Previously, based on a sequence conservation analysis of P-type PPR motifs, we identified the most highly conserved amino acids at each position and used this sequence (VVTYNTLIDGLCKAGKLDEALKLFEEMVEKGIKPD) as a scaffold on which to synthesize dPPRs to determine the RNA base specificity of the PPR codes ‘ND’, ‘NS’, ‘SN’ and ‘TD’ (27,30). However, commercial synthesis of a large number of PPRs that contain a series of repeat domains with different PPR codes is difficult, time consuming and costly. The repetitive nature of the PPR motif leads to challenges in the construction of custom PPRs by ordinary PCR-based cloning and ligation. Moreover, such constructs cannot be generated by high-throughput PPR synthesis.
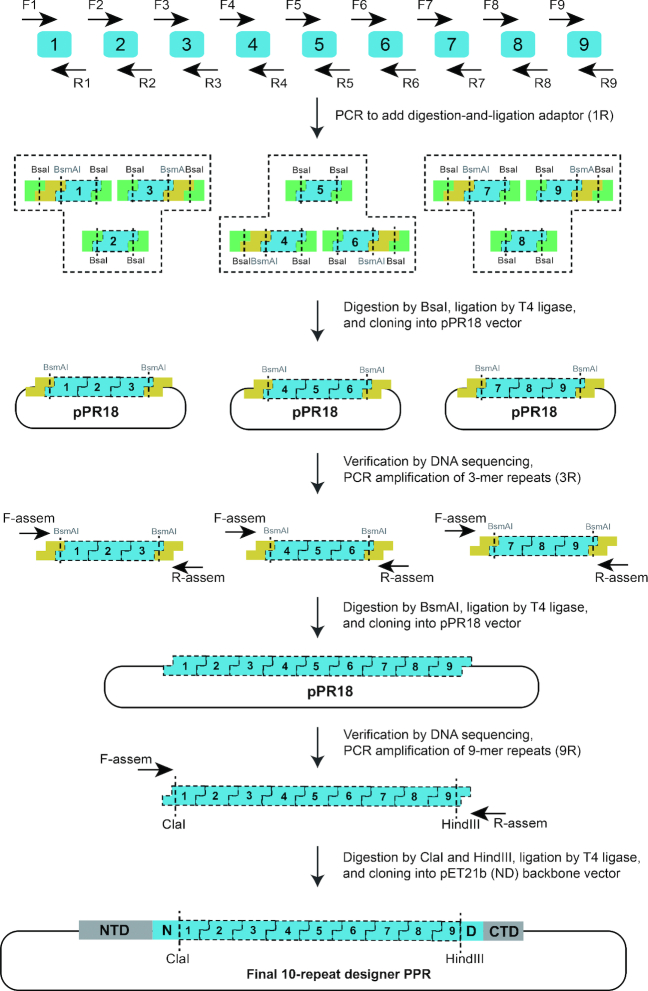
In this study, we developed a hierarchical ligation-based strategy for high-throughput construction of modular PPRs. We utilized the codon degeneracy of the adjacent GL residues because of the versatile codon combinations of these residues. To facilitate the assembly of the individual monomers in a specific order, the different codons at the GL junction between each pair of ligated monomers were used to construct a series of unique sticky-end ligation adaptors. Briefly, the basic modules encoded 25 C-terminal residues (LCKAGKLDEALKLFEEMVEKGIKPD) of repeat X and 10 N-terminal residues (VVTYNTLIDG) of repeat X+1 (Figure 1A). To maintain the integrity of PPR motifs in the final modular PPRs assembled, two fragments (VVTYNTLIDG and LCKAGKLDEALKLFEEMVEKGIKPD) were preintegrated into the amino-terminal domain (NTD) and carboxy-terminal domain (CTD) in the backbone vector (Figure 1A). The initial 35-amino-acid scaffold monomer was encoded by 105-bp DNA sequences constructed by PCR amplification.
Figure 1.
Modular assembly efficiency was explored using different numbers of monomers. (A) PPR motif conservation analysis and scaffold sequence selection. The sequence conservation analysis was performed with WebLogo (http://weblogo.berkeley.edu/logo.cgi). The most highly conserved residues at each position were VVTYNTLIDGLCKAGKLDEALKLFEEMVEKGIKPD. To facilitate the cloning and hierarchical ligation of PPR repeats, the codon degeneracy of GL was utilized to construct monomers with different sticky ends. The scaffold module was composed of 25 C-terminal residues (LCKAGKLDEALKLFEEMVEKGIKPD) of repeat X and 10 N-terminal residues (VVTYNTLIDG) of repeat X+1. Ligated residues G and L are colored in pink. Residues N and D are colored in light blue and red, respectively. (B) Schematic diagram of monomer ligation. Two to seven PPR repeats were tested by simultaneous ligation. (C) PCR screening of the ligated 2–7-repeat assemblies. The stars indicate bands with the expected molecular weights. The figures are representative of three replicates. (D) Statistical results for the positive rates of the ligated repeats. The clones were verified by DNA sequencing.
We first explored the effect of the number of ligated monomers (2R-7R) on the ligation efficiency (Figure 1B). The results showed that the ligation efficiency substantially decreased as the number of ligated monomers increased (Figure 1C and D). Simultaneous ligation with no more than four monomers resulted in a relatively high ligation efficiency (2R/3R/4R), the positive rate of which was more than 50% (Figure 1C and D). A previous study indicated a major distribution of 9–10 repeats of P-type PPR proteins (31). Moreover, the 10-repeat PPR proteins exhibited notable RNA-binding activity (27,30) and maximal specificity (34). Based on these findings, we assembled 10-repeat dPPRs with distinct PPR codes. Specifically, three monomers were ligated simultaneously to form 3-mer tandem repeats (Figure 2). Likewise, the 3-mer tandem repeats were ligated to obtain the desired 9-mer tandem repeats and subsequently cloned into the pET21b (ND) backbone vector that already contained a whole repeat but was divided at the GL junction (VVTYNTLIDG||LCKAGKLDEALKLFEEMVEKGIKPD) to achieve 10-mer tandem PPR tracts (Figure 2). To improve the solubility of the engineered proteins, parts of PPR10 from Zea mays were fused to the NTD and CTD of the dPPRs (27). Taking the 10-repeat dPPR containing ‘ND’ codes (referred to here as dPPR-(ND)10) as an example, size-exclusion chromatography (SEC) analysis revealed that the protein exhibited excellent solubility and very good solution behavior (Supplementary Figure S2A). To examine the effectiveness of the constructed PPR protein, we tested the RNA-binding activity of dPPR-(ND)10 by EMSA using a specific 5′-FAM-labeled RNA substrate. The protein exhibited notable RNA-binding activity with a Kd value <100 nM (Supplementary Figure S2B), which was comparable to the results of our previous study involving the use of commercially synthesized dPPR and a 32P-labeled RNA substrate (30).
Figure 2.
Flowchart of hierarchical ligation and modular assembly for the construction of custom PPRs. Nine individual PCRs with specific primers were performed to obtain the basic assembly units. Each monomer was customized with specific PPR codes, and each PCR product had a unique linker specifying the position of the PCR product in the assembly. After enzymatic digestion with a type II restriction endonuclease, orthogonal overhangs were derived at the junction position using distinct codons to preserve the same amino acids. The unique overhangs facilitated the positioning of each monomer in the ligation product. The fragments were cloned into pPR18 and verified by sequencing during assembly. The final fragments were cloned into a modified pET21b (ND) vector containing NTD and CTD sequences from PPR10 and a whole repeat divided at the GL junction to form a 10-repeat dPPR.
To expand the application potential of dPPR proteins, it is usually necessary to synthesize PPR proteins that contain different numbers of repeats. By randomly ligating the 2R/3R/4R assembly modules, dPPRs with the desired number of tandem repeats could be obtained. Consequently, a series of dPPRs containing increasing numbers of tandem repeats were obtained (Supplementary Figure S2C and D). In summary, we developed an effective modular assembly method that can be used for high-throughput construction of desired PPRs.
Systematic investigation of correlations between PPR codes and RNA bases
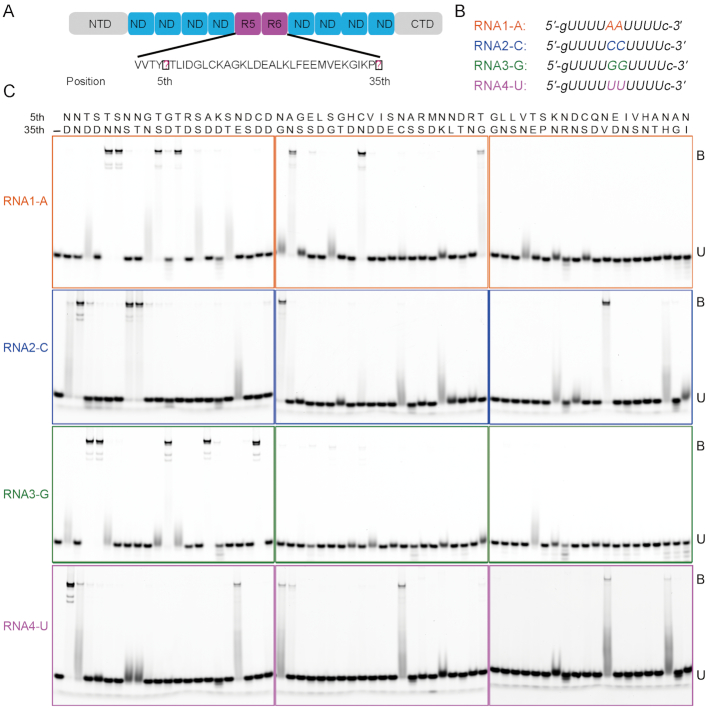
We previously designed four types of dPPR proteins dPPR-U8N2, consisting of 10 repeats harboring ND codes at all except the fifth and sixth repeats, at which the proteins harbored four different PPR codes (‘ND’, ‘NS’, ‘SN’ and ‘TD’) and investigated the RNA-binding selectivity of these dPPRs by EMSA using four corresponding RNA substrates with different nucleotides (A, C, G or U) at the fifth and sixth positions. These four PPR codes could specifically distinguish the four nucleotides (27). To gain a comprehensive understanding of PPR codes, we adopted an identical strategy to examine the base selectivity of the above-mentioned 62 PPR codes derived from land plants (Supplementary Figure S1). Therefore, 62 dPPRs containing 10-PPR tracts with different PPR codes at the fifth and sixth repeats were constructed via the modular assembly method described above. These proteins were all purified to homogeneity (Supplementary Figure S3A), and SEC analyses revealed that these proteins exhibited similar behaviors in solution (Supplementary Figure S3B). We screened the RNA-binding activity of these 62 dPPRs against four 5′ FAM-labeled RNA substrates by EMSA (Figure 3A and B). The binding landscapes can be generally categorized into three types: (i) sharply shifted bands (ii) smeared bands and (iii) weak bands (Supplementary Figure S4).
Figure 3.
The RNA-binding landscape of the designer PPR was examined by EMSA. (A) Schematic diagram of the modularly assembled designer PPRs. The constructed designer PPRs consisted of 10 consecutive PPR repeats containing the ‘ND’ code at the 5th and 35th positions, except repeat five and six, in which desired PPR codes were used. Sixty-two designer PPRs were constructed. (B) RNA substrates were used to determine the RNA-binding specificity of the designer PPRs. These RNAs were 5′ FAM labeled. (C) The RNA-binding specificity of designer PPRs was examined by EMSA. The final protein concentration for each sample was 1 μM. The 5th and 35th di-residues above each lane represent a designer PPR protein containing the corresponding PPR code at repeats five and six. The di-residues are arranged by distribution frequency decreasing from left to right, corresponding to Supplementary Figure S1. B: bound. U: unbound. This figure is representative of three replicates.
The PPR codes ‘TN’ and ‘SN’ bind base A (2). In this study, several new PPR codes, including ‘TS’, ‘TT’, ‘AN’, ‘CN’ and ‘TG’ were identified as being able to recognize base A, similar to ‘TN’ and ‘SN’ (Figure 3C). In addition, the amino acid combinations ‘TD’, ‘GN’, ‘SS’, ‘ST’, ‘NG’, ‘SG’ and ‘VN’ exhibited smeared bands, suggesting the correlation of these combinations with base A. In addition, weak bands were observed for several combinations, such as ‘NS’, ‘NT’, ‘CS’, ‘GD’, ‘VD’ and ‘ES’ (Figure 3C and Supplementary Figure S4). Base G is correlated with PPR codes ‘TD’ and ‘SD’ (2). Herein, we identified three additional PPR codes, namely, ‘AD’, ‘CD’ and ‘GD’ that bind to base G strongly, yielding sharp bands, similar to ‘TD’ and ‘SD’ (Figure 3C). In addition, several other codes, including ‘ND’, ‘TN’, ‘TS’, ‘TT’ and ‘TE’ exhibited severely smeared bands, indicating the correlation of these codes with base G. In previous studies, serine and threonine at position 5 were reported to be correlated with purines: ‘TN’ and ‘SN’ were correlated with A, and ‘TD’ and ‘SD’ were correlated with G (2). Herein, we discovered three additional amino acids, namely, alanine, cysteine and glycine, at position 5, that were strongly correlated with purines (Figure 3). Under these circumstances, asparagine or aspartate at position 35 (Asn35, Asp35) play a determinant role in the recognition of base A or G.
For base C, we identified ‘NT’, ‘NG’ and ‘NV’ as combinations that strongly bind to C, similar to ‘NN’ and ‘NS’ (Figure 3C). In addition, ‘ND’, ‘NE’, ‘NC’, ‘NK’, ‘KN’ and ‘NH’ exhibited severely smeared bands, suggesting the correlation of these codes with base C. For base U, ‘ND’ and ‘NN’ were observed to be the two most frequently distributed PPR codes within the PPR motifs (Supplementary Figure S1) and are known to bind U (2). In this study, ‘NE’, ‘NG’, ‘NC’, ‘NV’ and ‘NH’ exhibited shifted bands, similar to ‘NN’, and smeared bands of ‘NS’ and ‘NT’ were observed (Figure 3C), suggesting the correlation of these codes with base U.
In addition to the aforementioned PPR codes, it is clear that many of the amino acid combinations at the 5th and 35th positions are not involved in RNA base recognition (Figure 3C). For example, ‘RD’, ‘DS’, ‘MD’ and ‘RS’ are frequently distributed combinatorial amino acids at positions 5 and 35 within PPR motifs (Supplementary Figure S1); however, these combinations do not bind any of the four nucleotides (Figure 3C). Therefore, these PPR codes might play a minor role in RNA base coordination.
Quantitative determination of the RNA-binding affinity of dPPRs by ITC
We further measured the RNA-binding affinity of these 62 dPPRs by ITC experiments. Each dPPR protein was titrated against four RNA substrates. The titration curves for all the ITC assays are shown in Supplementary Figure S5. Kd values reflecting the binding affinity of each PPR–RNA pair were calculated. The ‘ND’, ‘SD’, ‘TN’ and ‘NS’ codes, for example, exhibited binding preferences for U, G, A and C, respectively (Figure 4A), which was consistent with the EMSA results (Figure 3C) and with the results of previous investigations of these codes (2,28). For further analysis, the binding affinity derived from ITC was categorized into three levels: strong, medium and weak. ‘Strong’ indicates that the Kd value is in the range of 10–100 nM; ‘medium’ indicates a range of 100–1000 nM; ‘weak’ indicates a range of 1–10 μM. In addition, several PPR codes exhibited no detectable binding. Based on these criteria, we constructed a heat map of all the binding events tested by ITC to reveal the binding landscape of each PPR code (Figure 4B).
Figure 4.
The RNA-binding affinity of the designer PPR was examined by ITC. (A) ITC binding curves show the distinct binding affinity of the dPPR against four RNA substrates. The results are exemplified by four known PPR codes. (B) Binding affinity of each designer PPR against four RNA substrates. The figure was constructed using R. The binding affinities are categorized into three levels (strong, medium and weak); ‘Strong,’ shown in dark blue, represents Kd values in the range of 10–100 nM; ‘Medium,’ shown in slate, represents Kd values in the range of 100–1000 nM; ‘Weak,’ shown in seashell, represents Kd values larger than 1 μM. ‘Null,’ shown in Alice blue, represents no detectable binding by ITC.
Subsequently, we aligned the results from both EMSA and ITC based on the binding affinities (Supplementary Figure S6). All the sharply shifted (type I) or smeared (type II) bands observed by EMSA exhibited a binding affinity within a range of 10–1000 nM by ITC (Supplementary Figure S6), indicating strong correlations between these PPR codes and the corresponding bases. However, the weak binding events (type III) shown by EMSA exhibited varied results determined by ITC (Supplementary Figure S6). Some weak binding events exhibited affinities in the range of 1–10 μM (e.g. ‘NS’ or ‘NT’ to A), suggesting weak correlation or tolerance, whereas some other binding events were undetectable. For example, ‘AD’ and ‘GD’ were not observed to bind base A. Taken together, the binding events that were confirmed by both EMSA and ITC suggested the correlations between these PPR codes and the respective bases. Based on these results, the bases correlated with each PPR code were identified (Supplementary Table S4).
Target RNA prediction for PPR proteins
PPR proteins, which constitute a very large protein family, are involved in RNA metabolism, but only a limited number of these proteins have been functionally characterized because most of the RNA targets have not been identified. Systematic investigation of the correlations between PPR codes and RNA bases greatly enriches the pool of PPR codes, which will contribute to the prediction of the RNA targets of a large number of PPR proteins. For target RNA prediction, we developed the PPRCODE web server by using the ScanProsite program (33). After a certain PPR protein is submitted to the server, a set of results is provided, including the PPR motif, PPR code and correlated RNA bases. Using this server, we predicted the corresponding RNA targets of all P-type PPR proteins of the 65 land plants (Supplementary Table S5). The effectiveness of the server was validated by comparison of the predicted RNA targets of SOT1 (AT5G46580) with the functionally identified RNA sequence (Figure 5A) (3,35). For PPR proteins with unidentified target RNA sequences, such as EMP12 (36), the server could provide the correlated RNA bases for the PPR motifs (Figure 5B). Moreover, the PPRCODE server could predict new correlations between PPR codes and RNA bases. Take the two PPR proteins GLYMA11G11880.1 from Glycine max and BV7U_180180_ANIA.T1 from Beta vulgaris (Supplementary Figure S7) as examples; these two proteins have not been functionally characterized, and most codes within the PPR motifs of these proteins were previously unidentified. Based on the correlations of PPR codes and RNA bases identified in this study, the PPRCODE server could predict the correlated RNA bases for each PPR motif (Figure 5C), thereby narrowing the query scope for the bona fide RNA target in organelle transcripts. To determine whether the identified PPR codes can be generally applied beyond land plants, we analyzed the code distribution frequency in other species. Interestingly, a similar PPR code distribution pattern was observed among algae, protists, fungi and metazoans (Supplementary Figure S8 and Supplementary Tables S6–9), suggesting the general usability of the identified codes in these species.
Figure 5.
RNA target prediction by the PPRCODE web server. (A) RNA target prediction and comparison of a functionally identified PPR, namely, SOT1 from Arabidopsis. (B) RNA target prediction of a functionally investigated PPR with an unidentified RNA sequence, namely, EMP12 from maize. (C) RNA target prediction of PPR proteins with unknown function, namely, GLYMA11G11880.1 from Glycine max and BV7U_180180_ANIA.T1 from Beta vulgaris.
DISCUSSION
The Pumilio and fem-3 messenger RNA-binding factor (PUF) and PPR proteins are two types of α-helical repeat proteins that can recognize ssRNA in a modular manner with one repeat corresponding to one base. Canonical PUF proteins contain eight tandem repeats; each repeat comprises 36 residues that form three α-helices, and the amino acids at positions 12, 13 and 16 within the second α-helix determine the RNA-binding specificity (37). The base recognition specificity of serial combinations of three amino acids was identified by high-throughput sequencing using a large random RNA library (38). Several previous studies have provided insight into the application of engineered PUF for recognition of specific RNA targets or for conjugation with functional modules to catalyze various RNA metabolic processes (39–42). In contrast to PUF, PPR proteins contain 2–30 PPR repeats; each repeat contains two antiparallel α-helices, and the amino acids at positions 5 and 35 determine the RNA-binding specificity. dPPRs can be tailored to new RNA targets (35), inducing specific RNA cleavage in plants (43). To comprehensively understand PPR codes, we developed a high-throughput modular assembly method to synthesize 62 designer PPRs and delineated the RNA-binding landscapes of 62 PPR codes (Figures 3C and 4B). Along with the increased number of identified PPR codes and improved custom design of artificial or natural PPR motifs, we expect dPPRs with tailored specificity toward various RNA manipulations to have broad application potential (41,44,45).
Previously, based on the crystal structures of dPPRs in complex with the corresponding RNAs, we revealed that different types of hydrogen bond networks mediate the interaction between the PPR code and RNA base (27). In this study, we identified a number of PPR codes that were correlated with specific bases. The PPR code ‘SN’ was found to be associated with base A. Both serine at position 5 and asparagine at position 35 were attached to base A via a hydrogen bond (27). Similarly, it could be inferred that the identified codes ‘TS’, ‘TT’, ‘SS’ and ‘ST’ could bind base A by forming a pair of hydrogen bonds. The PPR code ‘TD’ binds to base G (2). Threonine at position 5 (Thr5) and aspartate at position 35 (Asp35) form one and two hydrogen bonds with base G, respectively (27), suggesting that Asp35 might play a more important role in mediating the interaction than the amino acid at position 5. Thus, it can be speculated that several amino acids, such as alanine, cysteine or glycine, at position 5 combined with Asp35 (‘AD’, ‘CD’ or ‘GD’) could coordinate base G (Figure 3C). However, we also observed that combinations of some other amino acids at position 5 with Asp35, such as ‘RD’, ‘DD’, ‘LD’, ‘ID’, ‘MD’ and ‘QD’, were unable to recognize G (Figure 3C). We hypothesize that the large side chains of arginine, aspartate, leucine, isoleucine, methionine and glutamine could lead to steric clashes that hinder the interaction of these amino acids with base G, whereas small side chains (e.g., alanine, cysteine and glycine) do not hinder these interactions. Moreover, we also identified several PPR codes, such as ‘NG’, ‘NE’, ‘NC’, ‘NV’ and ‘NH’, that could bind both bases C and U, similar to ‘NN’. In addition, ‘NT’ exhibited binding affinity for base C, similar to ‘NS’ (Figure 3C). These identified PPR codes conformed to the finding that asparagine at position 5 was correlated with pyrimidines (2). We verified the selectivity of the identified ‘GD’, ‘AD’, ‘CD’, ‘TT’, ‘NG,’ ‘NE’ and ‘NC’ codes via eight-repeat dPPRs with varied 5′-end PPR motifs (Supplementary Figure S9A–D). However, we failed to detect selective binding of ‘CN’ to base A. PPR codes at different positions exhibiting discrepant base preferences have been occasionally observed in natural PPR proteins (46), indicating the effects of code positions on binding. These limited findings are quite enlightening. Further crystal structure determination of these PPR codes in complex with the corresponding RNA bases will reveal the molecular basis of recognition.
Nearly half of the PPR proteins of land plants belong to the PLS subfamily, members of which consist of repeating triplets of P, L and S motifs, in which the L (35–38 amino acids) and S (31 amino acids) motifs are variants of the canonical P-type motif. Recently, Harrison et al. developed an HMM-based method, named aPPRove, to predict the RNA target of a PLS-type PPR protein. The method is based on the known P-type PPR binding code (47). Our PPRCODE server utilizes the ScanProsite program for P-type motif analysis to predict the bases correlated with individual PPR codes on the basis of code delineation. Actually, the L-type motif exhibited distinct motif contexts compared with the P- and S-type motifs, with the codes of L-type repeats remaining unknown. Previous bioinformatics analysis has suggested that the L-type repeats can form contacts with RNA, although the nucleotide–amino acid correlations for L-type motifs were weaker than those for P- and S-type motifs (29,48). Furthermore, our recent structural analyses revealed that the L-type motif exhibited conformations different from those of the P- and S-type motifs. The difference in conformations led to unfavorable base targeting. However, in the presence of an editing factor, namely, multiple organellar RNA editing factor (MORF), the L-type motifs underwent a striking conformational change and became capable of coordinating the base (49). Therefore, further elucidation of the codes and the binding contribution of L-type motifs will improve target prediction for PLS-type PPR proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the research associates at the Centre for Protein Research, Huazhong Agricultural University, for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2015CB910900]; Fok Ying-Tong Education Foundation [151021]; Fundamental Research Funds for the Central Universities [2662017PY031]; China Postdoctoral Science Foundation [2017T100561 to J.Y., 2018M640712 to C.S.]; National Natural Science Foundation of China [31722017 to P.Y., 31700203 to J.Y.]. Funding for open access charge: National Natural Science Foundation of China [31722017 to P. Y.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B. et al.. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004; 16:2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 3. Wu W., Liu S., Ruwe H., Zhang D., Melonek J., Zhu Y., Hu X., Gusewski S., Yin P., Small I.D. et al.. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016; 85:607–621. [DOI] [PubMed] [Google Scholar]
- 4. Hsieh W.Y., Liao J.C., Chang C.Y., Harrison T., Boucher C., Hsieh M.H.. The slow growth3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in arabidopsis. Plant Physiol. 2015; 168:490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yap A., Kindgren P., Colas des Francs-Small C., Kazama T., Tanz S.K., Toriyama K., Small I.. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 2015; 81:661–669. [DOI] [PubMed] [Google Scholar]
- 6. Zoschke R., Watkins K.P., Miranda R.G., Barkan A.. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016; 85:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hammani K., Takenaka M., Miranda R., Barkan A.. A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 2016; 44:4278–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Z., Dong F., Wang X., Wang T., Su R., Hong D., Yang G.. A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus. J. Exp. Bot. 2017; 68:4115–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu J., Wang K., Huang W., Liu G., Gao Y., Wang J., Huang Q., Ji Y., Qin X., Wan L. et al.. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell. 2012; 24:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uyttewaal M., Arnal N., Quadrado M., Martin-Canadell A., Vrielynck N., Hiard S., Gherbi H., Bendahmane A., Budar F., Mireau H.. Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell. 2008; 20:3331–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim Y.J., Zhang D.. Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 2018; 23:53–65. [DOI] [PubMed] [Google Scholar]
- 12. Tang H., Luo D., Zhou D., Zhang Q., Tian D., Zheng X., Chen L., Liu Y.G.. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant. 2014; 7:1497–1500. [DOI] [PubMed] [Google Scholar]
- 13. Huang W., Yu C., Hu J., Wang L., Dan Z., Zhou W., He C., Zeng Y., Yao G., Qi J. et al.. Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14984–14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chateigner-Boutin A.L., Ramos-Vega M., Guevara-Garcia A., Andres C., de la Luz Gutierrez-Nava M., Cantero A., Delannoy E., Jimenez L.F., Lurin C., Small I. et al.. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008; 56:590–602. [DOI] [PubMed] [Google Scholar]
- 15. Sosso D., Mbelo S., Vernoud V., Gendrot G., Dedieu A., Chambrier P., Dauzat M., Heurtevin L., Guyon V., Takenaka M. et al.. PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell. 2012; 24:676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu Q., Dugardeyn J., Zhang C., Takenaka M., Kuhn K., Craddock C., Smalle J., Karampelias M., Denecke J., Peters J. et al.. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012; 71:836–849. [DOI] [PubMed] [Google Scholar]
- 17. Liu Y.J., Xiu Z.H., Meeley R., Tan B.C.. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell. 2013; 25:868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aryamanesh N., Ruwe H., Sanglard L.V., Eshraghi L., Bussell J.D., Howell K.A., Small I., des Francs-Small C.C.. The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 2017; 173:1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu D., Jiang L., Gong H., Liu C.M.. Embryonic factor 19 encodes a pentatricopeptide repeat protein that is essential for the initiation of zygotic embryogenesis in Arabidopsis. J. Integr. Plant Biol. 2012; 54:55–64. [DOI] [PubMed] [Google Scholar]
- 20. Li X.J., Zhang Y.F., Hou M., Sun F., Shen Y., Xiu Z.H., Wang X., Chen Z.L., Sun S.S., Small I. et al.. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 2014; 79:797–809. [DOI] [PubMed] [Google Scholar]
- 21. Tan J., Tan Z., Wu F., Sheng P., Heng Y., Wang X., Ren Y., Wang J., Guo X., Zhang X. et al.. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant. 2014; 7:1329–1349. [DOI] [PubMed] [Google Scholar]
- 22. Mei C., Jiang S.C., Lu Y.F., Wu F.Q., Yu Y.T., Liang S., Feng X.J., Portoles Comeras S., Lu K., Wu Z. et al.. Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling. J. Exp. Bot. 2014; 65:5317–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W. et al.. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013; 504:168–171. [DOI] [PubMed] [Google Scholar]
- 24. Ke J., Chen R.Z., Ban T., Zhou X.E., Gu X., Tan M.H., Chen C., Kang Y., Brunzelle J.S., Zhu J.K. et al.. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 2013; 20:1377–1382. [DOI] [PubMed] [Google Scholar]
- 25. Coquille S., Filipovska A., Chia T., Rajappa L., Lingford J.P., Razif M.F., Thore S., Rackham O.. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014; 5:5729. [DOI] [PubMed] [Google Scholar]
- 26. Gully B.S., Shah K.R., Lee M., Shearston K., Smith N.M., Sadowska A., Blythe A.J., Bernath-Levin K., Stanley W.A., Small I.D. et al.. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D. Biol. Crystallogr. 2015; 71:196–208. [DOI] [PubMed] [Google Scholar]
- 27. Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., Wang X., Wang Q., Zhang Q., Fan S. et al.. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016; 7:11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I.. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012; 8:e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T.. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013; 8:e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen C., Wang X., Liu Y., Li Q., Yang Z., Yan N., Zou T., Yin P.. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol. Plant. 2015; 8:667–670. [DOI] [PubMed] [Google Scholar]
- 31. Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M.F., Bai F., Castleden I., Song Y., Song B., Huang J. et al.. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016; 85:532–547. [DOI] [PubMed] [Google Scholar]
- 32. Huang J., Dong X., Gong Z., Qin L.Y., Yang S., Zhu Y.L., Wang X., Zhang D., Zou T., Yin P. et al.. Solution structure of the RNA recognition domain of METTL3-METTL14 N(6)-methyladenosine methyltransferase. Protein &Cell. 2018; doi:10.1007/s13238-018-0518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Castro E., Sigrist C.J., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N.. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006; 34:W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miranda R.G., McDermott J.J., Barkan A.. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 2018; 46:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou W., Lu Q., Li Q., Wang L., Ding S., Zhang A., Wen X., Zhang L., Lu C.. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E1554–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun F., Xiu Z., Jiang R., Liu Y., Zhang X., Yang Y.Z., Li X., Zhang X., Wang Y., Tan B.C.. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J. Exp. Bot. 2019; 70:963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., McLachlan J., Zamore P.D., Hall T.M.. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 38. Campbell Z.T., Valley C.T., Wickens M.. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 2014; 21:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei H., Wang Z.. Engineering RNA-binding proteins with diverse activities. Wiley Interdiscip. Rev. RNA. 2015; 6:597–613. [DOI] [PubMed] [Google Scholar]
- 40. Abil Z., Zhao H.. Engineering reprogrammable RNA-binding proteins for study and manipulation of the transcriptome. Mol. Biosyst. 2015; 11:2658–2665. [DOI] [PubMed] [Google Scholar]
- 41. Yagi Y., Nakamura T., Small I.. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014; 78:772–782. [DOI] [PubMed] [Google Scholar]
- 42. Hall T.M. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016; 36:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colas des Francs-Small C., Vincis Pereira Sanglard L., Small I.. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun Biol. 2018; 1:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015; 113:93–99. [DOI] [PubMed] [Google Scholar]
- 45. Filipovska A., Rackham O.. Pentatricopeptide repeats: modular blocks for building RNA-binding proteins. RNA Biol. 2013; 10:1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miranda R.G., Rojas M., Montgomery M.P., Gribbin K.P., Barkan A.. RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA. 2017; 23:586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrison T., Ruiz J., Sloan D.B., Ben-Hur A., Boucher C.. aPPRove: an HMM-Based method for accurate prediction of RNA-Pentatricopeptide repeat protein binding events. PLoS One. 2016; 11:e0160645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takenaka M., Zehrmann A., Brennicke A., Graichen K.. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013; 8:e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yan J., Zhang Q., Guan Z., Wang Q., Li L., Ruan F., Lin R., Zou T., Yin P.. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants. 2017; 3:17037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.