Abstract
Cockayne syndrome group B (CSB, also known as ERCC6) protein is involved in many DNA repair processes and essential for transcription-coupled repair (TCR). The central region of CSB has the helicase motif, whereas the C-terminal region contains important regulatory elements for repair of UV- and oxidative stress-induced damages and double-strand breaks (DSBs). A previous study suggested that a small part (∼30 residues) within this region was responsible for binding to ubiquitin (Ub). Here, we show that the Ub-binding of CSB requires a larger part of CSB, which was previously identified as a winged-helix domain (WHD) and is involved in the recruitment of CSB to DSBs. We also present the crystal structure of CSB WHD in complex with Ub. CSB WHD folds as a single globular domain, defining a class of Ub-binding domains (UBDs) different from 23 UBD classes identified so far. The second α-helix and C-terminal extremity of CSB WHD interact with Ub. Together with structure-guided mutational analysis, we identified the residues critical for the binding to Ub. CSB mutants defective in the Ub binding reduced repair of UV-induced damage. This study supports the notion that DSB repair and TCR may be associated with the Ub-binding of CSB.
INTRODUCTION
During transcription, bulky DNA lesions such as UV-induced adducts can arrest the progression of RNA polymerases. The arrested polymerases serve as signals to recruit DNA repair machinery to remove the obstructing lesions. This molecular mechanism is termed transcription-coupled repair (TCR) (1,2). Defects in the TCR pathway are associated with the hereditary disorders Cockayne syndrome (CS) and UV-sensitive syndrome (UVSS) (1,3). CS is an autosomal recessive genetic disorder characterized by growth retardation, progressive neurological degeneration, severe photosensitivity, and premature aging (4). CS cells are deficient in TCR, leading to hypersensitivity to UV irradiation and delay in the recovery of RNA synthesis after DNA damage. About 80% of CS cases are caused by a defect in the CS group B gene (CSB; also called ERCC6) with the remaining cases caused by a defect in the CS group A gene (CSA; also called ERCC8) and other less common genetic defects (5,6). UVSS is another genetic disorder associated with the invalid TCR. A main symptom of UVSS patients is photosensitivity with no neurologic or somatic abnormalities. UVSS is caused by a mutation in the CSA, CSB or UVSSA gene (3,7–9).
CSB is a protein that is recruited in an early stage of TCR induced by the arrest of RNA polymerase II (Pol II). In addition to DNA damage-dependent arrests, Pol II can also be stopped by naturally occurring non-canonical DNA structures. A recent study based on the cryo-electron microscopy of yeast CSB homologue Rad26 in complex with Pol II showed that CSB promotes the forward movement of Pol II to bypass certain translocation barriers (10). Pol II cannot bypass bulky DNA lesions such as UV-induced cyclobutane pyrimidine dimers (CPD), leading to the stabilization of CSB on the DNA (11). CSB can then recruit nucleotide excision repair (NER) enzymes to remove the DNA damage (12). Interestingly, CSB is also important to promote cellular survival after oxidative damage (13–15). Major oxidation products of DNA, in particular 8-oxoguanine (8-oxoG), do not block transcription elongation and should not be associated with transcription elongation arrests (16). However, several studies indicate that intermediate products of 8-oxoG repair including a single strand gap left after excision of 8-oxoG may lead to transcription arrest (17,18). Less frequent DNA lesions associated with oxidative stress, such as a single strand break (SSB) and a double strand break (DSB), may also be associated with TCR (19). Consistently, CSB has been shown to be involved in the repair of DSB, notably through its interaction with the RIF1 protein (20). Additionally, CSB has a function in transcriptional arrest after genotoxic stress that induces the polyubiquitylation of ATF3 and its further proteasomal degradation (21).
The C-terminal region of CSB has been found to bind to Ub (22). A putative Ub-binding domain (UBD) has been assigned on the basis of the sequence conservation and secondary structure prediction (22). Cells expressing a truncated CSB without this UBD exhibited hypersensitivity to UV irradiation (22,23). The mechanism involved in this DNA damage response remains unknown, since ubiquitylated proteins recognized by CSB UBD have not been identified yet. A recent study showed that the ubiquitylation of CSB on Lys991 is associated with oxidative stress (24). The K991R mutant of CSB (CSBK991R) exhibits a transcription profile similar to the UBD-lacking CSB (CSBΔUBD), indicating some link between CSB ubiquitylation and Ub binding. However, CSBΔUBD is hypersensitive to both UV irradiation and oxidative stress, whereas CSBK991R only affects the cellular response to the latter. Another study showed that the C-terminally truncated mutation of CSB such as ΔUBD or truncation of the C-terminal 30 residues (Δ30) affects cell survival after UV irradiation (23). In the same study, both CSBΔUBD and CSBΔ30 failed to interact with CSA or Pol II in vivo. In addition, the UBD and C-terminal extremity were required for sumoylation of CSB on Lys205. The K205R mutant of CSB was hypersensitive to UV irradiation, indicating functional importance of this posttranslational modification (23). Recently, a computational modeling suggested that the last 76 amino acids of CSB resembled a winged-helix domain (WHD) overlapping with the UBD. This WHD has been shown to modulate the RIF1–CSB interaction to mediate the repair of DSBs (20).
In this study, we biochemically characterize the interaction between CSB and Ub and identify CSB WHD as the minimal Ub-binding domain. We further present the crystal structure of the CSB WHD in complex with Ub at 2.6 Å resolution. CSB WHD forms a single globular domain. CSB WHD represents a unique Ub-binding domain, which is different from the previously identified 23 types of UBDs. The interaction of CSB WHD with Ub shares similarities with FAAP20 UBZ: one α-helix mediates most of the interactions, whereas the C-terminal extension stabilizes this interaction (25,26). Since CSB WHD was shown to mediate the recruitment of CSB to DSBs, our structural data and accompanying in vitro and in vivo mutational analyses suggest an important function of the Ub binding in the CSB-mediated DSB repair and TCR.
MATERIALS AND METHODS
Sample preparation
The genes encoding the C-terminal regions of human CSB (residues 1220–1493, 1386–1493, 1401–1493, 1412–1493, 1426–1493 and 1386–1491) were cloned into the pCold-GST (Takara Bio) vector using NdeI and XhoI. Escherichia coli strain Rosetta (DE3) cells (Invitrogen) were transformed with each expression vector and cultured in LB medium containing 100 mg l−1 ampicillin at 37°C. When the optical density of the culture at 600 nm reached ∼0.5, protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. The culture was continued for 24 h at 15°C. The cells were disrupted by sonication in phosphate buffered saline (PBS) containing 1 mM dithiothreitol (DTT) and 0.5% Triton X-100. The lysate was clarified by centrifugation at 30 000 g for 60 min. The supernatant was then loaded onto a Glutathione Sepharose FF (GE Healthcare) column pre-equilibrated with PBS containing 1 mM DTT and 0.5% Triton X-100. The column was washed with PBS containing 1 mM DTT and 0.5% Triton X-100 and then with PBS containing 1 mM DTT. The GST fusion sample was eluted with 50 mM Tris–HCl buffer (pH 8.0) containing 200 mM NaCl, 1 mM DTT, and 15 mM reduced glutathione. For crystallization, the GST tag was cleaved by PreScission protease (GE Healthcare). The sample was dialyzed against 50 mM Tris–HCl buffer (pH 8.0) containing 1 mM DTT and then loaded onto a ResourceQ anion exchange column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl buffer (pH 8.0) containing 1 mM DTT. The sample was eluted with a linear gradient of 0–1 M NaCl. The peak fractions were collected, concentrated, and subjected onto a Superdex75 16/60 (prep grade) column in 10 mM Tris–HCl buffer (pH 8.0) containing 200 mM NaCl and 5 mM β-mercaptoethanol. The fractions abundant in the purified protein were collected and concentrated to ∼10 g l−1 using an Amicon Ultra-4 10 000 MWCO filter (Millipore). Ub and M1-linked diUb (M1-Ub2) were produced at 20°C in E. coli strain Rosetta (DE3) cells (Invitrogen) transformed with the pET26b (Novagen) expression vector harboring mouse Ub and human M1-Ub2 genes, respectively (27). K6-, K11-, K29-, K33-, K48- and K63-linked diUb samples (K6-, K11-, K29-, K33-, K48- and K63-Ub2, respectively) were synthesized enzymatically (27). The samples were purified by a ResourceS cation exchange column (GE Healthcare) and a HiLoad Superdex75 16/60 or 26/60 size-exclusion column (GE Healthcare). The purified Ub2 species were concentrated to ∼30 g l−1 using an Amicon Ultra-4 10 000 MWCO filter (Millipore) and stored at −80°C until use.
Crystallization and data collection
To prepare the complex between CSB WHD (residues 1401–1493) and K48-Ub2, a 1.5-fold molar excess of CSB WHD was incubated with K48-Ub2 for 1 h at 4°C. For crystallization, K48-Ub2 was synthesized from equimolar amounts of K48R Ub and D77 Ub, instead of wild-type Ub. Both K48R Ub and D77 Ub were prepared in a manner similar to wild-type Ub. The complex sample was loaded onto a HiLoad 16/60 Superdex75 (prep grade) column (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl buffer (pH 8.0) containing 50 mM NaCl and 5 mM β-mercaptoethanol to remove the unbound CSB WHD. The purified complex was concentrated to ∼10 g l−1 by using an Amicon Ultra-4 10 000 MWCO filter (Millipore). Initial crystallization screening was performed using the sitting drop vapor diffusion method at 20°C with a Mosquito liquid-handling robot (TTP Lab Tech). We tested about 600 conditions, using crystallization reagent kits supplied by Hampton Research. Crystals of the complex between CSB WHD and K48-Ub2 were grown at 20°C with the sitting drop vapor diffusion method by mixing 200 nl of the protein solution with an equal amount of precipitant solution containing 28% PEG1000, 10% glycerol, 0.1 M tricine (pH 8.0), and 350 mM MgCl2. For data collection, crystals were transferred to 28% PEG1000, 10% glycerol, 0.1 M tricine (pH 8.0), 350 mM MgCl2 and 25.5% 2-methyl-2,4-pendanediol for cryoprotection. The cryoprotected crystals were flash frozen in liquid nitrogen.
Structural determination and refinement
Diffraction data sets were collected at 100 K at the beamline BL41XU in SPring-8 (Hyogo, Japan) and were processed with the program HKL2000 (28) and Phenix program suite (29). The complex structures reported in this study were determined by molecular replacement using the program Phaser (30) and Phenix AutoBuild wizard (31). The crystal structure of ubiquitin (PDB 1UBQ) (32) was used as the search model. Structure refinement was carried out by using the program Phenix with iterative correction and refinement of the atomic model. The atomic model was built to fit 2Fo – Fc electron density map by using the program Coot (33) with careful inspection. The final model has excellent stereochemistry with Rfree of 23.5% at 2.6 Å resolution. Data collection and refinement statistics are shown in Table 1. All molecular graphics were prepared with PyMOL (DeLano Scientific; http://www.pymol.org). Protein-protein interface analysis between UBDs and Ub were analyzed using Proteins, Interfaces, Structures and Assemblies (PISA) database (34). Electrostatic surface potential was calculated using the program APBS tool (35).
Table 1.
Data collection and refinement statistics
| CSB WHD–K48-Ub2 | |
|---|---|
| Data collection | |
| Beamline | SPring-8 BL41XU |
| Space group | P21 |
| Cell constants | |
| a, b, c (Å) | 54.2, 101.7, 66.0 |
| α, β, γ (°) | 90.0, 101.8, 90.0 |
| Resolution | 40–2.6 (2.64–2.6) |
| R sym | 0.210 (0.583) |
| I/σI | 7.4 (1.2) |
| Redundancy | 5.4 (2.6) |
| Completeness (%) | 99.6 (95.3) |
| Refinement | |
| Resolution | 40–2.6 |
| No. reflections | 21,345 |
| R work/Rfree | 0.179/0.236 |
| No. atoms | |
| Protein | 5,282 |
| Ligand | 19 |
| Water | 105 |
| Average B (Å2) | |
| Protein | 44.3 |
| Ligand | 59.6 |
| Water | 35.3 |
| Rmsds | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.677 |
| Ramachandran plot (%) | |
| Favored | 98.6 |
| Outliers | 0.0 |
Values in parentheses are for the highest resolution shell.
SPR analysis
SPR measurements were carried out using a Biacore T200 instrument (GE Healthcare) with HBS-P buffer (10 mM HEPES-Na (pH 7.4), 150 mM NaCl and 0.05% surfactant P20) at 25°C. Anti-GST antibodies (GE Healthcare) were covalently immobilized on the CM5 sensor chip (GE Healthcare) at a density of about 13 000 resonance units (RU). The GST-fused CSB WHD was then captured on the sensor chip at a density of 1000–1500 RU. Ub was injected for 60 s at a flow rate of 10 μl per min. Equilibrium dissociation constants (Kd) were computed by fitting steady state binding level (Req) to a 1:1 interaction model using the Biacore T200 evaluation software (GE Healthcare). All assays were carried out thrice for each sample. The data are presented as mean ± standard deviation.
Expression constructs and stable cell lines
To construct epitope-tagged CSB expression plasmids, the PCR-amplified wild-type CSB (CSBWT) cDNA was cut with XhoI at the 5′ end and with XbaI at the 3′ end, and then cloned in-frame and downstream of the sequence encoding the FLAG epitope, followed by the HA epitope in pcDNA3.1 (Invitrogen). To introduce mutations, the DNA fragment flanked by two ApaI sites (one is located within the CSB cDNA, while the other is derived from pcDNA3.1) was replaced with the mutated fragment, which was generated by PCR using mutated primers. Gibson assembly (New England Biochem) was used for this replacement. To isolate stable transfectants, SV40 immortalized CS1AN (CS1ANSV) cells were transfected with each CSB expression construct using Effectene transfection reagent (Qiagen). Stable transfectants were selected in the presence of 500 μg ml−1 G418 (Nacalai Tesque).
All cell lines used in this study (WI38VA13 (normal), CS1ANSV (CS-B), and CS1ANSV cells stably expressing CSBWT or each CSB mutant) were cultured in DMEM containing 10% FBS, 100 units ml−1 penicillin, and 100 μg ml−1 streptomycin at 37°C in an incubator containing 5% CO2. The protein expression level of CSBWT or mutant CSB in each stable cell line was examined by Western blotting using monoclonal anti-HA tag antibody (catalog no. 11867423001; Roche) and polyclonal anti-lamin B antibody (loading control; catalog no. sc-6216; Santa Cruz Biotechnology). Whole cell extracts for Western blotting were prepared from 1.0 × 106 cells, which were washed once with PBS and lysed with 200 μl of SDS-PAGE sample buffer (62.5 mM Tris–HCl (pH 6.8), 2% SDS, 10% glycerol, 0.01% bromphenol blue and 2.5% mercaptoethanol) by boiling for 5 min.
Cell survival after UV irradiation
Cells (7.0 × 102) were seeded into 10-cm dishes and incubated overnight. The cells were washed once with PBS and exposed to UV-C light irradiation with the indicated dose, immediately followed by addition of culture medium. The irradiated cells were cultured for 10–12 days, fixed with 3% formaldehyde prepared in PBS, and stained with 0.1% crystal violet solution. Colonies were counted using a stereo microscope.
Recovery of RNA synthesis after UV irradiation
Two sets of cells (1.0 × 105) were seeded into 35-mm dishes and incubated overnight. The cells were washed once with PBS and exposed to UV-C light irradiation with 10 J m−2 or no irradiation, followed by incubation in 1 ml of culture medium. After 24 h of incubation, the cells in one set were counted. To measure RNA synthesis, [3H] uridine (PerkinElmer Life Sciences) was supplemented to the other set of cells to a concentration of 370 kBq ml−1. After incubation for 30 min, labeling was terminated by adding NaN3 to a final concentration of 200 μg ml−1. The cells were solubilized with 0.8% SDS, supplemented with an equal volume of 10% TCA containing 0.1 M sodium diphosphate, and incubated on ice. Acid-insoluble material was collected on glass microfiber filters (Whatman GF/C). Radioactivity was measured in Insta-Fluor Plus mixture (PerkinElmer Life Sciences) with a liquid scintillation counter (PerkinElmer Life Sciences Tri-Carb 2810TR). Radioactivity was normalized to cell number. The ratio of the radioactivity of UV-irradiated cells to that of non-irradiated cells was considered to reflect the recovery of RNA synthesis after UV irradiation.
RESULTS
Ub-interacting region of CSB
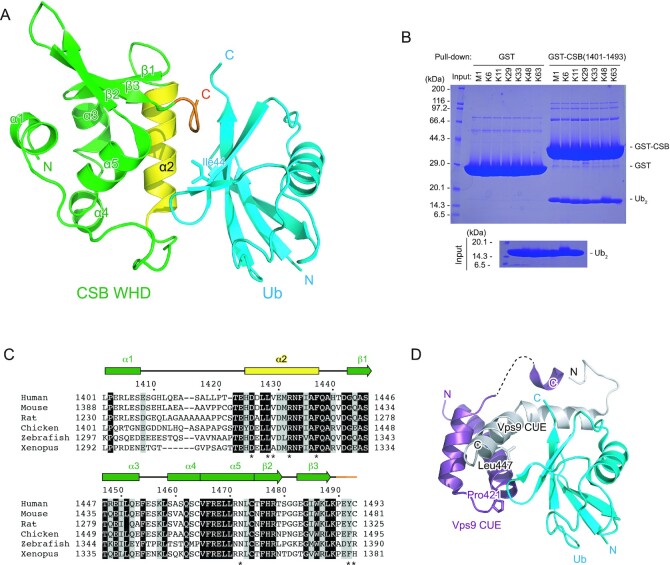
CSB is a 168-kDa protein belonging to the SWI2/SNF2 family, which is usually involved in chromatin remodeling. CSB contains a central ATPase domain consisting of seven conserved helicase motifs (36,37) (Figure 1A). The acidic patch in the N-terminal part of CSB is involved in the regulation of the chromatin remodeling activity (38). In addition, the C-terminal 274 residues of CSB (residues 1220–1493) have been shown to bind to Ub. Some sequence homology has been suggested between a part of the C-terminal region (residues 1400–1428) and a CUE domain, one of the well-studied UBDs (22). Based on the secondary structure prediction and conserved proline residue (Pro1420) and dileucine motif (Leu1427-Leu1428), it has been suggested that residues 1400–1428 of CSB bind to Ub in a manner similar to the CUE domain (22,39) (Figure 1B). However, the GST fusion protein containing the putative CSB UBD (residues 1389–1431) cannot bind to Ub, indicating that this region is devoid of residues critical for protein folding and/or Ub interaction (22). Other part of the C-terminal region of CSB has been shown to be important for UV-induced damage and DSB repair (20,23). In particular, the last 76 residues of CSB (residues 1418–1493) were predicted to fold as a winged-helix domain (WHD) and are required for the recruitment of CSB to DSBs in vivo (20) (Figure 1B). To determine which part of the C-terminal region of CSB is sufficient for binding to Ub, we prepared several truncated forms of this region and assessed their Ub-binding abilities by pull-down assays. Although we were not able to purify the full region (residues 1220–1493) owing to its poor stability in solution, the region encompassing residues 1386–1493 was successfully purified and efficiently bound to Ub, indicating that residues 1220–1385 are dispensable for binding to Ub (Figure 1C). In contrast, a truncation of the last two residues (residues 1386–1491) abolishes Ub binding, revealing that CSB WHD is critical for binding to Ub. Among three N-terminally truncated forms of the putative UBD, two forms (residues 1401–1493 and 1412–1493) can bind to Ub, whereas another form (residues 1426–1493) cannot, indicating that CSB WHD can function as the minimal Ub-binding domain of CSB.
Figure 1.
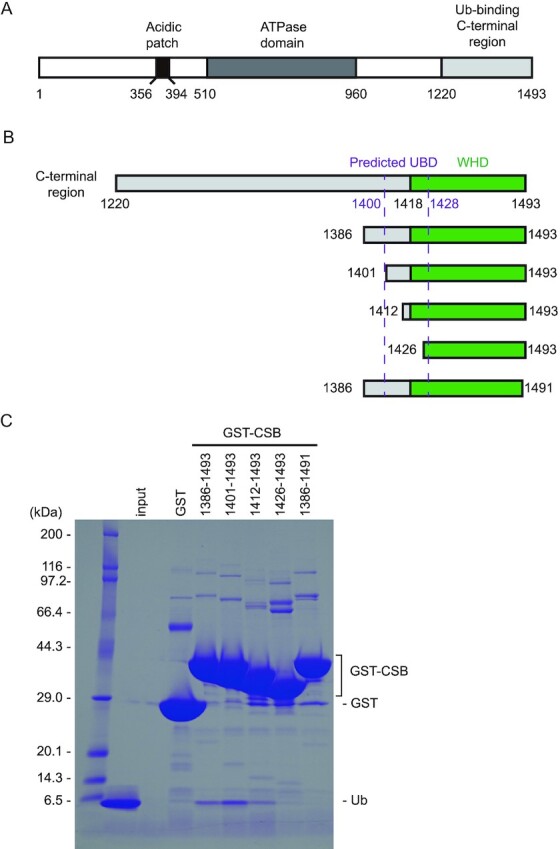
Identification of the minimal Ub-binding domain of CSB. (A) Schematic view of the domain composition of human CSB. The N-terminal acidic patch, ATPase domain, and Ub-binding C-terminal region are shown as black, dark gray and light grey boxes, respectively. (B) Schematic view of CSB constructs used for identification of the minimal Ub-binding domain of CSB. WHD is shown as green boxes. The predicted UBD is delimited by dotted purple lines. (C) Pull-down assays between Ub and the CSB constructs shown in B. The bound Ub molecules were analyzed by SDS-PAGE with Coomassie brilliant blue staining.
Overall structure of CSB WHD in complex with Ub
To understand how CSB WHD binds to Ub, we determined the crystal structure of CSB WHD in complex with K48-Ub2 (Table 1). For crystallization, we used residues 1401–1493 of CSB with the Q1413R variant allele (refSNP rs2228529). The dissociation constants determined by surface plasmon resonance (SPR) spectroscopy are comparable between the two alleles (36.1 ± 2.9 μM for wild-type WHD and 51.5 ± 9.1 μM for the allele variant (Q1413R)) (Table 2 and Supplementary Figure S1). K48-Ub2 (instead of monoUb) was used just for yielding diffraction-quality crystals. The asymmetric unit of the crystal contains four CSB WHD and two K48-Ub2 molecules (Figure 2A). Each WHD molecule interacts with one Ub moiety to exhibit four WHD–Ub complexes, which resemble each other (Figure 2B). The Cα rmsd between two CSB WHDs is 0.24 Å on average. CSB WHD does not interact simultaneously with two Ub moieties, suggesting no binding preference to specific Ub chain types (Figures 2A and 3A). Consistently, WHD bound similarly to M1-, K6-, K11-, K29-, K33-, K48- and K63-Ub2 in our pull-down analysis (Figure 3B). A previous work also showed that this region displayed no chain specificity between Lys48- and Lys63-linked Ub chains (22).
Table 2.
Binding affinity of CSB CTD for Ub
| K d (μM) | |
|---|---|
| WT | 35.6 ± 2.9 |
| Q1413R (variant allele) | 51.5 ± 9.1 |
| D1425A | ND |
| L1428A | ND |
| R1432A | ND |
| F1437D | ND |
| Y1492A | ND |
ND, not detectable.
Data are presented as mean ± standard deviation; n = 3 independent experiments.
Figure 2.
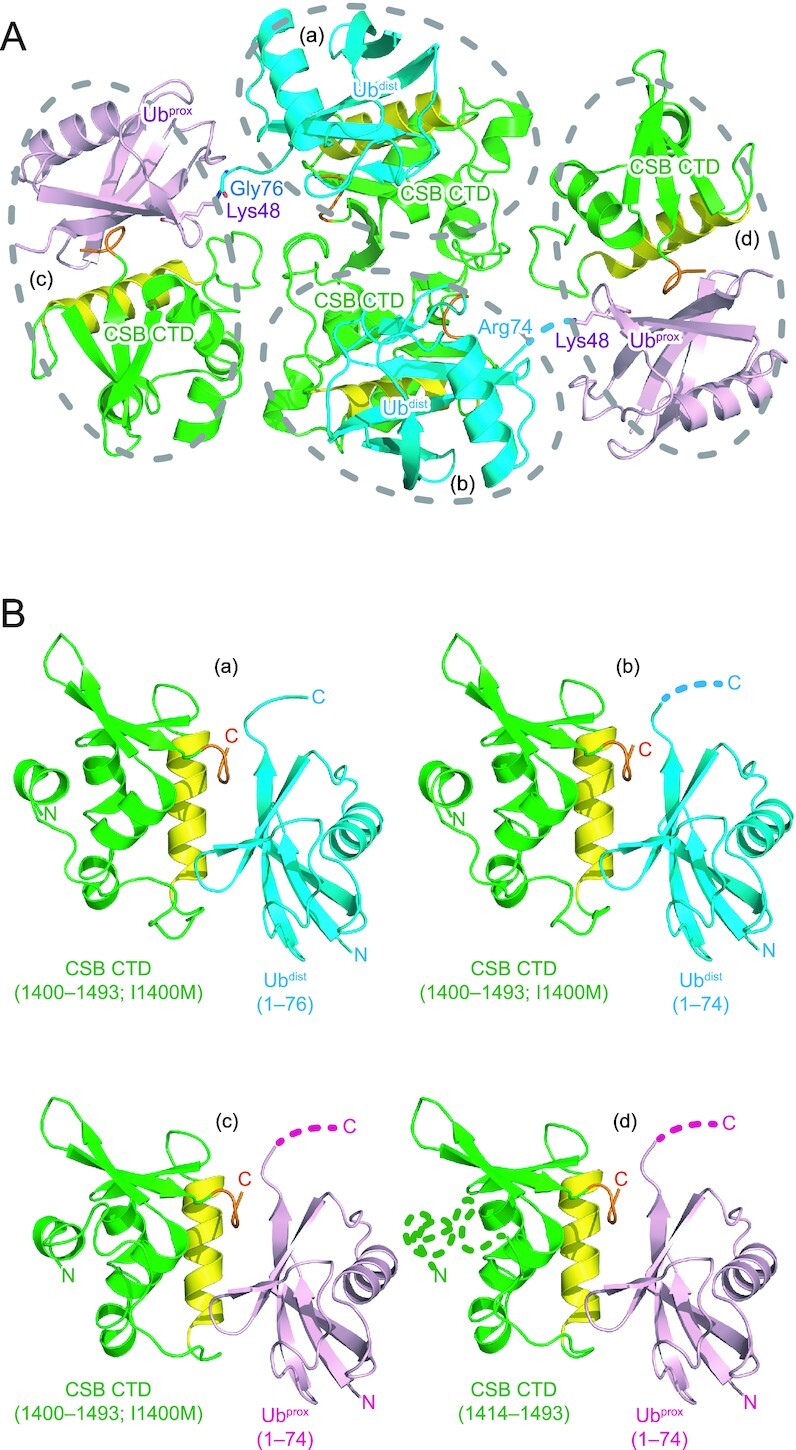
Crystal packing of the CSB WHD–K48-Ub2 complex. CSB WHD is colored green, except that the Ub-interacting α2 and C-terminal extremity are colored yellow and orange, respectively. The proximal and distal Ub moieties of K48-Ub2 (Ubprox and Ubdist, respectively) are colored pink and cyan, respectively. The linkage between Gly76 of Ubdist and Lys48 of Ubprox is shown as sticks. The disordered linkage is indicated by a dotted line. (A) Four CSB WHD and two K48-Ub2 molecules in the asymmetric unit. Four CSB WHD–Ub pairs (a–d) are encircled by dotted lines. (B) Structural comparison of the four CSB WHD–Ub complexes (a–d) in the asymmetric unit. The disordered regions are indicated by dotted lines.
Figure 3.
Structure of the complex between CSB WHD and Ub. (A) Crystal structure of one CSB WHD in complex with Ub. The coloring scheme is the same as that in Figure 2. (B) Pull-down assays between CSB WHD (1401–1493) and M1-, K6-, K11-, K29-, K33-, K48- or K63-Ub2. The bound Ub2 species were analyzed by SDS-PAGE with Coomassie brilliant blue staining. (C) Sequence alignment of CSB WHD from human (NP_000115.1), mouse (NP_001074690.1), rat (NP_001100766.1), chicken (XP_421656.2), zebrafish (XP_688972.2) and Xenopus (NP_001016056.1). Multiple sequence alignment was performed using the program ClustalW (53). Fully conserved residues are colored white with black backgrounds, whereas residues with similar properties (scoring > 0.5 in the Gonnet matrix (54)) are marked with grey backgrounds. Asterisks represent the residues whose side chains interact with Ub. (D) Structure of the complex between Ub and the Vps9 CUE dimer. One Vps9 CUE molecule in the dimer is colored white, whereas the other molecule is colored purple. The side chains of Pro421 and Leu447 in Vps9 CUE are shown as sticks.
CSB WHD folds as a globular domain containing five α-helices (α1–α5) and three β-strands (β1–β3) (Figure 3A, C), although the N-terminal 14 residues (residues 1400–1413) including α1 were not visible in one of the four CSB WHDs in the asymmetric unit (Figure 2B panel (d)). In the other three CSB WHDs, α1 appears to be anchored to an aliphatic patch between α3 and α4 in part by the N-terminal Met, which is a cloning artifact, and the loop connecting α1 and α2 (residues 1401–1420) displays different conformations (Figure 2B). These structural features raise the possibility that the globular part of CSB WHD starts from α2. However, CSB (1422–1493) was poorly expressed in E. coli, suggesting that the proper folding of CSB WHD requires α1 and the following α1–α2 loop. The N-terminal residue of α1 is originally Ile, which likely binds to the aliphatic patch between α3 and α4 in a manner similar to Met in the present CSB WHD structure.
The interaction of CSB WHD with Ub relies on α2 and the C-terminal extremity of CSB WHD (Figure 3A). The amino acid sequences of these two regions are highly conserved among representative vertebrates (Figure 3C). We confirmed that Gln1413 of CSB, which was mutated to Arg in the CSB allele used for the present crystallographic analysis, is located between α1 and α2 and does not interact with Ub. Previously, the region containing residues 1401–1428 of CSB WHD was predicted to interact with Ub in a manner similar to the UBA or CUE domains (22). The dimerized CUE domains of Vps9 bind to Ub through the interactions mediated by Met and Phe in the MFP (Met419 and Phe420 in Vps9 CUE) motif of one CUE domain and Leu in the LL motif (Leu447 in Vps9 CUE) of the other CUE domain (Figure 3D) (39). Although CSB comprises the highly conserved Pro1402 and Leu1428 in the positions corresponding to Pro421 and Leu447 of Vps9, respectively, at the primary sequence level, the Ub-interacting α-helices of CSB display different orientations from those of Vps9 CUE (Figure 3A, D).
Binding interface between CSB WHD and Ub
In the N-terminal part of α2, Asp1425 of CSB forms hydrogen bonds with the main chain NH groups of Ala46 and Gly47 of Ub (Figure 4A). Asn1472 of CSB further forms a hydrogen bond with the main chain CO group of Ub Gly47. In the middle of α2, the side chain of CSB Val1429 is inserted in the canonical hydrophobic pocket of Ub formed by Ile44, Val70, and the aliphatic portion of His68 (Figure 4A). This binding is stabilized by Leu1428 and Tyr1492 in CSB. In the C-terminal extremity of α2, Phe1437 of CSB intercalates between Leu71 and Leu73 of Ub (Figure 4B). The terminal carboxyl group of CSB Cys1493 forms hydrogen bonds with Arg42 and the main chain NH group of Leu73 in Ub. In addition, Arg1432 of CSB also forms a hydrogen bond with the main chain CO group of Ub Leu71 (Figure 4B). Furthermore, the main chain CO group of CSB Pro1490 forms a hydrogen bond with Arg42 of Ub. These Ub-interacting residues in α2 of CSB WHD are conserved or replaced by functionally equivalent residues among representative vertebrates, except that Asn1472 and Val1429 are replaced by Arg and Ala, respectively, in Xenopus CSB (Figure 3C). Xenopus CSB WHD may display different binding interface with Ub from those of other vertebrates.
Figure 4.
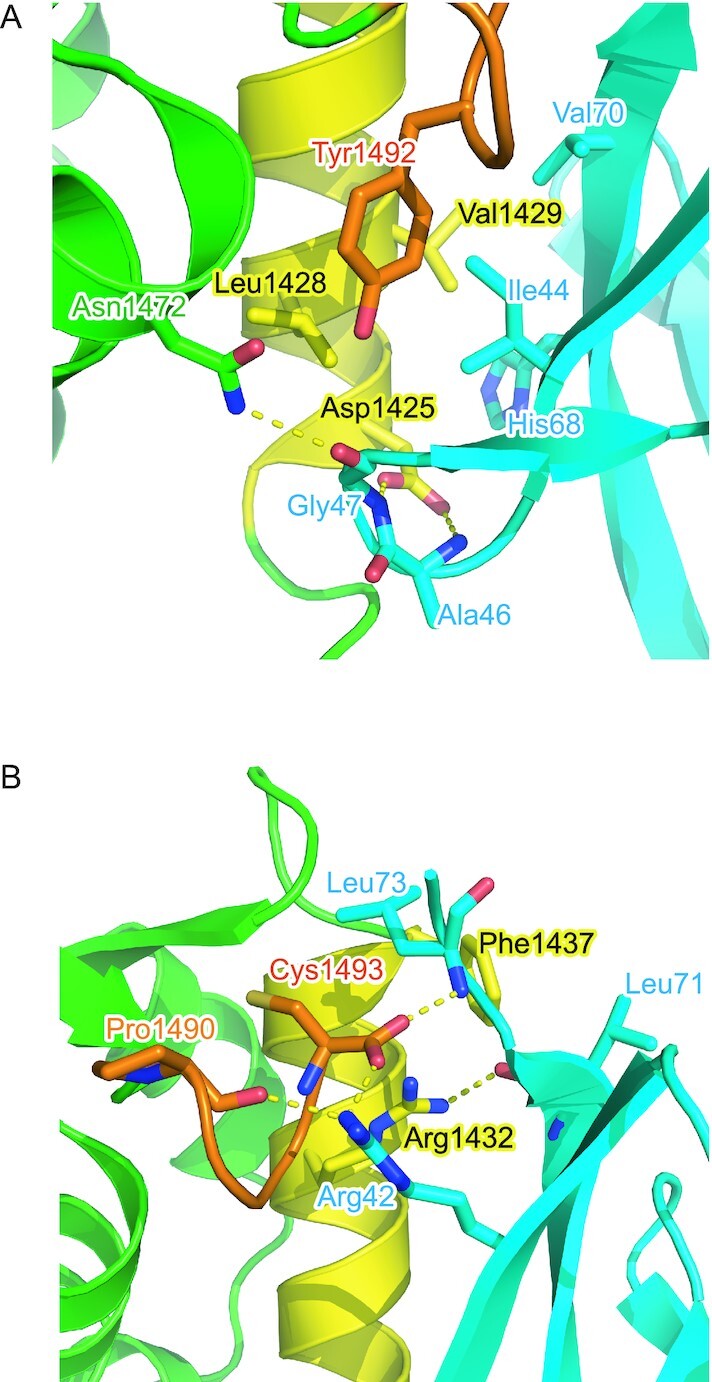
Interfaces between CSB WHD and Ub. The interfaces around the N-terminal (A) and C-terminal (B) regions of α2 are shown. The residues involved in the interaction between CSB WHD and Ub are shown as sticks. Hydrogen bonds are shown as dotted lines. The coloring scheme is the same as that in Figure 2.
To assess the contribution of the Ub-interacting residues in α2 and the C-terminal extremity of CSB WHD to the binding affinity, we investigated the binding between site-directed CSB WHD mutants and Ub by surface plasmon resonance (SPR) spectroscopy. No binding was detected for the D1425A mutant, which should be defective in interacting with the Ala46-Gly47 loop of Ub (Figure 4A, Table 2 and Supplementary Figure S1). Similarly, the L1428A and Y1492A mutations of CSB, which should disrupt the interaction with the Ile44-centered patch of Ub, eliminated Ub binding (Figure 4A, Table 2 and Supplementary Figure S1). Furthermore, no binding could be detected for the R1432A or F1437D mutant, which should be defective in hydrogen bonding to the main chain of Ub Leu71 or hydrophobic interactions with Leu71 and Leu73 of Ub, respectively. Taken together, the Ub-interacting residues in α2 and the C-terminal extremity of CSB WHD, which we found in the present structure of the CSB WHD–Ub complex, are critical for Ub binding.
Assessment of the CSB WHD–Ub interaction in vivo
To further assess the functional significance of the CSB WHD–Ub interaction in vivo, the effects of mutations disrupting the CSB WHD–Ub interaction were examined in the context of cell survival and recovery of RNA synthesis after UV irradiation (Figure 5). We generated stable cell lines expressing CSBWT or site-directed CSB mutants defective in binding to Ub (i.e., CSBD1425A, CSBL1428A, CSBR1432A, CSBF1437D and CSBY1492A) from the CSB-deficient cell line (CS1ANSV) (Figure 5A), and then compared the rates of their cell survival and recovery of RNA synthesis after UV irradiation (RRS). The wild-type cell line (WI38) and CS1ANSV were also examined as normal and UV-sensitive controls, respectively. CS1ANSV was highly sensitive to UV irradiation, whereas CS1ANSV expressing CSBWT was as tolerant to UV irradiation as WI38 (Figure 5B). Consistently, CS1ANSV exhibited the failure of RRS (∼5%), which was recovered to the normal level (WI38; ∼90%) by the expression of CSBWT. These results are in agreement with previous studies (23). On the other hand, all CSB mutant-expressing CS1ANSV cell lines examined in this study exhibited lower UV-survival rates than WI38 or CS1ANSV expressing CSBWT but higher rates than CS1ANSV (Figure 5B). Correspondingly, the failure of RRS in CS1ANSV was partly recovered (∼15–50%) by the expression of the CSB mutants but not to the level of WI38 or CS1ANSV expressing CSBWT (Figure 5C). A similar result has been reported for CS1ANSV expressing CSBL1427G/L1428G, which is analogous to CSBL1428A tested in this study (23). Taken together, defects in the Ub binding of CSB obviously decrease the rates of RRS and cell survival after UV irradiation, suggesting that the CSB WHD–Ub interaction we characterized by crystallography and SPR analysis has a functional relevance in vivo, at least for the repair of UV-induced damage.
Figure 5.
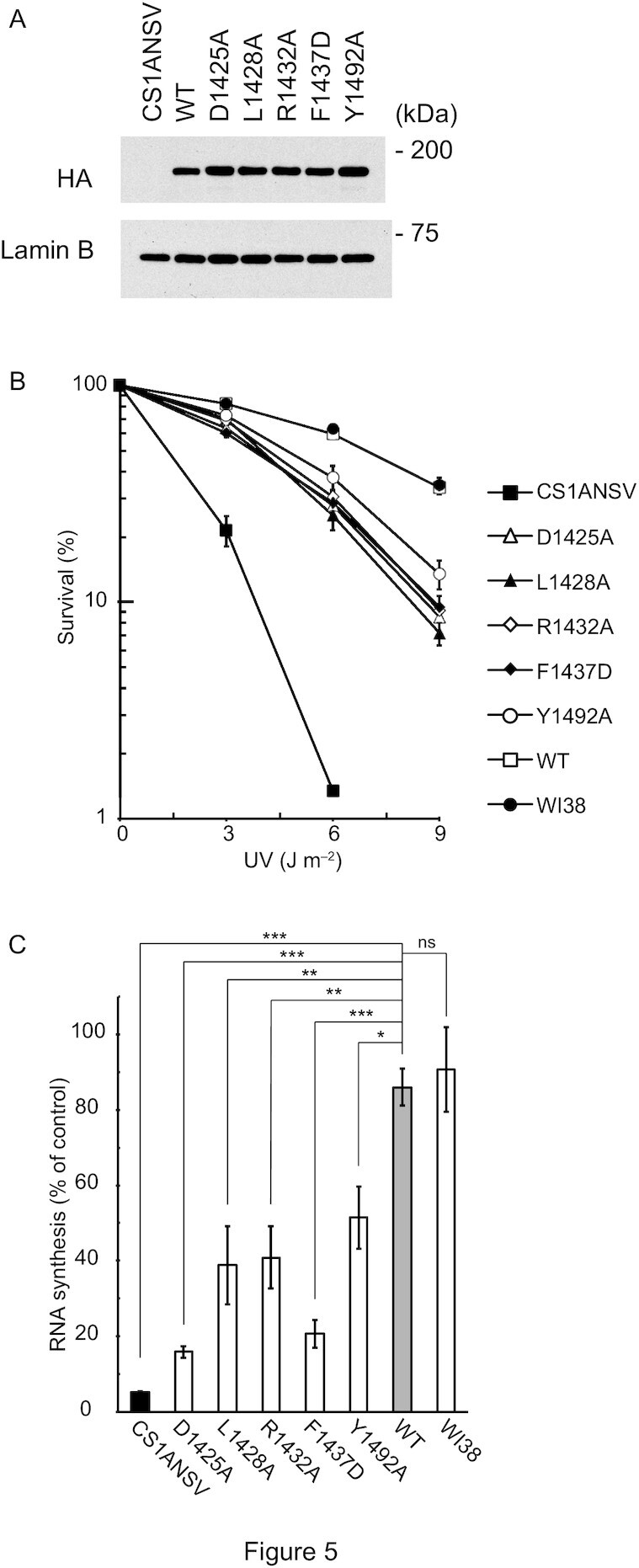
Assessment of the CSB WHD–Ub interaction in vivo. (A) Expression levels of CSBWT and mutant CSB proteins in the CS1ANSV-derived stable cell lines used in this study. The HA tag of each protein was detected by Western blotting using anti-HA tag antibody. Lamin B was also detected as a loading control by anti-lamin B antibody. (B) UV survival of WI38, CS1ANSV, and the CS1ANSV-derived stable cell lines expressing CSBWT or the indicated mutant CSB. The percentage of surviving cells is plotted against UV dose. Error bars indicate standard error from three independent experiments. (C) RNA synthesis recovery after UV irradiation in WI38, CS1ANSV, and the CS1ANSV-derived stable cell lines expressing CSBWT or the indicated mutant CSB. The ratio of the incorporation of [3H] uridine in 10 J m−2-irradiated cells to that of non-irradiated cells was considered to reflect the recovery of RNA synthesis after UV irradiation. Error bars indicate standard error from three independent experiments. Data were statistically analyzed by one-way analysis of variance (ANOVA) with Dunnett's post hoc tests (***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant).
Comparison of Ub-binding architecture
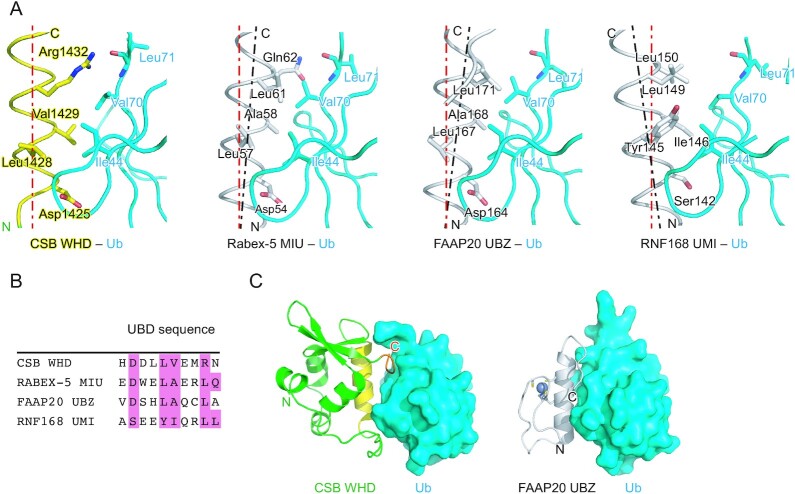
Among the WHDs identified so far, CSB WHD is the first one reported as a UBD. Previous studies on Ub-binding proteins and Ub-related enzymes have identified 23 different types of UBDs (40–45). These UBD structures are clearly different from that of CSB WHD (Supplementary Figure S2). Among the UBDs, UIM (Ub-interacting motif), MIU (motif interacting with Ub), UMI (UIM and MIU-related UBD), and UBZ (Ub-binding zinc finger) involve a single Ub-interacting α-helix (25,26,43,46–48). The orientation of the Ub-interacting helix of CSB WHD (i.e. α2) is the same as those of MIU, UMI, and UBZ (Figure 6A). Asp1425 of CSB is spatially conserved in both Rabex-5 MIU and FAAP20 UBZ, whereas it is replaced by Ser in RNF168 UMI (Figure 6A). The interaction with the Ile44-centered hydrophobic patch of Ub is shared among these four UBDs, although their Ile44 patch-interacting residues are divergent: Val1429 in CSB WHD, Ala58 in Rabex-5, Ala168 in FAAP20 UBZ, and Ile146 in RNF168 UMI (Figure 6B). This difference affects the orientation of each UBD relative to Ub. In Rabex-5 MIU and FAAP20 UBZ, the short side chain of the Ile44 patch-interacting Ala allows the C-terminal end of the Ub-interacting helix to position in close proximity to Ub so that Leu61 of Rabex5 MIU and Leu171 of FAAP20 UBZ can hydrophobically interact with Val70 of Ub (Figure 6A). In contrast, the longer side chain of the Ile44 patch-interacting Ile or Val in CSB WHD or RNF168 UMI, respectively, pushes out the C-terminal end of the Ub-interacting helix. As a result, Leu149 of RNF168 UMI does not interact with Val70 of Ub (Figure 6A). Intriguingly, this conserved Leu of these UBDs is replaced by Arg1432 in CSB WHD (25,26,43,46–48). The aliphatic portion of Arg1432 of CSB WHD interacts with Val70, and the guanidino group forms a hydrogen bond with the main chain CO group of Leu71 in Ub. Hydrogen bonds are also observed between Gln62 of Rabex-5 MIU and the main chain NH and CO groups of Leu71 in Ub in the spatially equivalent position (Figure 6A, B).
Figure 6.
Comparison of CSB WHD, Rabex-5 MIU, FAAP20 UBZ, and RNF168 UMI. (A) Structure comparison of the Ub-binding interfaces of CSB WHD (this study), Rabex-5 MIU (PDB 2C7M), FAAP20 UBZ (PDB 3WWQ), and RNF168 UMI (PDB 5XIS). The coloring scheme of the CSB WHD–Ub complex is the same as that in Figure 2. Rabex-5 MIU, FAAP20 UBZ, and RNF168 UMI are colored white. The residues involved in the Ub-binding interfaces are shown as sticks. The helical axis of α2 in CSB WHD is shown as a red dotted line in each panel to highlight the difference in the orientation of the Ub-interacting helix relative to Ub. The helical axes of α2 in Rabex-5 MIU, FAAP20 UBZ, and RNF168 UMI are shown as black dotted lines. (B) Sequence alignment of CSB WHD, Rabex-5 MIU, FAAP20 UBZ, and RNF168 UMI. The residues involved in the interaction with Ub are highlighted in pink. (C) Comparison between the CSB WHD–Ub and FAAP20 UBZ–Ub (PDB 3WWQ) complexes. The coloring scheme is the same as that in A. The bound Zn2+ and Zn2+-coordinating residues in FAAP20 UBZ are shown as a sphere and sticks, respectively.
The C-terminal extremity of CSB WHD is involved in binding to Ub, as also observed in the FAAP20 UBZ–Ub complex. In FAAP20 UBZ, the flexible C-terminal tail expands the Ub-binding interface beyond the canonical UBZ module (25,26) (Figure 6C). These additional interactions increase the buried surface areas in the FAAP20 UBZ–Ub and CSB WHD–Ub complexes to 621 and 691 Å2, respectively, which are comparable to those in the Rabex-5 MIU–Ub, Vps27 UIM–Ub and RNF168 UMI–Ub complexes (709, 555 and 579 Å2, respectively) (all these values are averaged in the asymmetric unit, except for Vps27 UIM–Ub). At the primary sequence level, the distance between α2 and the C-terminal extremity in CSB WHD (50 residues) is longer than that between the Ub-interacting helix and C-terminal tail in FAAP20 UBZ (seven residues). This difference expands the length of CSB WHD as a UBD. To our knowledge, the essential role of the C-terminal extremity for Ub binding has been described only for these two UBDs. However, other unidentified UBDs might also contain additional structural elements for Ub binding in their C-terminals.
DISCUSSION
The winged-helix motif is known as a helix-turn-helix DNA-binding motif consisting of three α helices (H1–H3) and three β strands (S1–S3), which are connected in order of H1–S1–H2–H3–S2–S3 (49). The long loop between S2 and S3 and that following S3 are called wings W1 and W2, respectively, for their similarities with the appendages of a butterfly. In most WHDs such as HNF-3γ WHD (Figure 7A), H3 plays a central role in the DNA recognition with the contribution of W2 (50). H3 corresponds to α4 and α5 in CSB WHD, which appear to form a single continuous helix but are separated at Val1465. The surface of α4 and α5 corresponding to the DNA-interacting surface of the DNA-binding winged-helix motif is covered with the loop connecting α1 and α2 in CSB WHD (Figure 7A), although one cannot exclude the possibility that the loop conformation could be changed, considering the above-mentioned flexibility of this loop and α1. The truncation of W2 in CSB WHD might favor the interaction with Ub. Other WHDs such as RFX1 (Figure 7A, right) display a different DNA-binding mechanism mainly through the interaction between W1 and DNA (51). In CSB WHD, the 3-residue loop (1480SGG1482) corresponding to W1 should not be able to interact with the DNA. Electrostatically, the surfaces of HNF-3γ and RFX1 WHDs contain positively charged regions that are widely distributed for the interaction with the negatively charged phosphate backbone of DNA (Figure 7B). In contrast, positive charges on the surface of CSB WHD are relatively weak and sparsely distributed, suggesting that CSB WHD is incapable of binding to DNA (or RNA) (Figure 7B). Reversely, to our knowledge, no other WHDs have been reported (or predicted) as UBDs. To clearly distinguish between the DNA-binding and Ub-binding WHDs, we propose to name the latter UBW (Ub-binding WHD) as a new class of UBDs.
Figure 7.
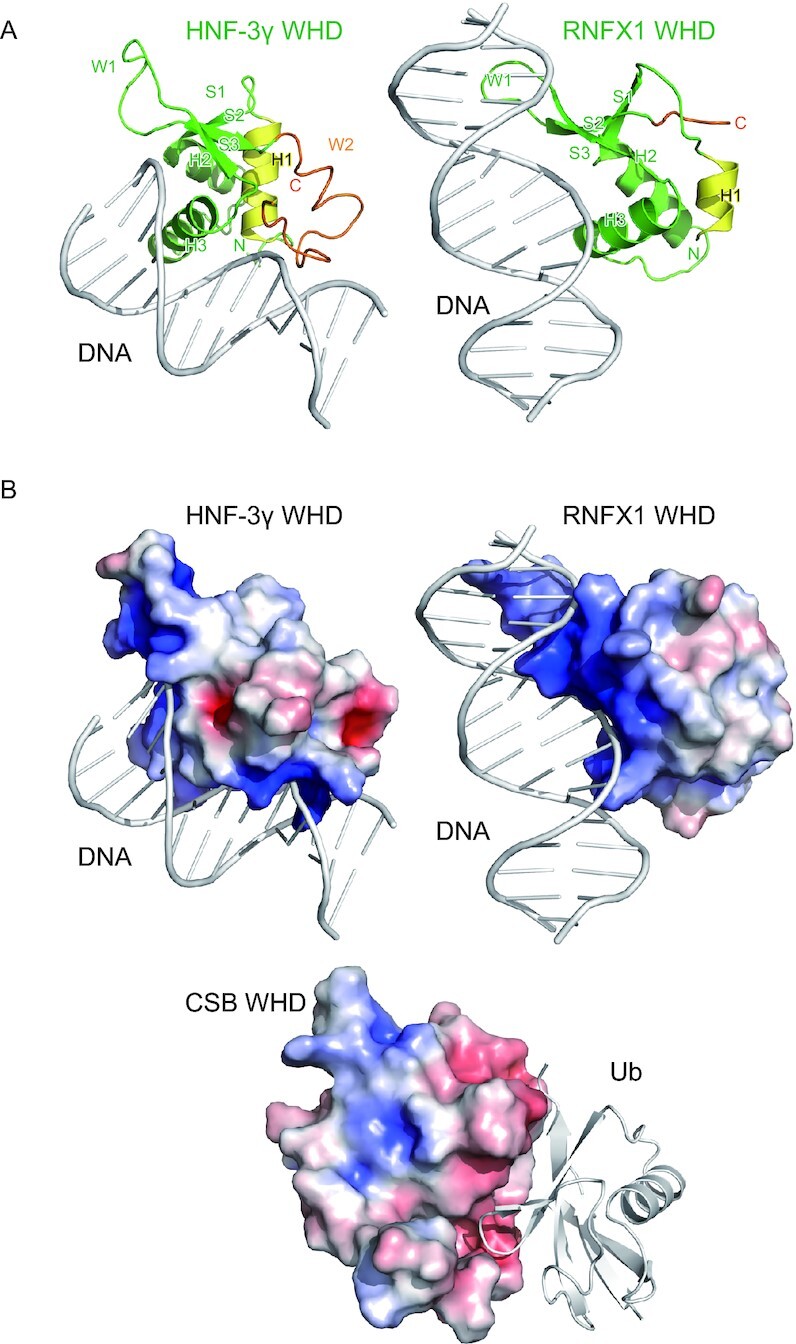
Comparison with DNA-binding WHDs. (A) Crystal structures of the DNA-bound HNF-3γ WHD (PDB 1VTN) and RFX1 WHD (PDB 1DP7). The coloring scheme of the WHDs is based on that in Figure 2. The DNA molecules are colored white. (B) Electrostatic surface potentials of CSB WHD, HNF-3γ WHD and RFX1 WHD on a scale from −5 kBT/e (red) to +5 kBT/e (blue). Interacting DNA or Ub are also shown as white cartoon models.
The crystal structure of the CSB WHD–Ub complex shows the α-helix-mediated Ub-binding mechanism, which is unexpectedly similar to those of other UBDs such as UIM, MIU, UBZ, and UMI, despite divergent primary sequences (Figure 6B). The C-terminal extremity of CSB WHD is also involved in Ub binding (20,23). Therefore, the role of CSB WHD on DSB repair and SUMO-dependent NER may be associated with ubiquitylation events. Deletion of CSB UBD or WHD has been performed to understand their functional roles in DNA repair. However, such deletion may affect not only a Ub-binding property of CSB but also other unknown functions (19,20,23,24). Point mutations are beneficial to assess a specific function of CSB (20,23). Our crystal structure-based analysis revealed point mutations that can more specifically inactivate the CSB interaction with Ub. The truncation of the last two residues of CSB is also sufficient to prevent CSB from binding to Ub with little effect on the rest of CSB. Overall, the crystal structure of the CSB WHD–Ub complex presented in this study should be helpful for understanding of the CSB function in DNA repair including Ub-dependent degradation of ATF3 (21,52).
DATA AVAILABILITY
Coordinates and structure factors of the CSB WHD–K48–Ub2 complex have been deposited in the Protein Data Bank under accession code 6A6I. Other data are available from the corresponding authors upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline staff of macromolecular crystallography beamlines of Photon Factory (Tsukuba, Japan) and BL41XU of SPring-8 (Hyogo, Japan) for technical help during data collection.
Authors contributions: D.T. designed the work, performed the crystallography and interaction analyses, and wrote the paper. Y.S., A.Y. and S.G.-I. assisted with the crystallography and interaction analyses. M.S. performed the experiments with mutant CSB transfectants. All authors discussed the results. S.F. supervised the work and wrote the paper.
Contributor Information
Diane T Takahashi, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan; Synchrotron Radiation Research Organization, The University of Tokyo, Tokyo 113-0032, Japan.
Yusuke Sato, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan; Synchrotron Radiation Research Organization, The University of Tokyo, Tokyo 113-0032, Japan; Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-8562, Japan.
Atsushi Yamagata, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan; Synchrotron Radiation Research Organization, The University of Tokyo, Tokyo 113-0032, Japan; Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-8562, Japan.
Sakurako Goto-Ito, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan; Synchrotron Radiation Research Organization, The University of Tokyo, Tokyo 113-0032, Japan.
Masafumi Saijo, Graduate School of Frontier Biosciences, Osaka University, Yamadaoka 1-3, Suita, Osaka 565-0871, Japan.
Shuya Fukai, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan; Synchrotron Radiation Research Organization, The University of Tokyo, Tokyo 113-0032, Japan; Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-8562, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science [16H04750 to Y.S., 15F15386 to D.T.]; Japan Science and Technology Agency [JPMJCR1XMX5 to S.F.]. Funding for open access charge: Japan Society for the Promotion of Science [18H05501 to S.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 2. Laine J.P., Egly J.M.. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006; 25:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spivak G. UV-sensitive syndrome. Mutat. Res. 2005; 577:162–169. [DOI] [PubMed] [Google Scholar]
- 4. Nance M.A., Berry S.A.. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 1992; 42:68–84. [DOI] [PubMed] [Google Scholar]
- 5. Laugel V., Dalloz C., Durand M., Sauvanaud F., Kristensen U., Vincent M.C., Pasquier L., Odent S., Cormier-Daire V., Gener B.et al.. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum. Mutat. 2010; 31:113–126. [DOI] [PubMed] [Google Scholar]
- 6. Mallery D.L., Tanganelli B., Colella S., Steingrimsdottir H., van Gool A.J., Troelstra C., Stefanini M., Lehmann A.R.. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998; 62:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakazawa Y., Sasaki K., Mitsutake N., Matsuse M., Shimada M., Nardo T., Takahashi Y., Ohyama K., Ito K., Mishima H.et al.. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 2012; 44:586–592. [DOI] [PubMed] [Google Scholar]
- 8. Schwertman P., Lagarou A., Dekkers D.H., Raams A., van der Hoek A.C., Laffeber C., Hoeijmakers J.H., Demmers J.A., Fousteri M., Vermeulen W.et al.. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 2012; 44:598–602. [DOI] [PubMed] [Google Scholar]
- 9. Zhang X., Horibata K., Saijo M., Ishigami C., Ukai A., Kanno S., Tahara H., Neilan E.G., Honma M., Nohmi T.et al.. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat. Genet. 2012; 44:593–597. [DOI] [PubMed] [Google Scholar]
- 10. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M.A., Chong J., Hare A.A., Dervan P.B., DiMaio F., Leschziner A.E.et al.. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017; 551:653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brueckner F., Hennecke U., Carell T., Cramer P.. CPD damage recognition by transcribing RNA polymerase II. Science. 2007; 315:859–862. [DOI] [PubMed] [Google Scholar]
- 12. Fousteri M., Vermeulen W., van Zeeland A.A., Mullenders L.H.. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 2006; 23:471–482. [DOI] [PubMed] [Google Scholar]
- 13. de Waard H., de Wit J., Gorgels T.G., van den Aardweg G., Andressoo J.O., Vermeij M., van Steeg H., Hoeijmakers J.H., van der Horst G.T.. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst.). 2003; 2:13–25. [DOI] [PubMed] [Google Scholar]
- 14. Maddukuri L., Speina E., Christiansen M., Dudzinska D., Zaim J., Obtulowicz T., Kabaczyk S., Komisarski M., Bukowy Z., Szczegielniak J.et al.. Cockayne syndrome group B protein is engaged in processing of DNA adducts of lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat. Res. 2009; 666:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tuo J., Muftuoglu M., Chen C., Jaruga P., Selzer R.R., Brosh R.M. Jr, Rodriguez H., Dizdaroglu M., Bohr V.A.. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001; 276:45772–45779. [DOI] [PubMed] [Google Scholar]
- 16. Charlet-Berguerand N., Feuerhahn S., Kong S.E., Ziserman H., Conaway J.W., Conaway R., Egly J.M.. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006; 25:5481–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khobta A., Kitsera N., Speckmann B., Epe B.. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair (Amst.). 2009; 8:309–317. [DOI] [PubMed] [Google Scholar]
- 18. Kitsera N., Stathis D., Luhnsdorf B., Muller H., Carell T., Epe B., Khobta A.. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011; 39:5926–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei L., Nakajima S., Bohm S., Bernstein K.A., Shen Z., Tsang M., Levine A.S., Lan L.. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3495–E3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Batenburg N.L., Walker J.R., Noordermeer S.M., Moatti N., Durocher D., Zhu X.D.. ATM and CDK2 control chromatin remodeler CSB to inhibit RIF1 in DSB repair pathway choice. Nat. Commun. 2017; 8:1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Epanchintsev A., Costanzo F., Rauschendorf M.A., Caputo M., Ye T., Donnio L.M., Proietti-de-Santis L., Coin F., Laugel V., Egly J.M.. Cockayne's syndrome A and B proteins regulate transcription arrest after genotoxic stress by promoting ATF3 degradation. Mol. Cell. 2017; 68:1054–1066. [DOI] [PubMed] [Google Scholar]
- 22. Anindya R., Mari P.O., Kristensen U., Kool H., Giglia-Mari G., Mullenders L.H., Fousteri M., Vermeulen W., Egly J.M., Svejstrup J.Q.. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell. 2010; 38:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sin Y., Tanaka K., Saijo M.. The C-terminal region and SUMOylation of Cockayne syndrome group B protein play critical roles in transcription-coupled nucleotide excision repair. J. Biol. Chem. 2016; 291:1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ranes M., Boeing S., Wang Y., Wienholz F., Menoni H., Walker J., Encheva V., Chakravarty P., Mari P.O., Stewart A.et al.. A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2016; 44:5246–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toma A., Takahashi D.T., Sato Y., Yamagata A., Goto-Ito S., Nakada S., Fukuto A., Horikoshi Y., Tashiro S., Fukai S.. Structural basis for ubiquitin recognition by ubiquitin-binding zinc finger of FAAP20. PLoS One. 2015; 10:e0120887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wojtaszek J.L., Wang S., Kim H., Wu Q., D’Andrea A.D., Zhou P.. Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain. Nucleic Acids Res. 2014; 42:13997–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sato Y., Okatsu K., Saeki Y., Yamano K., Matsuda N., Kaiho A., Yamagata A., Goto-Ito S., Ishikawa M., Hashimoto Y.et al.. Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30. Nat. Struct. Mol. Biol. 2017; 24:911–919. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Echols N., Headd J.J., Hung L.W., Jain S., Kapral G.J., Grosse Kunstleve R.W.et al.. The Phenix software for automated determination of macromolecular structures. Methods. 2011; 55:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D.. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vijay-Kumar S., Bugg C.E., Cook W.J.. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 1987; 194:531–544. [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 35. Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A.. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Selby C.P., Sancar A.. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997; 272:1885–1890. [DOI] [PubMed] [Google Scholar]
- 37. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 38. Lake R.J., Geyko A., Hemashettar G., Zhao Y., Fan H.Y.. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell. 2010; 37:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prag G., Misra S., Jones E.A., Ghirlando R., Davies B.A., Horazdovsky B.F., Hurley J.H.. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003; 113:609–620. [DOI] [PubMed] [Google Scholar]
- 40. Dikic I., Wakatsuki S., Walters K.J.. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009; 10:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He F., Wollscheid H.P., Nowicka U., Biancospino M., Valentini E., Ehlinger A., Acconcia F., Magistrati E., Polo S., Walters K.J.. Myosin VI contains a compact structural motif that binds to ubiquitin chains. Cell Rep. 2016; 14:2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pinato S., Gatti M., Scandiuzzi C., Confalonieri S., Penengo L.. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol. Cell. Biol. 2011; 31:118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi D.T., Hirade Y., Toma A., Sato Y., Yamagata A., Goto-Ito S., Tomita A., Nakada S., Fukai S.. Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168. Nat. Commun. 2018; 9:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peisley A., Wu B., Xu H., Chen Z.J., Hur S.. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014; 509:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J.. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010; 141:315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S., Tsai Y.C., Mattera R., Smith W.J., Kostelansky M.S., Weissman A.M., Bonifacino J.S., Hurley J.H.. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. 2006; 13:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Penengo L., Mapelli M., Murachelli A.G., Confalonieri S., Magri L., Musacchio A., Di Fiore P.P., Polo S., Schneider T.R.. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006; 124:1183–1195. [DOI] [PubMed] [Google Scholar]
- 48. Swanson K.A., Kang R.S., Stamenova S.D., Hicke L., Radhakrishnan I.. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003; 22:4597–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harami G.M., Gyimesi M., Kovacs M.. From keys to bulldozers: expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem. Sci. 2013; 38:364–371. [DOI] [PubMed] [Google Scholar]
- 50. Clark K.L., Halay E.D., Lai E., Burley S.K.. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993; 364:412–420. [DOI] [PubMed] [Google Scholar]
- 51. Gajiwala K.S., Chen H., Cornille F., Roques B.P., Reith W., Mach B., Burley S.K.. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000; 403:916–921. [DOI] [PubMed] [Google Scholar]
- 52. Caputo M., Balzerano A., Arisi I., D’Onofrio M., Brandi R., Bongiorni S., Brancorsini S., Frontini M., Proietti-De-Santis L.. CSB ablation induced apoptosis is mediated by increased endoplasmic reticulum stress response. PLoS One. 2017; 12:e0172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R.et al.. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 54. Gonnet G.H., Cohen M.A., Benner S.A.. Exhaustive matching of the entire protein sequence database. Science. 1992; 256:1443–1445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors of the CSB WHD–K48–Ub2 complex have been deposited in the Protein Data Bank under accession code 6A6I. Other data are available from the corresponding authors upon reasonable request.