Abstract
The optimal root system architecture (RSA) of a crop is context dependent and critical for efficient resource capture in the soil. Narrow root growth angle promoting deeper root growth is often associated with improved access to water and nutrients in deep soils during terminal drought. RSA, therefore is a drought-adaptive trait that could minimize yield losses in regions with limited rainfall. Here, GWAS for seminal root angle (SRA) identified seven marker-trait associations clustered on chromosome 6A, representing a major quantitative trait locus (qSRA-6A) which also displayed high levels of pairwise LD (r2 = 0.67). Subsequent haplotype analysis revealed significant differences between major groups. Candidate gene analysis revealed loci related to gravitropism, polar growth and hormonal signaling. No differences were observed for root biomass between lines carrying hap1 and hap2 for qSRA-6A, highlighting the opportunity to perform marker-assisted selection for the qSRA-6A locus and directly select for wide or narrow RSA, without influencing root biomass. Our study revealed that the genetic predisposition for deep rooting was best expressed under water-limitation, yet the root system displayed plasticity producing root growth in response to water availability in upper soil layers. We discuss the potential to deploy root architectural traits in cultivars to enhance yield stability in environments that experience limited rainfall.
Keywords: root angle, seminal roots, root architecture, GWAS, QTL, haplotype, drought adaptation
Introduction
Durum wheat (Triticum durum Desf.) is a major staple crop in the Mediterranean region (Shewry and Hey, 2015) and other semi-arid regions of the world (Araus et al., 2002). The crop is typically grown under rain-fed conditions where water scarcity is a major limiting factor for productivity, particularly when drought occurs during the flowering or grain filling period (Loss and Siddique, 1994; Belaid, 2000; Mohammadi et al., 2011; Bassi and Sanchez-Garcia, 2017). Due to climate change, rainfall patterns are predicted to change in most durum production regions worldwide, particularly in the Mediterranean region (Christensen et al., 2007; Carvalho et al., 2014). Therefore, breeding durum for water-limiting environments is a priority (Cattivelli et al., 2008; Boutraa, 2010).
Until recently, breeding programs have focused on above ground traits and direct selection for yield per se, while the crop’s “hidden-half,” i.e., the roots have been largely overlooked. Plant roots are important organs in determining grain productivity driven by water uptake and nutrient acquisition (Sharma et al., 2009; Ehdaie et al., 2012; Shen et al., 2013; Palta and Yang, 2014). Hence, improving RSA in breeding programs is a promising strategy to increase the resilience of durum wheat genotypes in drought-prone environments (Sanguineti et al., 2007; Manschadi et al., 2008). RSA has been recognized as one of the foundations for crop adaptation under water stress conditions (Manschadi et al., 2006; Christopher et al., 2008; Gregory et al., 2009; Asif and Kamran, 2011). Root length, density and root depth are the main components of RSA influencing water extraction in deep soils (King et al., 2003; Asif and Kamran, 2011; Carvalho et al., 2014). These adaptive features determining the root distribution in the soil profile have been associated with root growth angle (Nakamoto et al., 1991; Oyanagi et al., 1993; Oyanagi, 1994; Borrell et al., 2014). In durum wheat, seminal root angle (SRA) is representative of the mature RSA and provides a useful proxy because the trait can be easily phenotyped at the seedling stage (Tuberosa et al., 2002a,b, 2007; de Dorlodot et al., 2007; Fang et al., 2017; El Hassouni et al., 2018). For instance, a narrow SRA is associated with a higher proportion of roots at depth at the mature stage in wheat (Nakamoto and Oyanagi, 1994; Bengough et al., 2004; Manschadi et al., 2008), similar to root growth angle reported in other major crops like sorghum and rice (Omori and Mano, 2007; Uga et al., 2011; Mace et al., 2012). A narrow SRA can improve access to residual moisture in deep soils, particularly under terminal drought conditions (Manschadi et al., 2006; Reynolds et al., 2007; Christopher et al., 2008; Acuña and Wade, 2012; Hamada et al., 2012) and can prolong the grain filling period to improve yield (Blum et al., 1983; Lynch, 1995; Kashiwagi et al., 2005). On the other hand, wide SRA is associated with a shallow root system that may be beneficial for exploring the superficial soil layers and capturing in-season rainfall. Therefore, identifying the optimal RSA in each target environment is critical to guide breeding efforts (El Hassouni et al., 2018). Minor differences in the distribution of roots in the soil space can lead to major impacts on yield. For instance, results from modeling studies suggest that wheat yield would increase by 55 kg.ha-1 for each additional millimeter of water extracted from the soil during the critical grain filling stage (Manschadi et al., 2006; Kirkegaard et al., 2007; Christopher et al., 2013). Furthermore, a recent study examining RSA in durum wheat suggests that genotypes with deep root systems could increase grain yield up to 35% and thousand kernel weight by 9% in environments with limited moisture, compared to genotypes with shallow root systems (El Hassouni et al., 2018). The availability of large genetic variability in terms of rooting patterns and the high heritability of SRA (Manschadi et al., 2006, 2008; Maccaferri et al., 2016; Alahmad et al., 2018; El Hassouni et al., 2018) are two key factors suggesting that optimization of the roots could potentially deliver high yielding durum cultivars in water-limiting environments.
In comparison to aboveground traits, studying root traits have been a challenge for plant breeders (Zhang et al., 2009), largely due to lack of efficient and reliable root phenotyping methods and limited knowledge of the genetic control of root development (Tuberosa et al., 2002b; Zhang et al., 2009; Richards et al., 2010; Mace et al., 2012; Shen et al., 2013; Carvalho et al., 2014; Wang et al., 2014). Recently, a high-throughput, affordable and scalable phenotyping method for screening seminal root angle under controlled conditions has been developed, known as the ‘clear pot’ method (Richard et al., 2015), and has been successfully applied to durum wheat, barley, and bread wheat. The technique has facilitated direct phenotypic selection of SRA (Alahmad et al., 2018; Richard et al., 2018), phenotyping cultivars and breeding lines to investigate yield trends (El Hassouni et al., 2018; Robinson et al., 2018), and phenotyping of mapping populations required for QTL discovery (Robinson et al., 2016). While evaluation of mature RSA in the field is challenging, moderately efficient techniques have been developed, such as ‘shovelomics’ (Trachsel et al., 2011), soil coring (Wasson et al., 2014) and the ‘pasta strainer’ method (El Hassouni et al., 2018). Despite the challenges, good progress has been made to identify some of the genomic regions influencing RSA in durum wheat, with several bi-parental and association mapping studies published to date (Sanguineti et al., 2007; Cane et al., 2014; Maccaferri et al., 2016). A recent prioritization analysis of QTL detected in bi-parental and association mapping studies identified nine main QTL clusters on chromosomes 2A, 2B, 4B, 6A, 7A, and 7B, which appear to be most valuable for breeding applications (Maccaferri et al., 2016). However, further research is required to dissect the genetics of RSA in durum wheat that is relevant to breeders, along with the discovery of large effect QTL that are most desirable for marker-assisted breeding.
This study applied the ‘clear pot’ method to phenotype elite durum populations derived from crosses between Australian and ICARDA germplasm pools and performed a genome-wide association study (GWAS) using DArT-seq markers. A major QTL was identified on chromosome 6A that modulates growth angle, but not root biomass. This major QTL could be exploited and combined with root biomass, thus facilitating the development of new varieties with designer root systems that optimize resource capture in the soil profile targeting different environments.
Materials and Methods
Plant Material
A panel of 14 genotypes (Table 1) was evaluated for SRA under controlled conditions and nodal root angle in the field to investigate correlation between these traits. This included eight genotypes imported into Australia in 2015 from ICARDA’s durum wheat breeding program in Morocco (Fastoz2, Fastoz3, Fastoz6, Fastoz7, Fastoz8, Fastoz10, Outrob4, and Fadda98). The lines were preselected for drought adaptation and used as parents in breeding programs targeting marginal rainfall regions of West Asia and North Africa. Three Australian durum commercial varieties were also included (DBA Aurora, Jandaroi, Yawa) which are preferred by growers and the pasta industry due to high yield potential and protein content. In addition, bread wheat varieties Mace, Wylie and Scout were included with Mace and Scout used as standards of known root angle phenotype (Table 1).
Table 1.
Details for the panel of 14 durum wheat and bread wheat standards examined in this study.
| Genotype ID | Ploidy | Origin | Pedigree |
|---|---|---|---|
| DBA-Aurora | Tetraploid | Australia | Tamaroi∗2/Kalka//RH920318/Kalka///Kalka∗2/Tamaroi |
| Jandaroi | Tetraploid | Australia | 110780/111587 |
| Yawa | Tetraploid | Australia | Westonia/Kalka//Kalka/Tamaroi///RAC875/Kalka//Tamaroi |
| Outrob4 | Tetraploid | ICARDA | Ouassel1/4/GdoVZ512/Cit//Ruff/Fg/3/Pin/Gre//Trob |
| Fadda98 | Tetraploid | ICARDA | Awl2/Bit |
| Fastoz2 | Tetraploid | ICARDA | T.polonicumTurkeyIG45272/6/ICAMORTA0463/5/Mra1/4/Aus1/3/Scar/GdoVZ579//Bit |
| Fastoz3 | Tetraploid | ICARDA | Msbl1//Awl2/Bit/3/T.dicoccoidesSYRIG117887 |
| Fastoz6 | Tetraploid | ICARDA | Azeghar1/6/Zna1/5/Awl1/4/Ruff//Jo/Cr/3/F9.3/7/Azeghar1//Msbl1/Quarmal |
| Fastoz7 | Tetraploid | ICARDA | CandocrossH25/Ysf1//CM829/CandocrossH25 |
| Fastoz8 | Tetraploid | ICARDA | MorlF38//Bcrch1/Kund1149/3/Bicrederaa1/Miki |
| Fastoz10 | Tetraploid | ICARDA | Younes/TdicoAlpCol//Korifla |
| Mace | Hexaploid | Australia | Wyalkatchem/Stylet//Wyalkatchem[3798] |
| Scout | Hexaploid | Australia | Sunstate/QH-71-6//Yitpi[4113][4174][4177] |
| Wylie | Hexaploid | Australia | QT-2327/Cook//QT-2804[3596][3784] |
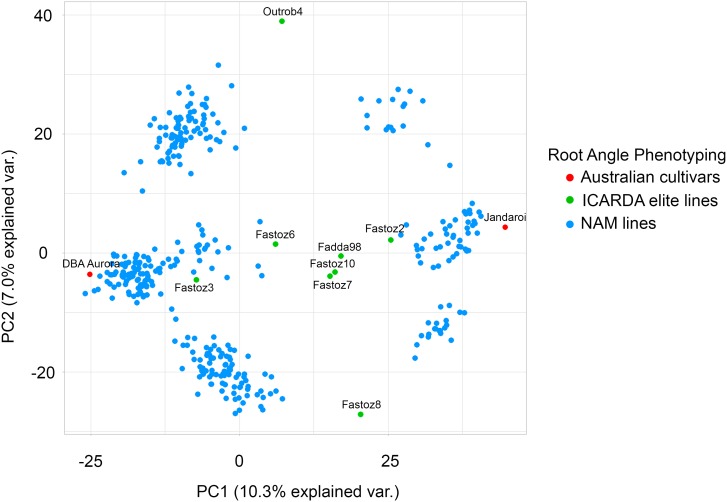
A subset of 393 durum recombinant inbred lines from a nested association mapping (NAM) population were evaluated for SRA and used for GWAS. The NAM population was generated by crossing the eight ICARDA lines listed above as ‘founders’ to the ‘reference’ Australian durum varieties Jandaroi and DBA Aurora. The speed breeding facility at The University of Queensland was used to rapidly progress through six generations of spring durum wheat in a year (Ghosh et al., 2018; Watson et al., 2018). The NAM resource comprises 10 donor × reference sub-populations of 92 F6 lines each (Figure 1). The subset of 393 lines evaluated for SRA was selected from the ten families based on agronomic appearance in the field.
FIGURE 1.
Population structure for the durum NAM lines evaluated for seminal root angle using the clear pot method. A principal component analysis based on pairwise modified Roger’s distances calculated from 2,541 polymorphic DArTseq markers was performed for the 393 NAM lines. The ten families NAM lines are derived from two Australian reference varieties (red) and ICARDA elite lines (green).
Phenotyping Seminal Root Angle Under Controlled Conditions
The panel of 14 genotypes including NAM parents and bread wheat standards (Table 1) were phenotyped for SRA, using the ‘clear pot’ method which is suitable for screening small grain crops (Richard et al., 2015; Robinson et al., 2016; Alahmad et al., 2018). In this experiment, clear pots were filled with composted fine, black-colored pine bark, consisting of 70% particles 0–5 mm in size, pre-mixed with 30% coco peat to increase the water-holding capacity. A randomized complete block design (RCBD) was adopted using R V3.4.3 (R Core Team, 2017), with 15 replicates per genotype and 24 positions per 4 L pot. Pots were placed on the bench in a distinct column/row grid according to the RCBD design. Seeds were planted in the pots carefully positioning the embryo facing the wall of the pot and vertically with the radical pointed downward. This allows enhanced visibility of the seminal roots following germination. Plants were grown in the glasshouse under diurnal natural light conditions and constant temperature (17 ± 2°C) as recommended by Richard et al. (2015). Images were captured 5 days after sowing (seminal roots 3–5 cm in length) using a Canon PowerShot SX600 HS 16MP Ultra–Zoom Digital camera. The angle between the first pair of seminal roots was measured from the images using ImageJ software1. Two bread wheat genotypes were tested as standards, including Mace for wide and Scout for narrow SRA (Alahmad et al., 2018). The subset of 393 NAM lines, parents and standards were subsequently phenotyped for SRA using the same procedure as described above.
Phenotyping Nodal Root Angle in the Field
The panel of 14 genotypes was evaluated for nodal root angle in the field using a ‘shovelomics’ approach (Trachsel et al., 2011). The field experiment was conducted at The University of Queensland Gatton Research Station (27°32′45″ S; 152°19′44″ E) known for summer dominant rainfall and clay soils, Queensland, Australia, in 2017. RCBD was adopted using R V3.4.3 (R Core Team, 2017) where each genotype was replicated 3 times in 7.5 m2 yield plots (5 rows spaced 0.3 m × 5 m long). Once all genotypes had reached anthesis, 10 plants per genotype were randomly selected and excavated from the internal two rows. Plants were manually removed to a depth of 20 cm. Excavated plants were then vigorously shaken to remove the loose dry soil before images of the crown roots were captured using a smart-phone camera in the field. These images were then analyzed and the outer angle of the nodal roots was measured using Image J software.
Analysis of Phenotype Data
All phenotypic data analyses were performed in R V3.4.3 (R Core Team, 2017). The root growth angle phenotypes for parental lines (n = 14) in the glasshouse and the field, as well as the subset of NAM lines (n = 393 F6 lines) in the glasshouse, were measured using ImageJ (Schneider et al., 2012). Statistical analyses of the root growth angle measurements were performed using ASReml-R (Butler et al., 2009). Best linear unbiased estimates (BLUEs) were calculated for each individual, including the parental lines and the NAM lines, based on root angle data generated in the glasshouse experiment using the ‘clear pot’ method. To account for spatial variation a mixed linear model was fitted. In this model, the genotypes were fitted as fixed effect, while the replicate, pot and position were fitted as random terms. The field experiment of the parental lines was conducted to investigate the correlation of mature root growth angle under field conditions with measurements of roots from plants grown under glasshouse conditions at early growth stage. BLUEs for the field data were obtained by fitting a linear mixed model with genotype as a fixed effect and the plot coordinates (row and column) as random effects. The BLUEs for the subset of NAM lines were used as phenotypes in the GWAS analyses. Significance of differences in root biomass between genotypes and between SRA haplotypes were tested using Tukey’s test for general linear hypothesis testing based on the linear models described above. Data derived from image analysis of rhizoboxes, and the anatomical traits from the cross sections were also analyzed and differences between the means of genotypes were tested for significance based on a Fisher’s Least Significant Difference (LSD) test for multiple comparison with a family-wise error rate 5%.
Genotyping and Curation of Marker Data
All 10 families of the durum NAM population were genotyped using the Diversity Arrays Technology (DArT) genotyping-by-sequencing platform (DArTseq; Figure 1). Leaf tissues were sampled from F6 plants and genomic DNA was extracted according to the protocol provided by DArT. Genotyping resulted in a total of 13,395 DArTseq markers which were ordered according to their genetic positions in the consensus map (version 4.0), provided by Diversity Arrays Technology Pty Ltd., Canberra, Australia. Markers with a frequency of heterozygotes of ≥0.1 and missing calls of ≥20% were omitted. Markers with ≥30% missing data and a minor allele frequency of <3% were omitted and only genotypes with ≤20% missing marker information were considered, resulting in a selection of 2,541 high-quality, polymorphic DArTseq markers in 393 durum wheat lines which were used for the subsequent genetic analyses.
Genome-Wide Association Mapping
The 2,541 high-quality genome-wide markers were used to investigate marker-trait-associations (MTA) for SRA. Significances for MTAs were calculated in a two-step mixed linear model approach that increases detection power without increasing the empirical type I error (Stich, 2009). We used a mixed model implemented in the R package GenABEL (Aulchenko et al., 2007) which adjusted for population stratification by including identity-by-state estimates for genotype pairs (as a kinship matrix) and a principal component adjustment that uses the first four principal components as fixed covariates. To reduce the type I error rate, we applied a stringent Bonferroni cut-off threshold of –log10(p-value) = 4.67 (α = 0.05) for SRA (Bland and Altman, 1995). The major SRA QTL exceeding this threshold was then compared with previously identified drought-related and yield component QTL in an alignment approach.
Local LD of the significant markers on chromosome 6A for SRA was calculated and used to group markers into one QTL. Markers with pairwise r2 values > 0.60 were assigned to an LD block and included in the haplotype analysis, resulting in eight haplotype variants which were observed in the population. Haplotype networks, showing TCS genealogies between haplotype variants (Clement et al., 2000), were calculated using PopART2 (Leigh and Bryant, 2015). The network nodes were colored according to the average SRA in the respective haplotype groups. To investigate the effect of the SRA QTL on the growth angle measurements while correcting for variability due to genetic background, we selected three sub-NAM populations, segregating for the SRA allele combinations associated with narrow and wide SRA including DBA Aurora × Outrob4, DBA Aurora × Fastoz8, DBA Aurora × Fastoz3. We compared the mean SRA of lines that carried the two most frequent haplotypes hap1 and hap2 within the families separately. A Tukey’s test was performed to test phenotypic differences in SRA between the haplotype groups within each family. The haplotype effects on root angle phenotypes, were visualized using GraphPad Prism V6 (Graphpad Software Inc.).
Evaluating Root and Shoot Biomass Effects of Root Angle QTL
To investigate whether the identified major SRA QTL is also associated with pleiotropic differences in root or shoot biomass, a glasshouse experiment was conducted under controlled conditions. A total of 40 closely related genotypes segregating for SRA QTL were evaluated, including 20 lines carrying hap1 and 20 lines carrying hap2. The panel was phenotyped for root biomass using the method reported by Voss-Fels et al., 2017 with some modifications. Here, ANOVA pots (ANOVApot®, 137 mm diameter, 140 mm height) were filled with 1,650 g of sand (with particle size ranging from 0.075–4.75 mm) to facilitate efficient root washing. An RCBD was used for the experimental design, with four plants per genotype in each 1.4 L pot, in three replicates. Fifteen pots were placed in a container fitted with capillary mats to provide sufficient water and nutrient supply. A hydroponic solution was added to each container (1.50 mL of Cultiplex per L of deionized water) and was maintained at the same level over the course of the experiment. The concentration of the nutrient solution was gradually increased as the plants developed and required additional nutrient supply (days 1–10: 1.50 mL/L, days 11–17: 2 mL/L, days 18–22: 2.50 mL/L, days 23–26: 3 mL/L).
Seeds were germinated using a cold treatment (4°C) for 3 days to promote synchronous germination. The germinated seeds were transplanted to the sand-filled plastic pots and grown under diurnal (12 h) photoperiod in a temperature-controlled glasshouse (22/17°C; day/night). At 26 days after sowing (early tillering stage) plants were extracted with minimum disruption to the roots by placing the pot in a water-filled container and carefully washing off the remaining sand in clean water. The roots and shoots from each pot were separated and placed in a dehydrator at 65°C for 72 h before dry weight was measured.
Evaluation of Root Ideotypes Under Well-Watered and Drought Conditions
To investigate the potential for breeding cultivars with different root ideotypes, durum NAM lines representative of four distinct root ideotypes (root angle-root biomass; wide-low, wide-high, narrow-high, and narrow-low) were evaluated using rhizoboxes, similar to those described by Singh et al. (2010). Representative lines were selected based on extreme root angle and biomass phenotypes, as well as haplotype information for the major SRA QTL. Briefly, germinated seeds were sown in rhizoboxes (4 cm × 26 cm × 60 cm) at a depth of 3 cm and maintained under diurnal photoperiod (12 h) and a temperature of 22/17°C (day/night). An RCBD design was adopted in three replicates as blocks in two treatments (well-watered and drought). Four plants per ideotype were planted in each rhizobox. Four rhizoboxes were placed in a container filled with 300 mL water to supply plants with water from the bottom of the rhizoboxes in both treatments. Following sowing, all chambers were watered daily until 1 week after sowing. The well-watered (control) treatment received daily watering from the top of the rhizobox while the drought treatment received no additional water and was subjected to severe water-limitation in the upper layer of the soil. The percentage of soil moisture was measured weekly over the course of the experiment using a soil moisture meter (PMS-714; Lutron Electronic; probe length 22 cm and probe diameter 1 cm) at a depth of 50 cm. Images of the rhizoboxes were captured after 5 weeks and analyzed using GIA Roots software (Galkovskyi et al., 2012). The images were cropped into three equal sections at 0–20, 20–40, and 40–60 cm to evaluate root distribution at various soil depths.
To investigate differences in root anatomy associated with the root angle QTL or root ideotype, the stele diameter (SD) and metaxylem area (MXA) was measured for root tissue sampled 10 cm from the seminal root apex in both well-watered and drought treatments. Roots were hand sectioned with a razor blade using a dissecting microscope. The sections were stained with Toluidine Blue O. Images of the root sections were processed using a Zeiss Axio Microscope (Scope.A1 with 100× magnification). All image analyses were processed using ZEN lite 2012 software (blue edition, Jena, Germany).
Alignment of Previously Reported QTL for Root and Yield Component Traits
The QTL reported in this study was positioned on the Svevo durum physical map (Maccaferri et al., 2019). The previously reported QTL associated with RSA, distribution and growth angle (Maccaferri et al., 2016) were also projected onto the map using MapChart V2.3 (Voorrips, 2002). In addition, the previously reported QTL associated with the yield components (TKW, grain yield per spike) and the quality parameter yellow pigment concentration were aligned on the chromosomal region of interest (Golabadi et al., 2011; Roncallo et al., 2012; Maccaferri et al., 2016; Mengistu et al., 2016).
Candidate Gene Analysis
Mapping of Marker Genes in the Bread Wheat Reference Genome IWGSC RefSeq v.1.0
Identified peak markers were mapped onto the homologous bread wheat pseudochromosome 6A using the recently published RefSeq v1.0 annotations (Appels et al., 2018). High confidence (HC) and low confidence (LC) RefSeq v1.0 gene models were extracted from the identified region and used in further analyses. Similarity searches were carried out using BLASTn with high stringency settings (with an e-value cut-off of 1e-100). Collinearity analysis of Chromosome 6AL between T. durum and Triticum aestivum regions were performed using Pretzel3. Mapped markers and genes expressed in root tissues in seedling stage were used for the analysis.
Gene Expression Analysis and Functional Predictions
Gene expression patterns of the selected bread wheat gene homologs on chromosome 6A were analyzed using the developmental gene expression atlas of polyploid wheat (Ramírez-González et al., 2018); Wheat eFP Browser at http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi and visualized in R using the Morpheus package4.
Translated sequences of selected durum gene models were subjected to functional KEGG pathway analysis using blastKOALA (Kanehisa et al., 2016). Potential interacting proteins were analyzed in STRING (Szklarczyk et al., 2015) using the reference genomes of Brachypodium distachyon, Hordeum vulgare, Oryza sativa, Zea mays, and Arabidopsis thaliana as data background.
Results
Variation for Root Angle: From Glasshouse to Field
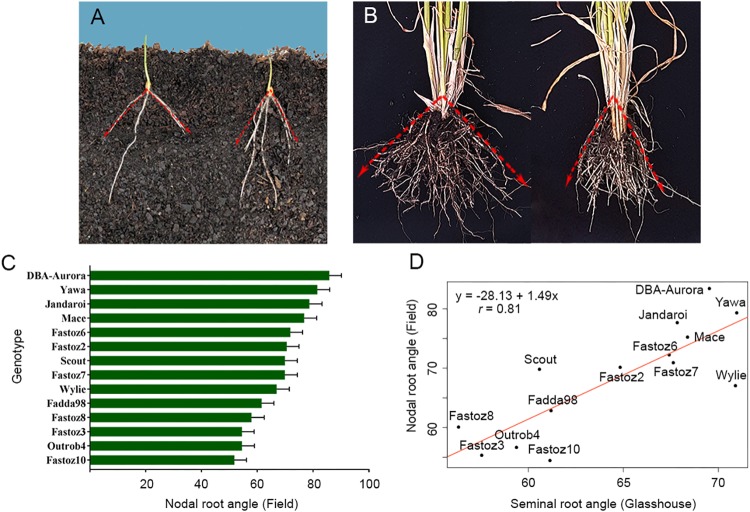
In this study, a panel consisting of the parents of the NAM population and standard lines with previously analyzed root characteristics was evaluated for SRA under controlled conditions in the glasshouse (Figure 2A) and nodal root angle under field conditions (Figure 2B). Phenotypes displayed by standards were as expected under glasshouse and field conditions, however, less variation under field conditions was observed. For example, the SRA for standard lines under glasshouse conditions were 110.1° (Mace) and 62.6° (Scout) compared to 76.8° (Mace) and 69.9° (Scout) for nodal roots under field conditions (Figure 2C). Although the absolute values varied between glasshouse and the field, Mace consistently displayed a wider root angle than Scout across both experiments.
FIGURE 2.
Root growth angle phenotypes measured in important durum wheat cultivars from Australia and ICARDA: (A) seminal root angle for Australian variety DBA Aurora (left, wide root angle) and an ICARDA elite founder line Outrob4 (right, narrow root angle) screened using the clear pot method, and (B) in the field using shovelomics method. (C) Nodal Root growth angle field measurements of 14 parental lines used for NAM population development. Correlation between seminal root angle in the glasshouse and mature roots in the field, r = 0.81, P = 0.00038 (D).
In both experiments, the ICARDA founder lines generally displayed a narrow root growth angle in comparison to the Australian durum cultivars. For example, the SRA for ICARDA founder lines ranged from 50.2–60.7° under glasshouse conditions and 51.8–61.6° in the field. Australian cultivars ranged from 83.0–97.8° under glasshouse conditions and 78.7–85.8° in the field. A strong correlation between seminal root angle in the glasshouse and mature root angle in the field was observed as shown in the regression analysis in Figure 2 (r = 0.81, P = 0.00038), wherein the panel of 14 lines showed consistent root growth angle phenotypes (Figure 2D).
Segregation for Root Angle in the NAM Lines
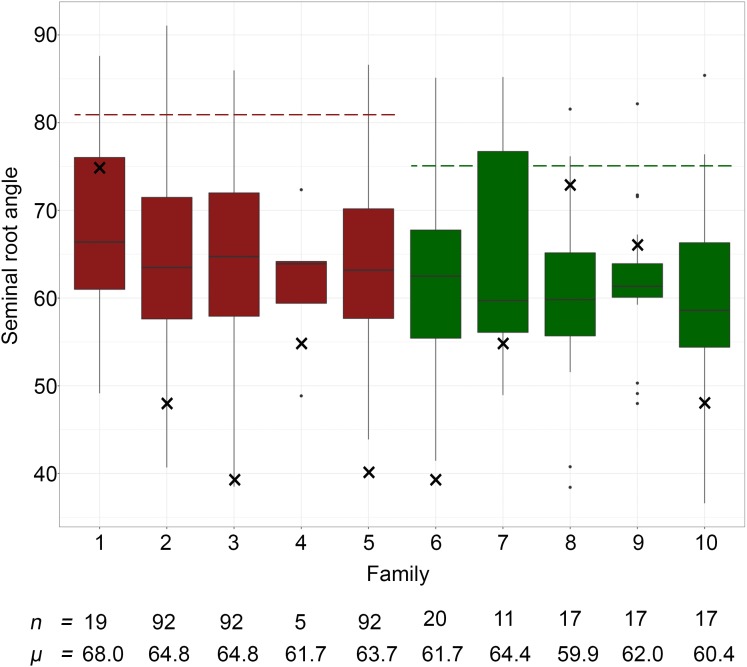
A high degree of variation for SRA was observed among the 393 NAM lines, with adjusted means ranging from 36.6–91.1° (Figure 3). In families derived from DBA Aurora (SRA = 81.1°), the SRA ranged from 38.5–91.1° and in the families derived from Jandaroi (SRA = 75.5°), the SRA ranged from 36.6–85.4°. In particular, three families (Family 2, Family 3 and Family 5) derived from crosses between DBA Aurora (widest root angle) and three ICARDA founder lines with the narrowest root angle (Outrob4 = 48.7°, Fastoz8 = 39.7° and Fastoz3° = 41.8°, respectively) displayed little transgressive segregation, with a number of lines showing slightly narrower or wider SRA phenotypes than the respective parents. For example, SRA of the individuals ranged from 40.7–91.1°, 38.5–86.0°, and 43.9–86.6° for Families 2, 3, and 5, respectively. Family 1 displayed a higher degree of transgressive segregation. In addition, two families (Jandaroi × Fastoz8 and Jandaroi × Outrob4, i.e., families 6 and 10, respectively) also displayed transgressive segregation, ranging from 41.5–85.1° (Family 6) and 36.6–85.4° (Family 10).
FIGURE 3.
Seminal root growth angle measurements of the 10 NAM families. Families 1 to 5 (red) share DBA Aurora as the common parent, and families 6–10 (green) share Jandaroi as the common parent. Family 1 = DBA Aurora × Fastoz7; Family 2 = DBA Aurora × Outrob4; Family 3 = DBA Aurora × Fastoz8; Family 4 = DBA Aurora × Fadda98; Family 5 = DBA Aurora × Fastoz3, Family 6 = Jandaroi × Fastoz8; Family 7 = Jandaroi × Fastoz10; Family 8 = Jandaroi × Fastoz6; Family 9 = Jandaroi × Fastoz2; Family 10 = Jandaroi × Outrob4. Boxplots display the quartile range and median SRA (horizontal line) of individuals within each of the 10 sub-NAM populations. The broken red line displays the mean SRA value of DBA Aurora and the broken green line displays the mean SRA value of Jandaroi; × represents the mean SRA value of ICARDA founder lines; n represents the number of individuals in each family; μ represents the mean SRA value of each family.
A Major QTL for Root Growth Angle Is Located on Chromosome 6A
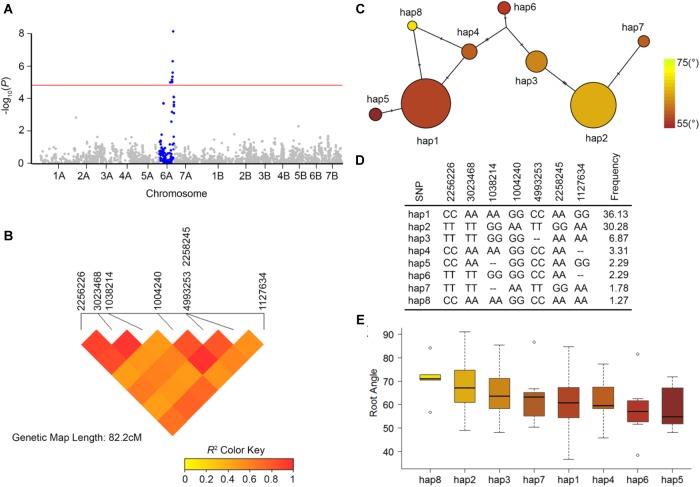
A total of seven highly significant markers for SRA were detected on chromosome 6A [at Bonferroni threshold = –log10(P) 4.67; Figure 4A]. A single major QTL region was defined based on high LD (r2 > 0.60) between pairwise markers, resulting in a QTL interval defined by the outer flanking markers 2256226 (86.46 cM DArTseq V4 consensus map) and 1127634 (94.68 cM DArTseq V4 consensus map) (Figure 4B). For this QTL, eight main haplotypes were detected (Figure 4C). Hap1 and hap2 were the most frequent allelic variants in the subset of NAM lines (frequency = 36.1 and 30.3%, respectively) (Figure 4D). The mean SRA for genotypes in the eight defined haplotype groups ranged from 57.8–71.0° (Figure 4E). Comparison of SRA between the most frequent haplotypes hap1 and hap2 revealed a highly significant difference of 7.7° (SE = 1.2, P = <0.001) across all families segregating for the QTL in both genetic reference backgrounds DBA Aurora and Jandaroi.
FIGURE 4.
Genome-wide association mapping for seminal root angle in 393 durum lines using 2,541 high quality DArTseq markers (minor allele frequency > 5%). (A) Manhattan plot showing chromosome 6A (blue) with significant marker-trait association at Bonferroni significant threshold 4.67 (red horizontal line). The x-axis displays the DArTseq markers on 14 chromosomes; y-axis is the –log10(P). (B) Heat map showing pairwise linkage disequilibrium (LD) between 7 significant markers representing major seminal root angle QTL on chromosome 6A (qSRA-6A). Color gradient represents LD as r2. (C) Haplotype network of 8 haplotype variants of the qSRA-6A that were found in the 393 NAM lines. Size of the circles represents the frequency of each haplotype in the population. Node color indicates mean seminal root angle for lines carrying the haplotype. (D) Allelic marker-combination of the 8 haplotypes for the 7 DArTseq markers and the frequency value of each haplotype. (E) Seminal root angle variation in each haplogroup.
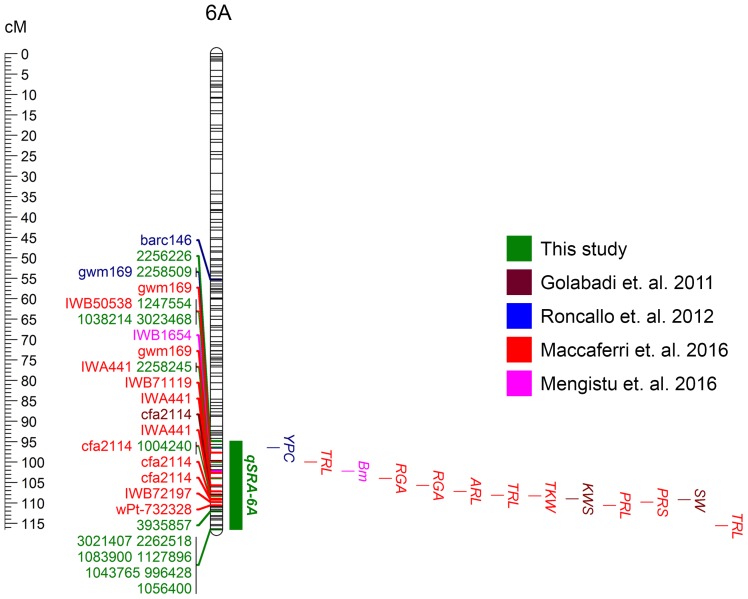
The QTL detected in this study and previously reported QTL in the same chromosomal region (Golabadi et al., 2011; Roncallo et al., 2012; Mengistu et al., 2016; Maccaferri et al., 2016) were positioned onto the durum reference genome (Svevo physical map, Maccaferri et al., 2019; Figure 5). The major QTL found in our study (qSRA-6A) was found to be co-located with previously reported durum QTL for root growth angle, total root length and root biomass, as well as QTL for yield components and quality traits (Figure 5).
FIGURE 5.
Major QTL for seminal root angle (qSRA-6A) positioned on the Svevo durum physical map (Mbp), along with QTL reported in previous mapping studies including root system architecture traits (TRL, total root length; RGA, root growth angle; ARL, average root length; TRL, total root length; PRL, primary root length; PRS, primary root surface), yield component traits (Bm, biomass; TKW, thousand kernel weight; KWS, grain yield per spike; SW, spike dry matter) and a quality trait (YPC, yellow pigment concentration).
qSRA-6A Influences Root Angle but Not Root Biomass in Different Genetic Backgrounds
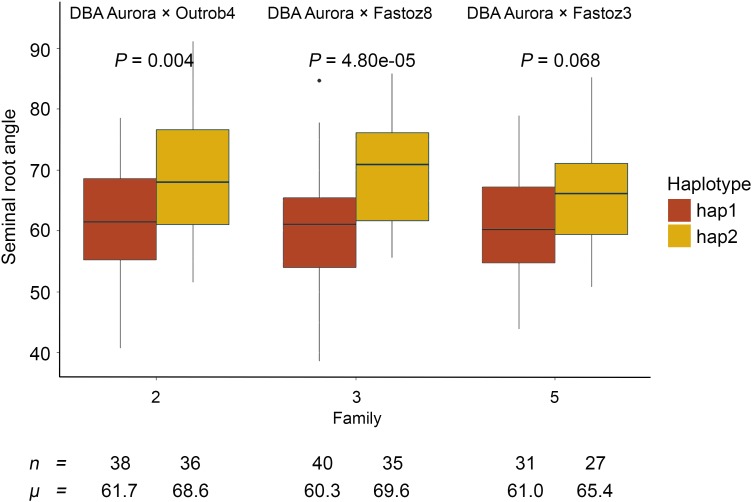
To evaluate haplotype effects for qSRA-6A, we compared the most common haplotypes in three families that were segregating for the QTL. The three families derived from crossing Outrob4, Fastoz8 and Fastoz3 with the common reference parent DBA Aurora were tested as these families segregated for hap1 and hap2 of the major QTL (Figure 6). The phenotypic differences in SRA between individuals carrying hap1 and hap2 were significant for family 2 and 3 and followed a similar trend in family 5. Amongst the families, the difference in SRA for lines carrying hap1 versus hap2 ranged between 4.4–9.3°. The largest effect was evident in the DBA Aurora × Fastoz8 family (60.3 and 69.6° for hap1 and hap2, respectively).
FIGURE 6.
Seminal root angle measurements in three NAM families segregating for the most common haplotype of the QTL qSRA-6A. Families were derived from crosses between DBA Aurora to three ICARDA lines (Outrob4, Fastoz3, and Fastoz8). In each family, mean SRA value of individuals carrying hap1 and hap2 was compared. The colors represent the two haplotype groups, n represents the number of individuals carrying different haplotype groups, μ represents the mean SRA value of each haplotype group, and P represents significance from an Honestly Significant Difference (HSD) test for the difference between the two haplotype groups within each family.
To investigate if the contrasting main haplotypes for qSRA-6A were only associated with root architectural differences or with overall plant development we conducted a subsequent experiment in which we assayed root and shoot biomass for a subset of 40 genotypes that represented hap1 (n = 20) and hap2 (n = 20). Comparing dried total root biomass, total shoot biomass and the root/shoot ratio of this subset showed no significant differences between the two main haplotype groups. Average values for hap1 and hap2 were 0.641 g/line and 0.638 g/line for total root biomass, 1.007 g/line and 1.023 g/line for total shoot biomass and 0.643 and 0.631 for root/shoot ratio.
Root Distribution and Anatomy of Four Root System Ideotypes
Root distribution under well-watered and drought conditions was investigated for four root system ideotypes in rhizoboxes (Supplementary Figure S1). Soil moisture of the rhizoboxes decreased dramatically with significant differences between treatments from 3 weeks after sowing (Supplementary Figure S2). Overall, plants in the drought treatment had less total root area (area of the roots in the images) (Figure 7A) and significantly reduced crown root growth (Figure 7B,C). Unexpectedly, the lines carrying the narrow allele were responsive to localized water availability in the upper strata (Figure 7B), while in the drought treatment root proliferation shifted deeper into the strata in response to soil moisture at depth (Figure 7C). For example, root ideotype ‘narrow-high’ produced significantly higher root area (23.54 cm2) in comparison to wide ideotypes (P < 0.05, wide-high = 21.15 cm2; wide-low = 15.11 cm2) in the upper soil layer (0–20 cm) of the rhizobox under well-watered conditions. In addition, the ‘narrow-high’ ideotype produced a significantly higher root area distribution under drought conditions in the middle and deepest soil layers (20–40 cm = 18.15 cm2; 40–60 cm = 12.13 cm2) when compared to both wide ideotypes (20–40 cm; 10.95–11.60 cm2, P < 0.05–0.1 and 40–60 cm; 2.60–9.35 cm2, P < 0.01–0.4; Figure 7).
FIGURE 7.
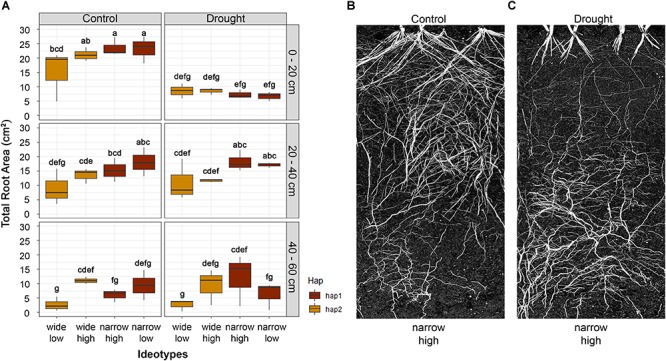
Root area distribution of the four root ideotypes wide-low, wide-high, narrow-high and narrow-low at different depths of the growth chamber. (A) Boxplots display root distribution of the four ideotypes under control (well-watered) and drought conditions. The colors represent the two haplotype groups of the root angle qSRA-6A. Letters above boxplots indicate significance difference between the four root ideotypes using least significant difference (LSD) test at α = 0.05. Visualization of a narrow-high root ideotype under (B) controlled (well-watered) and (C) drought conditions is shown.
To investigate associations between root architecture and root anatomical features, stele diameter (SD) and metaxylem area (MXA) were measured for the four ideotypes in the rhizobox experiment. Results suggested a strong link between root angle QTL qSRA-6A and SD (Figure 8A,B), as well as MXA at depth under well-watered conditions. Under well-watered conditions, the mean MXA for wide-low and wide-high root ideotypes were 11,930 ± 2,100 μm2 and 11,781 ± 3,287 μm2, respectively, in comparison to 3,137 ± 353 μm2 and 3,821 ± 794 μm2 for the narrow-high and narrow-low, respectively. However, the link was not evident under drought conditions. Under drought conditions, the mean MXA for wide-low and wide-high root ideotypes were 6,627 ± 1,203 μm2 and 4,543 ± 1,076 μm2, respectively, in comparison to 3,986 ± 256 μm2 and 5,577 ± 765 μm2 for the narrow-high and narrow-low, respectively (Figure 8C). Overall, wide root angle genotypes showed significantly reduced SD and MXA under drought conditions (P < 0.05). In addition, the ‘narrow-high’ ideotype which displayed the highest proportion of roots at depth, also showed smaller MXA under drought conditions at depth.
FIGURE 8.
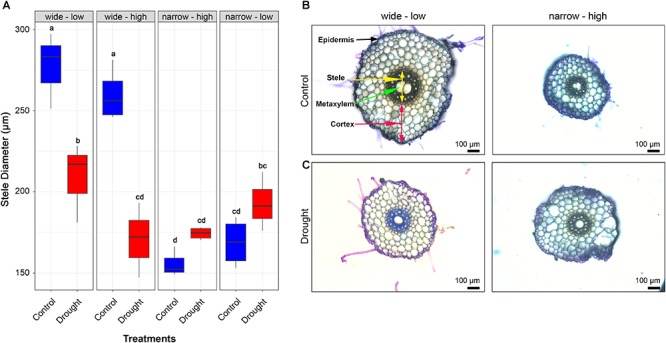
Anatomical features of roots sampled from durum wheat genotypes representative of distinct root system ideotypes. (A) Stele diameter of the four root ideotypes of samples collected 10 cm from root apex, under well-watered (control; boxplot colored in blue) and drought conditions (boxplot colored in red). Mean stele diameter with different letters above the boxplot are significantly different. Radial root cross sections on seminal root at 10 cm from root apex displaying anatomical variation in the root ideotype wide root angle with low biomass (B) and narrow root angle with high root biomass (C) under well-watered and drought conditions, scale bars in the cross sections = 100 μm.
Candidate Genes Underpinning the 6A QTL
Markers that were found to be significantly associated with SRA mapped to the distal end of chromosome 6A in durum and bread wheat. The length of the marked region was 22.82 Mbp in durum and 22.81 Mbp in bread wheat. These regions contain 393 gene models in durum, while 515 HC and 34 LC gene models were identified in bread wheat. The homologous genes had a high level of collinearity between the terminal regions of chromosome 6A in the T. durum reference cultivar Svevo and the bread wheat reference genome RefSeq v1.0 (Supplementary Figure S3).
Gene expression patterns were analyzed using the high-resolution tissue and stage-specific RNAseq data of Azurhnaya spring wheat (Winter et al., 2007; Ramírez-González et al., 2018). Altogether 206 genes show root specific expression during the plant life cycle, from which 76 genes show significant expression during early root development stages (radicle and roots at the seedling stage, one leaf and three leaf stage roots and root apical meristem tissues; Supplementary Figure S3).
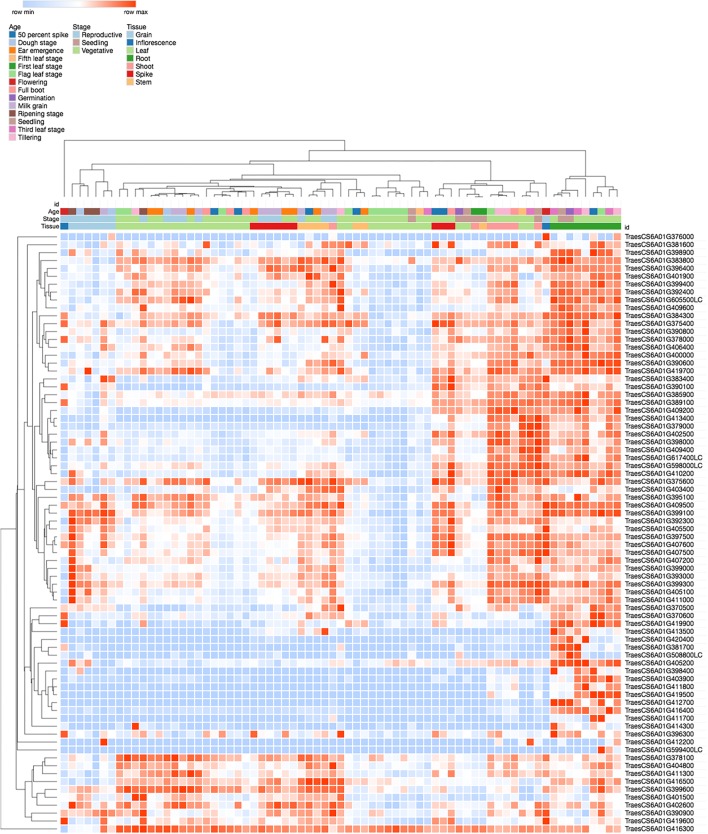
Transcript expression patterns from various tissues both at seedling stage, vegetative and reproductive stages are represented in Figure 9. Of these, 15 genes were primarily enriched in the root tissues during early root development (Figure 9 and Table 2).
FIGURE 9.
Expression patterns of bread wheat homologs at different age, stage, and tissue specific RNAseq libraries. Heatmap (blue = low; red = high) displayed high expression in root tissues (green) during seedling (pink) and vegetative stage (light green). Genes primarily expressed in the root tissues at seedling and vegetative stages are listed in Table 2.
Table 2.
List of 15 candidate genes identified using the homologous chromosome 6A genomic region of bread wheat through functional analysis of qSRA-6A.
| Gene ID | Homologous RefSeq v1.0 Gene ID | Molecular function | KEGG pathway |
|---|---|---|---|
| TRITD6Av1G217760 | TraesCS6A01G381700 | Cinnamoyl CoA reductase | Monolignol biosynthesis |
| TRITD6Av1G218270 | TraesCS6A01G383800 | Glutathione cytosolic | Glutathione metabolism |
| TRITD6Av1G218390 | TraesCS6A01G384300 | desumoylating isopeptidase 1-like | |
| TRITD6Av1G220070 | TraesCS6A01G392400 | 3-ketoacyl- thiolase peroxisomal | Fatty acid metabolism, Jasmonic acid biosynthesis, Beta-oxidation |
| TRITD6Av1G221500 | TraesCS6A01G396400 | Serine/threonine-protein phosphatase | Translation/mRNA surveillance pathway |
| TRITD6Av1G222970 | TraesCS6A01G605500LC | Membrane protein | |
| TRITD6Av1G223490 | TraesCS6A01G407600 | F-box family protein | |
| TRITD6Av1G223520 | TraesCS6A01G407600 | F-box family protein | |
| TRITD6Av1G223760 | TraesCS6A01G409500 | Transmembrane protein | |
| TRITD6Av1G223780 | TraesCS6A01G409600 | Electron transfer flavoprotein beta-subunit | leucine catabolism and in phytol degradation |
| TRITD6Av1G224460 | TraesCS6A01G412700 | Protein kinase, Wall-associated receptor kinase 2 | MAPK signaling |
| TRITD6Av1G224970 | TraesCS6A01G413500 | Ripening-related protein, RIPER1 | |
| TRITD6Av1G225080 | TraesCS6A01G508800LC | 12-oxophytodienoate reductase 2 | Fatty acid metabolism, Jasmonic acid biosynthesis |
| TRITD6Av1G225140 | TraesCS6A01G414300 | Disease resistance protein RPM1 | Plant/pathogen interaction |
| TRITD6Av1G225520 | TraesCS6A01G414300 | Disease resistance protein RPM1 | Plant/pathogen interaction |
In the bread wheat genome (RefSeq v1.0 chr 6A region) the mapped markers overlap with gene models representing a NAC transcription factor (TraesCS6A1G386700), a fatty acid hydroxylase family protein (TraesCS6A1G384600), a PRONE protein (TraesCS6A1G405000) and a SAWADEE homeodomain protein 2 (TraesCS6A1G420700). The position of the peak marker from the SRA QTL qSRA-6A mapped to the exon region both in the NAC domain-containing protein (3023468) and the PRONE protein (3935857). The observed SNP caused 8:T > A and 7:C > G nucleotide changes, respectively, which resulted in an amino acid change to the translated protein.
Translated durum protein sequences mapped to the QTL region were subjected to KEGG pathway analysis. From the 387 T. durum sequences, 114 proteins had significant blast hits in the KEGG database. The following KEGG pathways were enriched: 13 proteins involved in pathogen defense mechanisms, seven proteins in secondary metabolite biosynthesis (monolignol biosynthesis) and five proteins in fatty acid metabolism [fatty acid biosynthesis, jasmonic acid (JA) biosynthesis and beta-oxidation]. Genes significantly expressed in the radicle and roots at the seedling stage as well as roots and root apical meristem at the three leaf stage were analyzed in more detail, using the STRING database to predict potential interacting protein networks. Studies using related monocot species (B. distachyon, H. vulgare, O. sativa, and Z. mays) as well as the model dicot species A. thaliana indicated the conserved patterns of interacting proteins enriched in functions involved in monolignol biosynthesis, fatty acid metabolism, jasmonic acid metabolism and beta-oxidation. Proteins belonging to plant–pathogen interaction pathways were also detected using both the moncot and dicot data backgrounds. However, homologous proteins of Arabidopsis were also related to fatty acid metabolism pathways. Extended interaction networks in all backgrounds also highlighted proteins that are identified in auxin metabolism.
Discussion
A Major QTL on 6A Determines Seminal Root Growth Angle
Here, we report a QTL on chromosome 6A (qSRA-6A) that has a significant effect on root growth angle in a subset of 393 durum NAM lines. The co-location of qSRA-6A with previously mapped QTL in durum wheat for various root traits, including root length, root surface and root biomass (Maccaferri et al., 2016; Mengistu et al., 2016) suggests that this chromosomal region has a major impact on root development. In addition, qSRA-6A also aligned with genomic regions influencing yield components and quality parameters, such as thousand kernel weight and yellow pigment content (Golabadi et al., 2011; Roncallo et al., 2012; Mengistu et al., 2016). Therefore, this region appears important for root system development, which may impact other agronomically important traits including grain yield and end-use quality parameters. Analysis of local LD around the main QTL peak showed high levels of pairwise LD between seven SRA-associated markers. Similar to reported observations for root traits in bread wheat (Voss-Fels et al., 2017) this suggests strong directional selection for this chromosomal block, resulting in a block-wise co-inheritance of markers in tight LD due to strong allelic fixation in important durum wheat germplasm (Tuberosa et al., 2002a; Hayes et al., 2007). Since root architecture directly affects the Source-sink relationship, it is likely that the underlying genetic mechanisms for drought-adaptive traits, such as root growth characteristics, also influence above ground traits like spike grain weight, TKW, spike dry weight and grain quality, which facilitates detection of similar QTL in segregating populations.
A recent study on SRA in bread wheat suggests that root angle is under complex genetic control with multiple small effect QTL involved (Richard et al., 2018). In barley, similar to our study, seminal root traits were reported as being affected by a major QTL on chromosome 5H (Robinson et al., 2016). In maize, a major QTL was reported as constitutive and was associated with root growth angle, root branching and root thickness. This QTL exhibited consistent strong effects under glasshouse and field conditions with different water treatments (Giuliani et al., 2005). In sorghum, four QTL for nodal root angle were mapped, two of which had a major effect and appeared to co-locate with previously identified QTL for stay-green expressed under low moisture conditions (Mace et al., 2012).
On the other hand, VERNALIZATION1 (VRN1), which controls flowering time in cereals like wheat and barley, was found to modulate RSA in bread wheat and barley (Voss-Fels et al., 2018a). The QTL identified in this study provides the opportunity to introgress novel diversity into durum and bread wheat to modulate RSA, potentially leading to improved performance under specific environmental conditions.
Promising Candidate Genes for qSRA-6A Have Root Growth-Related Functions
Analyses revealed that the position of the mapped markers of qSRA-6A overlaps with a genomic region enriched with genes related to gravitropism, polar growth and hormonal signaling. Notably, genes that are expressed only in root tissues at early stages of plant development (e.g., seedling and one-leaf stage) are involved in pathways such as fatty acid metabolism, jasmonic acid biosynthesis, and monolignol biosynthesis, that might be related to root angle variations in the analyzed phenotypes. Fatty acid metabolism and beta-oxidation play a significant role in the early germination steps when reserve lipids are mobilized to serve as respiratory substrates and to sustain the growth of the seedling. Among the identified early root development-related genes, genes encoding a 3-ketoacyl-CoA thiolase-like protein, Electron transfer flavoprotein beta-subunit and 12-oxophytodienoate reductase 2 and glutathione reductase are related to fatty acid metabolism and beta-oxidation in barley, rice, maize.
During triacylglycerol degradation, fatty acids are released and channeled into gluconeogenesis. Beta-oxidation is also essential to produce secondary metabolites in oxylipin signaling such as the 12-oxophytodienoic acid (OPR2) and jasmonic acid, which serve as signaling compounds in plant growth and pathogen defense mechanisms (Christine and Clifford, 2009; Wasternack and Hause, 2013). An inhibiting role of OPR2 on seed germination has been described (Dave et al., 2011; Dave and Graham, 2012), showing the interaction between 12-oxophytodienoic acid and abscisic acid that leads to increased ABA isensitive5 (ABI5) gene expression and suppressed germination. The identified durum OPR2 shows close homology to OPR5 in maize and OPR2 in barley; both of which are known to be involved in Jasmonic-acid biosynthesis pathways (Helmut et al., 2000). In roots, the growth inhibition by JA and OPR2 occurs via cross-talk with auxin and possibly other hormones, such as gibberelic acid (GA) and brassinoteroids (BR), mostly as an indirect effect via auxin. Jasmonic acid also regulates root gravitropism through affecting the biosynthesis of auxin. It directly influences the gradient formation by modulating its polar distribution of auxin (Singh et al., 2017). JA induced gene expression is most characteristic in the outer layers of the roots (Gasperini et al., 2015).
The monolignol biosynthesis primarily regulated by Cinnamoyl CoA reductases plays an essential role in cell wall lignification in the casparian strips of the root. In the analyzed genomic region of the durum chromosome 6A, there are five cinnamoyl reductase genes encoded, and additional genes (e.g., ABC transporter, laccase) related to monolignol biosynthesis were also found in the region. Drought conditions were reported to enhance the monolignol biosynthesis in the root elongation zone of the seedlings by the inhibition of the cell wall extensibility and root growth (Ma, 2007). Similarly, lignin-related phenolics biosynthesis was also reported during biotic stress (Silva et al., 2010). It is plausible to therefore suggest that these candidate genes may be having a role to play in the constitiution of the durum root ideotype.
The concentration of genes functioning in fatty acid metabolism, monolignol biosynthesis and jasmonic acid biosynthesis pathways in the identified QTL region highlight the importance of jasmonic acid-auxin crosstalk in gravitropism perception and primary root angle formation. The potential target genes include enzymes involved in cell wall expansion (monolignol biosynthesis) and JA biosynthetic pathways. JA signaling in young root tissues acts contrary to auxin signaling effects, is also related to gravitropism and therefore might be directly related to root angle variations. Genetic variations observed in the encoding genes or their regulating cis-promoter elements can help to identify phenotypes where the coordinated negative impact of increasing JA levels and lignification is controlled. Next to the growth-related functions, these genes are also involved in abiotic and biotic stress responses, including drought stress.
The Context-Dependency of Root System Ideotypes in Different Environments
The architecture of roots has great importance for sourcing underground water and nutrients which is essential for plant growth, particularly in marginal environments characterized by water limitation (Manschadi et al., 2006; Asif and Kamran, 2011). In barley, it has been hypothesized that shallow root growth as characterized by a wide root growth angle may be advantageous for accessing nutrients in the upper soil surface under environments where plants experience sporadic rainfall throughout the growing season (Robinson et al., 2016). However, studies showed that this may not be always the case. For example, in the Mediterranean climate of South Australia, which experiences high in-season precipitation, narrow root angle seems advantageous and tends to be associated with higher grain yield (McDonald, 2010). On the other hand, ‘steep, deep, and cheap’ ideotypes with longer roots or more root branching at depth are most desirable for enhanced access to nutrients and water stored in deep layers of the soil under environments experiencing terminal drought (Manschadi et al., 2008; Christopher et al., 2013; Lynch, 2013). Moreover, deep roots could be ideal to reduce between-plants competition for resources under high-density planting in high-input conditions (Manske and Vlek, 2002). Plants that express drought-adaptive traits under water-limited environments have been shown to sustain increased yield (Manschadi et al., 2010). This is likely due to increased water access post-anthesis which can be through deeper and more efficient root systems (Mace et al., 2012). Manschadi et al. (2006) demonstrated in their modeling study a yield increase of an extra 55 kg/ha for each millimeter of water extracted from the soil after anthesis and during the grain filling stage. The key reason for that was an increase in marginal water use efficiency to almost three times after anthesis, due to enhanced access to water available in deep soils (Kirkegaard et al., 2007; Christopher et al., 2013). The qSRA-6A QTL identified in this study is highly associated with root growth angle. This suggests deployment of the narrow (hap1) allele for qSRA-6A could be beneficial in breeding programs targeting production environments with deep soils that often experience water stress. The root plasticity under drought suggests that durum genotypes carrying the narrow allele may not have a yield penalty in high rainfall seasons because root growth appears to respond and take advantage of resource availability in the upper soil layers. The G × E for root development and utility of this feature should be further explored.
Interestingly, no association between qSRA-6A QTL with root and shoot biomass was found when comparing contrasting haplotype groups with similar genetic backgrounds. This implies that root growth angle and root biomass are under separate genetic control, opening up the possibility to create customized root systems, e.g., by using marker-assisted introgression approaches. Results of our study also highlighted that the ‘narrow-high’ ideotype produced the highest root proliferation at the deepest soil level with the smallest MXA under drought. This suggests the mechanism for accumulation of root biomass may not only be related to root branching at depth but also associated with an adaptive mechanism involving reduced water use uptake during early stages of crop development. If the loci controlling root biomass are deployed with loci influencing the direction of root growth, root proliferation could be directed and concentrated at desired soil depths. Such allelic combinations assembled through plant breeding could give rise to improved commercial varieties with designer roots tailored for specific target environments (Voss-Fels et al., 2018b). A similar observation was made in a rice study where a major RSA gene called DEEPER ROOTING 1 (DRO1) was cloned (Uga et al., 2013). They showed that DRO1 is involved in gravitropic response of root cells, thereby influencing root growth direction, but without a significant effect on root biomass. The qSRA-6A QTL is unlikely to be DRO1 because the ortholog is located on the group 5 chromosomes of wheat and wild emmer. Additional QTL related to root growth angle (DRO2, DRO3, DRO4, and DRO5) have also been reported (Kitomi et al., 2015). The QTL region DRO4 is located on the long arm of chromosome 2 in rice and Aux/IAA8, OsPIN1, and SAUR were reported as major contributors to the measured phenotypes (Kitomi et al., 2015; Lou et al., 2015). Using these rice orthologs we found that the corresponding genes are not overlapping with the genomic region determined in our study and are located closer to the centromere of chromosome 6A both in hexaploid and tetraploid wheat. Recently, another QTL has been reported by Wang et al. (2018), showing a significant role of OsPIN2 gene in root growth angle in rice. OsPIN2 encodes an Auxin-efflux carrier (Os06g0660200) that showed the highest sequence homology to the homologous gene group on chromosome 7 in wheat (TraesCS7A02G492400, TraesCS7B01G398100, and TraesCS7D01G478800). Overall, the lack of alignment between known genes in rice and the QTL on 6A mapped in the present study implies the region likely contains novel or currently uncharacterized gene(s).
The genetically stable effects of the qSRA-6A haplotypes across three different families implies that marker-assisted backcrossing strategies using the marker sequences identified in this study could effectively modulate RSA in future breeding attempts. Numerous studies have reported that root growth angle at the seedling stage was predictive for root growth angle in the field (Tuberosa et al., 2002a; Landi et al., 2010; Li et al., 2015; Richard et al., 2015; Uga et al., 2015; Kanehisa et al., 2016). Furthermore, recent studies have shown that RSA can be manipulated through recurrent phenotypic selection at the seedling stage under glasshouse conditions in which root traits could be measured at high broad-sense heritabilities (ranging from 0.62 to 0.79), leading to significant shifts in population distributions after a few cycles of selection (Alahmad et al., 2018; Richard et al., 2018). This offers plant breeders different options to directly manipulate RSA.
One major limitation for the direct consideration of root traits in defined breeding goals is the high context-dependency of varying RSA in different environments and the interplay of roots with other key phenology traits like flowering time in the expression of the end-point trait such as grain yield (Voss-Fels et al., 2018b). It was recently shown in a comprehensive study involving multi-environment trials in barley that the genetic correlation of root growth angle and yield was highly context dependent, ranging from situations in which shallow roots were associated with increased yield performance and vice versa (Robinson et al., 2018). Multi-environment field trials are required to thoroughly evaluate the value of qSRA-6A and different root ideotypes to improve or stabilize durum grain yield in a range of environmental circumstances.
Author Contributions
SA and LH conceived and designed the experiments. SA, CY, AA, and GQ performed the experiments. SA, ED, CY, KV-F, EM, and AJ analyzed the data. SA, JA, FB, and LH provided germplasm. SA, KEH, ED, CY, EM, AJ, KV-F, JA, FB, JC, and LH wrote or reviewed the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. SA was supported by Monsanto’s Beachell-Borlaug International Scholars Program (MBBISP) Iowa, the United States and a Ph.D. scholarship from The University of Queensland, Australia (UQRS). LH was supported through an Early Career Discovery Research Award (DE170101296) from the Australian Research Council. Genotyping was supported by the Food and Agriculture Organization (FAO; Grant LoA/TF/W3B-PR-02/JORDAN/2016/AGDT).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00436/full#supplementary-material
References
- Acuña T. B., Wade L. (2012). Genotype × environment interactions for root depth of wheat. Field Crops Res. 137 117–125. 10.1093/aob/mcs251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahmad S., Dinglasan E., Leung K. M., Riaz A., Derbal N., Voss-Fels K. P., et al. (2018). Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 14:36. 10.1186/s13007-018-0302-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Araus J., Slafer G., Reynolds M., Royo C. (2002). Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. 89 925–940. 10.1093/aob/mcf049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Kamran A. (2011). Plant breeding for water-limited environments. Crop Sci. 51 2911–2912. 10.2135/cropsci2011.12.0004br [DOI] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., Van Duijn C. M. (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Bassi F., Sanchez-Garcia M. (2017). Adaptation and Stability Analysis of ICARDA Durum Wheat Elites across 18 Countries. Crop Sci. 57 2419–2430. 10.2135/cropsci2016.11.0916 [DOI] [Google Scholar]
- Belaid A. (2000). “Durum wheat in WANA: production, trade and gains from technological change,” in Durum Wheat Improvement in the Mediterranean Region: New challenges. Options Méditerranéennes, eds Royo C. M., Nachit M., Di Fonzo N., Araus J.L. (Zaragoza: CIHEAM; ),63–70. [Google Scholar]
- Bengough A., Gordon D., Al-Menaie H., Ellis R., Allan D., Keith R., et al. (2004). Gel observation chamber for rapid screening of root traits in cereal seedlings. Plant Soil 262 63–70. 10.1023/B:PLSO.0000037029.82618.27 [DOI] [Google Scholar]
- Bland J. M., Altman D. G. (1995). Multiple significance tests: the Bonferroni method. BMJ 310:170 10.1136/bmj.310.6973.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A., Mayer J., Gozlan G. (1983). Associations between plant production and some physiological components of drought resistance in wheat. Plant Cell Environ. 6 219–225. 27677299 [Google Scholar]
- Borrell A. K., Mullet J. E., George-Jaeggli B., Van Oosterom E. J., Hammer G. L., Klein P. E., et al. (2014). Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 65 6251–6263. 10.1093/jxb/eru232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutraa T. (2010). Improvement of water use efficiency in irrigated agriculture: a review. J. Agron. 9 1–8. 10.3389/fchem.2018.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D., Cullis B. R., Gilmour A., Gogel B. (2009). ASReml-R Reference Manual. Brisbane: Department of Primary Industries and Fisheries. [Google Scholar]
- Cane M. A., Maccaferri M., Nazemi G., Salvi S., Francia R., Colalongo C., et al. (2014). Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Mol. Breed. 34 1629–1645. 10.1007/s11032-014-0177-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Azam-Ali S., Foulkes M. J. (2014). Quantifying relationships between rooting traits and water uptake under drought in Mediterranean barley and durum wheat. J. Integr. Plant Biol. 56 455–469. 10.1111/jipb.12109 [DOI] [PubMed] [Google Scholar]
- Cattivelli L., Rizza F., Badeck F.-W., Mazzucotelli E., Mastrangelo A. M., Francia E., et al. (2008). Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Res. 105 1–14. 10.3389/fpls.2015.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J. H., Hewitson B., Busuioc A., Chen A., Gao X., Held R., et al. (2007). “Regional Climate Projections,” in Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds Solomon S., Qin D., Manning M., Chen Z., Marquis M., Averyt K. B., et al. (Cambridge: Cambridge University Press; ). [Google Scholar]
- Christine M., Clifford W. (2009). Influence of mitochondrial β-oxidation on early pea seedling development. New Phytol. 181 832–842. 10.1111/j.1469-8137.2008.02717.x [DOI] [PubMed] [Google Scholar]
- Christopher J., Christopher M., Jennings R., Jones S., Fletcher S., Borrell A., et al. (2013). QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor. Appl. Genet. 126 1563–1574. 10.1007/s00122-013-2074-0 [DOI] [PubMed] [Google Scholar]
- Christopher J., Manschadi A., Hammer G., Borrell A. (2008). Developmental and physiological traits associated with high yield and stay-green phenotype in wheat. Aust. J. Agric. Res. 59 354–364. 10.1071/AR07193 [DOI] [Google Scholar]
- Clement M., Posada D., Crandall K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9 1657–1659. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Dave A., Graham I. (2012). Oxylipin Signaling: a Distinct Role for the Jasmonic Acid Precursor cis-(+)-12-Oxo-Phytodienoic Acid (cis-OPDA). Front. Plant Sci. 3:42. 10.3389/fpls.2012.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Hernández M. L., He Z., Andriotis V. M. E., Vaistij F. E., Larson T. R., et al. (2011). 12-Oxo-Phytodienoic Acid Accumulation during Seed Development Represses Seed Germination in Arabidopsis. Plant Cell 23 583–599. 10.1105/tpc.110.081489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot S., Forster B., Pagès L., Price A., Tuberosa R., Draye X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12 474–481. 10.1016/j.tplants.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Ehdaie B., Layne A. P., Waines J. G. (2012). Root system plasticity to drought influences grain yield in bread wheat. Euphytica 186 219–232. 10.1007/s10681-011-0585-9 [DOI] [Google Scholar]
- El Hassouni K., Alahmad S., Belkadi B., Filali-Maltouf A., Hickey L., Bassi F. (2018). Root system architecture and its association with yield under different water regimes in durum wheat. Crop Sci. 58 1–16. 10.2135/cropsci2018.01.0076 [DOI] [Google Scholar]
- Fang Y., Du Y., Wang J., Wu A., Qiao S., Xu B., et al. (2017). Moderate drought stress affected root growth and grain yield in old, modern and newly released cultivars of winter wheat. Front. Plant Sci. 8:672. 10.3389/fpls.2017.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkovskyi T., Mileyko Y., Bucksch A., Moore B., Symonova O., Price C. A., et al. (2012). GiA Roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 12:116. 10.1186/1471-2229-12-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini D., Chételat A., Acosta I. F., Goossens J., Pauwels L., Goossens A., et al. (2015). Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 11:e1005300. 10.1371/journal.pgen.1005300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Watson A., Gonzalez-Navarro O. E., Ramirez-Gonzalez R. H., Yanes L., Mendoza-Suárez M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13 2944–2963. 10.1038/s41596-018-0072-z [DOI] [PubMed] [Google Scholar]
- Giuliani S., Sanguineti M. C., Tuberosa R., Bellotti M., Salvi S., Landi P. (2005). Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J. Exp. Bot. 56 3061–3070. 10.1093/jxb/eri303 [DOI] [PubMed] [Google Scholar]
- Golabadi M., Arzani A., Maibody S. M., Tabatabaei B. S., Mohammadi S. (2011). Identification of microsatellite markers linked with yield components under drought stress at terminal growth stages in durum wheat. Euphytica 177 207–221. 10.1007/s10681-010-0242-8 [DOI] [Google Scholar]
- Gregory P. J., Bengough A. G., Grinev D., Schmidt S., Thomas W. B. T., Wojciechowski T., et al. (2009). Root phenomics of crops: opportunities and challenges. Funct. Plant Biol. 36 922–929. 10.1071/FP09150 [DOI] [PubMed] [Google Scholar]
- Hamada A., Nitta M., Nasuda S., Kato K., Fujita M., Matsunaka H., et al. (2012). Novel QTLs for growth angle of seminal roots in wheat (Triticum aestivum L.). Plant Soil 354 395–405. 10.1007/s11104-011-1075-5 [DOI] [Google Scholar]
- Hayes B., Chamberlain A., Mcpartlan H., Macleod I., Sethuraman L., Goddard M. (2007). Accuracy of marker-assisted selection with single markers and marker haplotypes in cattle. Genet. Res. 89 215–220. 10.1017/S0016672307008865 [DOI] [PubMed] [Google Scholar]
- Helmut M., Bettina H., Ivo F., Jörg Z., Claus W. (2000). Allene oxide synthases of barley (Hordeum vulgare cv. Salome): tissue specific regulation in seedling development. Plant J. 21 199–213. 10.1046/j.1365-313x.2000.00669.x [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428 726–731. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Kashiwagi J., Krishnamurthy L., Upadhyaya H. D., Krishna H., Chandra S., Vadez V., et al. (2005). Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum L.). Euphytica 146 213–222. 10.1007/s10681-005-9007-1 [DOI] [Google Scholar]
- King J., Gay A., Sylvester-Bradley R., Bingham I., Foulkes J., Gregory P., et al. (2003). Modelling cereal root systems for water and nitrogen capture: towards an economic optimum. Ann. Bot. 91 383–390. 10.1093/aob/mcg033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard J., Lilley J., Howe G., Graham J. M. (2007). Impact of subsoil water use on wheat yield. Aust. J. Agric. Res. 58 303–315. 10.1071/AR06285 [DOI] [Google Scholar]
- Kitomi Y., Kanno N., Kawai S., Mizubayashi T., Fukuoka S., Uga Y. J. R. (2015). QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 8:16. 10.1186/s12284-015-0049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi P., Giuliani S., Salvi S., Ferri M., Tuberosa R., Sanguineti M. C. (2010). Characterization of root-yield-1.06, a major constitutive QTL for root and agronomic traits in maize across water regimes. J. Exp. Bot. 61 3553–3562. 10.1093/jxb/erq192 [DOI] [PubMed] [Google Scholar]
- Leigh J. W., Bryant D. (2015). Popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 1110–1116. 10.1111/2041-210X.12410 [DOI] [Google Scholar]
- Li P., Chen F., Cai H., Liu J., Pan Q., Liu Z., et al. (2015). A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 66 3175–3188. 10.1093/jxb/erv127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss S. P., Siddique K. (1994). Morphological and physiological traits associated with wheat yield increases in Mediterranean environments. Adv. Agron. 52 229–276. 10.1016/S0065-2113(08)60625-2 [DOI] [Google Scholar]
- Lou Q., Chen L., Mei H., Wei H., Feng F., Wang P., et al. (2015). Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66 4749–4757. 10.1093/jxb/erv246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995). Root architecture and plant productivity. Plant Physiol. 109 7–13. 10.1104/pp.109.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. P. (2013). Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112 347–357. 10.1093/aob/mcs293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q.-H. (2007). Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. J. Exp. Bot. 58 2011–2021. 10.1093/jxb/erm064 [DOI] [PubMed] [Google Scholar]
- Maccaferri M., El-Feki W., Nazemi G., Salvi S., Canè M. A., Colalongo M. C., et al. (2016). Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. Bot. 67 1161–1178. 10.1093/jxb/erw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M., Harris N. S., Twardziok S. O., Pasam R. K., Gundlach H., Spannagl M., et al. (2019). Durum wheat genome reveals past domestication signatures and future improvement targets. Nat. Genet. 10.1038/s41588-019-0381-3 [DOI] [PubMed] [Google Scholar]
- Mace E., Singh V., Van Oosterom E., Hammer G., Hunt C., Jordan D. (2012). QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor. Appl. Genet. 124 97–109. 10.1007/s00122-011-1690-9 [DOI] [PubMed] [Google Scholar]
- Manschadi A., Christopher J., Hammer G., Devoil P. (2010). Experimental and modelling studies of drought-adaptive root architectural traits in wheat (Triticum aestivum L.). Plant Biosystems 144 458–462. 10.1080/11263501003731805 [DOI] [Google Scholar]
- Manschadi A. M., Christopher J., Hammer G. L. (2006). The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant Biol. 33 823–837. 10.1071/FP06055 [DOI] [PubMed] [Google Scholar]
- Manschadi A. M., Hammer G. L., Christopher J. T., Devoil P. (2008). Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 303 115–129. 10.1007/s11104-007-9492-1 [DOI] [Google Scholar]
- Manske G. G., Vlek P. L. (2002). Root architecture–wheat as a model plant. Plant Roots 3 249–259. 10.1201/9780203909423.ch15 [DOI] [Google Scholar]
- McDonald G. (2010). “The effects of root angle on root growth and yield of wheat in the Australian cereal belt,” in Proceedings of 15th Agronomy Conference: Food Security from Sustainable Agriculture, Lincoln. [Google Scholar]
- Mengistu D. K., Kidane Y. G., Catellani M., Frascaroli E., Fadda C., Pè M. E., et al. (2016). High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveals high diversity and potential for wheat breeding. Plant Biotechnol. J. 14 1800–1812. 10.1111/pbi.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi R., Sadeghzadeh D., Armion M., Amri A. (2011). Evaluation of durum wheat experimental lines under different climate and water regime conditions of Iran. Crop Pasture Sci. 62 137–151. 10.1071/CP10284 [DOI] [Google Scholar]
- Nakamoto T., Oyanagi A. (1994). The direction of growth of seminal roots of Triticum aestivum L. and experimental modification thereof. Ann. Bot. 73 363–367. 10.1006/anbo.1994.1045 [DOI] [Google Scholar]
- Nakamoto T., Shimoda K., Matsuzaki A. (1991). Elongation angle of nodal roots and its possible relation to spatial root distribution in maize and foxtail millet. Jpn. J. Crop Sci. 60 543–549. 10.1626/jcs.60.543 [DOI] [Google Scholar]
- Omori F., Mano Y. (2007). QTL mapping of root angle in F2 populations from maize ‘B73’ × teosinte ‘Zea luxurians’. Plant Root 1 57–65. 10.3117/plantroot.1.57 [DOI] [Google Scholar]
- Oyanagi A. (1994). Gravitropic response growth angle and vertical distribution of roots of wheat (Triticum aestivum L.). Plant Soil 165 323–326. 10.1007/BF00008076 [DOI] [Google Scholar]
- Oyanagi A., Nakamoto T., Wada M. (1993). Relationship between root growth angle of seedlings and vertical distribution of roots in the field in wheat cultivars. Jpn. J. Crop Sci. 62 565–570. 10.1626/jcs.62.565 [DOI] [Google Scholar]
- Palta J. A., Yang J. (2014). Crop root system behaviour and yield. Field Crops Res. 165 1–4. 10.1016/j.fcr.2014.06.024 [DOI] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ramírez-González R., Borrill P., Lang D., Harrington S., Brinton J., Venturini L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:eaar6089. 10.1126/science.aar6089 [DOI] [PubMed] [Google Scholar]
- Reynolds M. P., Pierre C. S., Saad A. S., Vargas M., Condon A. G. (2007). Evaluating potential genetic gains in wheat associated with stress-adaptive trait expression in elite genetic resources under drought and heat stress. Crop Sci. 47 S-172–S-189 10.2135/cropsci2007.10.0022IPBS [DOI] [Google Scholar]
- Richard C., Christopher J., Chenu K., Borrell A., Christopher M., Hickey L. (2018). Selection in early generations to shift allele frequency for seminal root angle in wheat. Plant Genome 11:170071. 10.3835/plantgenome2017.08.0071 [DOI] [PubMed] [Google Scholar]
- Richard C. A., Hickey L. T., Fletcher S., Jennings R., Chenu K., Christopher J. T. (2015). High-throughput phenotyping of seminal root traits in wheat. Plant Methods 11:13. 10.1186/s13007-015-0055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R. A., Rebetzke G. J., Watt M., Condon A. G., Spielmeyer W., Dolferus R. (2010). Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment. Funct. Plant Biol. 37 85–97. 10.1071/FP09219 [DOI] [Google Scholar]
- Robinson H., Hickey L., Richard C., Mace E., Kelly A., Borrell A., et al. (2016). Genomic regions influencing seminal root traits in barley. Plant Genome 9 1–13. 10.3835/plantgenome2015.03.0012 [DOI] [PubMed] [Google Scholar]
- Robinson H., Kelly A., Fox G., Franckowiak J., Borrell A., Hickey L. (2018). Root architectural traits and yield: exploring the relationship in barley breeding trials. Euphytica 214:151 10.1007/s10681-018-2219-y [DOI] [Google Scholar]
- Roncallo P. F., Cervigni G. L., Jensen C., Miranda R., Carrera A. D., Helguera M., et al. (2012). QTL analysis of main and epistatic effects for flour color traits in durum wheat. Euphytica 185 77–92. 10.1007/s10681-012-0628-x [DOI] [Google Scholar]
- Sanguineti M., Li S., Maccaferri M., Corneti S., Rotondo F., Chiari T., et al. (2007). Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 151 291–305. 10.1111/j.1744-7348.2007.00198.x [DOI] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Bhat P. R., Ehdaie B., Close T. J., Lukaszewski A. J., Waines J. G. (2009). Integrated genetic map and genetic analysis of a region associated with root traits on the short arm of rye chromosome 1 in bread wheat. Theor. Appl. Genet. 119 783–793. 10.1007/s00122-009-1088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Li S., Shao M. (2013). Effects of spatial coupling of water and fertilizer applications on root growth characteristics and water use of winter wheat. J. Plant Nutr. 36 515–528. 10.1080/01904167.2012.717160 [DOI] [Google Scholar]
- Shewry P. R., Hey S. J. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4 178–202. 10.1002/fes3.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. J. C. M., Valencise B. C. A., Juliana D. O. F. V., Carnier D. M., Paulo M. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52 360–376. 10.1111/j.1744-7909.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Singh M., Gupta A., Laxmi A. (2017). Striking the right chord: signaling enigma during root gravitropism. Front. Plant Sci. 8:1304. 10.3389/fpls.2017.01304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Van Oosterom E. J., Jordan D. R., Messina C. D., Cooper M., Hammer G. L. (2010). Morphological and architectural development of root systems in sorghum and maize. Plant Soil 333 287–299. 10.1007/s11104-010-0343-0 [DOI] [Google Scholar]
- Stich B. (2009). Comparison of mating designs for establishing nested association mapping populations in maize and Arabidopsis thaliana. Genetics 183 1525–1534. 10.1534/genetics.109.108449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S., Kaeppler S. M., Brown K. M., Lynch J. P. (2011). Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341 75–87. 10.1007/s11104-010-0623-8 [DOI] [Google Scholar]
- Tuberosa R., Giuliani S., Parry M., Araus J. (2007). Improving water use efficiency in Mediterranean agriculture: What limits the adoption of new technologies? Ann. Appl. Biol. 150 157–162. 10.1111/j.1744-7348.2007.00127.x [DOI] [Google Scholar]
- Tuberosa R., Salvi S., Sanguineti M. C., Landi P., Maccaferri M., Conti S. (2002a). Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann. Bot. 89 941–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa R., Sanguineti M. C., Landi P., Giuliani M. M., Salvi S., Conti S. (2002b). Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol. Biol. 48 697–712. [DOI] [PubMed] [Google Scholar]
- Uga Y., Kitomi Y., Ishikawa S., Yano M. (2015). Genetic improvement for root growth angle to enhance crop production. Breed. Sci. 65 111–119. 10.1270/jsbbs.65.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Okuno K., Yano M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62 2485–2494. 10.1093/jxb/erq429 [DOI] [PubMed] [Google Scholar]
- Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45 1097–1102. 10.1038/ng.2725 [DOI] [PubMed] [Google Scholar]
- Voorrips R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Voss-Fels K. P., Qian L., Parra-Londono S., Uptmoor R., Frisch M., Keeble-Gagnère G., Appels R., Snowdon R. J. (2017). Linkage drag constrains the roots of modern wheat. Plant Cell Environ. 40 717—725. 10.1111/pce.12888 [DOI] [PubMed] [Google Scholar]
- Voss-Fels K. P., Robinson H., Mudge S. R., Richard C., Newman S., Wittkop B., et al. (2018a). VERNALIZATION1 modulates root system architecture in wheat and barley. Mol. Plant 11 226–229. 10.1016/j.molp.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Voss-Fels K. P., Snowdon R. J., Hickey L. T. (2018b). Designer roots for future crops. Trends Plant Sci. 23 957–960. 10.1016/j.tplants.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Wang C., Liu W., Li Q., Ma D., Lu H., Feng W., et al. (2014). Effects of different irrigation and nitrogen regimes on root growth and its correlation with above-ground plant parts in high-yielding wheat under field conditions. Field Crops Res. 165 138–149. 10.1016/j.fcr.2014.04.011 [DOI] [Google Scholar]
- Wang L., Guo M., Li Y., Ruan W., Mo X., Wu Z., et al. (2018). LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 69 385–397. 10.1093/jxb/erx427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson A., Rebetzke G., Kirkegaard J., Christopher J., Richards R., Watt M. (2014). Soil coring at multiple field environments can directly quantify variation in deep root traits to select wheat genotypes for breeding. J. Exp. Bot. 65 6231–6249. 10.1093/jxb/eru250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111 1021–1058. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A., Ghosh S., Williams M. J., Cuddy W. S., Simmonds J., Rey M.-D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4 23–29. 10.1038/s41477-017-0083-8 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen S., Sun H., Wang Y., Shao L. (2009). Root size, distribution and soil water depletion as affected by cultivars and environmental factors. Field Crops Res. 114 75–83. 10.1016/j.fcr.2009.07.006 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.