Abstract
The developmental asymmetry of fission yeast daughter cells derives from inheriting ‘older Watson’ versus ‘older Crick’ DNA strand from the parental cell, strands that are complementary but not identical with each other. A novel DNA strand-specific ‘imprint’, installed during DNA replication at the mating-type locus (mat1), imparts competence for cell type inter-conversion to one of the two chromosome replicas. The catalytic subunit of DNA Polymerase α (Polα) has been implicated in the imprinting process. Based on its known biochemical function, Polα might install the mat1 imprint during lagging strand synthesis. The nature of the imprint is not clear: it is either a nick or a ribonucleotide insertion. Our investigations do not support a direct role of Polα in nicking through putative endonuclease domains but confirm its indirect role in installing an alkali-labile moiety as the imprint. While ruling out the role of the primase subunit of Polα holoenzyme, we find that mutations in the Polα-recruitment and putative primase homology domain in Mcm10/Cdc23 abrogate the ribonucleotide imprint formation. These results, while confirming the ribonucleotide nature of the imprint suggest the possibility of a direct role of Mcm10/Cdc23 in installing it in cooperation with Polα and Swi1.
INTRODUCTION
In Schizosaccharomyces pombe, the mating-type region comprises three loci located on chromosome II: mat1M or P, mat2P and mat3M (Figure 1A). The mat1 cassette is expressed and it dictates the Plus or Minus sex/cell type to the cell. The mat2P and mat3M cassettes are transcriptionally silenced by an epigenetic mechanism and function as master copies for switching mat1. Switching occurs by highly regulated recombination through transposition/substitution of the mat1 allele with the opposite mating-type information, copied from either mat2P or mat3M cassettes (1–4). The mating-type switching is an exquisitely regulated process. First, of the four ‘granddaughter’ cells derived from a single cell, only one cell switches in nearly 90% of pedigrees (2,5,6). This pattern results from asymmetric cell division occurring in each of the two consecutive generations in the progeny of a single cell. The switching program is initiated by a novel ‘imprint,’ that is installed in specific DNA strand during replication of the mat1 locus on chromosome II (1,2). In the following cell division, the imprint consummates into a switch but only in one of the sister chromatids during mat1 replication. The mat1-switching event removes the imprint. The mat1 locus in the chromosome is replicated only in one direction. Chromosomal inversion of the mat1 ‘cassette’ abolishes imprinting, which is partially restored by genetic manipulations promoting mat1 replication in the opposite direction (7). Thus, replication of a specific mat1 strand specifically by the lagging-strand replication complex is critical for generating the imprint. The imprinting process requires three genes: swi1, swi3 and swi7/polα (8–11). Mechanistically, Swi1 and Swi3 create a replication pause at the imprint site. They also block replication forks originating from the centromeric side of mat1 (9). Furthermore, histone H3-Lys9 demethylases Lsd1/Lsd2 play a role in replication pause upstream of Swi1 and Swi3 (12). The single swi7-1/polα imprinting-deficient mutant, however, shows a normal pause and normal replication fork block, and is, therefore, defective at some other undefined step in the imprinting pathway (9,11).
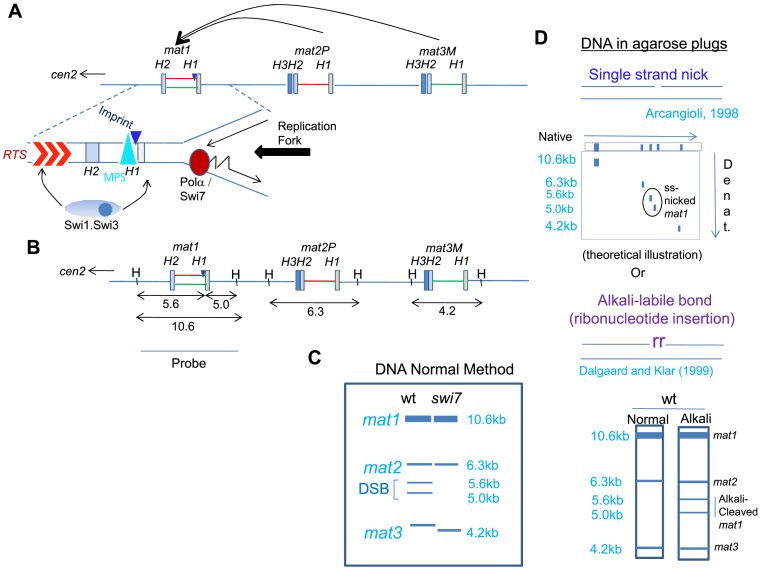
Figure 1.
Schematic diagram depicting the organization of mating-type loci in S. pombe. (A) The loci mat1, mat2 and mat3 are located in ∼30kb region of chromosome 2. They comprise short conserved homology regions H1 to H3, which flank the ∼1.1 kb allele-specific sequences. Following imprinting at the boundary of mat1 and the allele-specific region (dark blue triangle), a copy of the donor locus (mat2 or mat3) is transposed to the mat1 locus resulting in its switching by gene replacement. Replication fork progression from the centromere distal direction is met by a pause site (MPS1, blue triangle; 9), while fork from the left side encounters the replication termination sequence (RTS; 51). (B) A schematic diagram representing the HindIII restriction pattern of the mating type region with the corresponding result obtained following Southern blot hybridization using the mat1P or M fragment of 10.6 kb as a probe, schematically represented in (C), wherein mat1, mat2 and mat3 loci migrate at the positions of 10.6, 6.2 and 4.3 kb, respectively. Occurrence of imprint at mat1 generates a fragile site, which appears as a double strand break (DSB) generating the bands of 5.6 and 5.0 kb when DNA is prepared by the conventional method (left lane). Due to lack of imprint no DSB is seen in swi1, swi3 or swi7 mutants. (D) Schematic representation of the methodology used to detect a nick as the imprint, which can be visualized by 2-dimensional gel electrophoresis. (Top panel) DNA is prepared in plugs and then resolved by acrylamide gel electrophoresis in the first dimension, while the 2nd dimension is run in a denaturing acrylamide gel, as described earlier (13). Alternatively, samples embedded in agarose are digested with HindIII and then subjected to electrophoresis in native agarose gel without or with alkali-treatment (lower panel; 7), followed by Southern blotting and hybridization as in (C).
However, the nature of the imprint remains unresolved; it is thought to be either a site- and strand-specific nick (13) or an alkali-labile, RNase-sensitive modification, consisting of one or two ribonucleotides incorporated in the mat1 DNA (14,15). The imprint creates a DNA fragile site, which is artifactually converted into double-strand break (DSB) due to hydrodynamic shear during DNA extraction from cells (7,13). Therefore, the imprint level is usually determined by quantifying the DSB at the mat1 locus through Southern blot analysis.
The swi7 gene, encoding the catalytic subunit of Polα, is inherently required in initiating both leading and lagging strand replication at the replication origins and for Okazaki fragment synthesis during the lagging-strand replication (11). The biochemical role of Polα/Swi7 in generating the imprint has remained elusive. Being an essential gene limits its analysis; only one allele, swi7-1/polα (G1116E) in the catalytic subunit of Polα, is known to affect imprinting (11). Notably, the imprinting event occurs only on the newly synthesized lagging strand during S phase (16). Since the Polα/Primase complex can synthesize and extend an RNA moiety on DNA template, Polα/Swi7 is a plausible candidate for installing the imprint as a ribonucleotide(s) insertion at mat1, through the primase subunit. Alternatively, Polα-catalyzed DNA nicking may constitute the imprint (11). However, our results rule out the endonuclease function of Polα in generating the single strand nick. Instead, we show that even a catalytically dead Polα can install the ribonucleotide imprint. Furthermore, we identify Mcm10/Cdc23 as a new gene product required for installing the ribonucleotide imprint through its primase domain and interaction with Polα and Swi1.
MATERIALS AND METHODS
Strains and plasmids
The lists of strains, plasmids and oligos used in this study are provided in Supplementary Tables S1–S3, respectively (Supplementary Data). Media and growth conditions employed were as described (17). For viability assays, the cultures of the required strains were normalized to the same OD600, serially diluted 10-fold and 5μl of each dilution was spotted on the required plates. Plates were incubated at 25°C for 5 days or 30°C for 3–4 days and then photographed.
Southern hybridization
DNA was prepared by the normal method as described earlier (17). DNA was isolated from yeast cells, digested with HindIII and subjected to agarose gel electrophoresis, followed by Southern blotting and hybridization, as described earlier [11]. Alternatively, DNA was prepared from cells embedded in agarose plugs and subjected to restriction endonuclease digestion and alkali treatment, as previously described (7). The 10.6kb mat1M–containing HindIII fragment was used as a probe for Southern hybridization (Figure 1A). For experiments shown in Figure 2, donor deleted strain was used containing only the mat1 locus, with mat2 and mat3 loci being deleted (18). Normally, the level of DSB ranges around 25% of the mat1 DNA when DNA is prepared by the conventional method. However, we often observe a higher level of ∼35% because of partial shearing of the mat1 DNA. Furthermore, the level of DSB observed in alkali blots is ∼10–12 % of mat1 DNA. The 2–3-fold difference between the two methods of DNA preparation can be explained by the fact that blots of alkali-treated plugs detect only the fragments of the imprinted strand while in case of DNA prepared by the normal method both strands are detected as the mat1 DNA undergoes a DSB.
Figure 2.
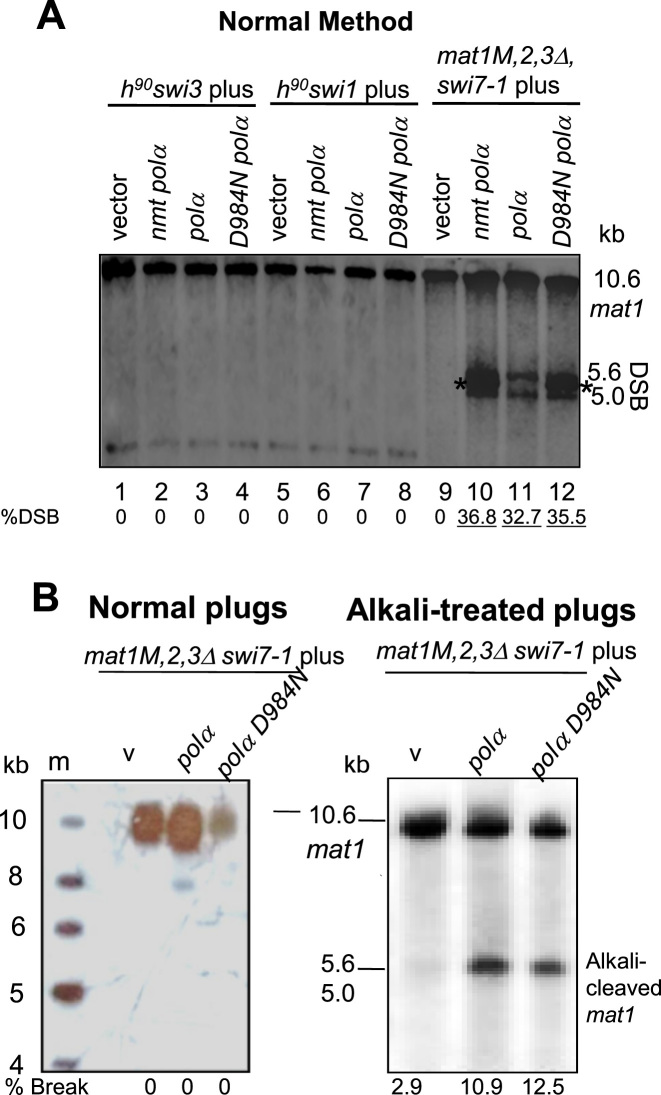
The catalytically dead polα (D984N) mutant complements the swi7-1 imprinting defect. (A) Southern blot analysis. polαD984N mutant complements the imprinting defect of swi7-1, but not that of the swi1 or swi3 mutant, in donor-deleted strains. Indicated strains were transformed with vector, nmtpolα and nmtpolαD984N plasmids and growth media plates containing thiamine. Asterisk (*) indicates the cross hybridization with the high copy vector pREP3. (B) DNA was prepared from the donor deleted swi7-1 mutant strains transformed with vector, nmtpolα and nmtpolαD984N plasmids and grown on selective plates containing thiamine. DNA was prepared in plugs (B) and digested with HindIII and Southern blotted followed by hybridization with radio-labelled mat1M probe (7). DNA was prepared in plugs and Southern blotted, without (left panel) or with alkali treatment (right panel), followed by hybridization.
Quantitation of switching efficiency
The homothallic (h90) cells, efficiently switch their mating type. Thereby, yeast colonies are composed of an equal proportion of P and M cells. Cells of opposite type mate under nitrogen starvation conditions and the resulting ‘zygotic’ diploid cell undergoes meiosis and sporulation to produce four haploid spores, called ascospores. The per cent switching efficiency was determined by using the equation: 100 × [{2(number of zygotes)}/{2(umber of zygotes) + number of vegetative cells}].
Also, the spores synthesize starch but the vegetative growing cells do not. Because starch readily reacts with iodine vapors, colonies of efficiently switching wild type strain stain black in colour while those of switching defective mutants stain lighter when exposed to iodine vapors (8,17). This procedure was used to test complementation of switching defective mutants. The transformant strains containing plasmid borne polα or cdc23 genes, expressed under the control of nmt1 or nmt41 promoters, were assayed for complementation on plates containing the repressor, thiamine. Under these conditions, leaky expression was observed from the nmt1 promoter (19; Ahmed and Singh, unpublished).
Quantitation of DSB as a measure of switching was done by using the formula: [A/(A + B)] × 100, where A represents the combined band intensity of 5.6 and 5.0 kb bands generated by DSB and B, the 10.6 kb band of the mat1 locus.
Pull-down experiments
Vector constructs encoding MBP-tagged polα or MBP-tagged swi7-1/polα gene, cloned in the expression vector pMALp2 were expressed by inducing with 1mM IPTG for 8hrs at 25°C. MBP-Polα or MBP-PolαSwi7-1were purified using amylose resin (NEB) according to manufacturer's instructions.
Genes encoding cdc23 and cdc23M36 were cloned into GST fusion vector pGEX2T and were expressed in Codon Plus strain by inducing with 1mM IPTG for 16 hrs at 18°C.For the pulldown assay, 2mg of MBP-Polα+ or MBP-PolαSwi7-1 extracts were first bound to the amylose resin and then extracts containing ultrafilter-concentrated GST-tagged Cdc23 or Cdc23-M36 were added at increasing concentration from 1 to 4 mg. After incubation at 4°C for 4 h, the amylose resin was washed with 10 column volumes of binding buffer (20mM Tris pH 7.4, 10 mM EDTA and 2 mM β-mercaptoethanol), boiled in 50 μl of SDS loading buffer for 5min and subjected to western blot analysis by using antibodies against MBP and GST at 1:1000 dilution.
Western blots were visualized by using the ECL kit (GE Healthcare).
Co-immunoprecipitation experiments
Cells of strains having the genotype cdc23-HA/swi1-TAP and cdc23HA/swi3-myc as well as untagged strains were harvested by centrifugation and washed once with ice-cold lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 50 mM NaF, 5 mM EDTA, 0.1% PMSF) containing 60 mM β-glycerophosphate. An equal volume of Zirconium beads (Sigma) was added and cells were broken by shaking in a mixer mill (Bead Beater) for 15 min at 4°C, intermittently for an hour. Zirconium beads and cell debris were removed by centrifugation for 30–35 min followed by centrifugation for another 10 min in a microfuge at 4°C.
For analysis of whole-cell extracts, 2 × Läemmeli sample buffer was added to the supernatant and the mix was boiled for 5 min at 94°C. For immunoprecipitations, extracts containing ∼1–1.5 mg of total protein were mixed with either α-CBP (Calmodulin Binding Protein; Millipore Sigma, 1μg/ml lysate), α-HA (Santacruz, 1 μg/ml lysate) or α-Myc (SantaCruz, 1.5 μg/ml lysate) specific antibodies. After overnight incubation at 4°C, 50 μl of protein A–agarose was added to total 1–1.5 mg of protein lysate and the mixtures incubated for another 4 h at 4°C. Beads were collected by centrifugation at 1000 rpm, washed three times with lysis buffer, resuspended in Läemmli sample buffer and boiled for 10 min. Western analyses were performed using Mouse monoclonal antibodies anti-HA (1:2000), anti-Myc (1:2500), and anti-CBP (1:2500) for detection of HA- tagged Cdc23 (from cells overexpressing cdc23 gene cloned in the vector pREP41HAN), myc-tagged Swi3 and TAP-tagged Swi1, respectively. For inputs, 100 μg protein (∼7.5–10%) was loaded on the SDS-PAGE before the transfer with 5–10% methanol and Tris–Cl buffer. Polyclonal antibody was raised against purified MBP-tagged Polα in rabbits and precleared by binding to MBP-Amylose resin before using for western blotting at dilution of 1:2000.
Data aquisition and analysis
The images were scanned for all the blots using the digital systems for enhanced chemiluminescence and autoradiograph, using the Biorad Pharos FX Plus Molecular Imager. The linear range was determined through the system by detection of saturation. The exposures for westerns were kept constant for each blot. The image quantitation was performed using Image J software. Equal concentrations of proteins were used and normalized with an internal control. Replicates and repeats were performed to ensure the data did not suffer experimental bias. The statistical analysis was done using one-way ANOVA followed by Tukey's multiple comparisons. Multiple comparisons with pairwise comparisons was performed within all columns using Graphed Prism software. For autorads, the automatic detection system during scanning, minimized the background and saturation. The levels of DNA and protein were constant in each experiment. The statistical analysis was done using one-way ANOVA followed by Tukey's multiple comparisons. Multiple comparisons with pairwise comparisons were performed within all columns using Graphpad Prism software.’
Directionality assay
The preferential mating type displayed by a strain was determined by PCR according to ref. 20). The reaction included 25ng of genomic DNA and primers, which included the common primer MT1, mat1P-specific primer MP and mat1M-specific primer MM; Table S3), which yielded mat1P specific product of 987 bp and mat1M-specific product of 729 bp. The extent of directionality was determined by calculating the ratio of signals of the Plus/Minus mating type, which is close to 1 in wt h90 strain and deviates significantly from 1 in the mutants displaying the directionality defect.
Far-western blotting
Far-western blotting was carried out as reported (21).
RESULTS
Polα catalytic subunit does not play a direct role in imprinting
It was reported earlier that the DNA prepared by the normal method causes hydrodynamic shearing, resulting in conversion of a nick into double strand break at the imprint site (13). As a result the mat1 HindIII fragment of 10.6kb is split into two bands of 5.6 and 5.0 kb (Figure 1B, C). According to this study, preparation of the DNA in agarose plugs avoids the hydrodynamic shear and mat1 DNA shows a nick in the top DNA strand as the putative imprint (13). Therefore, we first investigated whether the catalytic subunit of Polα may be directly involved in generating the imprint in the form of a nick at the mat1 locus. One family of homing endonucleases contains the LAGLIDADG motif (4). Interestingly, a region in Polα of S. pombe shares similarity with the LAGLIDADG motif (22–24; Supplementary Figure S1A). To ascertain the role of this motif in imprinting, both of the conserved Asp (D) residues at positions 1158 and 1160 labeled D1 and D2 (Supplementary Figure S1A) in the polα gene were individually substituted with Ala (A) residue. After cloning them into the pART1 vector containing the constitutive adh1 promoter, both wt and mutated genes were integrated into the resident swi7-1 allele by homologous recombination (Supplementary Figure S1B). Increased iodine staining of the transformant colonies showed that, like intact episomal polα, both wt and polα D to A single and double mutant constructs restored switching when chromosomally integrated by homologous recombination into the swi7-1 mutant (Supplementary Figure S1C). Thus, the putative DADG region is not involved in imprinting.
Polα also contains the restriction endonuclease (REase) signature sequence called PD-(D/E)XK (25) within the region-V (Supplementary Figure S1D). The region spanning the residues 1040 to 1056 with the sequence LDSQGKPNLDVKGLDMK contains two overlapping LD-(D/E)XK motifs: LD—X(7)—DVK and LD—X(12)—DMK. The critical Asp (D) residues are located at positions 1041 and 1049 and at residues 1041 and 1054, respectively (Supplementary Figure S1D). Each of the Asp residues at positions 1041, 1049 and 1054 was mutated to Ala. After cloning into pART1, the DNA of wt and mutant polα was transformed into swi7-1 mutant after linearization at BglII site. However, like the wt polα gene, all three mutants restored iodine staining/switching (Supplementary Figure S1E), ruling out the role of PD(D/E)XK motif in imprinting.
We further tested whether Polα exhibits a mat1-specific endonuclease activity. We observed that the largely supercoiled (sc) form of Bluescript vector containing the mat1M fragment was efficiently converted into open circular form (oc) upon incubation with MBP-Polα but not so effectively by MBP alone (Supplementary Data; Supplementary Figure S2A). No effect was observed upon incubation of the linearized mat1M plasmid DNA (Supplementary Figure S2B). However, sequencing of the purified oc DNA obtained upon incubation with MBP or MBP-Polα did not show any discontinuity or decrease in the sequencing signal coinciding with the known site of the imprint (inverted arrows; Supplementary Figure S2C, D). Thus, Polα may only be associated with a random endonuclease activity, which acts on supercoiled DNA but not specifically on mat1 DNA.
Next, we asked whether the integrity of Polα in the replication complex or its catalytic activity of Okazaki fragment synthesis establishes the imprint. Mutation of the conserved Asp residue in ‘region-I’ (the most conserved region of the α-like DNA polymerases) of human polα to Asn (D1004N) is known to abolish the catalytic activity without altering the structure or stability of the Polα–primase complex (26,27). The corresponding Asp residue (D984) in the fission yeast Polα exerts a dominant negative growth defect on cells similar to that of the human Polα D1004N mutation (26,27). We checked whether the polα D984N mutation affects imprinting. Transformants were plated on the thiamine repressor-containing medium to minimize the deleterious effect of the (D984N) mutation. Under these conditions, the wild type polα gene complemented the swi7-1 mutation (Ahmed and Singh, unpublished), likely due to leaky expression of the nmt1 promoter. To avoid the complication of donor loci alterations, and because mat1 locus is normally imprinted in the mat2 and mat3 donors-deleted cells (18), we also tested the effect of polα (D984N) on imprinting in the donors-deleted swi7-1 mutant by transforming it with either vector control, polα or polα (D984N) plasmid. The transformants were grown in the presence of the repressor thiamine. Southern blot analysis showed that the DSB level is restored in the donor-deleted swi7-1 cells by the D984N plasmid (Figure 2A, lanes 9–12) but not in swi3-157 or swi1-111 mutant (Figure 2A, lanes 1–8). Thus, the polα (D984N) mutation only complemented the imprinting defect of the swi7-1 mutant and not that of the other two known swi1 and swi3 imprinting-deficient mutants. These results also suggest the requirement of Swi1 and Swi3 for the complementation by polα D984N mutant.
Results so far failed to identify any endonuclease like function in Polα. Although this does not completely rule out the role of a Polα-associated nuclease, we also considered it unlikely as broken DNA molecules are subject to the action of nucleases and lead to lethality. Although appearance of break in DNA prepared in alkali-treated plugs does not distinguish between the presence of a nick or a ribonucleotide insertion, we noted that Dalgaard and Klar failed to detect a nick at mat1 locus when genomic DNA of wt cells was prepared in agarose plugs and subjected to denaturing gel electrophoresis in the presence of formaldehyde (7).
These considerations led us to the possibility of the existence of a ribonucleotide bond at the imprint site. This possibility was tested by performing the experiment according to Dalgaard and Klar (7), by preparing the DNA of the transformants of the swi7-1 mutant with polα and polα D984N in agarose plugs, followed by HindIII digestion and electrophoresis with and without treatment of plugs with alkali, Southern blotting and hybridization, as done earlier. The results show a lack of any band at 5–5.6 kb in DNA of swi7-1 mutant transformed with vector, polα and polα D984N plasmids prepared in plugs (Figure 2B, left panel). Surprisingly, a broad band was observed at 5–5.6kb when the DNA of cells transformed with wt polα as well as polα D984N plasmid was prepared in plugs and subjected to alkali treatment before electrophoresis (Figure 2B, right panel). These results led us to the conclusion that both polα and polαD984N generate an alkali-labile bond. Thus, the imprint is likely to be constituted by a mono- or di-ribonucleotide insertion in the mat1 DNA, as reported (7,15).
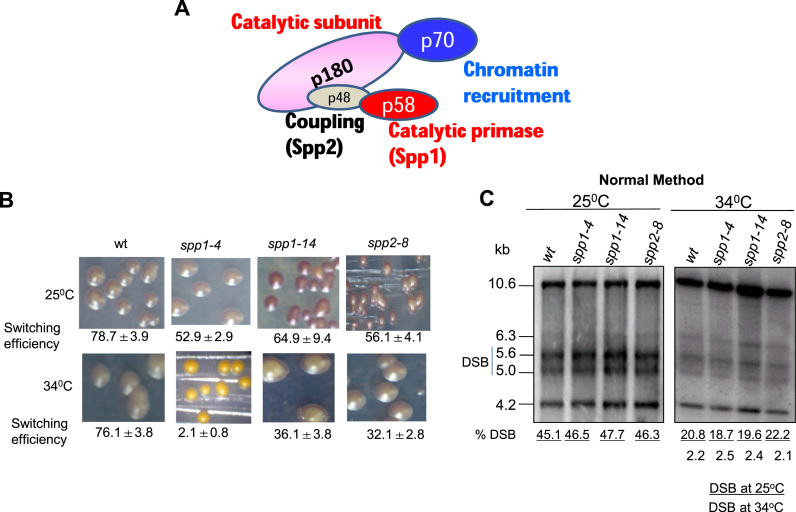
These results created a conundrum: How might the catalytically dead Polα complement the imprinting defect of swi-1 mutant? The polαD984N mutation in the evolutionarily conserved Asp residue does not alter either the stability or assembly of the mutant Polα–primase complex (26). Notably, the catalytic subunit mutant protein is unable to further extend RNA primers synthesized by the primase subunit (26). We envisaged two possibilities: either the PolαSwi7-1 mutant protein complex is defective in the primase activity or in utilizing the RNA primer synthesized by the primase subunit for DNA synthesis. To distinguish between these possibilities, we tested the imprinting and switching efficiency of temperature sensitive mutants in subunits of Polα: spp1-4, spp1-14 alleles of the primase spp1 and spp2-8 allele of spp2 gene (Figure 3A; 29). Although, somewhat reduced efficiency of switching was observed, especially in spp1-4 mutant (Figure 3B), the level of imprint/DSB was affected in these mutants to an extent similar to the wt strain when cultured at 34°C (Figure 3C). Thus, these results do not support a role of the Polα-primase subunit in mat1 imprinting. However, the extremely low rate of switching in the spp1-4 mutant at 34°C may be due to lower efficiency of utilization of the imprint for switching, as observed earlier in case of swi2, swi5 and swi6 mutants (8).
Figure 3.
Polα subunit spp1 and spp2 mutants are not defective in imprinting. (A) Sub unit structure of DNA Polα shows the largest catalytic sub unit (p180), primase sub unit Spp1 (p59), coupling subunit Spp2 (p48) and the p70 sub unit required for chromatin recruitment. (B) Picture of the iodine-staining phenotype of colonies of strains grown/sporulated at the indicated temperatures. Numbers denote the efficiency of switching represented as percent level of zygotic asci. (C) Southern blot analysis of strains described in (B). DNA was prepared by the normal method. Numbers indicate the level of DSB.
Interestingly, we find that, similar to swi6 mutant, the spp1-4 mutant shows a defect in directionality of switching; it contains nearly 20 fold more mat1M than mat1P DNA (Supplementary Data; Supplementary Figure S3A, B), indicating that like swi6, the spp1-4 mutant displays a defect in directionality of switching from Minus to Plus mating type.
Cdc23/SpMcm10 performs the imprinting function at mat1
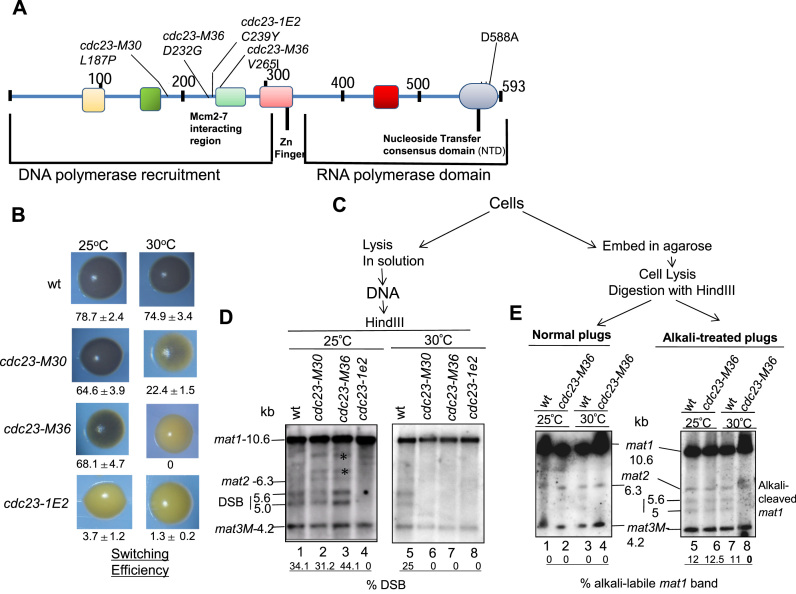
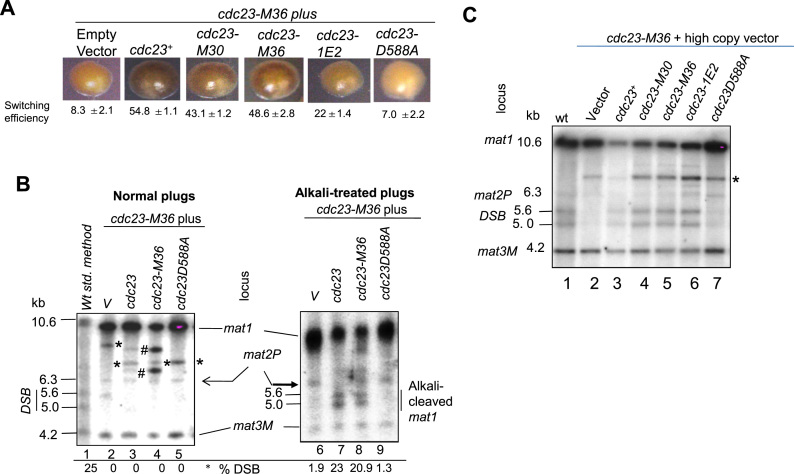
Because of a reported primase-like function of Mcm10/Cdc23, we next investigated the efficiency of switching and mat1 imprinting of cdc23 mutants (Figure 4A). Previous structure-function analysis of Cdc23 has revealed three functional domains: Polα- interaction domain, zinc domain and putative primase domain, having similarity to the bacteriophage T7 gene 4 primase (Figure 4A) (30). Notably, the cdc23-M30, cdc23-M36 and cdc23-1E2 mutations are located in its Mcm/Polα-interacting domain (30). Both cdc23-M30 and cdc23-M36 mutants showed normal iodine staining and switching (Figure 4B, left panel) at permissive temperature (25°C) but reduced iodine staining and switching at 30°C semi-permissive growth temperature (Figure 4B, right panel). In contrast, the cdc23-1E2 mutant (Figure 4A), showed low iodine staining and switching efficiency at both growth temperatures (Figure 4B). The effect on iodine staining was paralleled by the effect on level of haploid zygotic asci confirming that the low iodine staining was due to reduced switching (Supplementary Figure S4).
Figure 4.
Role of Cdc23/SpMcm10 in Imprinting at the mat1 locus. (A) Structure of Cdc23 protein, indicating the N-terminal domain required for binding with ssDNA and DNA Polα, the Zinc finger domain and the C-terminal domain performing the RNA polymerase function. Locations of mutations M30, M36 and 1E2 in the N-terminal domain and D588A in the C-terminal RNA polymerase domain are shown (adapted from ref. 30). (B) Iodine-staining phenotypes of the indicated strains in h90 background after growth on PMA+ plates at 25°C or 30°C. Numbers indicate switching efficiency, as represented by level of zygotic asci and vegetative cells. Experiment could not be performed with the cdc23-D588A mutant due to its lack of viability (ref. 30). (C) Schematic showing the process of preparation of DNA by normal method or in plugs for experiments shown in (D) and (E). *represents bands due to mating type rearrangements. (D) cdc23 mutants exhibit imprinting defect. DNA was prepared and analyzed by the standard method. (E) DNA was prepared in plugs from indicated cultures grown at 25°C or 30°C, without (left panel) and with alkali treatment (right panel) followed by Southern blotting and hybridization. Numbers represent the level of imprint in the mat1 DNA.
The effect on imprinting was assessed by determining the effect of temperature on the level of DSB; for ease we prepared the DNA by conventional method. Results show that both cdc23-30 and cdc23-36 mutants contained normal level of DSB at 25°C but reduced DSB at 30°C (Figure 4D, compare lanes 2 and 3 with lanes 6 and 7), while cdc23-1e2 mutant showed reduced DSB at both the temperatures, paralleling the effect of growth temperature on switching (Figure 4D, compare lanes 4 and 8). Importantly, results of experiments done with DNA prepared in plugs and subjected to electrophoresis with and without alkali treatment showed that, as indicated by the bands of 5.6 and 5.0 kb, the cdc23-M36 mutant contains an alkali-labile bond at mat1 when grown at 25°C but not at 30°C (Figure 4E, right panel; compare lanes 6 and 8). Thus, Mcm- and Polα-interacting domain of Cdc23 plays a role in installing the alkali-labile imprint.
Because the primase mutant is inviable (30), we explored the role of Cdc23 domains in imprinting by genetic complementation experiments. We tested the ability of the high copy number plasmids bearing different cdc23 mutant alleles to suppress the imprinting defect of the cdc23-M36 allele. While plasmids bearing wild-type cdc23+, cdc23-M30, cdc23-M36 or cdc23-1E2 complemented the switching defect of the cdc23-M36 mutant grown at 30°C, the primase defective mutant cdc23-D588A (30) did not (Figure 5A). Results with DNA prepared in plugs with and without alkali treatment showed that plasmids bearing cdc23+ and cdc23-M36 restored the alkali-labile bond at mat1 in cdc23-M36 mutants grown at 30°C (Figure 5B, compare lanes 7 and 8 with lanes 3 and 4 ), while the primase defective cdc23-D588A gene did not (Figure 5B, lane 9). Similar results were obtained when DNA prepared from the transformants by the normal method was subjected to Southern blot hybridization (Figure 5C). Surprisingly, all three cdc23 mutants in the Polα-interacting domain-M30, M36 and 1E2 complement the imprinting defect of cdc23-m36 mutant at 30°C (Figure 5C, lanes 4–6). Importantly, the mutant D588A did not restore the imprint/DSB in the cdc23-M36 mutant (Figure 5C, lane 7). It is possible that the effect of cdc23-D588A mutant gene on cdc23-M36 mutant may be due to a dominant negative effect on imprinting and switching. However, this possibility was ruled out as the D588A mutant gene had no effect on switching in the wt h90 strain (Supplementary Figure S5).
Figure 5.
Primase domain of Cdc23 is essential for imprinting. (A) Iodine-staining phenotypes of h90, cdc23-M36 mutant transformed with plasmids bearing indicated cdc23 gene mutations and grown on selective medium plates at 30°C. (B, C) cdc23 genes having mutation in primase domain fail to complement the imprinting defect of the cdc23-M36 mutant. DNA was prepared from transformants of cdc23-M36 mutant with the high copy plasmids containing the indicated genes and grown at 30°C. (B) DNA was prepared in agarose plugs. Lane labeled wt indicates DNA prepared from wild type strain. Vector only lane shows cross-reacting vectors backbone bands (marked by *) present in all lanes, while bands marked (#) correspond to mating-type rearrangements. DNA prepared in plugs from strains grown at 30°C was digested with HindIII, followed by agarose gel electrophoresis without (left panel) and with alkali treatment (right panel). (C) DNA prepared from the indicated transformant cells grown at 30°C by the normal method was digested with HindIII followed by agarose gel relectrophoresis. After Southern blotting membranes were subjected to hybridization. * represents cross reacting band of the vector DNA.
Genetic and biochemical interactions of Cdc23 with Swi1, Swi3 and Swi7
Polα physically interacts with Cdc23 (30–33). Since both cdc23 and swi7-1/polα mutants are defective in imprinting, we tested genetic interaction between their mutations. The double mutant showed much reduced switching efficiency and sporulation on minimal medium at 30°C (Supplementary Data, Supplementary Figure S6A). This result indicates that both these factors are required at the same step in imprinting. Surprisingly, unlike the single mutants, the double mutant failed to grow on rich media at 30°C (Supplementary Data, Supplementary Figure S6C).
We also investigated genetic interactions of cdc23 mutant with swi1 and swi3 mutants in switching and viability. Interestingly, cdc23-M36 mutant showed cumulative effect on switching efficiency in combination with swi1 and swi3 mutants on minimal medium at 30°C (Supplementary Figure S6B). The double mutants of cdc23 with swi1 and swi3 mutants also showed synthetic lethality on rich medium at 30°C (Supplementary Figure S6D). The discrepancy between growth on minimal medium and lack of growth on rich medium at 30°C is surprising. It may reflect that the double mutants of cdc23M36 with swi1, swi3 and swi7 may affect some important physiological function during vegetative growth but not during starvation.
Interestingly, co-immunoprecipitation experiments also showed that Cdc23 interacts with Swi1 but not Swi3 in vivo (Supplementary Data, Supplementary Figure S7A, B). Thus, Cdc23 may also act at a common step involving Swi1 and Polα.
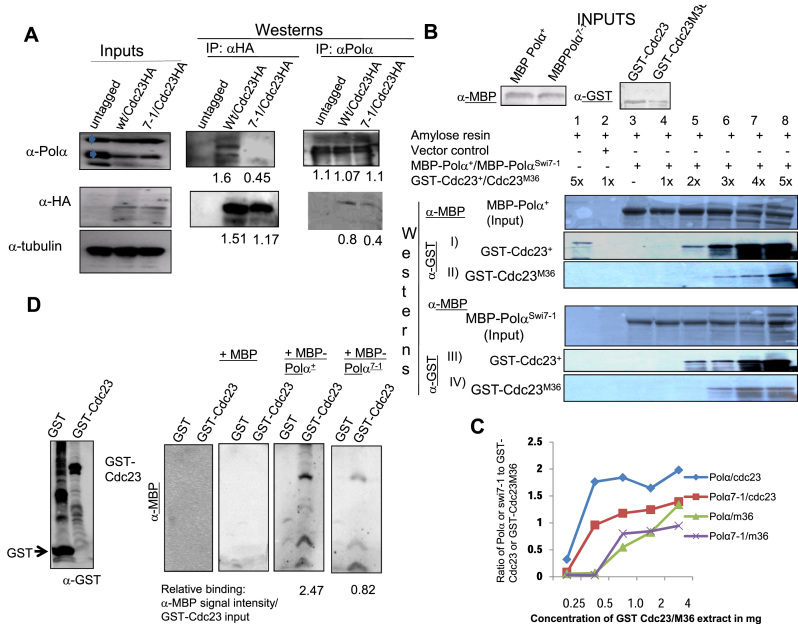
We further investigated direct biochemical interactions in vivo and in vitro. Polα+p could be readily co-immunoprecipitated with Cdc23+-HA but in comparison the mutant Polα7-1p was less efficiently co-immunoprecipitated (Figure 6A; arrowheads). In vitro pull-down experiments showed that MBP-Polα+ interacted more strongly with GST-Cdc23 than with GST-Cdc23M36 (Figure 6B, panels I and II; Figure 6C). Likewise, Polα7-1 interacted more strongly with Cdc23+ than with Cdc23M36 (Figure 6B, panels III and IV). Furthermore, Cdc23 interacted with Polα+ more strongly than with Polα7-1 (Figure 6B, compare panels I and III, panels II and IV; Figure 6C). The order of strength of interaction was Polα+-Cdc23+ > PolαSwi7-1-Cdc23+ > Polα+- Cdc23-M36 = PolαSwi7-1-Cdc23-M36. As Cdc23 recruits Polα to chromatin (34–36), our results suggest that the recruitment of Polα may be reduced in swi7-1 and cdc23-M36 mutants. Far-western analysis showed nearly 3-fold weaker interaction of Cdc23 with Polαswi7-1 as compared to Polα+ (Figure 6D).
Figure 6.
Reduced interaction of Cdc23 and Polα mutant proteins. (A) Co-IP of HA-tagged Cdc23 with Polα and Polαswi7-1. Inputs blot shows equivalent amounts of HA-tagged Cdc23, Polα and Polαswi7-1 was present in the indicated strains. Polα appears as a doublet, as indicated by arrowheads. IP was performed with anti-HA antibody followed by immunoblotting with anti-HA and anti-Polα antibodies. (B–D) In vitro interactions. (B, C) Pull-down assay. (B) Identical amounts of MBP-Polα (panels I and II) and MBP-Polαswi7-1 (panels III and IV) were bound to amylose beads and incubated with increasing concentrations of the normalized amounts of GST-Cdc23 (panels I and III) and GST-Cdc23M36 (panels II and IV), as indicated. Following SDS-PAGE of the bound proteins, the blots were probed with anti-MBP (panels I and II) and anti-GST antibodies (panels III and IV). (C) Quantitation of MBP-Polα and MPB-Polαswi7-1 binding with Cdc23 and Cdc23M36. X axis shows the concentration of Cdc23 or Cdc23M36 and the Y axis shows the ratio of Polα or Polαswi7-1 to input GST-Cdc23 or GST-Cdc23M36. (D) Far-western analysis. Partially purified GST and GST-tagged Cdc23 were subjected to SDS-PAGE and western blotted. The blots were subjected to progressive renaturing conditions, purified extracts from E. coli strains expressing MBP, MBP-Polα+ and MBP-PolαSwi7-1, as indicated. Blots were probed with anti-Polα (1:1000), anti-HA, anti-GST and anti α-tubulin (1:2,000) antibodies. Control indicates extract from E. coli control strain.
A trivial possibility to explain our genetic results could be reduction in the level of Polα in cdc23 mutants, as inactivation of Cdc23 homolog, Mcm10, is known to cause rapid loss of Polα in S. cerevisiae (34–36). However, western blot analysis shows that, compared to the wild type strain, the level of Polα is not reduced significantly in cdc23-M36 mutant grown at 25°C and 30°C (Supplementary Figure S8A, B).
MCM helicase subunits do not play a role in imprinting
Our findings indicated a role of DNA replication initiation in imprinting. Next, we queried whether the components of MCM-helicase complex, essential for DNA replication initiation, are required for imprinting (Supplementary Figure S9A). We observed normal iodine staining of colonies of mutants in mcm2 (cdc19-p1) (37), mcm4 (cdc21-M68) (38), mcm5 (nda4-108) (39) and mcm6 (mis5-268) (40), in homothallic (h90) background, indicative of normal switching at permissive temperature (25°C) and semi-permissive temperature (30°C) (Supplementary Data; Supplementary Figure S9B). This result argues against the role of earlier steps of replication initiation in imprinting.
DISCUSSION
This study addressed the mechanism of action of the catalytic subunit of Polα in strand-specific imprinting at the mat1 locus in S. pombe. While Swi1 and Swi3 play an indirect role by providing a paused replication fork, the enzymatic activity catalyzing the imprint is not known. Given its role in lagging strand synthesis (11,41), we investigated whether the catalytic subunit of Polα plays a direct role in generating a nick or ribonucleotide insertion. Results showing lack of a role of endonuclease motifs in imprinting as well as lack of a specific endonuclease activity of recombinant Polα argues against a nick being generated by Polα at the mat1 locus. Most surprisingly, even a catalytically dead polαD984N mutant gene was complementation proficient. Likewise, the spp1 and spp2 subunit mutants showed normal imprinting. None of the components of the MCM helicase complex was found to be involved in imprinting. Finally, mutants of the non-canonical primase mcm10/cdc23 gene were found to be defective in imprinting. These results support the ribonucleotide nature of the imprint. We show further that Cdc23 inserts the imprint through its primase domain and interaction with Swi1 and DNA Polα.
Mcm10/Cdc23 has been studied in S. cerevisiae, S. pombe and metazoans. However, primase activity has been demonstrated in detail in S. pombe but not in the vertebrate orthologs (30–32,42). In vitro studies have shown that the SpCdc23 makes 2–20nt long RNA primers, which are then transferred to and extended by the catalytic subunit of Polα (41). It is pertinent that even a short, 2-ribonucleotide primer synthesized by Cdc23 can be extended into a DNA chain by Polα (30,32). We suggest that primers as short as two ribonucleotides, that are synthesized by primase domain of Cdc23 at the mat1 locus, can be extended into a DNA chain by the catalytic domain of Polα; Polα is recruited through N-terminal domain of Cdc23 (4). It is puzzling how such an event could occur at the pause site. In this regard it has been shown that multiple rounds of primer synthesis occur at replication pause site (7,43). Furthermore, these two ribonucleotides may not be processed by RNaseH and could be ligated with the 3′-end of the adjacent Okazaki fragment, thus establishing the imprint.
Furthermore, the reduced binding of Cdc23-M36 to Polα+ and still poorer binding to Polα7-1, may impede extension of RNA primers by the Polα7-1. This idea can explain the cumulative reduction in imprinting/switching of cdc23-M36, polα7-1 double mutant on synthetic medium at 30°C. The ability of PolαD984N to restore imprinting in swi7-1 mutant may be ascribed to possible restoration of recruitment of Polαswi7-1 to a Cdc23-bound template to help extend the RNA primers. The interaction of Cdc23 with Swi1 also suggests that the unique primase activity may occur through interaction of Cdc23 with Polα and Swi1 at the pause site.
Another puzzling observation is the ability of the cdc23 mutant genes M30, M36 and 1E2 to complement the imprinting defect of the cdc23-M36 mutant. This can be explained by envisaging the dosage effect wherein extra amount of the above mutant proteins may compensate for the weaker interaction of the mutant protein with Polα. The inability of similarly overexpressed cdc23-D588A mutant gene to complement the imprinting defect of cdc23-M36 mutant also rules out an indirect effect and lends support to the role of primase domain of Cdc23 in inserting the ribonucleotide imprint.
How imprinting is caused uniquely at the mat1 locus remains a puzzle. One possibility is that nucleotide sequence and unique DNA/nucleoprotein architecture at the mat1 locus leads to a Swi1- and Swi3-dependent pause at the imprint site. This pause may facilitate localization of Polα to persist at the imprint site long enough to synthesize a 2-ribonucleotide primer, which is then extended by the catalytic subunit of Polα during the elongation stage of DNA replication (Figure 7). A weaker interaction of mutant Cdc23 with Polα protein may also cause reduced sequence specificity and/or efficiency to installing the imprint. Indeed, multiple break sites have been reported distal to the imprint site at the mat1 locus in the swi7-1 mutant (44). Again, possibly due to weaker interaction with Cdc23, the mutant PolαSwi7-1 may extend multiple ribonucleotide insertions near the imprint site. In sum, our results suggest that Cdc23 plays a more direct role in imprinting than the previously described factors.
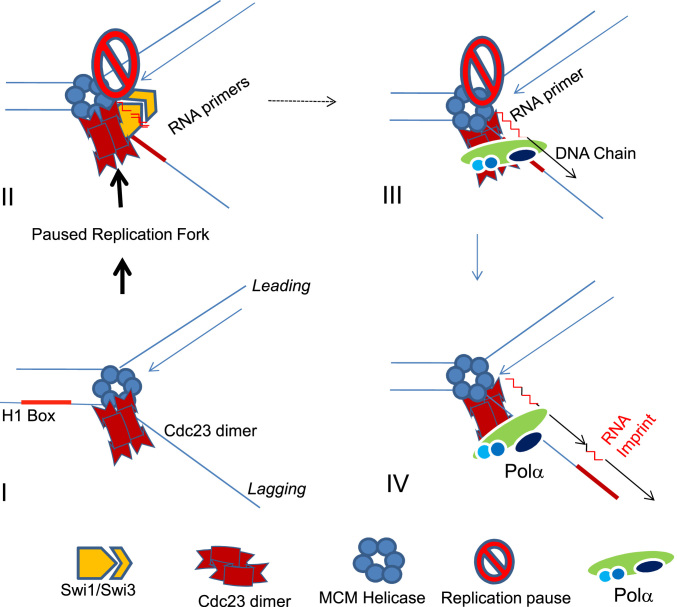
Figure 7.
A model visualizing the role of Cdc23 Primase and DNA Polα during lagging strand synthesis in generating the imprint at the pause site at the junction of the H1 box and allele-specific sequence at the mat1 locus. Pausing at the imprint site by Swi1 and Swi3 may lead to allow Cdc23 primase to interact with Swi1 and to linger at the pause site for prolonged period during elongation phase of DNA replication (II). This may allow synthesis of a short primer of two ribonucleotides length (II), which is subsequently extended by the catalytic subunit of Polα (III). The imprint may persist due to ligation with the next adjoining Okazaki fragment synthesized further upstream (IV).
Interestingly, the residue D588 is conserved between S. pombe and metazonas, including human, mice, Xenopus (30), though the metazoan homologs have been shown to lack primase activity in vitro (31). However, given the presence of an extra domain in the metazoan orthologs (30,42), a developmental control of primase activity through the extra domain may occur. Further studies are required to investigate such a possibility.
The idea of strand-specific incorporation of ribonucleotides as a part of asymmetric DNA replication contributing to generation of developmental asymmetry of sister cells appears to be unique (4). Indeed a similar mechanism was shown recently in the evolutionarily distant yeast, Schizosaccharomyces japonicus (45).
Widespread incorporation of ribonucleotides has indeed been reported during mitochondrial DNA replication (46) and replication by both Polα and Polδ in vitro (47). Thus, ribonucleotide insertion in DNA may play a role during differentiation or to facilitate recombination (48). Similarly, fragile sites occur in mammalian genome predominantly in AT-rich sequences with a potential to form stem-loop structures in response to the inhibitors of DNA replication by Polα (49). Interestingly, a homozygous mutation in Mcm10 in mice causes defect during morula to blastocyst stage, the stage when the inner cell mass (ICM) is formed, leading to formation of different organs (50). Thus, it would be interesting to investigate the role of Mcm10 in generation of DNA fragile sites, the incorporation of ribonucleotide insertions at these sites and their possible role in asymmetric cell differentiation during development in metazoans.
Supplementary Material
ACKNOWLEDGEMENTS
B.S. and K.K.B. were supported by Senior Research Fellowships from Council of Scientific and Industrial Research, New Delhi, India. We are grateful to P. Nurse, J. Hurwitz, T. Wang and S. Kearsey for gifts of strains and plasmids. We thank L. Kaur for editorial help. Help of students in the labs of A. Kumar, V. Bhalla, C. Sharma, R. Suri, D. Sharma, P. Gupta, P. Sen, J. Agrewala and D. Dutta is gratefully acknowledged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Council of Scientific and Industrial Research, New Delhi, India; A.K.’s research is supported by the Intramural Research Program of The National Institutes of Health, Frederick National Laboratory for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Klar A.J. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987; 326:466–470. [DOI] [PubMed] [Google Scholar]
- 2. Klar A.J. The developmental fate of fission yeast cells is determined by the pattern of Inheritance of parental and grand parental DNA strands. EMBO J. 1990; 9:1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egel R. Fission yeast mating-type switching: programmed damage and repair. DNA Repair (Amst.). 2005; 4:525–536. [DOI] [PubMed] [Google Scholar]
- 4. Klar A.J. Lessons learned from studies of fission yeast mating-type switching and silencing. Ann. Rev. Genet. 2007; 41:213–236. [DOI] [PubMed] [Google Scholar]
- 5. Egel R., Eie E.. Cell lineage asymmetry of Schizosaccharomyces pombe: unilateral transmission of a high-frequency mating-type switching in diploid pedigrees. Curr. Genet. 1987; 3:5–12. [Google Scholar]
- 6. Miyata H., Miyata M.. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Micro. 1981; 27:365–371. [Google Scholar]
- 7. Dalgaard J.Z., Klar A.J.. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature. 1999; 400:181–184. [DOI] [PubMed] [Google Scholar]
- 8. Egel R., Beach D.H., Klar A.J.. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dalgaard J.Z., Klar A.J.. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000; 102:745–751. [DOI] [PubMed] [Google Scholar]
- 10. Lee B.S., Grewal S.I., Klar A.J.. Biochemical interactions between proteins and mat1 cis-acting sequences required for imprinting in fission yeast. Mol. Cell Biol. 2004; 24:9813–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh J., Klar A.J.. DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature. 1993; 361:271–273. [DOI] [PubMed] [Google Scholar]
- 12. Holmes A., Roseaulin L., Schurra C., Waxin H., Lambert S., Zaratiegui M., Martienssen R.A., Arcangioli B.. Lsd1 and lsd2 control programmed replication fork pauses and imprinting in fission yeast. Cell Rep. 2012; 2:1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arcangioli B. A site-and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 1998; 17:4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vengrova S., Dalgaard J.Z.. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 2004; 18:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vengrova S., Dalgaard J.Z.. The wild type Schizosaccharomyces pombe mat1 imprint of two ribonucleoetides. EMBO Rep. 2006; 7:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes A.M., Kaykov A., Arcangioli B.. Molecular and cellular dissection of mating- type switching steps in Schizosaccharomyces pombe. Mol. Cell Biol. 2005; 25:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moreno S., Klar A.J., Nurse P.. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991; 194:795–823. [DOI] [PubMed] [Google Scholar]
- 18. Klar A.J., Miglio L.M.. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell. 1986; 46:725–731. [DOI] [PubMed] [Google Scholar]
- 19. Forsburg S. L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993; 21:2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu C., Bonaduce M.J., Klar A.. Going in the right direction: mating-type switching inSchizosacchamromycs pombeis controlled by judicious expression of two different swi2 transcripts. Genetics. 2012; 190:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Y., Li Q., Chen X-Z.. Detecting protein-protein interactions by far-western blotting. Nat.Str. Biol. 2007; 2:3278–3284. [DOI] [PubMed] [Google Scholar]
- 22. Matsumura H., Takahashi H., Inoue T., Yamamoto T., Hashimoto H., Fujiwara S., Takagi M., Imanaka T., Kai Y.. Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain. Proteins. 2006; 63:711–715. [DOI] [PubMed] [Google Scholar]
- 23. Stoddard B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005; 38:49–95. [DOI] [PubMed] [Google Scholar]
- 24. Bakhrat A., Jurica M.S., Stoddard B.L., Raveh D.. Homology Modeling and MutationalAnalysisof HO Endonuclease of Yeast. Genetics. 2004; 166:721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kniezewski L., Kinch L.N., Grishin N.V., Rychlewski L., Ginalski K.. Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive metaprofile searches. BMS Str. Biol. 2007; 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Copeland W.C., Wang T.S.F.. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem. 1993; 268:11028–11040. [PubMed] [Google Scholar]
- 27. Bhaumik D., Wang T.S.F.. Mutational effect of fission yeast Pol alpha in cell cycle events. Mol. Biol Cell. 1998; 9:2107–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan S., Wang T.. Analysis of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol. Cell. Biol. 2000; 20:7853–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffiths D.J.F., Liu V.F., Nurse P., Wang T.S.F.. Role of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol. Biol. Cell. 2001; 12:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fien K., Hurwitz J.. Fission Yeast Mcm10 Contains Primase ACtivity. J. Biol. Chem. 2006; 281:22248–22260. [DOI] [PubMed] [Google Scholar]
- 31. Robertson P.D., Warren E.M., Zhang H., Friedman D.B., Lary J.W., Cole J.L., Tutter A.V., Nalter J.C., Fanning E., Eichman E.F.. Domain architecture and biochemical characterization of vertebrate Mcm10. J. Biol. Chem. 2007; 283:3338–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fien K., Cho V-S., Lee J-K., Raychaudhary S., Tappin I., Hurwitz J.. Primer utilization by DNA polymerase a-primase is influence by its interaction with Mcm10. J. Biol. Chem. 2004; 229:16144–16153. [DOI] [PubMed] [Google Scholar]
- 33. Warren E.M., Huang H., Fanning E., Chazin W.J., Eichman B.F.. Physical interactions between Mcm10, DNA and DNA Polymerase a. J. Biol. Chem. 2009; 284:24662–24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ricke R.M., Bielinski A.K.. Mcm10 regulates the stability and chromatin association of DNA Pola. Mol. Cell. 2004; 16:173–185. [DOI] [PubMed] [Google Scholar]
- 35. Yang X., Gregan G., Lindner K., Young H., Kearsey S.E.. Nuclear distribution and chromatin association of DNA polymerase a-primase is affected by TEV protease clevage of Cdc23 (Mcm10) in fission yeast. BMC Mol. Biol. 2005; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel L. K., Kornbluth A., Dutta A.. Mcm10 and And-1/CTF4 recruitDNA polymerasea to chromatin for initiation of DNA replication. Genes Dev. 2007; 21:2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forsburg S. L., Nurse P.. The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J. Cell Sci. 1994; 107:2779–2788. [DOI] [PubMed] [Google Scholar]
- 38. Coxon A., Maundrell K., Kearsey K.. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992; 20:5571–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyake S., Okishio N., Samejima I., Kiraoka Y., Toda T., Saitoh I.. Fission yeast genes nda1+ and nda4+, mutations of which lead to S phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol. Biol. Cell. 1993; 4:1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takahashi K., Yamada Y., Yanagida M.. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy. Mol. Biol. cell. 1994; 10:1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masai H., Matsumoto S., You Y., Yoshizawa-Sugata N., Oda M.. Eukaryotic Chromosomal DNA replication: Where, when and how?. Ann. Rev. Biochem. 2010; 79:89–130. [DOI] [PubMed] [Google Scholar]
- 42. Thu Y.M., Bielinski A., K.. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem. Sci. 2007; 38:184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van C., Yan S., Michael W.M., Waga S., Cimprinch K.A.. Continued primer synthesis at stalled replication forms contributes to checkpoint activation. J. Cell Biol. 2010; 189:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen O., Egel R.. Mapping the double-strand breaks at the mating type locus in fission yeast by genome sequencing. EMBO J. 1989; 8:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu C., Bonaduce M.J., Klar A.J.S.. Defining the epigenetic mechanism of asymmetric cell division of Schizosaccharomyces japonicus yeast. Genetics. 2013; 193:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang M.Y., Bowmaker M., Reyes A., Vergani L., Angeli P., Gringeri E., Jacobs H.T., Holt I.J.. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetricDNA replication. Cell. 2002; 111:495–505. [DOI] [PubMed] [Google Scholar]
- 47. Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Angeli P., Gringeri E., Lundstrom E.M., Burgers P.M., Johansson E., Chabes A.. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010; 107:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dalgaard J.Z. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012; 28:592–597. [DOI] [PubMed] [Google Scholar]
- 49. Durkin S.G., Glover T.W.. Chromosome fragile sites. Ann. Rev. Genet. 2007; 41:169–192. [DOI] [PubMed] [Google Scholar]
- 50. Lim H.J., Jeon Y., Jeon C.H., Kim J.H., Lee H.. lethality Targeted disruption of Mcm10 causes defective embryonic cell prolieration and embryonic. Biohcim. biophys. Acta. 2011; 1813:1777–1783. [DOI] [PubMed] [Google Scholar]
- 51. Dalgaard J.Z., Klar A.J.S.. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication of mat1 in S. pombe. Genes Dev. 2001; 15:2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.