Abstract
To explore microRNA (miR)-193b expression and its potential role in colon cancer, reverse transcription-quantitative polymerase chain reaction was performed to detect the miR-193b expression levels in 62 colon cancer tissues and normal adjacent tissues. The miR-193b-overexpressed cell line SW620 was used to study the role of miR-193b in colon cancer. Subsequently, a Transwell assay and cell cycle assay were performed to observe the functional cell changes in the in vitro expression levels of miR-193b. Results indicated that miR-193b expression levels were significantly decreased in colon cancer tissues compared with adjacent normal tissue (P<0.001) and the expression of miR-193b was significantly correlated with TNM staging (P=0.03) and lymph node invasion (P=0.007). Furthermore, overexpression of miR-193b significantly decreased colon cancer cell cycle progression and its migration ability. In addition, the present findings suggested that the increased expression of miR-193b by RAB22A, inhibited downstream proteins involved in the Ras signaling pathway, including the Ras and extracellular signal-related kinase which may inhibit cancer proliferation and migration. In conclusion, the aim was to clarify the association of miR-193b expression with colon cancer, and to explore the mechanism of miR-193b in colon cancer proliferation and cell migration. The preliminary findings revealed that miR-193b may have an important role in the process in colon cancer cell cycle and migration by the RAB22A-Ras signaling pathway, thus providing a theoretical basis for miR-193b as a potential molecular target for colon cancer treatment.
Keywords: microRNA-193b, RAB22A, colon cancer, Ras signaling pathway, progression
Introduction
Colon cancer is a type of cancer that causes severe damage to human health, and its pathogenesis to date remains unclear. Colon cancer is a leading cause of mortality in developed Western countries, second only to lung cancer (1). In China, in addition to the development of social economy and the improvement in the standard of living, the dietary structure and way of life has also changed for decades; for example, the typical dietary structure of residents in China has transitioned from consisting predominantly of grain, vegetables, sugar and high-fiber, to meat, eggs, milk, fat, high-protein and low-fiber (2,3). Changes in the dietary structure have been indicated as a notable causes of colon cancer (4). Along with an increase in the aging population in China, the incidence of colon cancer and fatalities have continued to rise (5). According to the World Health Organization, the incidence of colon cancer and mortality in malignant tumors ranked third and fourth respectively for Chinese residents (4). Despite the fact that treatment with surgery, chemotherapy, radiotherapy and comprehensive biological treatment has progressed, the 5-year survival rate of advanced colon cancer has not improved (6). Colon cancer with early-onset symptoms is not typical and the primary causes of the poor prognosis in patients with colon cancer include the strong tendency of invasion and metastasis (7,8). In light of these factors, identification of novel tumor markers and research towards colon cancer prevention and control has vital practical importance.
The occurrence and development of colon cancer is a multi-factorial, multi-phase, complicated multi-step process. As the understanding of tumor molecular biology has increased, an increasing number of genes have been identified to participate in the complex process of colon cancer development (9). microRNAs (miRNAs or miRs) are small non-coding RNAs that exist widely in eukaryotic cells, are 19–24 nucleotides in length and inhibit the expression of target genes at transcriptional or translational levels (10,11). In recent years, research has revealed that miR-193b is a typical multi-function gene that is mediated by its downstream genes, and is associated with various physiological and pathological processes, including inflammation, immunodeficiency, and the occurrence and development of tumors (12,13). The miR-193b gene is located on human chromosome 16p13.12 and has an abnormal expression in prostate cancer, hepatocellular carcinoma and breast cancer (14–16). However, the specific biological functions of its invasion and metastasis in colon cancer have not yet been elucidated.
To explore the influences and mechanisms associated with miR-193b, mature miR-193b sequences (mimics) were synthesized and transfected into SW480 colon cancer cells. The present study aimed to detect cell migration, cell invasion and metastasis ability changes following miR-193b transfection. RAB22A, the predicted target gene regulated by miR-193b, may have an effect on invasion and metastasis of colon cancer, although its mechanism requires further study. The present research provides a novel theoretical basis for the treatment of colon cancer.
Materials and methods
Patient tissue samples
A total of 62 tumor tissues and adjacent counterparts (<2 cm from the tumor site) were extracted from patients (42 male, 20 female; age, 28–79 years; median age, 66 years; T1-T4 stage; no distant metastasis or preoperative chemotherapy) with colon cancer who received tumor resections between January 2010 and December 2012 at the Department of Anus and Intestine Surgery, Weifang People's Hospital (Weifang, China). Patients' basic information was recorded, including Tumor-Node-Metastasis (TNM) stage, the degree of differentiation, lymph node metastasis and distant metastasis, according to the 2002 International Cancer Alliance TNM staging criteria (17). All tissues were rapidly placed in liquid nitrogen within half an hour after tumor resection. The Ethics Committee of Weifang People's Hospital approved the present study and informed consent from all patients was received upon admission. The clinical features of all cases are indicated in Table I.
Table I.
Association between miR-193b distribution and clinicopathological characteristics of patients with colon cancer.
| miR-193b expression (n) | |||||
|---|---|---|---|---|---|
| Characteristics | Patients (n) | Low | High | χ2 | P-value |
| Total cases | 62 | 40 | 22 | ||
| Age (years) | 0.015 | 0.903 | |||
| ≥60 | 36 | 23 | 13 | ||
| <60 | 26 | 17 | 9 | ||
| Sex | 1.974 | 0.160 | |||
| Male | 30 | 22 | 8 | ||
| Female | 32 | 18 | 14 | ||
| Tumor size | 0.282 | 0.596 | |||
| ≥5 cm | 31 | 21 | 10 | ||
| <5 cm | 31 | 19 | 12 | ||
| TNM stage | 4.700 | 0.030 | |||
| I+II | 28 | 14 | 14 | ||
| III+IV | 34 | 26 | 8 | ||
| Lymph node metastasis status | 7.297 | 0.007 | |||
| Positive | 34 | 27 | 7 | ||
| Negative | 28 | 13 | 15 | ||
| Tumor differentiation | 0.448 | 0.799 | |||
| Well-differentiated | 20 | 12 | 8 | ||
| Moderately-differentiated | 20 | 14 | 6 | ||
| Poorly-differentiated | 22 | 14 | 8 | ||
| Tumors (n) | 0.518 | 0.472 | |||
| 1 | 30 | 18 | 12 | ||
| >1 | 32 | 22 | 10 | ||
miR-193b, microRNA-193b.
Cell transfection and cell invasion activity detection
The human colorectal cancer cell lines SW480, HT-29, LOVO, HCT116 and CACO-2 were purchased from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were cultured at 37°C in an atmosphere containing 5% CO2 until they reached a 60–80% confluence for subsequent experiments.
miR-193b was transfected in 6-well plates for 24 h at 37°C with 6×105 SW480 cell culture cells (70–80% confluence). An miR-193b mimic (transfected with miR-193b mimic; all obtained from Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), negative-scramble control (transfected with the negative mimic, NC) and blank control (untransfected cells, BC) were used for transfection. The sequences of the miRNAs used were as follows: miR-193b mimic, sense 5′-AACUGGCCCUCAAAGUCCCGCU-3′ and antisense 5′-AGCGGGACUUUGAGGGCCAGUU-3′; and miR-193b SC, sense 5′-UUCUCGGAACCUGUCCAGUTT-3′ and antisense 5′-ACGUCAGACGUUGCGACAATT-3′. The transfection procedure was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The following mixtures were prepared: A) 10 µl miR-193b mimic (50 pmol/µl) + 250 µl opti-MEM, as provided by the kit; and B) 10 µl Lipofectamine® 2000 + 250 µl opti-MEM. The A+B liquid was mixed together and plated for 20 min at room temperature. Subsequently, 500 µl cell culture was added drop-wise into the liquid media and 1.5 ml opti-MEM was added in each 6-well plate. The transfected cells were incubated at 37°C for 6 h and were left to culture with 2 ml RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS for 36 h at 37°C prior to further experiments. Each group was established in three replicate wells.
Transwell migration assay
Using the above-described transfection method, 1×105 cells/well were counted manually using a hemocytometer at 2 days following transfection. Transfected SW480 cells, which exhibited the lowest relative expression of miR-193b, were seeded at 8×103 cells/well in a 6-well plate in serum-free RPMI-1640 medium for 12 h at 37°C in the upper chamber of Transwell plates (Costar; Corning Incorporated, Corning, NY, USA). The lower chamber contained 500 µl 20% fetal bovine Dulbecco's modified Eagle's medium. Following incubation for 36 h at 37°C the cells in the upper chamber were wiped out, fixed with 0.1% ethanol at room temperature for 20 min, stained with crystal violet at room temperature for 20 min. All migrated cells from the upper chamber were observed under an optical microscope (magnification, ×400) with crystal violet staining. Cells were counted in 5–6 randomly visual fields. Experiments were performed in triplicate.
Wound healing assay
Following transfection, transfected cells were seeded in 12-well plates (4×105 cells/well) and cultured at 37°C. On reaching 100% confluence, a wound was created by scratching the cell surface with a 1 ml pipette tip. Cells were then washed three times with PBS and incubated in RPMI-1640 with 2% FBS. Wounds were observed under a light microscope (magnification, ×400) at 0 and 48 h. Cell migration was determined by subtracting the final wound width (µm) from the initial wound width (µm).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from tissues and all cell lines using TRIzol reagent extract (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA RT-qPCR TaqMan method was used to detect miR-193b expression level. Specific steps were followed using the TaqMan miRNA reverse transcription kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The following conditions were applied to conduct the RT reaction: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. The PCR product was preserved at 4°C. qPCR reaction system operation steps were performed according to the TaqMan Small RNA Assay instructions (Takara Bio, Inc., Otsu, Japan). The following thermocycling conditions were applied for qPCR: 95°C for 10 min; 95°C for 15 sec and 60°C for 60 sec for 42 cycles; and 72°C for 10 min using a Stratagene Mx3005P PCR system. The results were calculated using the 2−ΔΔCq method (18). The miR-193b and U6 Ct value in each sample was revised via U6 internal calibration, with 2−ΔΔCt relative quantitative method to compare the miR-193b expressed in colon cancer and matched normal tissues: ΔΔCq=Cqcarcinoma miR-193b-Cqcarcinoma U6-(Cqadjacent miR-193b-Cqadjacent U6). The relative levels of gene expression were represented as ΔCq-Cq gene-Cq reference, and the fold change of the gene expression was calculated by the 2-ΔΔCt method. Experiments were repeated in triplicate. The following primers were used: RAB22A, forward 5′-TTGTAGTTGCCATTGCAGGA-3′ and reverse 5′-AGGCTGTCTTCGGAGTTTGA-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Western blot analysis
Total SW480 cell protein was extracted using radioimmunoprecipitation assay lysis buffer (Pierce; Thermo Fisher Scientific, Inc.), centrifuged at 12,000 × g for 15 min at 4°C and quantified using the Bradford method (19). Protein (50 µg) was separated using 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA), which were then blocked for 1 h with 5% skimmed milk in Tris-buffered saline with Tween-20 at room temperature. Primary antibodies against RAB22A (cat. no. ab137093; 1:5,000, Abcam, Cambridge, UK), extracellular signal-related kinase (ERK; cat. no. AF1576; 1:800; Novus Biologicals Canada ULC; Oakville, ON, Canada), GAPDH (cat. no. sc-25778; 1:2,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), Ras (cat. no. sc-68743; 1:1,000; Santa Cruz Biotechnology, Inc.) and phosphorylated (p-)ERK l/2 (cat. no. sc-101761; 1:800; Santa Cruz Biotechnology, Inc.) were added and incubated at 4°C overnight. Following elution, mouse anti-rabbit horseradish peroxidase conjugated Immunoglobulin G (cat. no. sc-2357; 1:5,000; Santa Cruz Biotechnology, Inc.) was added and incubated for 2 h at room temperature. GAPDH was used as the reference protein. Enhanced chemiluminescent color (EMD Millipore) was added, and the blots were developed. The experiment was repeated in triplicate.
Ras separation and detection was performed using the Active Ras Pull-Down and Detection kit (cat. no. 16117; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. A total of 2×106 SW480 cells were seeded in 6-well plates and transfected with miR-193b mimic for 24 h at 37°C. Subsequently, cells were treated with 500 µl pyrolysis liquid with protease inhibitors for 5 min to lyse the cells, prior to incubation with 1 mg sample and 80 µg Glutathione S-transferase-Rafl-RBD (part of the detection kit) for 1 h at 4°C. Protein centrifugation (6,000 × g for 30 sec at 4°C) was performed followed by protein concentration determination (20). Following electrophoresis and transfer to the polyvinylidene fluoride membrane, the blot was incubated at 4°C overnight with primary antibody (anti-Ras antibody; 1:200), which was included in the Ras Pull-Down and Detection kit.
miR-193b target gene prediction and dual-luciferase assay
Targetscan (http://www.targetscan.org/), Pictar (http://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/microrna/home.do) were used to predict the downstream target gene of miR-193b, through bioinformatics analysis. The RAB22A 3′ untranslated region (UTR) was identified at miR-193b binding sites, and wild-type and mutant RAB22A 3′UTR luciferase reporter gene plasmids (Shanghai GenePharma Co., Ltd., Shanghai, China) were constructed for further study. Luc-RAB22A-3′UTR wild-type or mutant-type 3′pGL3 vector were constructed by Shanghai GeneChem Co., Ltd. (Shanghai, China). HEK293T cells were plated onto 96-well plates at density of 2×104 cells/ml for each well 24 h prior to transfection. The Luc-RAB22A-3′ UTR wild-type or mutant-type 3′ pGL3 vector and miR-193b mimics were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At the time of transfection, 50 µl RPMI-1640 medium, 50 µl Dual Glo Luciferase Reagent (premixed; Promega Corporation, Madison, WI, USA), and 100 µl Glo Stop & Glo Reagent were added to each well of a LockWell MaxiSorp test plate. After 48 h of transfection, the Firefly and Renilla luciferase fluorescence values were detected using a Dual-Luciferase® Reporter Assay System (Promega Corporation). Results were normalized to that of Renilla luciferase and the experiment was repeated in triplicate.
Flow cytometric analysis
SW480 Cells were harvested by trypsinization, washed with PBS and fixed in cold 70% EtOH at room temperature for 1 h. Then SW480 cells were washed with PBS. Staining was performed with propidium iodide (cat. no. 81845; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at room temperature for 15 min and cell cycle profiles were analyzed using a FACS Aria II flow cytometer (BD FACSAria II; BD Biosciences, San Jose, CA, USA). Data were acquired and analyzed using the FACS DiVa software program (BD Biosciences).
Statistical analysis
GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) were used for systemic analyses and plotting data. One-way Analysis of variance was used to determine significant differences between the test conditions. ImageJ 1.51p 22 software (National Institutes of Health, Bethesda, MD, USA) was used to analyze the grey ratios of western blotting bands. The association between miR-193b and clinicopathological parameters was compared using a χ2 test. Data are presented as the mean ± or + standard error of the mean as indicated. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-193b in clinical tissue samples and cell lines
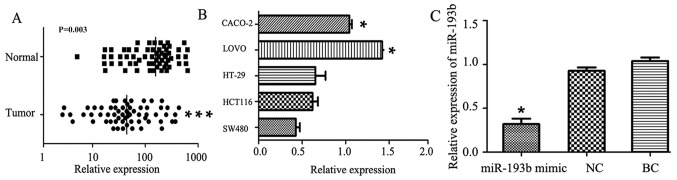
RT-qPCR was used to measure the expression levels of miR-193 in 62 colon cancer tissues and adjacent normal tissue. Of the 62 patients, 40 (64.5%) exhibited a significantly decreased level of miR-193b in colon cancer tissue specimens compared with adjacent normal tissue specimens (P=0.003; Fig. 1A), The median expression value in colon cancer tissues (64.32, normalized by the internal U6 gene) was significantly decreased compared with the median value (163.7, normalized to internal U6 gene) in the corresponding normal tissues (Fig. 1A). miR-193b was expressed at the lowest level (0.43±0.08) in the SW480 cell line compared with the other four cancer cell lines (HCT116, HT-29, LOVO and CACO-2) as indicated in Fig. 1B. The SW480 cell line was therefore selected for further study. The relative expression of miR-193b in the miR-193b mimic group was significantly downregulated compared with the NC group (P<0.05), with no significant differences between the NC and BC groups (Fig. 1C).
Figure 1.
miR-193b expression levels in colon cancer tissue and cancer cells. (A) miR-193b expression levels in colon cancer tissues were significantly decreased compared with that in normal adjacent tissues. (B) Expression of miR-193b in colon cancer cell line SW480 had the lowest expression levels compared with CACO-2, and LOVO colorectal cancer cell lines. (C) Reverse transcription-quantitative polymerase chain reaction analysis of miR-193b in SW480 cell lines compared with the NC or BC group. Data are presented as the mean + standard error of the mean. ***P<0.05 vs. normal adjacent tissue; *P<0.05 vs. the SW48 cell line, NC and BC. miR-193b, microRNA-193b, NC, negative-scramble control; BC, blank control.
Association between miR-193b expression and colon cancer clinical pathology parameters
miR-193b expression levels were significantly altered in accordance with TNM stage and lymph node metastasis status (Table I). Results indicated the miR-193b expression levels in patients with stage III+IV were significantly lower compared with those at stage I+II (P=0.030). Furthermore, patients with lymph node metastasis exhibited significantly decreased miR-193b expression levels compared with those with no lymph node metastasis (P=0.007). However, no significant differences were observed relating to age, histological type or tumor size (Table I).
miR-193b function in SW480 cancer cells following transfection
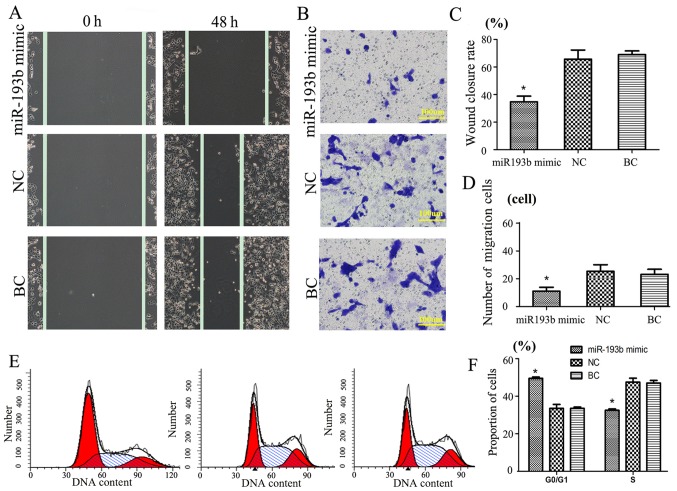
A Transwell migration assay at 48 h was performed to evaluate the function of miR-193b in SW480 cancer cells (Fig. 2). Results revealed that, following transfection of miR-193b mimic in SW480 cells group, the wound closure rate was 38.3±4.4%, whereas the wound closure rate of the NC and BC groups (compared with miR-193b mimic; P<0.05) were significantly increased in comparison (64.1±6.7 and 66.9±5.2%, respectively; Fig. 2A and C).
Figure 2.
Cell migration and migration ability following miR-193b transfection. (A and B) Following transfection, the cell migration ability in the miR-193b mimic group was markedly decreased compared with the NC and BC groups. (C) Histogram of wound closure rate indicating the rate was significantly reduced in the miR-193b mimic group compared with the NC and BC groups. (D) Histogram of cancer cell migration. (E and F) Flow cytometry analysis indicated that miR-193b mimic led to a higher G1 phase cell cycle arrest in SW480 cells compared with those of the NC or BC group. Data are presented as the mean + standard error of the mean. *P<0.05 vs. the NC and BC groups. miR-193b, microRNA-193b, NC, negative-scramble control; BC, blank control.
Additionally, Transwell assay results indicated that miR-193b may have a negative correlation with invasion. The results suggested that, compared with the NC (25±4) and BC groups (22±3), the miR-193b mimic group cell migration number (11±2) was significantly decreased (P<0.05, Fig. 2B and D). No significant difference was detected between the NC and BC groups. These results suggest that exogenous miR-193b may significantly inhibit SW480 cell migration.
SW480 cycle cell cycle results indicated that transfection with miR-193b mimic compared with the other groups was significantly different (P<0.05); the proportion of cells in the G0/G1 phase of the miR-193b mimic group was significantly increased, whereas the proportion of cells in the S phase was significantly reduced compared with the NC and BC groups, respectively (P<0.05; Fig. 2E and F). There was no significant difference between the groups in the G2/M phase (data not shown).
miR-193b biological function and pathway research in SW480 cells
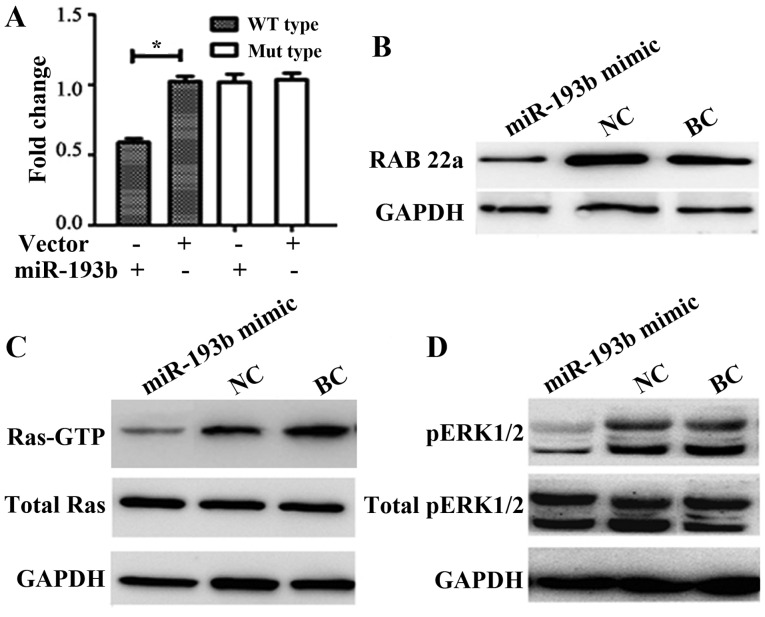
miR-193b mimic and pMIR-REPORT luciferase vector containing the RAB22A 3′UTR binding site fragment were co-transfected for 24 h. Results indicated that the luciferase activity compared with the NC or BC group, the enzyme activity was significantly reduced by ~40% in the mimic co-transfection group (P<0.05; Fig. 3A). The luciferase assay directly demonstrated that RAB22A may be a target of miR-193b (Fig. 3A).
Figure 3.
miR-193b targeted the RAB22A-Ras signaling pathway. (A) Relative luciferase activity analyses. The relative luciferase activity of the vector (+) group was set as 1. Western blot analysis was performed to determine the protein expression levels of (B) RAB22A, (C) Ras-GTP protein and (D) ERK1/2 following transfection with miR-193b mimic compared with NC and BC groups. Data are presented as the mean + standard error of the mean. *P<0.05. WT, wild-type; Mut, mutant; miR-193b, microRNA-193b, NC, negative-scramble control; BC, blank control; ERK, extracellular signal-related kinase.
The primary downstream target gene of miR-193b was predicted to be RAB22A according to bioinformatics analysis. Following transfection with miR-193b mimic into SW480 cells, RAB22A expression was significantly decreased compared with that of the NC group (0.35±0.11 vs. 1.11±0.19; P<0.01; BC group densitometry value=1) via western blot analysis. No marked differences were detected between the BC and NC groups (Fig. 3B).
The association of miR-193b and RAB22A was further analyzed by exploring the role of the Ras signaling pathway. Results indicated that the miR-193b mimic markedly reduced the protein expression levels of Ras-GTP compared with the NC and BC groups (Fig. 3C). Furthermore, ERK protein is a key Ras signaling factor (21), and the phosphorylation of ERK1/2 was evaluated by western blot analysis. In SW480 cells transfected with miR-193b, the protein expression levels of p-ERK l/2 were markedly decreased compared with the NC and BC groups (Fig. 3D).
Discussion
A number of previous studies have demonstrated that miR is associated with the occurrence of colon cancer (22,23). miRNA is important in the field of gene expression regulation and control, and is associated with apoptosis, cell cycle, cell differentiation and development (24,25). For example, in B-cell lymphoma, the miR-17–92 family is located on chromosome 13 q31 and the amplification of miR-17-92 may increase the degree of the malignant tumor formation (26). Currently, investigation into novel miR targets for cancer treatment is actively being conducted; some results have revealed the potential clinical significance of miRNA as therapeutic targets. For example, in bile duct cancer cells, miR-21, miR-141 and miR-200b were expressed at high levels, and inhibition of miR-21 and miR-200b was indicated to increase the sensitivity of the bile duct cancer cells to gemcitabine, and inhibition of miR-141 reduced cell proliferation (22,23,27). To facilitate gemcitabine treatment in transplanted tumors, a variety of changes in miR expression are required (28). Additionally, miR-21 may activate phosphatidylinositol 3 kinase signaling pathways to regulate cell apoptosis induced by gemcitabine (29). Tazawa et al (30) previously indicated that miR-34a downregulated E2F and upregulated the p53 signaling pathway, inducing an HCT116 and phenotypic RKO colon cancer cell senescence-like phenotype, which subsequently inhibited cell proliferation. These findings indicate that miR-34a has a critical role in the development of colon cancer (30). Additionally, Akao et al (31) identified that let-7 expression was reduced in colon tumors, and pre-miR let-7-1a transfected in the DLD colon cancer cell line significantly inhibited cancer cell growth.
RAB22A is a member of the RAS oncogene family that is amplified or expressed in specific tumors, including lung cancer, melanoma and osteosarcoma (32–34). Rabs serves as master regulator of transport between membrane compartments within cells, recruiting multiple effector molecules that recognize cargo, promote membrane fission and fusion, alter lipid composition, mediate transport along cytoskeletal elements and regulate other Rabs. (35,36). RAB22A is highly expressed in hepatocellular carcinoma (37). Furthermore, overexpression of RAB22A causes a prominent morphological enlargement of early and late endosomes, but hampers the transport between endosomes and the Golgi apparatus (38,39). RAB22A is associated with the co-localization of auto phagosomes, which is associated with another mechanism that leads to excessive RAB22A activation in tumor cells, including the interaction with early endosome antigen 1 and membrane transport (40). Previous research has indicated that RAB22A depletion may activate the RAF-mitogen-activated protein kinase-ERK signaling pathway, eventually leading to the occurrence of tumor development (41). The present study indicated that miR-193b targeted RAB22A through the Ras signaling pathway, resulting in the dephosphorylation of ERK1/2, repression of downstream factors and the inhibition of cell proliferation, migration, and other biological functions.
Through the study of molecular biology on tumor occurrence and development, the research suggests that the role of oncogenes and tumor suppressor genes are associated with the cell cycle mechanism by directly or indirectly regulating the cell cycle, which leads to disorder of cell cycle and uncontrolled growth even into cancerous (42). miR-193b abnormal expression is associated with human malignant tumor development (12). Xu et al (43) demonstrated that upregulated miR-193b expression targeted cyclin D1 (CCND1) and ETS1, resulting in blocking the cell cycle and suppression of liver cancer cell migration and invasion. Chen et al (44) suggested that increased expression of miR-193b inhibited malignant melanoma cell proliferation and suppressed CCND1 expression. CCND1, which is a type of oncogene that encodes protein, is the key factor in the G1 proliferation phase that has been identified to have an important role in the development of malignant tumors, and is therefore associated with tumor occurrence, metastasis and prognosis (45). A previous study has indicated that overexpression of CCND1 may shorten the G1 phase and promote cells to enter S phase, leading to uncontrolled cell proliferation and resulting in hyperplastic lesions or canceration (46). Therefore, it may be speculated that miR-193b regulates the cell cycle, not only by RAB22A-Ras signaling as indicated in the present study, but also by targeting the CCND1 gene.
The present study demonstrated that miR-193b expressed in colon cancer tissues was significantly decreased compared with that in the adjacent normal tissues and reduced expression was also observed in the SW480 colon cancer cell line. miR-193b mimic transfection into SW480 cells resulted in a reduction in migration, indicating the potential role that miR-193b has as a tumor suppressor gene in colon cancer. Furthermore, results indicated that miR-193b combined with RAB22A in the 3′UTR region, and that the specific binding of RAB22A is an miR-193b downstream target. Western blot analysis revealed that miR-193b may inhibit RAB22A protein expression levels in SW480 colon cancer cells.
These findings suggest that miR-193b has potential applications in the diagnosis and treatment of colon cancer and that it may have an important role in colon cancer development. Further studies into the function of miR-193b on the occurrence and development of colon cancer and further elucidation into the underlying molecular mechanism are required. In conclusion, these findings suggest the theoretical basis of miR-193b for the diagnosis and treatment of colon cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors’ contributions
ZMF and SCL performed the experiments and wrote the manuscript. CRL contributed significantly to the data analysis. All authors read and approved the manuscript and agree to be accountable for all aspects of the research and ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The Ethics Committee of Weifang People's Hospital (Weifang, China) approved the present study and informed consent from all patients was received upon admission.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Janakiram NB, Rao CV. The role of inflammation in colon cancer. Adv Exp Med Biol. 2014;816:25–52. doi: 10.1007/978-3-0348-0837-8_2. [DOI] [PubMed] [Google Scholar]
- 2.Deng M, Lan Y, Luo S. Quality of life estimate in stomach, colon, and rectal cancer patients in a hospital in China. Tumour Biol. 2013;34:2809–2815. doi: 10.1007/s13277-013-0839-3. [DOI] [PubMed] [Google Scholar]
- 3.Fengju S, Guanglin W, Kexin C. Incidence of colon cancer in Tianjin, China, 1981–2000. Asia Pac J Public Health. 2005;17:22–25. doi: 10.1177/101053950501700106. [DOI] [PubMed] [Google Scholar]
- 4.Hou L, Ji BT, Blair A, Dai Q, Gao YT, Chow WH. Commuting physical activity and risk of colon cancer in Shanghai, China. Am J Epidemiol. 2004;160:860–867. doi: 10.1093/aje/kwh301. [DOI] [PubMed] [Google Scholar]
- 5.Chiu BC, Ji BT, Dai Q, Gridley G, McLaughlin JK, Gao YT, Fraumeni JF, Jr, Chow WH. Dietary factors and risk of colon cancer in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2003;12:201–208. [PubMed] [Google Scholar]
- 6.Diao D, Wang L, Wan J, Chen Z, Peng J, Liu H, Chen X, Wang W, Zou L. MEK5 overexpression is associated with the occurrence and development of colorectal cancer. BMC Cancer. 2016;16:302. doi: 10.1186/s12885-016-2327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassmann S, Kreutz C, Schoepflin A, Hopt U, Timmer J, Werner M. A novel approach for reliable microarray analysis of microdissected tumor cells from formalin-fixed and paraffin-embedded colorectal cancer resection specimens. J Mol Med (Berl) 2009;87:211–224. doi: 10.1007/s00109-008-0419-y. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res. 2016;23:303–309. doi: 10.3727/096504016X14567549091305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren W, Shen S, Sun Z, Shu P, Shen X, Bu C, Ai F, Zhang X, Tang A, Tian L, et al. Jak-STAT3 pathway triggers DICER1 for proteasomal degradation by ubiquitin ligase complex of CUL4A(DCAF1) to promote colon cancer development. Cancer Lett. 2016;375:209–220. doi: 10.1016/j.canlet.2016.02.055. [DOI] [PubMed] [Google Scholar]
- 11.Mullany LE, Wolff RK, Herrick JS, Buas MF, Slattery ML. SNP regulation of microRNA expression and subsequent colon cancer risk. PLoS One. 2015;10:e0143894. doi: 10.1371/journal.pone.0143894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a novel biomarker for patients with ovarian cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feuermann Y, Kang K, Gavrilova O, Haetscher N, Jang SJ, Yoo KH, Jiang C, Gonzalez FJ, Robinson GW, Hennighausen L. MiR-193b and miR-365-1 are not required for the development and function of brown fat in the mouse. RNA Biol. 2013;10:1807–1814. doi: 10.4161/rna.27239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Kong F, Wu K, Song K, He J, Sun W. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol Med Rep. 2014;10:2613–2620. doi: 10.3892/mmr.2014.2558. [DOI] [PubMed] [Google Scholar]
- 15.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 16.Mao K, Zhang J, He C, Xu K, Liu J, Sun J, Wu G, Tan C, Zeng Y, Wang J, Xiao Z. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352:245–252. doi: 10.1016/j.canlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Hu H, Krasinskas A, Willis J. Perspectives on current tumor-node-metastasis (TNM) staging of cancers of the colon and rectum. Semin Oncol. 2011;38:500–510. doi: 10.1053/j.seminoncol.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, II, Cox AD, Philips MR. Ras signaling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Gao J, Du Z, Zhang X, Yang F, Gao W. Expression of factors and key components associated with the PI3K signaling pathway in colon cancer. Oncol Lett. 2018;15:5465–5472. doi: 10.3892/ol.2018.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YN, Chen ZH, Chen WC. Novel circulating microRNAs expression profile in colon cancer: A pilot study. Eur J Med Res. 2017;22:51. doi: 10.1186/s40001-017-0294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as regulator of signaling networks in metastatic colon cancer. Biomed Res Int. 2015;2015:823620. doi: 10.1155/2015/823620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Qin B, Liu BO. Delineating the effect of demethylating agent 5-aza-2′-deoxycytidine on human Caco-2 colonic carcinoma cells. Oncol Lett. 2016;12:139–143. doi: 10.3892/ol.2016.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagliara V, Saide A, Mitidieri E, d'Emmanuele di Villa Bianca R, Sorrentino R, Russo G, Russo A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-beta-synthase in colon cancer cells lacking p53. Oncotarget. 2016;7:50333–50348. doi: 10.18632/oncotarget.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider B, Nagel S, Ehrentraut S, Kaufmann M, Meyer C, Geffers R, Drexler HG, MacLeod RA. Neoplastic MiR-17~92 deregulation at a DNA fragility motif (SIDD) Genes Chromosomes Cancer. 2012;51:219–228. doi: 10.1002/gcc.20946. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen KN, Lindebjerg J, Nielsen BS, Hansen TF, Sørensen FB. MicroRNA-200b is downregulated in colon cancer budding cells. PLoS One. 2017;12:e0178564. doi: 10.1371/journal.pone.0178564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng L, Shi Y, Wang H, Yin B, Xia J, Wang Z. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–1749. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Chen J, Liu X, Guo Z, Sun X, Zhang J. The real-time dynamic monitoring of microRNA function in cholangiocarcinoma. PLoS One. 2014;9:e99431. doi: 10.1371/journal.pone.0099431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Wu B, Li JH, Nan G, Jiang JL, Chen ZN. Rab22a enhances CD147 recycling and is required for lung cancer cell migration and invasion. Exp Cell Res. 2017;357:9–16. doi: 10.1016/j.yexcr.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Su F, Chen Y, Zhu S, Li F, Zhao S, Wu L, Chen X, Su J. RAB22A overexpression promotes the tumor growth of melanoma. Oncotarget. 2016;7:71744–71753. doi: 10.18632/oncotarget.12329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Yang D, Liu G, Wang K. miR-203 acts as a tumor suppressor gene in osteosarcoma by regulating RAB22A. PLoS One. 2015;10:e0132225. doi: 10.1371/journal.pone.0132225. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Opdam FJ, Kamps G, Croes H, van Bokhoven H, Ginsel LA, Fransen JA. Expression of Rab small GTPases in epithelial Caco-2 cells: Rab21 is an apically located GTP-binding protein in polarised intestinal epithelial cells. Eur J Cell Biol. 2000;79:308–316. doi: 10.1078/S0171-9335(04)70034-5. [DOI] [PubMed] [Google Scholar]
- 36.Ngalame NN, Tokar EJ, Person RJ, Xu Y, Waalkes MP. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci. 2014;138:268–277. doi: 10.1093/toxsci/kfu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He H, Dai F, Yu L, She X, Zhao Y, Jiang J, Chen X, Zhao S. Identification and characterization of nine novel human small GTPases showing variable expressions in liver cancer tissues. Gene Expr. 2002;10:231–242. doi: 10.3727/000000002783992406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesa R, Salomón C, Roggero M, Stahl PD, Mayorga LS. Rab22a affects the morphology and function of the endocytic pathway. J Cell Sci. 2001;114:4041–4049. doi: 10.1242/jcs.114.22.4041. [DOI] [PubMed] [Google Scholar]
- 39.Mesa R, Magadán J, Barbieri A, López C, Stahl PD, Mayorga LS. Overexpression of Rab22a hampers the transport between endosomes and the Golgi apparatus. Exp Cell Res. 2005;304:339–353. doi: 10.1016/j.yexcr.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Kauppi M, Simonsen A, Bremnes B, Vieira A, Callaghan J, Stenmark H, Olkkonen VM. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- 41.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 42.Slattery ML, Herrick JS, Mullany LE, Samowitz WS, Sevens JR, Sakoda L, Wolff RK. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56:769–787. doi: 10.1002/gcc.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, Xing R, Jin Y, Sun Z, Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fusté NP, Castelblanco E, Felip I, Santacana M, Fernández-Hernández R, Gatius S, Pedraza N, Pallarés J, Cemeli T, Valls J, et al. Characterization of cytoplasmic cyclin D1 as a marker of invasiveness in cancer. Oncotarget. 2016;7:26979–26991. doi: 10.18632/oncotarget.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.