Abstract
Molecular-dynamics simulations were performed on the structures of the water channel aquaporin-1. The results provide an atomistic description of the interactions involved in the water permeation. Two major curvilinear pathways were identified. The simulations confirmed that the water selectivity is due primarily to the size-exclusion effect; i.e., maximally, one water molecule is allowed to pass through the narrow constriction in the aqueous pathway. Most importantly, in contrast to previous proposals, the hydrogen-bonding interactions of water molecules with the polar side chains of Asn-76 and Asn-192 on the strictly conserved Asn-Pro-Ala sequence motifs were found to be essential for maintaining the connectivity of water flow in the narrow constriction region. When Asn-76 and Asn-192 were replaced with near-isosteric hydrophobic residues in the simulation, the aqueous pathways were broken completely. Additionally, the size of the narrow constriction fluctuates significantly during the simulation, which frequently breaks the flow of water and, thus, breaks the single-file water network necessary for proton translocation. Moreover, mutations based on the simulation also have been suggested for further experimental investigation of the water-permeation mechanism of aquaporin-1.
Keywords: molecular-dynamics simulation‖membrane protein‖water permeation‖proton exclusion
Water channels belong to a recently discovered family of membrane channel proteins that allow living cells to adjust to osmotic and hydrostatic pressure changes in their environment. They are involved in numerous physiological processes (1). The first water channel that is functionally (2) and structurally (3–6) characterized is aquaporin-1 (AQP1) from human red blood cells. This channel allows a high flux of water to pass through by lowering the activation barrier of a membrane for water permeation from 10–20 kcal/mol to less than 5 kcal/mol (1). The rate of water permeation of the channel is in the range of 109-1010 molecules per second per channel (1). However, while allowing such a high rate of water permeation, AQP1 excludes small chemical species such as H+ and NH3. This exquisite selectivity remains a longstanding puzzle in physiology, because small ions like protons are known to be able to translocate through a hydrogen-bonded single file of water molecules (7).
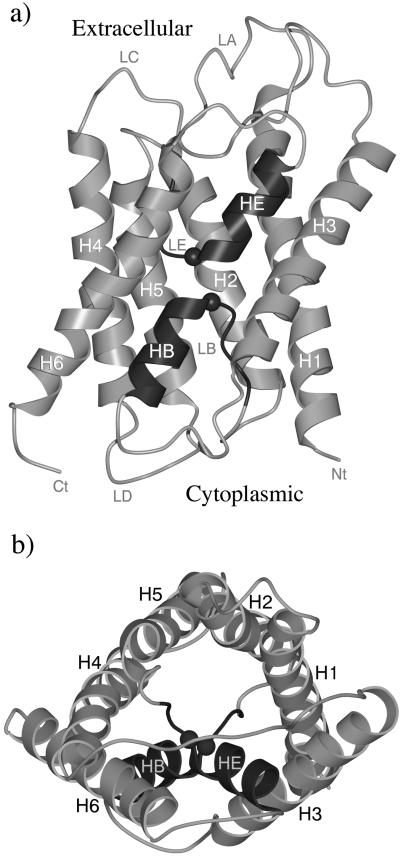
Two moderately resolved three-dimensional structures of AQP1 are available, one at 3.8 Å resolution (3) and the other at 3.7 Å resolution (4). Both were solved by electron crystallography but under slightly different conditions. AQP1 is a tetramer in vivo (8). Each monomer, which is functional alone (9, 10), consists of 269 residues (11) and contains six tilted, membrane-spanning α-helices that form a right-handed bundle with their N and C termini located on the cytoplasmic side of the membrane (12–14). Both structures suggest that the aqueous pathway is predominantly lined with conserved hydrophobic residues. A narrow constriction, with a core diameter of ≈3–4 Å over a span of one residue, is located near the middle of the aqueous pathway. Beside the constriction, loop B (LB) and loop E (LE) each contains an absolutely conserved Asn-Pro-Ala sequence motif (NPA motif) arranged in two nearly orthogonal planes. Fig. 1 shows the overall structure of an AQP1 monomer.
Figure 1.
Schematic representation of the structure of an AQP1 monomer. Side view (a) and top view from the extracellular side (b). Key secondary structural elements are labeled. The figure is based on the structure of 1FQY (3). The regions containing the two NPA motifs (Asn-76–Pro-77–Ala-78 and Asn-192–Pro-193–Ala-194) are LB-HB and LE-HE, respectively. They are highlighted by darker color. The positions of the Cα atoms of Asn-76 and Asn-192 are indicated by spheres. The figures were generated by the graphic packages of MOLSCRIPT (26) and RASTER3D (27).
Some features of the transport selectivity of AQP1 have been suggested based on functional and structural studies (3, 4, 15). However, the mechanism of water selectivity and proton discrimination remains highly controversial. Possible factors that have been proposed to contribute to the selectivity include the small size of the constriction (3, 4), the arrangement of helical dipole moment near the constriction (3, 4), and the energetic cost of bringing positively charged H3O+ ions into the cell membrane (4). The breakage of hydrogen bonds in the water flow, caused either by the competing interactions of the conserved residues Asn-76 and Asn-192 (3), or caused by the curvilinear nature of the aqueous pathway (4), also has been suggested to prohibit the formation of the single-file water network that would be required for efficient proton translocation (7).
To examine all of these proposed possibilities that contribute to the water selectivity of AQP1 and to obtain a detailed atomistic picture of the interactions involved, we performed molecular-dynamics simulations on the two structures of AQP1(3, 4). The results from the simulations provide insights into how AQP1 works and highlight the role of the conserved NPA motifs.
Methods
We performed molecular-dynamics simulations on both structures of AQP1. The initial coordinates were taken from the Protein Data Bank, PDB codes 1FQY (3) and 1HW0 (4). To reduce the size of the simulation system and to focus on the dynamics of the regions near the NPA motifs, we used the stochastic boundary molecular-dynamics (SBMD) method (16). The CHARMM package (17) was used for the simulations. The polar-hydrogen potential function (18) was used for the protein, and a modified TIP3P water model (19) was used for the water molecules. Each simulation system was divided into a reaction zone and a reservoir region; the reaction zone was divided further into a reaction region and a buffer region (16). The reference point for partitioning the system in SBMD was chosen to be near the center of the two NPA motifs. The reaction region was defined as a sphere of radius (r) 14 Å, the buffer region was 14 < r <16 Å, and the reservoir region was r > 16 Å; all atoms in the reservoir region were omitted. The final simulation systems contained 141 protein residues and 118 water molecules for 1FQY and 139 protein residues and 91 water molecules for 1HW0. Inside the reaction region, atoms were propagated by standard molecular dynamics, whereas atoms in the buffer region were propagated by the Langevin dynamics. Atoms inside the buffer region were restrained by harmonic restoring forces with force constants derived from the temperature factors in the crystal structure. Water molecules were confined to the vicinity of the active-site region by a deformable boundary potential (20). The friction constant in the Langevin dynamics simulation was set to 250 ps−1 for the protein atoms and 62 ps−1 for water molecules. All of the bonds involving hydrogen atoms were fixed by the SHAKE algorithm (21). The time step for the integration of the equations of motion was 1 fs. The initial velocities were randomly sampled from the Maxwell distribution (22). Initially, we performed 300 steps of minimizations by the steepest descent method and 500 steps of minimization by the Adapted Basis Newton Raphson method. After the minimization, the systems were dynamically equilibrated for 50 ps at 300 K. The time for the final production runs of both systems was 10 ns. For each structure, we ran a total of five simulation trajectories with different initial random velocities. For studying the role of the critical residues Asn-76 and Asn-192 on the two NPA motifs, we substituted these two Asn residues by two near-isosteric Leu residues and performed the same set of simulations.
In the simulations, we did not include lipid molecules. The portions of the structure near both the cytoplasmic and extracellular sides also were omitted in the SBMD simulation. Nevertheless, the simulation protocols we used permitted us to focus on the regions near the NPA motifs and, most importantly, to run multiple ns trajectories for more accurate statistics.
Results
We report our results primarily on the simulations of 1FQY (3). The comparisons of the original structures and the results of the simulations of 1FQY and 1HW0 are addressed at the end.
Overall Features of the Aqueous Pathway.
The water density distribution inside the channel, determined by a time average of a 10-ns simulation trajectory of 1FQY, is shown in Fig. 2. Starting from the extracellular side, the major aqueous pathway (the middle one) approaches the NPA motifs from the space between helices HE(LE), H1, and H2; after the NPA motifs, the pathway spreads into space between helices HB(LB), H2, and H5 at the cytoplasmic side. The hydrophobic lining of the pathway includes Phe-24 (H1), Val-25 (H1), Ile-29 (H1), Phe-56 (H2), Ile-60 (H2), Ala-73 (LB), and Leu-75 (LB). In the simulation, the side chain of Asn-192 faces this pathway. It is clear that the distribution of water density near the NPA motifs is significantly thinner. This observation confirms that the narrower space around that region imposes a constriction to the flow of water (4). The size of the narrow constriction fluctuates significantly during the simulation (Fig. 3a). Fig. 3b shows the overall distribution of the constriction size averaged over all five 10-ns trajectories, which peaks around 5.5 Å. Equally clear is the curvilinear nature of the pathway, as seen in Fig. 2, with a bend around the NPA motifs (4). Although AQP1 has a pseudo 2-fold symmetry in the middle of the structure, the aqueous pathways don't seem to have any symmetry.
Figure 2.
Stereo diagram of the water-density distribution. The figure is in the same orientation as that in Fig. 1a. The water density is represented by blue surface, and the positions of the Cα atoms of Asn-76 (lower) and Asn-192 (upper) are indicated by yellow spheres. The main aqueous pathway passes by the NPA motif. The water-density distribution beyond the regions immediately surrounding the NPA motifs were not simulated in stochastic boundary molecular-dynamics simulation. The remote regions, mostly surface loops that were omitted in the simulation, are drawn in thinner lines.
Figure 3.
(a) The size of the constriction as a function of time measured as the distance between Cζ of Phe-24 and Nδ of Asn-192. The data are shown in a 5-ns simulation window. (b) The distribution (histogram, ref. 28) of the constriction size averaged over all five 10-ns trajectories.
From the water distributions in Fig. 2, we also observed a second potential aqueous pathway. Although the precise boundary of each pathway is hard to define, significant water density was observed in the space near H4 and H5 as well. This pathway also is predominantly lined with hydrophobic residues such as Val-79 (HB), Leu-149 (H4), Val-153 (H4), Val-176 (H5), Ile-172 (H5), and Ile-191 (LE). In the simulation, the side chain of Asn-76 faces this pathway.
Important Interactions Involved.
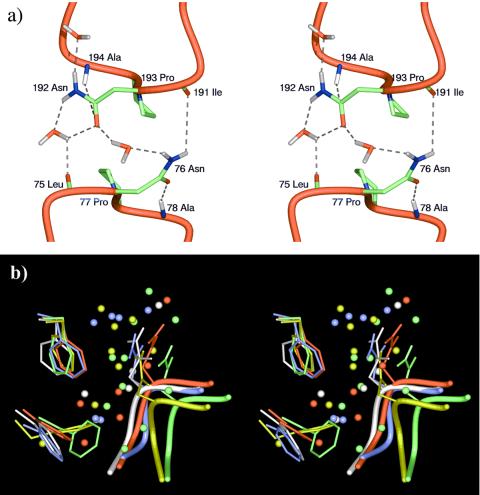
An unsolved issue in the mechanism of AQP1 is the exquisite selectivity of water over other small chemical species. In our simulation, we found that Asn-76 and Asn-192 on the NPA motifs are positioned in such a way that they more or less obey 2-fold symmetry in that region, roughly pointing to the two aqueous pathways, respectively. Pro-77 and Pro-193 on the NPA motifs are completely symmetric. As shown in Fig. 4a, the Oδ atom of Asn-76 side chain forms a hydrogen bond with the main chain N–H group of Ala-78, and the side chain Nδ–H group of Asn-76 forms a hydrogen bond with the main chain carbonyl group of Ile-191. Similarly, the Oδ of Asn-192 side chain forms a hydrogen bond with the main chain N–H group of Ala-194, and the side chain Nδ–H group of Asn-192 forms a hydrogen bond with the main chain carbonyl group of Leu-75. As the constriction opens up, water molecules can step in and form hydrogen bonds with Asn-76 and Asn-192. These interactions are stable throughout the entire duration of the simulation of 1FQY except the water-mediated interactions that can only form when there are water molecules in the constriction region.
Figure 4.
(a) Stereo diagram for the typical side chain interactions of two NPA motifs at the end of a 10-ns simulation trajectory. In this snapshot, the narrow constriction is open, and there are water molecules hydrogen-bonded to Asn-76 and Asn-192. (b) Stereo diagram for the interactions in the constriction region. The figure shows the superposition of five final structures at the end of five 10-ns trajectories (each in a different color). The side chains of Phe-24 are located at the upper left corner, and those of Phe-56 are at the lower left corner. The side chains of Asn-192 and LE are shown also. The spheres represent water molecules. The orientation is such that the cytoplasmic side is on the top. Only the yellow and green structures have enough space in the constriction region to allow water molecules to pass through.
An interesting observation is that Phe-24 and Phe-56 seem to play an important role in directing the water flow near the NPA motifs. Their highly mobile side chains, especially that of Phe-24, significantly influence the size of the constriction (Fig. 4b). Only when the distance between the side chains of Phe-24 and Asn-192 is larger than 5.0 Å (the green and yellow structures in Fig. 4b) are there water molecules in the constriction, hydrogen-bonded to the side chain and main chain of Asn-192. His-180, which is located close to Phe-56, has been found to be conserved among all of the water-selective aquaporins, but is frequently replaced with a Gly in glycerol-conducting channels (23). Very likely, His-180 plays a particular role in water conduction. In our simulation, it was observed that the side chain of His-180 participates in hydrogen-bonding interactions with water molecules in the constriction region and forms stable hydrogen bonds with the main chain carbonyl groups of Val-176 (H5) and Gly-190 (LE). Here, Gly-190 quickly flipped its backbone conformation from that in 1FQY within 50 ps of simulation so that its interaction with His-180 became possible. Given the fact that His-180 and Asn-192 are the only two polar residues near the constriction region, it would be very interesting to experimentally measure the water permeation rate for a system with a pair of swapped mutations, i.e., Asn-192 → His and His-180 → Asn.
Asn-76 → Leu and Asn-192 → Leu Double Mutations.
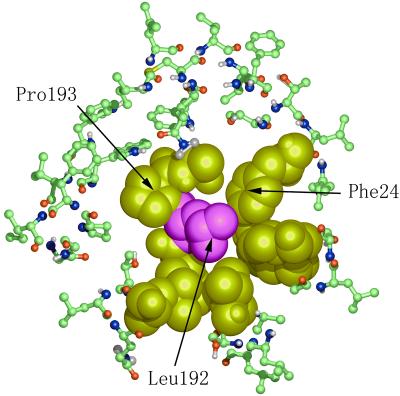
To probe further the role of the NPA motifs, we performed simulations of a system with Asn-76 and Asn-192 replaced with Leu, which has a side chain nearly isosteric to Asn but hydrophobic in chemical nature. The results show that the double mutant leads to a complete breakage of the aqueous pathway near the two NPA motifs. This breakage may be caused in part by the more extensive hydrophobic interactions in the mutant that result in a significantly increased probability of a complete closure of the constriction (Fig. 5). Also, the water density dramatically decreases around the totally hydrophobic constriction region in the mutant even when the constriction is opened up. Such results indicate that the polar side chains of Asn-76 and Asn-192 are essential for maintaining the water connectivity in the narrow constriction region. This result is contrary to earlier speculations (3) that the role of Asn-76 and Asn-192 is to compete for hydrogen bonds so as to break the single-file water network necessary for conducting protons.
Figure 5.
A cross section of the system of Asn-76 → Leu and Asn-192 → Leu double mutations. Leu-192 is in purple, and other key surrounding hydrophobic side chains are in yellow (shown as van der Waals spheres). The other residues are shown in ball-and-stick model. The side chain of Leu-192 completely breaks the water flow. The figure shows a view looking from the cytoplasmic side to the extracellular side.
It is important to point out that the wider regions of the aqueous pathway are lined with hydrophobic residues just like the pore of potassium channel KcsA (24). In a wider hydrophobic pathway, the cohesive forces among the polar water molecules are stronger than the adhesive forces between the polar water molecules and the hydrophobic wall, so that the water flow tends to form a “tube” that enables water molecules to flow quickly in the axial direction of the pathway without becoming stuck to the surrounding wall. However, in the region of the narrow constriction of AQP1, the water flow is as thin as a single water molecule. It is then essential to have polar side chains to maintain the hydrogen-bonding interactions (Fig. 4a) just like those in the gramicidin A channel (7). These observations provide a reasonable explanation for why a conserved Asn residue is required in the NPA motif. Ionic side chains would be energetically unfavorable in the middle of the cell membrane, hydrophobic residues (as shown in our simulation of the Asn-to-Leu double mutant) could not provide the required hydrogen-bonds to maintain the water flow, and other polar side chains might have size problems in maintaining the constriction. However, His, which has a similar size and chemical nature to Asn, could be an interesting candidate for substitution. Further experimental mutagenesis studies would be vital to verify our hypothesis.
Comparison of Simulations of the Two Available Structures.
Because the structures of AQP1 were determined only to moderate resolutions, inevitably there will be errors in details such as the positions of the side chains. To obtain more reliable information, we compared the original structures and the simulation results of 1FQY and 1HW0.
Although the overall features of the two structures are similar, there are several notable differences. The rms difference of Cα atoms between 1FQY and 1HW0 is 3.45 Å, which is relatively large for structures of an identical protein. This difference could be due to the moderate resolutions of the structures and/or due to the slightly different experimental conditions. Several helices in 1HW0 are not in canonical forms. The helical turns from residues 56 to 59 on H2 and residues 111 to 115 on H3 are deformed slightly. The helical turn from residues 216 to 220 on H6 is bulged out. Here the labeling of the secondary structural elements follows what was used in 1FQY. Another major difference is the orientation of the Asn-76 side chain on the first NPA motif. In 1FQY, the Asn-76 side chain roughly obeys a 2-fold symmetry with respect to that of Asn-192 on the second NPA motif. In contrast, the side chain of Asn-76 in 1HW0 seems to be positioned in such a way that it is too close to that of Asn-192 and points to an opposite direction from that of Asn-76 in 1FQY. The distance between the Nδ atoms of Asn-76 and Asn-192 is only 2.75 Å (3.67 Å in 1FQY). Several steps of simple energy minimization led to large displacements of the side chains due to strong repulsive forces between two Nδ atoms (data not shown). The local 2-fold symmetry is completely broken after only 50 ps simulations of 1HW0. Two other highly conserved residues, His-74 and Arg-195, have also drastically different (almost opposite) orientations in their side chains. Therefore, it is harder to realistically address the interactions involved with them. The conformations of His-180 are in a better agreement (see detailed discussion above). Other main features in the simulations of 1HW0 are consistent with what were observed in 1FQY.
Conclusions and Discussions.
One of the major implications of our simulation results is that the water selectivity of AQP1 is primarily caused by the size exclusion effect (3, 4, 25). The narrow constriction region imposes severe restriction so that there is usually only one water molecule allowed to pass through. Importantly, the size of the constriction fluctuates significantly, which frequently breaks the flow of water, thus breaking the single-file water network necessary for proton translocation (Fig. 4b). This observation is consistent with the fact that water molecules inside the channel diffuse three to five times slower than those in bulk water (1). The simulation also confirmed that the aqueous pathway is curvilinear with a bend around the NPA motifs (4). No symmetry of the aqueous pathways was observed, although the structure near the NPA motifs has a rough 2-fold arrangement. The existence of multiple aqueous pathways seems to be possible, although it is not an easy task to separate them completely. In the simulation, we identified two potential pathways; each has an Asn from the NPA motif pointing toward it. The strategic locations of Asn-76 and Asn-192 from the conserved NPA motifs seem to provide necessary hydrogen-bonding interactions to maintain the water connectivity in the narrow constriction region. Were Asn-76 and Asn-192 replaced with near-isosteric hydrophobic residues, such as Leu, the aqueous pathways would be broken completely. This result is in contrast to the previous speculation that Asn-76 and Asn-192 are competing for hydrogen bonding, so as to break the single-file water network necessary for proton translocation. In our simulation, the breakage of the water flow seems to arise from the diffusive fluctuation of the size of the constriction.
Based on the simulation results, we are able to propose some interesting mutations that can be used to study further the mechanism of AQP1. Of particular interest are Asn-76 → His and Asn-192 → His mutations. A pair of swapped mutations, Asn-192 → His and His-180 → Asn, also would be useful to probe the role of both highly conserved residues. Moreover, Pro-77 and Pro-193 in the NPA motifs may be important for maintaining the sharply curved conformations of loops LB and LE. Ala-78 and Ala-194 are just right for the size. There is not enough space to accommodate a larger side chain in the vicinity of the Cβ atoms of Ala-78 and Ala-194; a Gly there would make the loops too floppy.
Acknowledgments
We thank Professor Yoshinori Fujiyoshi for his encouragement and helpful discussion in the early stage of the project. We are grateful also for critical readings of the manuscript by Profs. Theodore G. Wensel and Florante A. Quiocho. We thank Dr. Yuxin Mao for his assistance with molecular graphics. This work was supported in part by a grant from the Robert A. Welch Foundation and in part by a grant from the American Heart Association (to J.M.).
Abbreviations
- AQP1
aquaporin-1
- LB
loop B
- LE
loop E
- NPA
Asn-Pro-Ala sequence motif
References
- 1.Heymann J B, Engel A. News Physiol Sci. 1999;14:187–193. doi: 10.1152/physiologyonline.1999.14.5.187. [DOI] [PubMed] [Google Scholar]
- 2.Preston G M, Carroll T P, Guggino W B, Agre P. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 3.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann J B, Engel A, Fujiyoshi Y. Nature (London) 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 4.Ren G, Reddy V S, Cheng A, Melnyk P, Mitra A K. Proc Natl Acad Sci USA. 2001;98:1398–1403. doi: 10.1073/pnas.041489198. . (First Published January 30, 2001; 10.1073/pnas.041489198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkman A S, Mitra A K. Am J Physiol. 2000;278:F13–F28. doi: 10.1152/ajprenal.2000.278.1.F13. [DOI] [PubMed] [Google Scholar]
- 6.Mitra A K. Vitam Horm (San Francisco) 2001;62:133–166. doi: 10.1016/s0083-6729(01)62003-x. [DOI] [PubMed] [Google Scholar]
- 7.Pomes R, Roux B. Biophys J. 1996;71:19–39. doi: 10.1016/S0006-3495(96)79211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verbavatz J M, Brown D, Sabolic I, Valenti G, Ausiello D A, Van Hoek A N, Ma T, Verkman A S. J Cell Biol. 1993;123:605–618. doi: 10.1083/jcb.123.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preston G M, Jung J S, Guggino W B, Agre P. J Biol Chem. 1993;268:17–20. [PubMed] [Google Scholar]
- 10.Shi L B, Skach W R, Verkman A S. J Biol Chem. 1994;269:10417–10422. [PubMed] [Google Scholar]
- 11.Preston G M, Agre P. Proc Natl Acad Sci USA. 1991;88:11110–11114. doi: 10.1073/pnas.88.24.11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wistow G J, Pisano M M, Chepelinsky A B. Trends Biochem Sci. 1991;16:170–171. doi: 10.1016/0968-0004(91)90065-4. [DOI] [PubMed] [Google Scholar]
- 13.Reizer J, Reizer A, Saier M H., Jr Crit Rev Biochem Mol Biol. 1993;28:235–257. doi: 10.3109/10409239309086796. [DOI] [PubMed] [Google Scholar]
- 14.Preston G M, Jung J S, Guggino W B, Agre P. J Biol Chem. 1994;269:1668–1673. [PubMed] [Google Scholar]
- 15.Jung J S, Preston G M, Smith B, Guggino W B, Agre P. J Biol Chem. 1994;269:14648–14654. [PubMed] [Google Scholar]
- 16.Brooks C L, 3rd, Karplus M. J Mol Biol. 1989;208:159–181. doi: 10.1016/0022-2836(89)90093-4. [DOI] [PubMed] [Google Scholar]
- 17.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 18.Neria E, Fischer S, Karplus M. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 19.Jorgensen W L. J Am Chem Soc. 1981;103:335–340. [Google Scholar]
- 20.Brooks C L, 3rd, Karplus M. J Chem Phys. 1983;79:6312–6325. [Google Scholar]
- 21.Ryckaert J P, Ciccotti G, Berendsen H J C. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 22.Allen M P, Tildesley D J. Computer Simulation of Liquids. Oxford: Clarendon; 1980. [Google Scholar]
- 23.Heymann J B, Engel A. J Mol Biol. 2000;295:1039–1053. doi: 10.1006/jmbi.1999.3413. [DOI] [PubMed] [Google Scholar]
- 24.Doyle D A, Cabral J M, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 25.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann J B, Engel A, Fujiyoshi Y. Kidney Int. 2001;60:399. [Google Scholar]
- 26.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 28.Kuang K, Haller J F, Shi G, Kang F, Cheung M, Iserovich P, Fischbarg J. Protein Sci. 2001;10:1627–1634. doi: 10.1110/ps.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]