Abstract
Non-coding antisense transcripts arise from the strand opposite the sense strand. Over 70% of the human genome generates non-coding antisense transcripts while less than 2% of the genome codes for proteins. Antisense transcripts and/or the act of antisense transcription regulate gene expression and genome integrity by interfering with sense transcription and modulating histone modifications or DNA methylation. Hence, they have significant pathological and physiological relevance. Indeed, antisense transcripts were found to be associated with various diseases including cancer, diabetes, cardiac and neurodegenerative disorders, and, thus, have promising potentials for prognostic and diagnostic markers and therapeutic development. However, it is not clearly understood how antisense transcription is initiated and epigenetically regulated. Such knowledge would provide new insights into the regulation of antisense transcription, and hence disease pathogenesis with therapeutic development. The recent studies on antisense transcription initiation and its epigenetic regulation, which are limited, are discussed here. Furthermore, we concisely describe how antisense transcription/transcripts regulate gene expression and genome integrity with implications in disease pathogenesis and therapeutic development.
Keywords: antisense transcription, long non-coding RNA, chromatin modification, GAL10, NuA4, TFIID, SAGA, RNA polymerase II
1. Introduction
Eukaryotic transcription of the protein-coding genes is a highly coordinated and complex process initiated by an assembly of general transcription factors and RNA Polymerase II at the promoter by an activator protein (activator), followed by elongation, and, finally, termination [1,2,3,4,5,6,7]. This process is tightly regulated by epigenetic factors and processes such as DNA methylation, histone modifications, and/or ATP-dependent chromatin remodeling [8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26]. These epigenetic events and transcriptions are further controlled by non-coding RNAs that include siRNAs (small interfering RNA), miRNAs (microRNA), piRNAs (Piwi-interacting RNA), lncRNAs (long non-coding RNA) or antisense non-coding transcripts [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41]. Non-coding antisense transcripts are generated from the strand opposite the sense strand and control sense transcription (and, therefore, gene expression). About 72% of the human genome generates antisense transcripts [42,43]. Antisense transcripts have important physiological and pathological significance [27,28,29,30,31,32,33,34,35,36,37,38,39,40,44,45,46,47,48,49,50,51,52,53,54,55,56]. Therefore, there are a number of studies on antisense oligonucleotide-based therapy for the regulation of gene expression, with clinical trials for treatment of various diseases including cancer, hypertension, respiratory illness, neurological and muscular disorders, and HIV infection [57,58,59,60,61,62]. Although antisense transcripts/transcription have great potential in disease pathogenesis and treatment, it remains unclear how antisense transcription is initiated and regulated by the chromatin structure. Such knowledge would provide additional avenues of regulation of antisense transcription/transcripts towards therapeutic development and disease pathogenesis. However, only a limited number of studies have focused on understanding the mechanisms of antisense transcription initiation and its regulation by the chromatin structure. These studies are discussed here. Furthermore, the roles of antisense transcription/transcripts in the regulation of gene expression and genomic stability, with implications in disease pathogenesis and therapeutic development, are also described below.
2. Antisense Transcription Initiation
Antisense transcripts were originally identified in bacteria [63]. Later, it was found that antisense transcripts are wide-spread throughout eukaryotic genomes [38,64]. More than 70% of the transcripts in humans and mice have antisense transcripts [42,43]. Antisense transcripts generally have a low abundance [65], and prefer to accumulate in the nucleus [66]. However, some antisense transcripts are found in the cytoplasm and the mitochondria [67]. Antisense transcripts are generated from independent promoters, bidirectional promoters of divergent transcription units or cryptic promoters [68,69,70,71,72,73,74,75,76]. Aside from their antisense orientation, antisense transcripts do not possess specific biochemical characteristics. Generally, antisense transcripts do not code for proteins, since the antisense transcript sequence is constrained by overlapping sense transcripts. However, there are examples of pairs of sense and antisense transcripts overlapping partially and both having protein-coding activity [77,78,79]. Independent of protein-coding potential/activity, antisense transcripts can contain specific domains that can interact with DNA, RNA or proteins to form specific functional complexes to execute cellular activities [80,81,82].
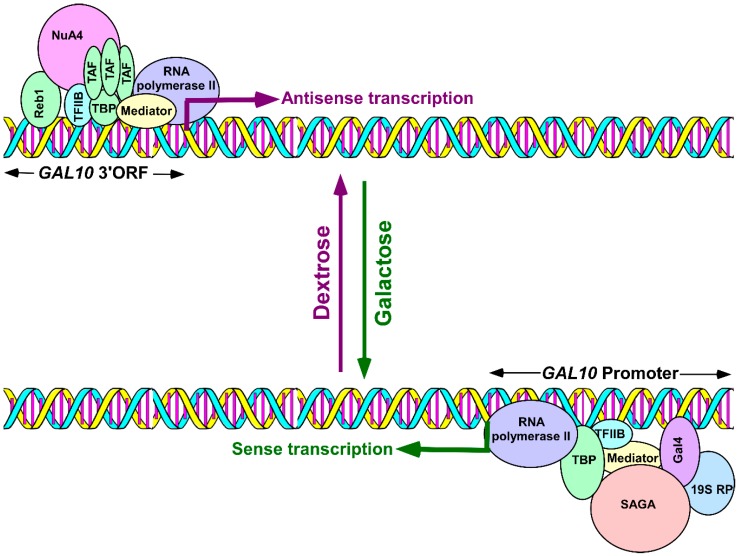
Although antisense transcription is wide-spread throughout eukaryotic genomes, it is not clearly understood how antisense transcription is initiated because it is technically difficult to study the mechanisms of antisense transcription in the background of sense transcription. Recently, we took advantage of the GAL gene cluster in yeast to study the mechanisms of antisense transcription initiation in a dextrose-containing growth medium that is permissive to antisense transcription but not to sense transcription [83,84]. GAL1, GAL7, and GAL10 constitute the GAL gene cluster, a galactose-inducible genetic unit. In this cluster, GAL1 and GAL10 are divergent genes with a bidirectional promoter, while GAL10 and GAL7 are tandem genes. Such organization has significant implications in gene regulation through transcriptional interference [85]. Previous studies [86,87] reported the existence of 2.6, 4, and 6 kb long non-coding antisense transcripts that initiated from the 3′-end of the GAL10 coding sequence in a dextrose-containing growth medium that represses GAL10 sense transcription. Such GAL10 antisense transcription attenuates GAL1-GAL10 sense transcription. Using this system, we analyzed the mechanisms of antisense transcription initiation, from the 3′-end of the GAL10 coding sequence, in a dextrose-containing growth medium [83,84]. GAL10 antisense transcription was found to be dependent on a Myb-related protein Reb1 that binds to the 3′-end of the GAL10 coding sequence [83,84,86,87]. The Reb1 binding site is located 158 bp upstream of the TATA-box at the 3′-end of the GAL10 coding sequence and 380 bp downstream of the translational stop codon [87]. However, there is another TATA-box 221 bp upstream of the Reb1 binding site [87]. Reb1 targets the recruitment of NuA4 (nucleosome acetyltransferase of histone H4) KAT (lysine (K) acetyltransferase) to the 3′-end of the GAL10 coding sequence for histone H4 acetylation targeting TBPs (TATA-box binding proteins) and TBP-associated factors (TAFs). This forms the pre-initiation complex (PIC) in recruiting RNA polymerase II (Figure 1). Consistently, NuA4 KAT, TBPs, TAFs, TFIIB (Transcription factor IIB) and a Mediator are required for the recruitment of RNA polymerase II to the 3′-end of the GAL10 coding sequence to initiate antisense transcription [83,84]. Under this growth condition (i.e., a dextrose-containing growth medium), RNA polymerase II, associated with sense transcription, was not found at the 3′-end of the GAL10 coding sequence [83]. This was because of a zinc finger protein Mig1-mediated repression as well as the masking the Gal4 activation domain by the repressor, Gal80, in a dextrose-containing growth medium [88,89,90,91,92,93,94]. Thus, our results [83,84] demonstrated, for the first time, the roles of various transcription factors, TBPs, TAFs, TFIIB, NuA4 and the Mediator, as well as the activator-binding site or Reb1 in facilitating the recruitment of RNA polymerase II to the antisense transcription initiation site at the 3′-end of the GAL10 coding sequence for antisense transcription initiation (Figure 1).
Figure 1.
Schematic diagram showing GAL10 sense and antisense transcriptions in galactose and dextrose-containing growth media, respectively. Bottom panel: The activator, Gal4, targets the co-activator, SAGA, to the GAL10 upstream activating sequence to enhance the formation of PIC, via the Mediator complex, independent of the TAFs that initiate GAL10 sense transcription a in galactose-containing growth medium [88,89]. The 19S proteasome subcomplex, or 19S RP, enhances the targeting of SAGA to Gal4 independent of the proteolytic activity of the 26S proteasome [97]. Top panel: Reb1 targets NuA4 KAT to the 3′-end of the GAL10 coding sequence (GAL10 3′ORF) for histone H4 acetylation and targeting of RNA polymerase II, via TFIID, to initiate GAL10 antisense transcription in dextrose-containing growth medium [83,84]. PIC—pre-initiation complex; SAGA—Spt-Ada-Gcn5-Acetyltransferase; TBP—TATA-box binding proteins; TAFs—TBP-associated factors; NuA4—Nucleosome acetyltransferase of histone H4; TFIID—Transcription factor IID, a complex of TBPs and a set of TAFs; TFIIB—Transcription factor IIB; and 19S RP—19S regulatory particle.
Intriguingly, we found that GAL10 antisense transcription is dependent on TFIID (transcription factor IID; an assembly of TBPs and TAFs), while its sense transcription does not require TFIID (Figure 1) [83,88,89,95,96]. Furthermore, GAL10 antisense transcription does not depend on the 19S proteasome complex or 19S regulatory particle (19S RP) that is required for sense transcription (Figure 1) [83,97]. Moreover, the Gal4 activator, which is essential for the sense transcription of GAL10, is dispensable for GAL10 antisense transcription (Figure 1) [83,88,89,96]. Furthermore, SAGA (Spt-Ada-Gcn5-acetyl-transferase) is required as a co-activator for GAL10 sense transcription but is dispensable for GAL10 antisense transcription (Figure 1) [83,88,89,96]. These results [83,84] supported the idea that GAL10 sense and antisense transcriptions are independent of each other and are regulated differently (Figure 1). Overall, our results [83,84] provided significant insights into the mechanisms of antisense transcription initiation from the 3′-end of the GAL10 coding sequence.
The initiation of antisense transcription from other loci might be regulated similarly to that of GAL10. However, it would be difficult to study the genome-wide mechanisms of antisense transcription initiation technically as antisense transcription is generally less frequent and occurs in the background of sense transcription. Furthermore, antisense promoters are less characterized and relatively weak. Nonetheless, our studies at the GAL locus, in the absence of sense transcription, clearly identified a distinct mechanism of antisense transcription initiation [83,84]. Additionally, it is technically challenging to track RNA polymerase II associated with antisense transcription in the background of sense transcription. Using the GAL system, we tracked RNA polymerase II, which is associated with GAL10 antisense transcription in the absence of sense transcription in a dextrose-containing growth medium [83]. Although our work at the GAL locus deciphered the mechanisms of antisense transcription initiation, it is poorly understood how antisense transcription is initiated from cryptic or bidirectional promoters.
3. Chromatin Regulation of Antisense Transcription
As DNA is packaged into chromatin within nucleus, chromatin structure is likely to play an important role in regulating antisense transcription. Indeed, our recent studies demonstrated that NuA4 KAT was targeted to the antisense transcription initiation site at the 3′-end of the GAL10 coding sequence for histone H4 acetylation [84]. Such targeting of NuA4 KAT or histone H4 acetylation promotes antisense transcription initiation by facilitating the recruitment of RNA polymerase II [84], similar to our results for sense transcription of the ribosomal protein genes and PHO84 [98,99,100,101,102,103]. Like NuA4 KAT, histone H3 K4 methyltransferase (Set1) and histone H3 K36 methyltransferase (Set2), which are required for histone H3 K4 and K36 methylation, respectively, also facilitate antisense transcription from the 3′-end of the GAL10 coding sequence [84]. Similarly, histone H2B ubiquitylation (which regulates histone H3 K4 methylation [104]) promotes GAL10 antisense transcription [84]. However, GAL sense transcription is facilitated by the histone H2B ubiquitylation independent of histone H3 K4 methylation [105,106]. Thus, the SAGA that possesses histone H2B deubiquitylation activity via its Ubp8 subunit [107] is likely to control antisense transcription. Intriguingly, SAGA was found to be dispensable for antisense transcription from the 3′-end of the GAL10 coding sequence but is required for the sense transcription of GAL genes [84,88,89,96,108,109,110,111]. In agreement with this, SAGA is not recruited to the 3′-end of a GAL10 coding sequence in a dextrose-containing growth medium [88,96] that is permissive for GAL10 antisense transcription but not sense transcription. However, in a galactose-containing growth medium that is permissive to GAL10 sense transcription, SAGA is targeted to the upstream activating sequence of GAL10 by the activator Gal4 [88,89,96]. Thus, the chromatin modification factor, SAGA, is differentially required for GAL10 sense and antisense transcriptions. Like SAGA, an ATP-dependent chromatin remodeling factor, SWI/SNF (switching–defective/sucrose non-fermenting) complex, is dispensable for GAL10 antisense transcription [84], but rather is required for sense transcription [112,113]. However, other ATP-dependent chromatin remodeling factor(s) may be involved in regulation of antisense transcription, which needs to be investigated further. Nonetheless, these recent studies demonstrate the roles of chromatin modification factors on the regulation of GAL10 antisense transcription. Antisense transcription from other loci is likely to be similarly epigenetically regulated, something which needs to be elucidated further.
4. Antisense Transcription in Regulation of Sense Transcription and Chromatin Structure
An antisense transcript can function by itself and/or by the act of its transcription in cis (which controls genes locally on the DNA strand involved in its origination) and/or in trans (which regulates genes on other DNA strands). The trans effect is usually mediated by the antisense transcript, while the cis effect is generally due to the act of antisense transcription [114]. Three-dimensional organization of chromatin can also allow the regions/sites of antisense transcription to interact with other loci for trans effects. Furthermore, an antisense transcript can be present at the place of its synthesis via stalled RNA polymerase, R-loops or triple helices, to exert its function in cis. It is suggested that antisense transcription/transcripts function more frequently in cis than in trans [80].
Antisense transcription regulates sense transcription by affecting DNA methylation at the CpG islands at the promoter [30,115]. For example, the hemoglobin α1 gene (HBA1) in α-thalassemia patients is repressed by antisense transcription, where an aberrant LUC7L (putative RNA-binding protein Luc7-like) RNA runs antisense to the HBA1 locus and methylates the CpG island at the promoter to repress the HBA1 gene expression [30]. Antisense transcription was also found to be involved in gene imprinting [116,117]. For example, antisense transcription of AIRN (antisense to insulin-like growth factor 2 receptor (IGF2R) non-coding RNA), but not AIRN transcript, represses IGF2R through transcriptional interference and DNA methylation in mice [117]. However, antisense transcription can also stimulate sense transcription by inhibiting de novo methylation at the promoter via R-loop formation [118,119]. Sense transcription is also regulated by antisense transcription via histone modifications. For example, X chromosome inactivation occurs through the regulation of histone modification by antisense expression. Antisense transcription also regulates histone modification in trans via antisense transcript. One classic example is mammalian HOTAIR (HOX transcript antisense intergenic RNA) that regulates histone modification, via PRC2 (polycomb repressive complex 2, required for histone H3 K27 methylation and a repressive mark), to control sense transcription [119,120]. In plants, a set of antisense transcripts to FLC (flowering locus C), namely COOLAIR (cold-assisted intronic non-coding RNA), increases histone H3 K27 methylation levels through recruitment of polycomb proteins to repress FLC expression in response to cold [121]. In addition, another antisense transcript, COLDAIR (cold-induced long antisense intragenic RNA; antisense to COOLAIR), is also responsible for the recruitment of polycomb proteins at the FLC locus [122]. In budding yeast, the antisense transcript to inorganic phosphate transporter gene PHO84 is upregulated upon chronological ageing and represses PHO84 sense transcription via histone deacetylation [123]. Furthermore, the act of antisense transcription itself regulates chromatin modifications. For example, antisense transcription from the internal cryptic promoters modifies the chromatin of the associated sense genes and, therefore, sense transcription [73,86,87]. In addition to the regulatory mechanisms of sense transcription and chromatin structure by antisense transcription discussed above, the formation of a triple RNA-DNA helix at the promoter in cis and in trans has been implicated in sense transcription repression [124,125,126]. Overall, antisense transcription/transcripts regulate gene expression by promoter methylation [30,117,127], histone modifications [81,128,129,130,131,132,133], or interfering/blocking sense transcriptional machinery [44,124,134,135].
Besides the functions discussed above, antisense transcription also controls mRNA splicing [136,137,138], mRNA stability [46] or translational efficiency through the recruitment of additional factors [139]. Thus, through these activities, antisense transcription/transcripts regulate gene expression. In addition to these gene regulatory functions, antisense transcription/transcripts are also involved in controlling the expression/generation of non-coding RNAs. For example, antisense transcription generates siRNAs from double-stranded sense-antisense hybrids [140,141]. Furthermore, an antisense transcript, namely lncTAM34a, was recently found to modulate the expression of miR34a that is associated with tumor suppression [142]. Thus, antisense transcription/transcripts play important roles in the expression/generation of coding as well as non-coding RNAs.
5. Antisense Transcription in Regulation of DNA Repair
In addition to controlling sense transcription, chromatin structure, mRNA splicing and stability, translation, and generation/expression of non-coding RNAs, antisense transcription/transcripts are involved in the regulation of DNA damage response and repair. Cells are continuously attacked by genotoxic factors, and DNA lesions and damage are repaired by various cellular mechanisms, including transcription-coupled DNA repair [143,144,145,146,147,148,149,150,151,152,153,154,155,156]. DNA damage response plays an important role in DNA repair. DNA damage activates checkpoints for cell cycle arrest and DNA repair. If DNA is not repaired or DNA repair fails, apoptosis will be triggered to remove cells with accumulated mutations [157]. Therefore, cell cycle arrest and apoptosis play crucial roles in handling detrimental genotoxic stress. These important processes of DNA repair are regulated by transcription factors and antisense transcripts/transcription, as described below.
A number of studies have indicated that the expression of antisense transcripts is altered in response to DNA damage in order to control downstream gene expression for DNA repair. For example, transcription of an antisense non-coding RNA is induced from the upstream region of the CCND1 (cyclin D1, a cell cycle regulator) gene in response to genotoxic stress [158]. Such damage-induced antisense transcripts establish a hypo-acetylated chromatin state upon binding to the RNA binding protein TLS (translocated in liposarcoma that inhibits CBP/p300 histone acetyltransferase activity), thus repressing CCND1 sense expression [159]. The reduced expression of CCND1 is associated with cell cycle arrest and check point activation for DNA repair. Another example is the long intergenic non-coding RNA-p21 (or lincRNA-p21) that is transcribed from the opposite strand of p21 (CDKN1A), a cell cycle regulator, in response to DNA damage [160]. Unlike the antisense RNA at CCND1 that acts locally, lincRNA-p21 functions globally to repress transcription of the genes that are associated with apoptosis and DNA repair [160]. Another antisense non-coding RNA at the INK locus is ANRIL (a 3.8 kb transcript in the opposite orientation of INK4B-ARF-INK4A gene cluster; also known as CDKN2B-AS1). This is also induced by the transcription factor E2F1 (E2F transcription factor 1) in an ATM (ataxia-telangiectasia mutated)-dependent fashion in response to DNA damage [161]. The transcriptional induction of ANRIL reduces the expression of INK4A (also known as CDKN2A and p16), INK4B (also known as CDKN2B and p15) and ARF (alternate reading frame; also known as p14) [162]. Such altered transcription of INK4A, INK4B and ARF permits cells to go back to their normal state following DNA repair, via the impediment of cell cycle checkpoints and stimulation of cell cycle progression [162]. In addition, ANRIL is also associated with homologous recombination-mediated DNA repair pathways [162]. Another antisense RNA, known as DLX6-AS1 (DLX6 antisense RNA 1 or Evf2 lncRNA), is involved in the regulation of DNA repair via interaction with the catalytic BRG1 (Brahma-related gene 1) subunit of the SWI/SNF chromatin remodeling complex through its ATM-dependent phosphorylation. Loss of BRG1 is associated with impaired homologous recombination [163,164,165]. Another antisense RNA, PANDA (p21-associated non-coding RNA, DNA damage activated) is located 4.5 kb upstream of the cell cycle regulator CDKN1A (p21) transcriptional start site. It is also induced in response to p53-dependent DNA damage [166]. PANDA interacts with the transcription factor NF-YA and suppresses the transcription of pro-apoptotic genes [166]. Thus, DNA damage induced PANDA prevents apoptosis via the recruitment of NF-YA. Furthermore, PANDA stabilizes p53 in response to DNA damage. Likewise, there are many examples of antisense transcripts associated with DNA repair [167,168,169]. Thus, antisense transcription/transcripts play important roles in the regulation of genomic integrity. Misregulation of antisense transcription/transcripts would alter genomic integrity, leading to cellular pathologies.
6. Antisense Transcription in Cancer
Antisense transcription/transcripts represent potential prognostic and diagnostic markers for therapeutic development for cancer. A promising candidate, HOTAIR, is significantly overexpressed in multiple tumors, including breast, colorectal, hepatocellular and pancreatic cancers [119,170,171,172,173,174,175,176]. HOTAIR is a 2.2 kb lncRNA that originates from the HOXC locus, antisense to the HOXC11 and HOXC12 genes [170]. This antisense RNA was found to interact with PRC2 for histone H3 K27 methylation and to silence chromatin [119]. HOTAIR enhances the occupancy of PRC2 at the HOXD locus, and silences transcription of the HOXD genes by altering the chromatin structure. Overexpression of HOTAIR induces genome-wide re-targeting of PRC2 to several hundred genes, leading to altered histone H3 K27 methylation, cancer progression and malignancy [119,170,171,172,173,174,175,176]. Importantly, the knockout of HOTAIR inhibits cell proliferation and migration, and induces apoptosis and cell cycle arrest in various cancer types [119,170,171,172,173,174,175,176]. Another widely studied antisense RNA is H19, one that is transcribed from H19/IGF2 on chromosome 11. Its overexpression is linked to cellular migration and invasion in various cancers including stomach, breast, liver, lung, and pancreas cancers [177]. Yoshimura et al. [178] reported that the inhibition of H19 antisense RNA could be an effective therapeutic strategy for the treatment of pancreatic cancer. Recently, another antisense RNA, MAPT-AS1 (MAPT antisense RNA 1), has been reported as a potential therapeutic target in ER (estrogen receptor)-negative breast cancers [179]. MAPT-AS1 was found to be highly expressed in breast cancer cells. Upregulation of this antisense RNA is associated with metastasis in breast cancer and other cancers, while its depletion reduces the proliferation and migration of cancer cells, thus implicating MAPT-AS1 as a therapeutic target for the treatment of ER-negative breast cancers [179].
There are other antisense RNAs that could be potential targets for cancer therapy. These include WRAP53 (WD repeat containing antisense to TP53), HOXA-AS2 (HOXA cluster antisense RNA 2), HOXA11-AS (HOXA11 Antisense RNA), PANDA, and ANRIL. WRAP53 regulates the tumor suppressor p53 and is overexpressed in a variety of tumor cell lines [180]. HOXA-AS2 is upregulated in breast cancer and its silencing inhibits the progression of breast cancer [181]. Thus, HOXA-AS2 can be a potential prognostic marker and therapeutic target for breast cancer. HOXA11-AS is upregulated in human gastric cancer cells [182]. PANDA is upregulated in gastric and breast cancers, and downregulated in non-small cell lung cancers [183,184,185]. Likewise, ANRIL is highly expressed in cancers including non-small cell lung cancer and cervical cancer, its depletion inhibits cell proliferation [186,187]. In addition to these antisense transcripts, there are other antisense transcripts involved in various cancers [188,189,190,191,192,193,194,195,196]; thus, it could serve as potential biomarkers and/or therapeutic targets for cancer therapy. Furthermore, a comprehensive dataset has been generated for a positive correlation of the differential expressions of sense–antisense transcripts with cancer [197,198].
7. Antisense Transcription in Neurological Disorders
In addition to their association with cancer, antisense transcripts are also involved in neurological disorders. The characterization of these transcripts and their modes of action may allow them to be used for diagnosis, monitoring disease progression and targeted therapies in neurological disorders. One important antisense transcript, BACE1-AS (β-site amyloid precursor protein-cleaving enzyme-antisense), is associated with Alzheimer’s disease. BACE1-AS is a 2 kb long transcript originating from BACE1 (β secretase 1) in the antisense orientation [46]. This antisense transcript plays an important role in enhancing the stability of BACE1 mRNA via the formation of the RNA duplex and, thus, leading to the elevated levels of BACE1 protein that are essential for the generation of β-amyloid [199]. The knockdown of this antisense transcript decreases the level of BACE1, thus reducing amyloid formation and aggregation in the brain. In Alzheimer’s disease, BACE1-AS is highly expressed and promotes amyloid formation via the enhanced stability of BACE1 [200], implicating BACE1-AS as an important biomarker and potential therapeutic target for the treatment of Alzheimer’s disease. Another antisense RNA, known as UCHL1-AS (ubiquitin carboxy-terminal hydrolase L1-antisense), is a 1.2 kb lncRNA that targets UCHL1 mRNA to heavy polysomes for efficient translation and to enhance UCHL1 protein level [139]. Overexpression of this UCHL1 was found to be associated with reduced amyloid β production and the delayed progression of Alzheimer’s disease [201]. Both UCHL1 and UCHL1-AS are downregulated in Parkinson’s disease [202]. Another antisense transcript, PINK1-AS (PINK1 antisense RNA), is transcribed from the antisense direction of the PINK1 gene that encodes PTEN (phosphatase and tensin homologue deleted on chromosome 10)-induced putative kinase 1. Mutation in the PINK locus causes the early onset of Parkinson’s disease. The PINK1-AS stabilizes the expression of a PINK1 splice variant, svPINK1, via a double strand RNA-mediated mechanism [203]. The silencing of PINK1-AS results in the reduced expression of svPINK1 in neuronal cells [203]. Thus, the modulation of the PINK1-AS expression may have a direct impact on Parkinson’s disease. Huntington’s disease (HD) is also associated with an antisense transcript, HTTAS (Huntingtin antisense). HTTAS is a natural antisense transcript at the Huntingtin CAG trinucleotide repeat locus. It is alternatively spliced into HTTAS-v1 (exons 1 and 3) and HTTAS-v2 (exons 2 and 3). Using cell systems, the HTTAS-v1 overexpression was found to correlate with reduced endogenous transcript levels of HTT (Huntingtin), while the knockdown of HTTAS-v1 positively influenced the HTT transcript level. The reduced expression of HTTAS-v1 was observed clinically in human HD frontal cortexes, suggesting the involvement of HTTAS-v1 in the regulation of HTT expression and the progression of HD [204,205]. Another antisense lncRNA, TUG1 (Taurine upregulated gene 1), is upregulated in HD patients [205]. TUG1 interacts with the EZH2 (enhancer of zeste homolog 2) component of PRC2 and, thus, epigenetically represses the expressions of the target genes [206,207]. The depletion of TUG1 induces apoptosis [208]. Thus, the dysregulation of TUG1 is associated with HD and other neurological disorders [205]. Likewise, there are other antisense transcripts associated with neurological disorders [209,210,211,212,213,214].
8. Antisense Transcription in Diabetes, Cardiovascular and Other Diseases
In addition to their involvement in cancer and neurological disorders, antisense transcripts are also associated with various other diseases including diabetes and cardiovascular disorders. Transcriptome-wide studies have revealed several antisense transcripts to be involved in diabetes mellitus, a metabolic disorder associated with high blood glucose levels. Misregulation of these antisense transcript expressions is linked to both type 1 and type 2 diabetes [215,216,217,218,219]. The antisense transcript ANRIL is a hot spot region associated with type 2 diabetes, diabetic nephropathy, diabetic retinopathy, diabetic cardiomyopathy and coronary artery disease (CAD). ANRIL expression is high in retina and retinal endothelial cells due to hyperglycemia. The elevated expression of ANRIL has been shown to regulate the transcription and function of VEGF (vascular endothelial growth factor) via interaction with epigenetic factors, namely p300 and PRC2, in diabetic mice [215]. The knockout of ANRIL in mice resulted in a low level of VEGF, as well as extracellular matrix proteins [215]. Thus, ANRIL controls the heart, kidneys and eyes in diabetes by regulating the expression of VEGF and the extracellular matrix proteins [215]. These findings suggest a novel therapeutic strategy to control diabetes and associated complications using an RNA-based approach. Furthermore, Qiu et al. [216] identified another antisense lncRNA, MEG3 (maternally expressed gene 3), in microvascular dysfunction, an important feature in diabetes complications [216]. In this study, the expression level of MEG3 was found to be significantly low in the retinas of streptozotocin-induced diabetic mice and in endothelial cells under high glucose stress. The knockdown of MEG3 significantly exacerbated retinal vascular abnormalities, resulting in endothelial cell proliferation, migration and tube formation [216]. Thus, MEG3 upregulation may serve as a new therapeutic approach in the treatment of diabetes-induced microvascular complications. Likewise, there are many antisense transcripts that are significantly misregulated in diabetes and its associated complications [217,218,219].
In addition to the involvement of antisense RNAs in cardiac diseases via diabetes, antisense transcription/transcripts are also directly associated with cardiovascular diseases. Recent studies have implicated antisense RNAs as new diagnostic markers with therapeutic potentials for cardiovascular diseases [210,211,212,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239]. For example, an antisense transcript H19, discussed above, is associated with cardiovascular diseases such as CAD. The knockdown of H19 induces cardiomyocyte hypertrophy [220], indicating H19′s role in maintaining cardiac health. Furthermore, H19 functions as a precursor for miR-675, which inhibits cardiac hypertrophy [221]. Importantly, CaMKIId (calcium/calmodulin-dependent protein kinase II delta), a downstream target of H19-miR-675, is a serine/threonine protein kinase that is associated with cardiac electrical conduction. Thus, misregulation of CaMKIId by H19-miR-675 has been linked to cardiac electrical conduction defects. Furthermore, H19 was found to be upregulated in atherosclerosis and to increase the levels of H19 in VSMC (vascular smooth muscle cells) and HUVSC (human umbilical vein endothelial cells), resulting in cellular proliferation and the suppression of apoptosis [222]. Similarly, another antisense RNA, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), is linked to cardiovascular diseases including hypertension and diabetic cardiomyopathy [223,224]. In addition, MALAT1 has been demonstrated to play a regulatory role in promoting angiogenesis via VEGFR2 (vascular endothelial growth factor receptor 2) [225]. Likewise, ANRIL is also associated with cardiac diseases including myocardial infarction (MI) and CAD. The promoter of ANRIL was found to be methylated in individuals with a high risk of CAD [226,227]. ANRIL variants (e.g., rs10965215 and rs10738605) resulting from single nucleotide polymorphism (SNP) are associated with the risk of MI in the Chinese Han population [228]. The ANRIL variant rs3217992 has been connected to adverse cardiovascular events [229]. Furthermore, another ANRIL SNP variant, rs10757278, was linked to the development of major adverse cardiovascular events in hemodialysis patients [230]. The expression level of the ANRIL transcript was found to be remarkably low in the blood of CAD patients, indicating a relationship between the expression level of ANRIL and the risk of disease. Like ANRIL, HOTAIR is also involved in AMI (acute MI) and its plasma concentration could be used to detect and monitor AMI [231]. HOTAIR is upregulated in the cardiac tissues and plasma of patients with AMI and CAD [231,232]. Together, these studies demonstrate the roles of antisense transcripts in the regulation of cardiac health and functions. These antisense transcripts could be diagnostic markers and therapeutic targets for cardiovascular diseases. In addition, many other antisense transcripts are correlated with cardiovascular diseases [210,211,212,233,234,235,236,237,238,239].
Antisense transcripts are also involved in regulating muscular diseases. For example, MALAT1 is linked to muscular dystrophy [240,241]. In proliferating myoblasts, MALAT1 recruits SUV39H1 (Su(var)3-9 homolog 1) to the binding site of the muscle differentiation regulator MyoD, and causes trimethylation of the histone H3 K9, thus repressing the expression of the MyoD target genes involved in muscle differentiation [240]. On the other hand, the knockdown of MALAT1 promotes myogenic differentiation in cultured cells [240]. In agreement with this, increased muscle regeneration was observed in MALAT1 knockout mice [240]. Recently, MALAT1 was also reported as a novel downstream target of myostatin [241], a negative regulator of muscle growth [242]. Thus, MALAT1 is associated with the regulation of myogenic differentiation and muscle regeneration [240], and the misregulation of MALAT1 is linked to muscular disorders [240,241]. Another antisense lncRNA, SIRT1 AS (sirtuin 1 antisense RNA), has also been shown to regulate myogenesis. The knockdown of SIRT1 leads to the differentiation of myoblasts in C2C12 and human skeletal muscle cells. The overexpression of SIRT1 AS increases the levels of the NAD+-dependent histone/protein deacetylase, SIRT1, via the formation of RNA duplexes and the facilitation of SIRT1 translation, by competing with miR34a (that can bind with SIRT1) to inhibit muscle formation [243,244]. Thus, dysfunction or misregulation of SIRT1 AS would alter myogenesis, leading to muscular diseases. Likewise, there are other antisense RNAs involved in muscle diseases [210,212,245,246]. Antisense transcripts were found to be involved in immune diseases [247,248,249]. For example, H19 is upregulated in rheumatoid arthritis patients [247]. HOTAIR was found to be expressed at high levels in rheumatoid arthritis [248]. MALAT1 is also overexpressed in rheumatoid arthritis fibroblast-like synoviocytes [249]. Thus, antisense transcripts have the potential to function as biomarkers in immune diseases.
Since gene expression is central to cellular processes, the regulation of protein-coding gene expression by antisense transcription/transcripts has a significant impact on cellular gene expression and health. The misregulation of antisense transcription or antisense transcripts would be associated with various diseases, some of which are discussed above. However, other diseases/conditions such as aging, metabolic disorders, stress, thalassemia and spinocerebellar ataxia are also associated with antisense transcription/transcripts [30,210,211,212].
9. Conclusions
Antisense transcripts are wide-spread throughout eukaryotic genomes and are generated from independent, bidirectional or cryptic promoters. Antisense transcription/transcripts regulate gene expression and genome integrity via transcriptional interference, histone modification, and/or DNA methylation. Antisense transcripts can bring different macromolecules together within the three-dimensional context of the cell to coordinately execute transcriptional, post-transcriptional, and epigenetic processes. Thus, antisense transcription/transcripts are involved in many biological processes and are misregulated in a variety of diseases including cancer, neurological diseases, diabetes and cardiovascular disorders. Therefore, an understanding of the regulatory mechanisms of antisense transcription, characterization of antisense transcripts and their modes of actions would be useful for the diagnosis, monitoring and targeted therapies of various diseases. Indeed, several antisense oligonucleotides are in clinical trials for the treatment of various diseases [62]. Two antisense oligonucleotide-mediated therapies are now available in clinics for the treatment of Duchenne muscular dystrophy and spinal muscular atrophy [62]. Thus, the rapid development of antisense transcription-based therapy holds great promise for the treatment of many diseases in the near future.
Although antisense transcription/transcripts play crucial roles in the regulation of gene expression and genomic integrity due to their involvement in various diseases, it remains unclear how antisense transcription is initiated and epigenetically regulated. Here, we began to develop an understanding of how antisense transcription is initiated and regulated by histone covalent modifications. Further studies are needed for a thorough understanding of antisense transcription initiation and its epigenetic regulation. Such knowledge will provide new insights into the regulation of antisense transcription/transcripts and will aid in understanding the etiologies of various diseases, therefore promoting the discovery of diagnostic markers and therapeutic interventions.
Funding
Work in the Bhaumik lab was supported by grants from the National Institute of Health (1R15GM088798-01, 2R15GM088798-02 and 3R15GM088798-03), the American Heart Association (15GRNT25700298), the Mallinckrodt Foundation, the American Cancer Society, and Southern Illinois University School of Medicine.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bhaumik S.R. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim. Biophys. Acta. 2011;1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik S.R., Malik S. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 2008;43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 3.Durairaj G., Malik S., Bhaumik S.R. Eukaryotic gene expression by RNA polymerase II. In: Mandal S.S., editor. Gene Regulation, Epigenetics, and Hormone Signaling. Volume 1. Wiley-Blackwell; Oxford, UK: 2017. pp. 1–28. [Google Scholar]
- 4.Shilatifard A., Conaway R.C., Conaway J.W. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 5.Karmakar S., Ponnusamy M.P., Bhaumik S.R., Batra S.K. RNA Polymerase II and Associated Transcription Factors. eLS. 2019 doi: 10.1002/9780470015902.a0003301.pub3. [DOI] [Google Scholar]
- 6.Karmakar S., Dey P., Vaz A.P., Bhaumik S.R., Ponnusamy M.P., Batra S.K. PD2/PAF1 at the crossroads of the cancer network. Cancer Res. 2018;78:313–319. doi: 10.1158/0008-5472.CAN-17-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla A., Natarajan A., Bhaumik S.R., El Shemy H., Lightfoot D. The interactions of the largest subunit of RNA polymerase II with other cellular proteins: A bioinformatic approach. Curr. Issues Mol. Biol. 2009;11:i65–i71. [PubMed] [Google Scholar]
- 8.Bhaumik S.R., Smith E., Shilatifard A. Histone covalent modifications in development and disease pathogenesis. Nat. Struct. Mo. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 9.Shukla A., Chaurasia P., Bhaumik S.R. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 2009;66:1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S., Bhaumik S.R. Mixed lineage leukemia: Histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 2010;277:1805–1821. doi: 10.1111/j.1742-4658.2010.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartholomew B. Regulating the chromatin landscape: Structural and mechanistic perspectives. Annu. Rev. Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorch Y., Kornberg R.D. Chromatin-remodeling for transcription. Q. Rev. Biophys. 2017;50:e5. doi: 10.1017/S003358351700004X. [DOI] [PubMed] [Google Scholar]
- 13.Sen R., Bhaumik S.R. Transcriptional stimulatory and repressive functions of histone H2B ubiquitin ligase. Transcription. 2013;4:221–226. doi: 10.4161/trns.26623. [DOI] [PubMed] [Google Scholar]
- 14.Svejstrup J.Q. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Lorch Y., Kornberg R.D. Chromatin-remodeling and the initiation of transcription. Q. Rev. Biophys. 2015;48:465–470. doi: 10.1017/S0033583515000116. [DOI] [PubMed] [Google Scholar]
- 16.Yaniv M. Chromatin remodeling: From transcription to cancer. Cancer Genet. 2014;207:352–357. doi: 10.1016/j.cancergen.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Narlikar G.J. A proposal for kinetic proof reading by ISWI family chromatin remodeling motors. Curr. Opin. Chem. Biol. 2010;14:660–665. doi: 10.1016/j.cbpa.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C.Y., Johnson S.L., Gamarra N.I., Narlikar G.J. Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 2016;45:153–181. doi: 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Li S., Wang X., Zhu J., Wei Y., Wang Y., Wen Y., Wang L., Huang Y., Zhang B., et al. DNA methylation dynamics: Identification and functional annotation. Brief Funct. Genomics. 2016;15:470–484. doi: 10.1093/bfgp/elw033. [DOI] [PubMed] [Google Scholar]
- 21.Wu S.C., Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla A., Stanojevic N., Duan Z., Shadle T., Bhaumik S.R. Functional analysis of H2B-K123 ubiquitination in regulation of H3-K4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J. Biol. Chem. 2006;281:19045–19054. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 23.Schneider J., Bajwa P., Johnson F.C., Bhaumik S.R., Shilatifard A. Rtt109 is required for proper H3K56 acetylation: A chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 24.Durairaj G., Chaurasia P., Lahudkar S., Malik S., Shukla A., Bhaumik S.R. Regulation of chromatin assembly/disassembly by Rtt109p, a histone H3 Lys56-specific acetyltransferase, in vivo. J. Biol. Chem. 2010;285:30472–30479. doi: 10.1074/jbc.M110.113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin S., Wang Q., Ray A., Wani G., Zhao Q., Bhaumik S.R., Wani A.A. Sem1p and Ubp6p orchestrate telomeric silencing by modulating histone H2B ubiquitination and H3 acetylation. Nucleic Acids Res. 2009;37:1843–1853. doi: 10.1093/nar/gkn1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood A., Shukla A., Schneider J., Lee J.-S., Stanton J.D., Dzuiba T., Swanson S.K., Florens L., Washburn M.P., Wyrick J., et al. Ctk complex regulation of histone methylation by COMPASS. Mol. Cell. Biol. 2007;27:709–720. doi: 10.1128/MCB.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su W.Y., Xiong H., Fang J.Y. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem. Biophys. Res. Commun. 2010;396:177–181. doi: 10.1016/j.bbrc.2010.04.147. [DOI] [PubMed] [Google Scholar]
- 28.Imamura T., Yamamoto S., Ohgane J., Hattori N., Tanaka S., Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 29.Werner A., Berdal A. Natural antisense transcripts: Sound or silence? Physiol. Genomics. 2005;23:125–131. doi: 10.1152/physiolgenomics.00124.2005. [DOI] [PubMed] [Google Scholar]
- 30.Tufarelli C., Stanley J.A., Garrick D., Sharpe J.A., Ayyub H., Wood W.G., Higgs D.R. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 31.Ohhata T., Hoki Y., Sasaki H., Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein E., Allis C.D. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein E., Duncan E.M., Masui O., Gil J., Heard E., Allis C.D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werner A., Sayer J.A. Naturally occurring antisense RNA: Function and mechanisms of action. Curr. Opin. Nephrol. Hypertens. 2009;18:343–349. doi: 10.1097/MNH.0b013e32832cb982. [DOI] [PubMed] [Google Scholar]
- 35.Lapidot M., Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavorgna G., Dahary D., Lehner B., Sorek R., Sanderson C.M., Casari G. In search of antisense. Trends Biochem. Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Werner A. Natural antisense transcripts. RNA Biol. 2005;2:53–62. doi: 10.4161/rna.2.2.1852. [DOI] [PubMed] [Google Scholar]
- 38.Vanhee-Brossollet C., Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/S0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 39.Sun M., Hurst L.D., Carmichael G.G., Chen J. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 2005;33:5533–5543. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X.J., Gaasterland T., Chua N.H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:1–11. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tisseur M., Kwapisz M., Morillon A. Pervasive transcription: Lessons from yeast. Biochimie. 2011;93:1889–18896. doi: 10.1016/j.biochi.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 43.He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hongay C.F., Grisafi P.L., Galitski T., Fink G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 45.Wery M., Kwapisz M., Morillon A. Noncoding RNAs in gene regulation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:728–738. doi: 10.1002/wsbm.148. [DOI] [PubMed] [Google Scholar]
- 46.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent III G., Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seitz A., Gourevitch D., Zhang X.M., Clark L., Chen P., Kragol M., Levenkova N., Rux J., Samulewicz S., Heber-Katz E. Sense and antisense transcripts of the apolipoprotein E gene in normal and ApoE knockout mice; their expression after spinal cord injury and corresponding human transcripts. Hum. Mol. Genet. 2005;14:2661–2670. doi: 10.1093/hmg/ddi296. [DOI] [PubMed] [Google Scholar]
- 48.Kumar M., Carmichael G.G. Antisense RNA: Function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reis E.M., Nakaya H.I., Louro R., Canavez F.C., Flatschart A.V., Almeida G.T., Egidio C.M., Paquola A.C., Machado A.A., Festa F., et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 50.Grinchuk O.V., Motakis E., Yenamandra S.P., Ow G.S., Jenjaroenpun P., Tang Z., Yarmishyn A.A., Ivshina A.V., Kuznetsov V.A. Sense-antisense gene-pairs in breast cancer and associated pathological pathways. Oncotarget. 2015;6:42197–42221. doi: 10.18632/oncotarget.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aartsma-Rus A., van Ommen G.J. Progress in therapeutic antisense applications for neuromuscular disorders. Eur. J. Hum. Genet. 2010;18:146–153. doi: 10.1038/ejhg.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooke R.M. Antisense oligonucleotides as therapeutics for hyperlipidaemias. Expert Opin. Biol. Ther. 2005;5:907–917. doi: 10.1517/14712598.5.7.907. [DOI] [PubMed] [Google Scholar]
- 53.Luther H.P. Role of endogenous antisense RNA in cardiac gene regulation. J. Mol. Med. 2005;83:26–32. doi: 10.1007/s00109-004-0613-5. [DOI] [PubMed] [Google Scholar]
- 54.Ulanova M., Schreiber A.D., Befus A.D. The future of antisense oligonucleotides in the treatment of respiratory diseases. BioDrugs. 2006;20:1–11. doi: 10.2165/00063030-200620010-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popescu F.D. Antisense- and RNA interference-based therapeutic strategies in allergy. J. Cell. Mol. Med. 2005;9:840–853. doi: 10.1111/j.1582-4934.2005.tb00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 57.Ferrari N., Seguin R., Renzi P. Oligonucleotides: A multi-targeted approach for the treatment of respiratory diseases. Future Med. Chem. 2011;3:1647–1662. doi: 10.4155/fmc.11.108. [DOI] [PubMed] [Google Scholar]
- 58.Lu X., Yu Q., Binder G.K., Chen Z., Slepushkina T., Rossi J., Dropulic B. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J. Virol. 2004;78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeVos S.L., Miller T.M. Antisense oligonucleotides: Treating neurodegeneration at the level of RNA. Neurotherapeutics. 2013;10:486–497. doi: 10.1007/s13311-013-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Z., Cooper T.A. Antisense oligonucleotides: Rising stars in eliminating RNA toxicity in myotonic dystrophy. Hum. Gene Ther. 2013;24:499–507. doi: 10.1089/hum.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visser M.E., Witztum J.L., Stroes E.S., Kastelein J.J. Antisense oligonucleotides for the treatment of dyslipidaemia. Eur. Heart J. 2012;33:1451–1458. doi: 10.1093/eurheartj/ehs084. [DOI] [PubMed] [Google Scholar]
- 62.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 63.Wagner E.G., Simons R.W. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 64.Brown T., Howe F.S., Murray S.C., Wouters M., Lorenz P., Seward E., Rata S., Angel A., Mellor J. Antisense transcription-dependent chromatin signature modulates sense transcript dynamics. Mol. Syst. Biol. 2018;14:e8007. doi: 10.15252/msb.20178007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozsolak F., Kapranov P., Foissac S., Kim S.W., Fishilevich E., Monaghan A.P., John B., Milos P.M. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C., et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 70.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim T., Xu Z., Clauder-Munster S., Steinmetz L.M., Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 75.Kaplan C.D., Laprade L., Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 76.Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 77.Su W.Y., Li J.T., Cui Y., Hong J., Du W., Wang Y.C., Lin Y.W., Xiong H., Wang J.L., Kong X., et al. Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res. 2012;22:1374–1389. doi: 10.1038/cr.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilkening S., Pelechano V., Järvelin A.I., Tekkedil M.M., Anders S., Benes V., Steinmetz L.M. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:1–8. doi: 10.1093/nar/gkt364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pelechano V., Wei W., Steinmetz L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malik S., Durairaj G., Bhaumik S.R. Mechanisms of antisense transcription initiation from the 3′-end of the GAL10 coding sequence in vivo. Mol. Cell. Biol. 2013;33:3549–3567. doi: 10.1128/MCB.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uprety B., Kaja A., Ferdoush J., Sen R., Bhaumik S.R. Regulation of antisense transcription by NuA4 histone acetyltransferase and other chromatin regulatory factors. Mol. Cell. Biol. 2016;36:992–1006. doi: 10.1128/MCB.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gullerova M., Proudfoot N.J. Transcriptional interference and gene orientation in yeast: Noncoding RNA connections. Cold Spring Harb. Symp. Quant. Biol. 2010;75:299–311. doi: 10.1101/sqb.2010.75.048. [DOI] [PubMed] [Google Scholar]
- 86.Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pinskaya M., Gourvennec S., Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhaumik S.R., Green M.R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhaumik S.R., Raha T., Aiello D.P., Green M.R. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnston M. A model fungal gene regulatory mechanism: The GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnston M., Carlson M. Regulation of carbon and phosphate utilization: The molecular and cellular biology of the yeast Saccharomyces. In: Jones E.W., Pringle J.R., Broach J.R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Volume II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1992. pp. 193–281. [Google Scholar]
- 92.Ozcan S., Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell. Biol. 1996;16:5536–5545. doi: 10.1128/MCB.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Campbell R.N., Leverentz M.K., Ryan L.A., Reece R.J. Metabolic control of transcription: Paradigms and lessons from Saccharomyces cerevisiae. Biochem. J. 2008;414:177–187. doi: 10.1042/BJ20080923. [DOI] [PubMed] [Google Scholar]
- 94.Sen R., Malik S., Frankland-Searby S., Uprety B., Lahudkar S., Bhaumik S.R. Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response. Nucleic Acids Res. 2014;42:9892–9907. doi: 10.1093/nar/gku703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li X.Y., Bhaumik S.R., Green M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 96.Larschan E., Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malik S., Shukla A., Sen P., Bhaumik S.R. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 2009;284:35714–35724. doi: 10.1074/jbc.M109.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uprety B., Lahudkar S., Malik S., Bhaumik S.R. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 2012;40:1969–1983. doi: 10.1093/nar/gkr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uprety B., Sen R., Bhaumik S.R. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol. Cell. Biol. 2015;35:2947–2964. doi: 10.1128/MCB.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uprety B., Kaja A., Bhaumik S.R. TOR facilitates the targeting of the 19S proteasome subcomplex to enhance transcription complex assembly at the promoters of the ribosomal protein genes. Mol. Cell. Biol. 2018;38:e00469-17. doi: 10.1128/MCB.00469-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferdoush J., Sen R., Kaja A., Barman P., Bhaumik S.R. Two Distinct Regulatory Mechanisms of Transcriptional Initiation in Response to Nutrient Signaling. Genetics. 2018;208:191–205. doi: 10.1534/genetics.117.300518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen W.C., Bhaumik S.R., Causton H.C., Simon I., Zhu X., Jennings E.G., Wang T.H., Young R.A., Green M.R. Systematic analysis of essential yeast TAFs in genome-wide transcription and pre-initiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li X.Y., Bhaumik S.R., Zhu X., Li L., Shen W.C., Dixit B.L., Green M.R. Selective recruitment of TAFs by upstream activating sequences: Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/S0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 104.Lee J.S., Shukla A., Schneider J., Florens L., Swanson S.K., Washburn M.P., Bhaumik S.R., Shilatifard A. Translating histone crosstalk between H2B monoubiquitination and H3 methylation by COMPASS and Dot1. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 105.Shukla A., Bhaumik S.R. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem. Biophys. Res. Commun. 2007;359:214–220. doi: 10.1016/j.bbrc.2007.05.105. [DOI] [PubMed] [Google Scholar]
- 106.Sen R., Lahudkar S., Durairaj G., Bhaumik S.R. Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo. J. Biol. Chem. 2013;288:9619–9633. doi: 10.1074/jbc.M113.450403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S.R. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shukla A., Bajwa P., Bhaumik S.R. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shukla A., Lahudkar S., Durairaj G., Bhaumik S.R. Sgf29p facilitates the recruitment of TATA-box-binding protein, but does not alter SAGA’s global structural integrity in vivo. Biochemistry. 2012;51:706–714. doi: 10.1021/bi201708z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Durairaj G., Sen R., Uprety B., Shukla A., Bhaumik S.R. Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation. J. Mol. Biol. 2014;426:2928–2941. doi: 10.1016/j.jmb.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bhaumik S.R., Green M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schwabish M.A., Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malik S., Chaurasia P., Lahudkar S., Uprety B., Bhaumik S.R. Rad26p regulates the occupancy of histone H2A-H2B dimer at the active genes in vivo. Nucleic Acids Res. 2012;40:3348–3363. doi: 10.1093/nar/gkr1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 115.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lyle R., Watanabe D., Vruchte D., Lerchner W., Smrzka O.W., Wutz A., Schageman J., Hahner L., Davies C., Barlow D.P. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 117.Latos P., Pauler F.M., Koerner M.V., Şenergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E., et al. Airn transcriptional overlap; but not its lncRNA products; induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 118.Guil S., Esteller M. cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 119.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP–seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Swiezewski S., Liu F., Magusin A., Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 122.Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 123.Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L.M., Stutz F. trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 125.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 126.Schmitz K.M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 129.Modarresi F., Faghihi M.A., Lopez-Toledano M.A., Fatemi R.P., Magistri M., Brothers S.P., van der Brug M.P., Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsui M., Chu Y., Zhang H., Gagnon K.T., Shaikh S., Kuchimanchi S., Manoharan M., Corey D.R., Janowski B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 132.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X., Li H., Burnett J.C., Rossi J.J. The role of antisense long noncoding RNA in small RNA-triggered gene activation. RNA. 2014;20:1916–1928. doi: 10.1261/rna.043968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xue Z., Ye Q., Anson S.R., Yang J., Xiao G., Kowbel D., Glass N.L., Crosthwaite S.K., Liu Y. Transcriptional interference by antisense RNA is required for circadian clock function. Nature. 2014;514:650–653. doi: 10.1038/nature13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stojic L., Niemczyk M., Orjalo A., Ito Y., Ruijter A.E., Uribe-Lewis S., Joseph N., Weston S., Menon S., Odom D.T., et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Munroe S.H., Lazar M.A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 137.Hastings M.L., Ingle H.A., Lazar M.A., Munroe S.H. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J. Biol. Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- 138.Beltran M., Puig I., Pena C., Garcia J.M., Alvarez A.B., Pena R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 140.Piatek M.J., Henderson V., Fearn A., Chaudhry B., Werner A. Ectopically expressed Slc34a2a sense-antisense transcripts cause a cerebellar phenotype in zebrafish embryos depending on RNA complementarity and Dicer. PLoS ONE. 2017;12:1–17. doi: 10.1371/journal.pone.0178219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Serviss J.T., Andrews N., Van den Eynden J., Richter F.C., Houtman M., Vesterlund M., Schwarzmueller L., Johnsson P., Larsson E., Grander D., Pokrovskaja T.K. An antisense RNA capable of modulating the expression of the tumor suppressor microRNA-34a. Cell Death Dis. 2018;9:1–15. doi: 10.1038/s41419-018-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Friedberg E.C., Walker G.C., Siede W., Wood R.D. DNA Repair and Mutagenesis. American Society for Microbiology Press; Washington, DC, USA: 2005. DNA Damage. [Google Scholar]
- 144.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 145.Seeberg E., Eide L., Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/S0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 146.Cromie G.A., Connelly J.C., Leach D.R. Recombination at Double-Strand Breaks and DNA Ends: Conserved Mechanisms from Phage to Humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/S1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 147.Kanaar R., Hoeijmakers J.H., van Gent D.C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/S0962-8924(98)01383-X. [DOI] [PubMed] [Google Scholar]
- 148.Hartwell L.H., Weinert T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 149.Hartwell L.H. Role of yeast in cancer research. Cancer. 1992;69:2615–2621. doi: 10.1002/1097-0142(19920515)69:10<2615::AID-CNCR2820691036>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 150.Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 151.Malik S., Bagla S., Chaurasia P., Duan Z., Bhaumik S.R. Elongating RNA polymerase II is disassembled through specific degradation of its largest but not other subunits in response to DNA damage in vivo. J. Biol. Chem. 2008;283:6897–6905. doi: 10.1074/jbc.M707649200. [DOI] [PubMed] [Google Scholar]
- 152.Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., Bhaumik S.R. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 2010;38:1461–1477. doi: 10.1093/nar/gkp1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Malik S., Bhaumik S.R. Rad26p, a transcription-coupled repair factor, promotes the eviction and prevents the reassociation of histone H2A-H2B dimer during transcriptional elongation in vivo. Biochemistry. 2012;51:5873–5875. doi: 10.1021/bi3005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Malik S., Bhaumik S.R. Regulation of active genome integrity and expression. Nucleus. 2014;5:520–526. doi: 10.4161/nucl.36230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chaurasia P., Sen R., Pandita T.K., Bhaumik S.R. Preferential DNA double-strand break repair at the active gene. J. Biol. Chem. 2012;287:36414–36422. doi: 10.1074/jbc.M112.364661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chaurasia P., Sen R., Bhaumik S.R. Functional analysis of Rad14p, a DNA damage recognition factor in nucleotide excision repair, in regulation of transcription in vivo. J. Biol. Chem. 2013;288:793–806. doi: 10.1074/jbc.M112.413716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 158.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X.T., Song X.Y., Glass C.K., Rosenfeld M.G. The Long Arm of Long Noncoding RNAs: Roles as Sensors Regulating Gene Transcriptional Programs. Cold Spring Harb. Perspect. Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M., et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Congrains A., Kamide K., Ohishi M., Rakugi H. ANRIL: Molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013;14:1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wan G., Mathur R., Hu X., Liu Y., Zhang X., Peng G., Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Qi W., Wang R., Chen H., Wang X., Xiao T., Boldogh I., Ba X., Han L., Zeng X. BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci. 2015;128:317–330. doi: 10.1242/jcs.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cajigas I., Leib D.E., Cochrane J., Luo H., Swyter K.R., Chen S., Clark B.S., Thompson J., Yates J.R., Kingston R.E., et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142:2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kwon S.J., Park J.H., Park E.J., Lee S.A., Lee H.S., Kang S.W., Kwon J. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene. 2015;34:303–313. doi: 10.1038/onc.2013.556. [DOI] [PubMed] [Google Scholar]
- 166.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P., et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sharma V., Misteli T. Noncoding RNAs in DNA damage and repair. FEBS Lett. 2013;587:1832–1839. doi: 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Y., Lu X. Non-coding RNAs in DNA damage response. Am. J. Cancer Res. 2012;2:658–675. [PMC free article] [PubMed] [Google Scholar]
- 169.Dianatpour A., Ghafouri-Fard S. The role of Long non-coding RNAs in the repair of DNA double strand breaks. Int. J. Mol. Cell Med. 2017;6:1–12. [PMC free article] [PubMed] [Google Scholar]
- 170.Rinn R.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Avazpour N., Hajjari M., Birgani M.T. HOTAIR: A Promising Long Non-coding RNA with Potential Role in Breast Invasive Carcinoma. Front. Genet. 2017;8:170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA HOTAIR regulates polycomb dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 173.Geng Y., Xie S., Li Q., Ma J., Wang G. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 174.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang L., Liao L.M., Liu A.W., Wu J.B., Cheng X.L., Lin J.X., Zheng M. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch. Gynecol. Obstet. 2014;290:717–723. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 176.He X., Bao W., Li X., Chen Z., Che Q., Wang H., Wan X.P. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int. J. Mol. Med. 2014;33:325–332. doi: 10.3892/ijmm.2013.1570. [DOI] [PubMed] [Google Scholar]
- 177.Alahari S.V., Eastlack S.C., Alahari S.K. Role of long noncoding RNAs in Neoplasia: Special emphasis on prostate cancer. Int. Rev. Cell Mol. Biol. 2016;324:229–254. doi: 10.1016/bs.ircmb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 178.Yoshimura H., Matsuda Y., Yamamoto M., Kamiya S., Ishiwata T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front. Biosci. 2018;23:614–625. doi: 10.2741/4608. [DOI] [PubMed] [Google Scholar]
- 179.Pan Y., Pan Y., Cheng Y., Yang F., Yao Z., Wang O. Knockdown of LncRNA MAPT-AS1 inhibits proliferation and migration and sensitizes cancer cells to paclitaxel by regulating MAPT expression in ER-negative breast cancers. Cell Biosci. 2018;8:1–13. doi: 10.1186/s13578-018-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mahmoudi S., Henriksson S., Farnebo L., Roberg K., Farnebo M. WRAP53 promotes cancer cell survival and is a potential target for cancer therapy. Cell Death Dis. 2011;2:e114. doi: 10.1038/cddis.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fang Y., Wang J., Wu F., Song Y., Zhao S., Zhang Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget. 2017;8:46090–46103. doi: 10.18632/oncotarget.17552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu Z., Chen Z., Fan R., Jiang B., Chen X., Chen Q., Nie F., Lu K., Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer. 2017;16:1–9. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ma P., Xu T., Huang M., Shu Y. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed. Pharmacother. 2016;78:172–176. doi: 10.1016/j.biopha.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 184.Sang Y., Tang J., Li S., Tang X., Cheng C., Luo Y., Qian X., Deng L., Liu L., Lv X. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16INK4A expression. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Han L., Zhang E., Yin D., Kong R., Xu T., Chen W., Xia R., Shu Y., De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:1–12. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang J.J., Wang D.D., Du C.X., Wang Y. Long Noncoding RNA ANRIL Promotes Cervical Cancer Development by Acting as a Sponge of miR-186. Oncol. Res. 2018;26:345–352. doi: 10.3727/096504017X14953948675449. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 187.Naemura M., Murasaki C., Inoue Y., Okamoto H., Kotake Y. Long Noncoding RNA ANRIL Regulates Proliferation of Non-small Cell Lung Cancer and Cervical Cancer Cells. Anticancer Res. 2015;35:5377–5382. [PubMed] [Google Scholar]
- 188.Bhan A., Soleimani M., Mandal S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Arun G., Diermeier S.D., Spector D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lin C., Yang L. Long noncoding RNA in Cancer: Wiring signaling circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bolha L., Ravnik-Glavac M., Glavac D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Parsons C., Tayoun A.M., Benando B.D., Ragusa G., Dorvil R.F., Rourke E.A., Connor K.O., Reed I.G., Alexander A., Willetts L., et al. The role of long noncoding RNAs in cancer metastasis. J. Cancer Metastasis Treat. 2018;4:1–23. doi: 10.20517/2394-4722.2018.11. [DOI] [Google Scholar]
- 193.Tano K., Akimitsu N. Long non-coding RNAs in cancer progression. Front. Genet. 2012;3:1–6. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Deva Magendhra Rao A.K., Arvinden V.R., Rajkumar T., Samson M. Functions and Therapeutic Applications of Antisense RNAs in Cancer. Austin J. Cancer Clin. Res. 2017;4:1–3. [Google Scholar]
- 195.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Renganathan A., Felley-Bosco E. Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv. Exp. Med. Biol. 2017;1008:199–222. doi: 10.1007/978-981-10-5203-3_7. [DOI] [PubMed] [Google Scholar]
- 197.Grigoriadis A., Oliver G.R., Tanney A., Kendrick H., Smalley M.J., Jat P., Neville A.M. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics. 2009;10:324. doi: 10.1186/1471-2164-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Maruyama R., Shipitsin M., Choudhury S., Wu Z., Protopopov A., Yao J., Lo P.K., Bessarabova M., Ishkin A., Nikolsky Y., et al. Altered antisense-to-sense transcript ratios in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2820–2824. doi: 10.1073/pnas.1010559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Faghihi M.A., Zhang M., Huang J., Modarresi F., Van der Brug M.P., Nalls M.A., Cookson M.R., St-Laurent III G., Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:1–13. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Modarresi F., Faghihi M.A., Patel N.S., Sahagan B.G., Wahlestedt C., Lopez-Toledano M.A. Knockdown of BACE1-AS nonprotein-coding transcript modulates betaamyloid- related hippocampal neurogenesis. Int. J. Alzheimers Dis. 2011;2011:1–11. doi: 10.4061/2011/929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang M., Cai F., Zhang S., Zhang S., Song W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci. Rep. 2015;4:1–6. doi: 10.1038/srep07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Carriieri C., Forrest A.R.R., Santoro C., Persichetti F., Carninci P., Zucchelli S., Gustincich S. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 2015;9:114. doi: 10.3389/fncel.2015.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Scheele C., Petrovic N., Faghihi M.A., Lassmann T., Fredriksson K., Rooyackers O., Wahlestedt C., Good L., Timmons J.A. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics. 2007;8:74. doi: 10.1186/1471-2164-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chung D.W., Rudnicki D.D., Yu L., Margolis R.L. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet. 2011;20:3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 206.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang E.B., Yin D.D., Sun M., Kong R., Liu X.-H., You L.-H., Han L., Xia R., Wang K.-M., Yang J.-S., et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer; partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:1–12. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 209.Wu P., Zuo X., Deng H., Liu X., Liu L., Ji A. Roles of long noncoding RNAs in brain development; functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 210.Kim J., Kim K.M., Noh J.H., Yoon J.H., Abdelmohsen K., Gorospe M. Long noncoding RNAs in diseases of aging. Biochim. Biophys. Acta. 2016;1859:209–221. doi: 10.1016/j.bbagrm.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cipolla G.A., de Oliveira J.C., Salviano-Silva A., Lobo-Alves S.C., Lemos D.S., Oliveira L.C., Jucoski T.S., Mathias C., Pedroso G.A., Zambalde E.P., et al. Long non-coding RNAs in multifactorial diseases: Another Layer of complexity. Noncoding RNA. 2018;4:13. doi: 10.3390/ncrna4020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim C., Kang D., Lee E.K., Lee J.S. Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxid. Med. Cell Longev. 2017;2017:1–21. doi: 10.1155/2017/2062384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pereira F.D., Bitar M., Jacobs F.M.J., Barry G. Long non-coding RNAs in Neuronal aging. Noncoding RNA. 2018;4:E12. doi: 10.3390/ncrna4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wei C.-W., Luo T., Zou S.-S., Wu A.-S. The role of Long noncoding RNAS in central nervous system and neurodegenerative diseases. Front. Behav. Neurosci. 2018;12:1–11. doi: 10.3389/fnbeh.2018.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thomas A.A., Feng B., Chakrabarti S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Invest Ophthalmol. Vis Sci. 2017;58:470–480. doi: 10.1167/iovs.16-20569. [DOI] [PubMed] [Google Scholar]
- 216.Qiu G.Z., Tian W., Fu H.T., Li C.P., Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016;471:135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 217.Ma C., Luo H., Liu B., Li F., Tschope C., Fa X. Long noncoding RNAs: A new player in the prevention and treatment of diabetic cardiomyopathy? Diabetes Metab. Res. Rev. 2018;34:e3056. doi: 10.1002/dmrr.3056. [DOI] [PubMed] [Google Scholar]
- 218.Gong Q., Su G. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci. Rep. 2017;37:1–18. doi: 10.1042/BSR20171157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Raut S.K., Khulla M. The big entity of new RNA world: Long non-coding RNAs in microvascular complications of diabetes. Front. Endocrinol. 2018;9:1–10. doi: 10.3389/fendo.2018.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang Z., Gao W., Long Q.-Q., Zhang J., Li Y.-F., Liu D.-C., Yan J.-J., Yang Z.-J., Wang L.-S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Liu L., An X., Li Z., Song Y., Li L., Zuo S., Liu N., Yang G., Wang H., Cheng X., et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovascular Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 222.Pan J.X. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21:322–328. [PubMed] [Google Scholar]
- 223.Zhuo Y., Zeng Q., Zhang P., Li G., Xie Q., Cheng Y. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin. Chem. Lab. Med. 2017;55:38–46. doi: 10.1515/cclm-2016-0056. [DOI] [PubMed] [Google Scholar]
- 224.Zhang M., Gu H., Chen J., Zhou X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int. J. Cardiol. 2016;202:753–755. doi: 10.1016/j.ijcard.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 225.Zhang X., Tang X., Hamblin M.H., Yin K.-J. Long non-coding RNA Malat1 regulates angiogenesis in hindlimb ischemia. Int. J. Mol. Sci. 2018;19:1723. doi: 10.3390/ijms19061723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circulation Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 227.Murray R., Bryant J., Titcombe P., Barton S.J., Inskip H., Harvey N.C., Cooper C., Lillycrop K., Hanson M., Godfrey K.M. DNA methylation at birth within the promoter of ANRIL predicts markers of cardiovascular risk at 9 years. Clin. Epigenetics. 2016;8:1–6. doi: 10.1186/s13148-016-0259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Cheng J., Cai M.-Y., Chen Y.-N., Li Z.-C., Tang S.-S., Yang X.-L., Chen C., Liu X., Xiong X.-D. Variants in ANRIL gene correlated with its expression contribute to myocardial infarction risk. Oncotarget. 2017;8:12607–12619. doi: 10.18632/oncotarget.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schulz S., Seitter L., Werdan K., Hofmann B., Schaller H.-G., Schlitt A., Reichert S. Single nucleotide polymorphisms in long noncoding RNA, ANRIL, are not associated with severe periodontitis but with adverse cardiovascular events among patients with cardiovascular disease. J. Periodontal Res. 2018;53:714–720. doi: 10.1111/jre.12555. [DOI] [PubMed] [Google Scholar]
- 230.Arbiol-Roca A., Padró-Miquel A., Vidal-Alabró A., Hueso M., Fontova P., Bestard O., Rama I., Torras J., Grinyó J.M., Alía-Ramos P., et al. ANRIL as a genetic marker for cardiovascular events in renal transplant patients—An observational follow-up cohort study. Transpl. Int. 2018;31:1018–1027. doi: 10.1111/tri.13276. [DOI] [PubMed] [Google Scholar]
- 231.Gao L., Liu Y., Guo S., Yao R., Wu L., Xiao L., Wang Z., Liu Y., Zhang Y. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol. Biochem. 2017;44:1497–1508. doi: 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- 232.Jiang Y., Mo H., Luo J., Zhao S., Liang S., Zhang M., Yuan J. HOTAIR is a potential novel biomarker in patients with congenital heart diseases. BioMed Res. Int. 2018;2018:2850657. doi: 10.1155/2018/2850657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Haemmig S., Simion V., Yang D., Deng Y., Feinberg M.W. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr. Opin. Cardiol. 2017;32:776–783. doi: 10.1097/HCO.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Shen S., Jiang H., Bei Y., Xiao J., Li X. Long Non-Coding RNAs in Cardiac Remodeling. Cell Physiol. Biochem. 2017;41:1830–1837. doi: 10.1159/000471913. [DOI] [PubMed] [Google Scholar]
- 235.Gomes C.P.C., Spencer H., Ford K.L., Michel L.Y.M., Baker A.H., Emanueli C., Balligand J.L., Devaux Y. The Function and Therapeutic Potential of Long Non-coding RNAs in Cardiovascular Development and Disease. Mol. Ther Nucleic Acids. 2017;8:494–507. doi: 10.1016/j.omtn.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Scheuermann J.C., Boyer L.A. Getting to the heart of the matter: Long non-coding RNAs in cardiac development and disease. EMBO J. 2013;32:1805–1816. doi: 10.1038/emboj.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Uchida S., Dimmeler S. Long Noncoding RNAs in Cardiovascular diseases. Circulation Research. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 238.Archer K., Broskova Z., Bayoumi A.S., Teoh J.P., Davila A., Tang Y., Su H., Kim I.M. Long Non-coding RNAs as Master regulators in cardiovascular diseases. Int. J. Mol. Sci. 2015;16:23651–23667. doi: 10.3390/ijms161023651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sallam T., Sandhu J., Tontonoz P. Long noncoding RNA discovery in cardiovascular disease decoding form to function. Circ. Res. 2018;122:155–166. doi: 10.1161/CIRCRESAHA.117.311802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Chen X., He L., Zhao Y., Li Y., Zhang S., Sun K., So K., Chen F., Zhou L., Lu L., et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017;3:1–23. doi: 10.1038/celldisc.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Watts R., Johnsen V.L., Shearer J., Hittel D.S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Cell Physiol. 2013;304:C995–C1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 242.Thomas M., Langley B., Berry C., Sharma M., Kirk S., Bass J., Kambadur R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 243.Wang Y., Pang W.J., Wei N., Xiong Y., Wu W.J., Zhao C.Z., Shen Q.W., Yang G.S. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. 2014;539:117–124. doi: 10.1016/j.gene.2014.01.037. [DOI] [PubMed] [Google Scholar]
- 244.Wang G., Wang Y., Xiong Y., Chen X., Ma M., Cai R., Gao Y., Sun Y., Yang G., Pang W. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lim Y.-H., Kwon D.-H., Kim J., Park W.J., Kook H., Kim Y.-K. Identification of long noncoding RNAs involved in muscle differentiation. PLoS ONE. 2018;13:1–15. doi: 10.1371/journal.pone.0193898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li Y., Chen X., Sun H., Wang H. Long non-coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Lett. 2018;417:58–64. doi: 10.1016/j.canlet.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 247.Stuhlmüller B., Kunisch E., Franz J., Martinez-Gamboa L., Hernandez M.M., Pruss A., Ulbrich N., Erdmann V.A., Burmester G.R., Kinne R.W. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003;163:901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Song J., Kim D., Han J., Kim Y., Lee M., Jin E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015;15:121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 249.Pan F., Zhu L., Lv H., Pei C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int. J. Mol. Med. 2016;38:1507–1514. doi: 10.3892/ijmm.2016.2755. [DOI] [PubMed] [Google Scholar]